Significance
Accumulating evidence shows that type I interferons (IFNs) induce an antiviral response. Retinoic acid-inducible gene I-like receptors detect Sindbis virus (SINV) RNA and mediate signaling to IFN-β promoter stimulator-1, an adaptor that functions on the surface of healthy mitochondria, to induce transcriptional activation of IFNs. Zinc finger antiviral protein (ZAP) is induced by IFNs and binds with SINV RNA to promote RNA degradation. We recently found an organelle stress-induced IFN-independent antiviral response. On SINV infection, BCL2-associated X protein and BCL2 antagonist/killer 1 cause mitochondria damage, leading to a nuclear-to-cytoplasmic redistribution of tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly(ADP-ribose) polymerase (TIPARP), a ZAP-like protein. TIPARP binds with SINV RNA to promote RNA degradation. Therefore, mitochondrial-nuclear crosstalk plays a vital role in the host defense against RNA viruses.
Keywords: innate immunity, antiviral response, mitochondrial damage, RNA degradation, reactive oxygen species
Abstract
The innate immune system senses RNA viruses by pattern recognition receptors (PRRs) and protects the host from virus infection. PRRs mediate the production of immune modulatory factors and direct the elimination of RNA viruses. Here, we show a unique PRR that mediates antiviral response. Tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly(ADP ribose) polymerase (TIPARP), a Cysteine3 Histidine (CCCH)-type zinc finger-containing protein, binds to Sindbis virus (SINV) RNA via its zinc finger domain and recruits an exosome to induce viral RNA degradation. TIPARP typically localizes in the nucleus, but it accumulates in the cytoplasm after SINV infection, allowing targeting of cytoplasmic SINV RNA. Redistribution of TIPARP is induced by reactive oxygen species (ROS)-dependent oxidization of the nuclear pore that affects cytoplasmic-nuclear transport. BCL2-associated X protein (BAX) and BCL2 antagonist/killer 1 (BAK1), B-cell leukemia/lymphoma 2 (BCL2) family members, mediate mitochondrial damage to generate ROS after SINV infection. Thus, TIPARP is a viral RNA-sensing PRR that mediates antiviral responses triggered by BAX- and BAK1-dependent mitochondrial damage.
Pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) sense viral RNA and induce an antiviral innate immune response. After recognition of viral RNA, TLRs, and RLRs trigger activation of intracellular signaling pathways to produce immune modulatory factors such as type I interferons (IFNs), cytokines, and chemokines (1). RLRs detect viral double-stranded RNA (dsRNA) and triphosphate RNA in the cytoplasm to mediate signaling to mitochondrial adaptor protein IFN-β promoter stimulator-1 (IPS-1) (also called mitochondrial antiviral signaling, MAVS). IPS-1 functions on the outer membrane of healthy mitochondria to activate IFN regulatory factor 3 and 7, which in turn induces expression of type I IFNs (2). Type I IFNs promote activation of an acquired immune response and direct the elimination of viruses, resulting in the protection of hosts from lethal virus infection (3).
Mitochondria are multifunctional organelles that regulate energy production, cellular metabolism, cell death, and signal transduction (4). Mitochondria play a critical role in antiviral responses because they mediate the induction of apoptosis as well as expression of type I IFNs. Apoptosis is controlled by members of the B-cell leukemia/lymphoma 2 (BCL2) family, which mainly localize on mitochondrial surfaces (5). Proapoptotic BCL2 family members such as BCL2-associated X protein (BAX) and BCL2 antagonist/killer 1 (BAK1) induce mitochondrial outer membrane permeabilization to activate caspase-9, leading to the induction of cell death, whereas the anti-apoptotic BCL2 family members inhibit BAX- and BAK1-dependent induction of mitochondrial outer membrane permeabilization. Apoptosis of virus-infected cells is beneficial in limiting further viral replication and propagation. However, apoptosis of terminally differentiated, nondividing cells such as neuronal cells and cardiac myocytes is harmful. Hence, apoptosis is a double-edged sword that protects and damages hosts during virus infection. BAX and BAK1 also mediate the expression of type I IFNs. BAX and BAK1 generate mitochondrial outer membrane permeabilization to promote the release of mitochondrial DNA, leading to activation of cyclic GMP–AMP synthase (cGAS), a cytosolic DNA sensor (6, 7). However, caspase-9 potently inhibits cGAS-triggered expression of type I IFNs to prevent the development of autoimmune diseases.
Zinc finger antiviral protein (ZAP, also called ZC3HAV1), a member of the Cysteine3 Histidine (CCCH)-type zinc finger domain-containing protein family, was originally identified as a cellular factor capable of inhibiting the replication of murine leukemia virus (8). ZAP inhibits the replication of not only murine leukemia virus but also various otherviruses, including Sindbis virus (SINV) (9, 10). ZAP senses RNA of target viruses through N-terminal CCCH-type zinc finger domains and functions as a PRR that induces degradation of targeted viral RNA by the exosome complex (11, 12). Recently, we generated Zc3hav1−/− mice and demonstrated that ZAP restricts the replication of SINV to protect hosts from lethal infection (10). In addition to ZAP, regnase-1, and ZFP36, members of the CCCH-type zinc finger domain-containing protein family are known to inhibit the replication of flaviviruses and HIV-1, respectively (13, 14). Hence, these family members are candidate intracellular PRRs that sense viral RNA and directly eliminate RNA viruses. However, the roles of the remaining family members in antiviral responses are poorly understood.
In this study, we identified tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly(ADP ribose) polymerase (TIPARP), a CCCH-type zinc finger domain-containing protein, as a viral RNA-sensing PRR and identified the critical role of mitochondria in TIPARP-mediated antiviral responses.
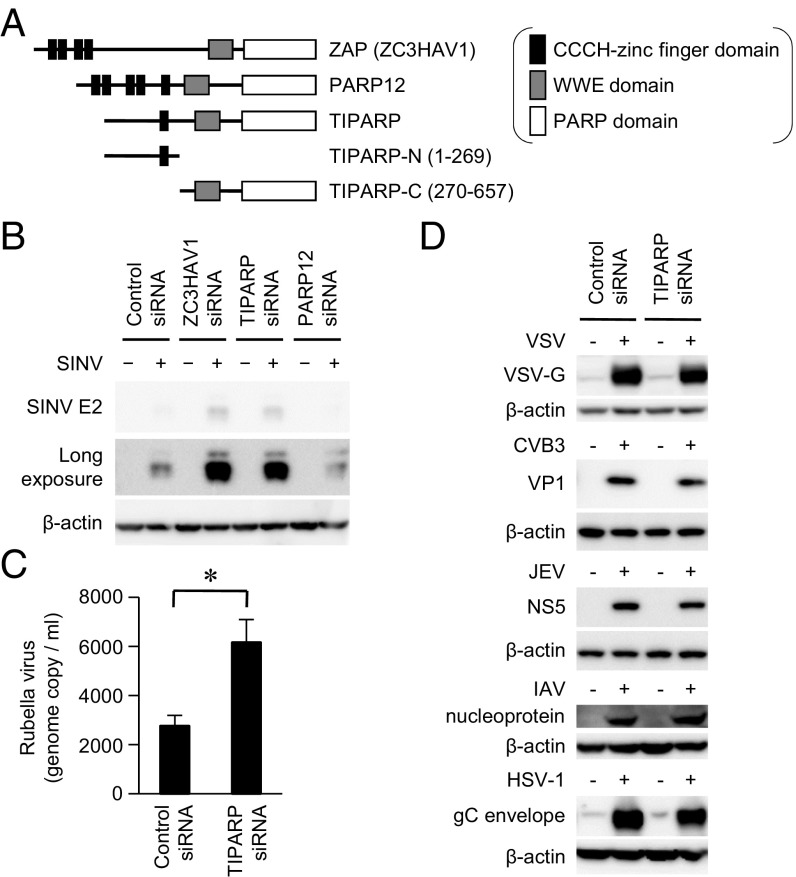
Results
TIPARP Mediates Host Defense Responses to Viruses in the Family Togaviridae.
We have studied PRRs that are involved in the antiviral response to SINV, a positive-sense single-stranded RNA virus, belonging to the genus Alphavirus in the family Togaviridae (10, 15). ZAP functions as a PRR that detects and eliminates SINV, but the function of ZAP-like proteins in an antiviral response is still incompletely understood. Therefore, we focused on the role of ZAP-like proteins in an antiviral response. ZAP and ZAP-like proteins TIPARP and PARP12 have CCCH-type zinc finger domain(s) at the N terminus (Fig. 1A). They also have PARP and tryptophan–tryptophan–glutamate (WWE) domains at the C terminus. Small interfering RNA (siRNA)-mediated knockdown of TIPARP mRNA, but not PARP12 mRNA, enhanced replication of SINV in a U373 human astrocyte cell line (Fig. 1B and Fig. S1A). These findings prompted us to investigate the involvement of TIPARP in host defenses against other RNA viruses. Knockdown of TIPARP mRNA also enhanced replication of rubella virus, which is also a member of the Togaviridae family, whereas knockdown of TIPARP mRNA failed to enhance replication of vesicular stomatitis virus (VSV), coxsackie virus B3 (CVB3), Japanese encephalitis virus (JEV), influenza A virus (IAV), and herpes simplex virus type 1 (HSV-1) (Fig. 1 C and D). Hence, TIPARP mediates antiviral responses to viruses in the family Togaviridae in a human astrocyte cell line.
Fig. 1.
TIPARP limits replication of viruses belonging to the family Togaviridae. (A) Domain architecture of ZAP, PARP12, and TIPARP. (B) U373-CD14 cells were treated with the indicated siRNAs and then infected with SINV (multiplicity of infection [MOI] = 1) for 24 h. The levels of viral protein and β-actin in the cell lysates were determined by immunoblotting analysis. (C and D) U373-CD14 cells were treated with control or TIPARP siRNA. (C) The cells were infected with rubella virus (MOI = 0.1) for 96 h. The levels of rubella virus RNA in the culture supernatants were measured by quantitative reverse transcription-PCR. (D) The cells were infected with VSV (MOI = 1), CVB3 (MOI = 1), JEV (MOI = 1), IAV (PR8, 100 hemagglutinin unit), or HSV-1 (MOI = 1) for 24 h. The levels of viral proteins and β-actin in the cell lysates were determined by immunoblotting analysis. Experiments were performed three times, and representative data are shown (means ± SD of three independent samples). *P < 0.05.
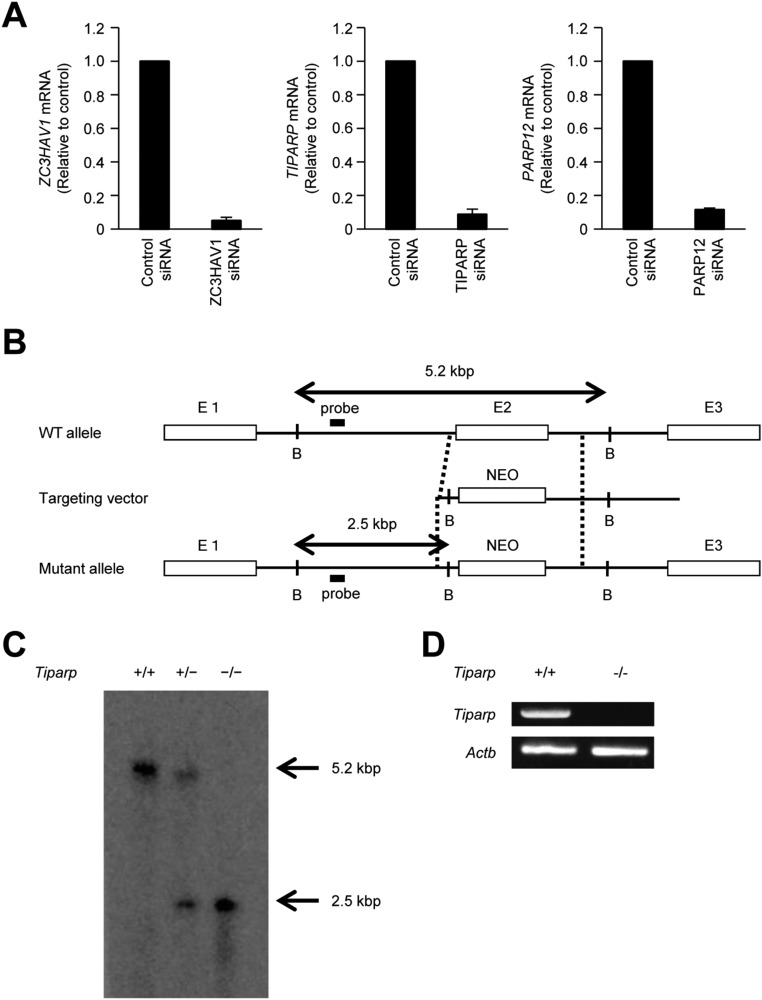
Fig. S1.
Generation of Tiparp−/− mice. (A) U373-CD14 cells were transiently transfected with the indicated siRNA for 48 h. The levels of ZC3HAV1, TIPARP, and PARP12 mRNA were measured by quantitative reverse transcription-PCR. (B) Schematic representation of Tiparp gene targeting. The targeting vector was constructed by replacing exon 2 of Tiparp with a neomycin resistance gene. E, exon; NEO, phosphoglycerate kinase promoter-driven neomycin-resistance gene cassette; B, BamH1. (C) Southern blot analysis of genomic DNA from Tiparp+/+, Tiparp+/−, and Tiparp−/− mice. Genomic DNA was separated by electrophoresis after digestion with BamH1 and hybridized with the radiolabeled probe shown in B. (D) Total RNAs from Tiparp+/+ and Tiparp−/− MEFs were subjected to RT-PCR analysis of the expression of Tiparp and Actb mRNA.
TIPARP Protects Mice from Lethal SINV Infection.
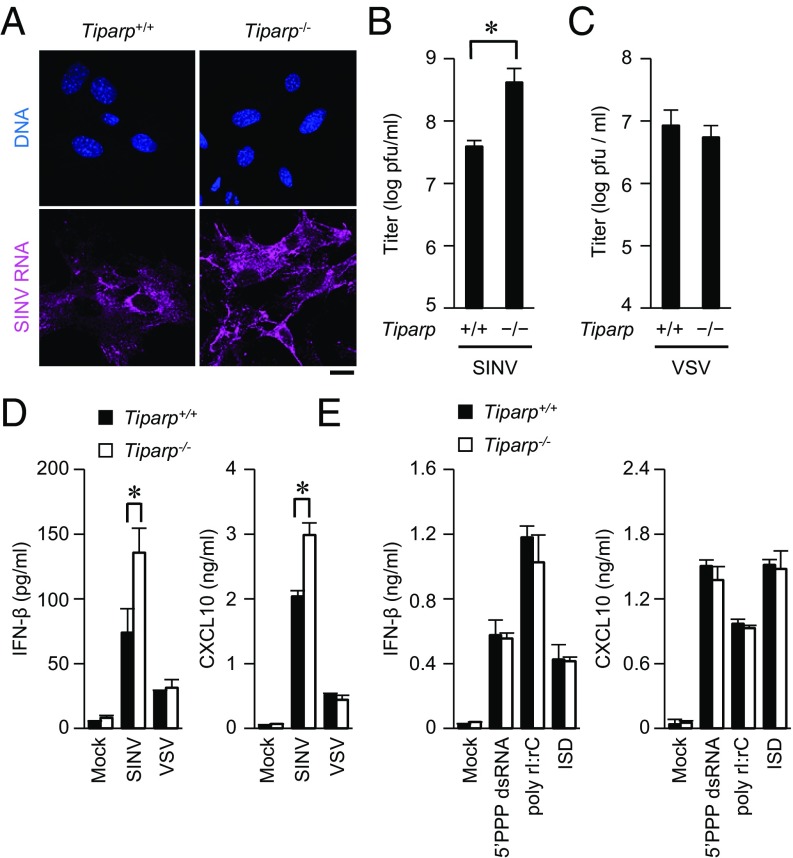
To investigate the pathophysiological role of TIPARP in an innate immune response, we generated Tiparp−/− mice by replacing exon 2 of mouse Tiparp with a neomycin resistance gene (Fig. S1 B and C). Abrogation of Tiparp mRNA expression in mouse embryonic fibroblasts (MEFs) was confirmed by reverse transcription-PCR (Fig. S1D). The number of T cells, B cells, neutrophils, macrophages, and dendritic cells were normal in the spleens of Tiparp−/− mice (Fig. S2A). Using Tiparp−/− mice, we examined the role of TIPARP in an antiviral response. The replication of SINV was significantly enhanced in primary Tiparp−/− MEFs (Fig. 2 A and B). However, the replication of VSV was normal in primary Tiparp−/− MEFs (Fig. 2C). These findings suggested that TIPARP specifically regulates the replication of SINV in MEFs.
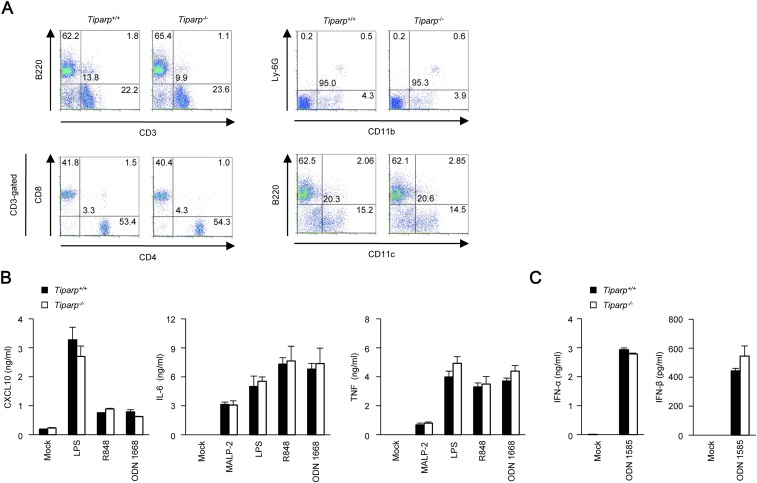
Fig. S2.
Tiparp−/− dendritic cells respond normally to TLR ligands. (A) Flow cytometric analysis of splenoctyes from Tiparp+/+ and Tiparp−/− mice. Cell surface expression of the indicated maker proteins was measured. (B) ELISA of CXCL10, IL-6, and TNF in culture supernatants from primary Tiparp+/+ and Tiparp−/− granulocyte-macrophage colony-stimulating factor-induced bone marrow-derived dendritic cells stimulated with macrophage-activating lipopeptide-2 (MALP-2) (TLR2 ligand, 10 ng/mL), lipopolysaccharide (LPS) (TLR4 ligand, 100 ng/mL), R848 (TLR7 ligand, 10 μg/mL), or ODN 1668 (TLR9 ligand, 1 μM) for 24 h. (C) ELISA of IFN-α and IFN-β in culture supernatants from primary Tiparp+/+ and Tiparp−/− FMS-like tyrosine kinase 3 ligand-induced bone marrow-derived dendritic cells stimulated with ODN 1585 (TLR9 ligand, 5 μM) for 24 h. Experiments were performed three times, and representative data are shown (means ± SD of three independent samples).
Fig. 2.
Loss of TIPARP enhances SINV replication. (A) Primary Tiparp+/+ and Tiparp−/− MEFs were infected with SINV (MOI = 1) for 12 h. Fixed samples were subjected to an RNA fluorescence in situ hybridization analysis of SINV RNA and to Hoechst 33342 staining of genomic DNA. (B and C) Primary Tiparp+/+ and Tiparp−/− MEFs were infected with SINV (MOI = 0.1) (B) or VSV (MOI = 0.1) (C) for 24 h. The viral titers in culture supernatants were determined by 50% tissue culture infectious dose assay. (D and E) ELISA of IFN-β and CXCL10 in culture supernatants of primary Tiparp+/+ and Tiparp−/− MEFs. (D) Cells were infected with SINV (MOI = 1) or VSV (MOI = 1) for 24 h. (E) Cells were stimulated with 5′PPP dsRNA (1 μg/mL), poly rI:rC (1 μg/mL), and IFN stimulatory DNA (ISD) (1 μg/mL), together with Lipofectamine 2000, for 24 h. (Scale bar, 20 μm.) Experiments were performed at least three times, and representative data are shown (means ± SD of three independent samples). *P < 0.05.
Type I IFN plays an important role in the induction of an antiviral response (3). However, SINV-induced production of IFN-β and C-X-C motif chemokine ligand 10 (CXCL10) was enhanced in primary Tiparp−/− MEFs compared with in Tiparp+/+ MEFs (Fig. 2D). In contrast, VSV-induced production of IFN-β and CXCL10 was normal in primary Tiparp−/− MEFs. Production of IFN-β and CXCL10, induced by synthetic nucleic acids such as 5′PPP dsRNA, polyinosinic–polycytidylic acid (poly rI:rC), and IFN stimulatory DNA, was normal in primary Tiparp−/− MEFs (Fig. 2E). Furthermore, production of CXCL10, interleukin (IL)-6, and tumor necrosis factor (TNF), induced by TLR ligands such as macrophage-activating lipopeptide-2, lipopolysaccharide, R848, and ODN1668, was normal in primary Tiparp−/− conventional dendritic cells (Fig. S2B). Production of IFN-α and IFN-β induced by ODN1585 was normal in primary Tiparp−/− plasmacytoid dendritic cells (Fig. S2C). These findings indicate that TIPARP does not regulate RLR-, cGAS-, or TLR-mediated production of IFNs, cytokines, or chemokine.
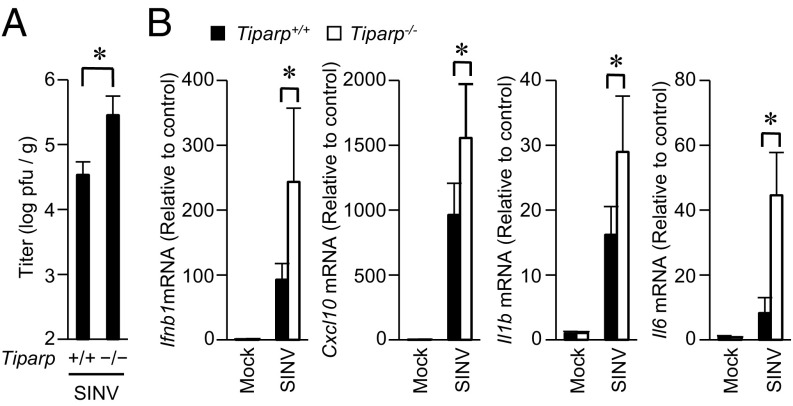
SINV infection causes encephalomyelitis in infant mice and diverse diseases that include fever, rash, and arthritis in humans (10, 16). To reveal the role of TIPARP in antiviral responses in vivo, we challenged infant Tiparp−/− mice with SINV. SINV efficiently replicated in the brains of infant Tiparp−/− mice compared with in Tiparp+/+ mice (Fig. 3A). The expression of Ifnb1 and Cxcl10 mRNA was up-regulated by SINV infection in the brains of infant Tiparp−/− mice relative to that in Tiparp+/+ mice (Fig. 3B). Furthermore, the expression of Il1b and Il6 mRNA, markers of encephalomyelitis, was also up-regulated by SINV infection in the brains of infant Tiparp−/− mice relative to that in Tiparp+/+ mice. These findings indicate that TIPARP protects infant mice from SINV infection.
Fig. 3.
TIPARP deficiency renders mice susceptible to SINV infection. (A and B) Ten-day-old Tiparp+/+ and Tiparp−/− mice (n = 5 each) were s.c. inoculated with SINV (100 plaque-forming units per mouse). At day 5 postinfection, the brains of SINV-infected Tiparp+/+ and Tiparp−/− mice were isolated. (A) The viral titers (plaque-forming units per tissue weight) in brains were determined by 50% tissue culture infectious dose. (B) The levels of Ifnb1, Cxcl10, Il1b, and Il6 mRNA in brains were measured by quantitative reverse transcription-PCR. Experiments were performed five times, and representative data are shown (means ± SD of five independent samples). *P < 0.05.
CCCH-Type Zinc Finger Domain Is Required for the Antiviral Action of TIPARP.
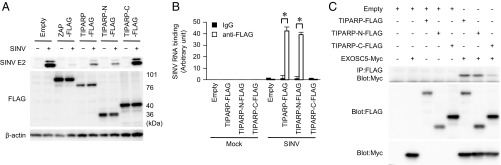
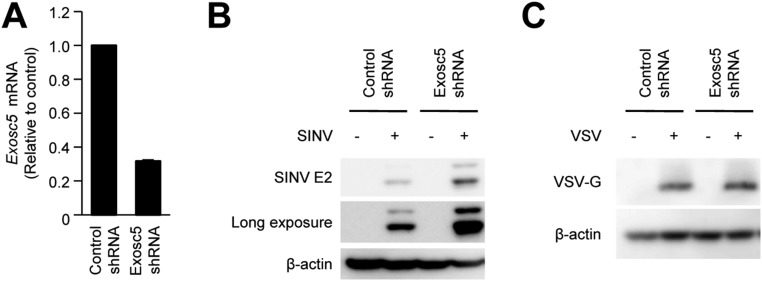
To reveal the mechanism underlying the TIPARP-dependent antiviral response to SINV, we determined the domain structure of TIPARP. Similar to ZAP, TIPARP required the N-terminal CCCH-type zinc finger domain for its antiviral action (Fig. 4A). The N-terminal CCCH-type zinc finger domain of TIPARP bound to SINV RNA and EXOSC5, a component of the exosome complex that is involved in the antiviral response to SINV (Fig. 4 B and C and Fig. S3 A–C). These findings indicate that TIPARP detects SINV RNA via its N-terminal portion and induces RNA degradation by recruiting the exosome complex.
Fig. 4.
TIPARP binds to SINV RNA and induces SINV RNA degradation. (A) Wild-type MEFs stably expressing the indicated FLAG-tagged proteins were infected with SINV (MOI = 0.1) for 24 h. The levels of viral E2 protein, FLAG-tagged protein, and β-actin in cell lysates were determined by immunoblotting analysis. (B) Wild-type MEFs stably expressing the indicated expression plasmids were infected with SINV for 24 h. The levels of SINV RNA binding to the indicated FLAG-tagged proteins were measured by RNA immunoprecipitation, coupled with quantitative reverse transcription-PCR. (C) U373-CD14 cells were transiently transfected with the indicated expression plasmids. Cell lysates were subjected to immunoprecipitation with anti-FLAG antibody and to immunoblot analysis with anti-FLAG and anti-Myc antibodies. The experiments were performed three times, and representative data are shown (means ± SD of three independent samples). *P < 0.05.
Fig. S3.
TIPARP recruits EXOSC5 to degrade Sindbis virus (SINV) RNA. (A) Wild-type MEFs were stably expressed with control shRNA or Exosc5 shRNA. The levels of Exosc5 mRNA expression were measured by quantitative reverse transcription-PCR. (B and C) Wild-type MEFs stably expressing control shRNA or Exosc5 shRNA were infected with SINV (MOI = 0.05) (B) or VSV (MOI = 0.05) (C) for 24 h. Levels of viral protein and β-actin in cell lysates were measured by immunoblotting. Experiments were performed three times, and representative data are shown (means ± SD of three independent samples). *P < 0.05.
SINV Induces Cytoplasmic Accumulation of TIPARP.
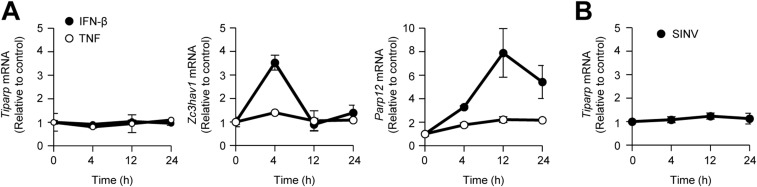
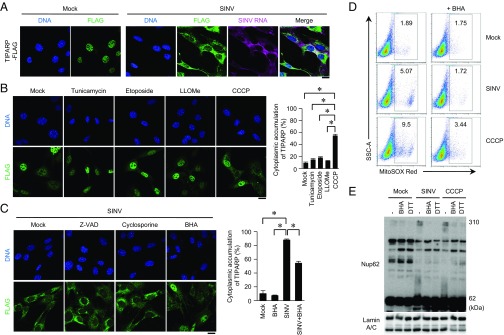
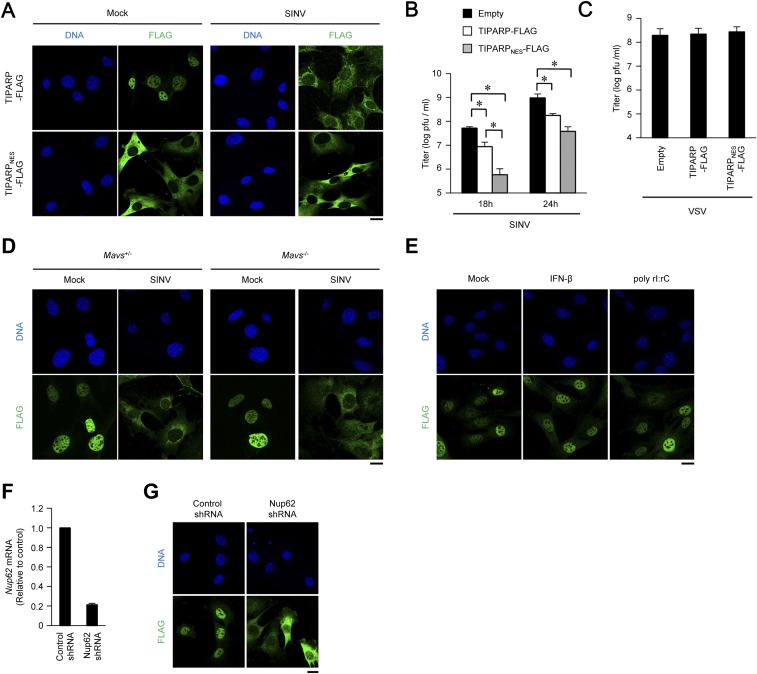
Type I IFNs are known to induce the expression of a set of antiviral proteins. Although IFN-β induced expression of Zc3hav1 and Parp12 mRNA, it failed to induce expression of Tiparp mRNA (Fig. S4A). TNF, an inflammatory cytokine, and SINV also failed to induce the expression of Tiparp mRNA (Fig. S4 A and B). Therefore, we focused on the subcellular localization of TIPARP. Consistent with a previous observation (17), TIPARP localized in the nuclei of uninfected cells (Fig. 5A). However, TIPARP localized in the cytoplasm on SINV infection (Fig. 5A). Enforced expression of TIPARP fused with a nuclear export signal induced enhanced elimination of SINV, but not VSV (Fig. S5 A–C). These findings indicate that cytoplasmic localization enables TIPARP to detect the RNA of replicating SINV.
Fig. S4.
Neither SINV nor IFN-β induces expression of Tiparp mRNA. (A) Wild-type MEFs were stimulated with IFN-β (10 ng/mL) or TNF (10 ng/mL) for the indicated durations. Levels of Tiparp, Zc3hav1, and Parp12 mRNA expression were measured by quantitative reverse transcription-PCR. (B) Wild-type MEFs were infected with SINV (MOI = 1) for the indicated durations. Levels of Tiparp mRNA expression were measured by quantitative reverse transcription-PCR. Experiments were performed three times, and representative data are shown (means ± SD of three independent samples).
Fig. 5.
Cytoplasmic accumulation of TIPARP promotes elimination of SINV. (A) Wild-type MEFs stably expressing TIPARP-FLAG were infected with SINV (MOI = 5) for 12 h. Fixed samples were subjected to RNA fluorescence in situ hybridization analysis of SINV RNA, immunocytochemistry analysis of TIPARP-FLAG, and Hoechst 33342 staining of genomic DNA. (B and C) Wild-type MEFs stably expressing TIPARP-FLAG were treated with tunicamycin (5 μg/mL), etoposide (10 μM), Leu-Leu methyl ester hydrobromide (LLOMe) (100 μM), or CCCP (10 μM) for 6 h (B). Wild-type MEFs stably expressing TIPARP-FLAG were infected with SINV (MOI = 5), together with Z-VAD (10 μM), cyclosporin A (5 μM), or BHA (5 μM) for 12 h (C). Fixed samples were subjected to immunocytochemistry analysis of FLAG-tagged protein and Hoechst 33342 staining of genomic DNA. Frequencies of MEFs with cytoplasmic accumulation of TIPARP were determined. (D) Wild-type MEFs were infected with SINV (MOI = 5) or stimulated with CCCP (10 μM), with or without BHA (5 μM), for 12 h, and then stained with MitoSOX Red. Samples were subjected to flow cytometric analysis to measure the level of mitochondrial reactive oxygen species. SSC-A, side scatter area. (E) U373-CD14 cells were infected with SINV (MOI = 5) or stimulated with CCCP (50 μM) in the presence or absence of BHA (5 μM) for 24 h. Cellular extracts were treated with or without 100 mM DTT. Samples were suspended in 2-mercaptoethanol-free loading buffer and were subjected to immunoblot analysis of Nup62 and lamin A/C. (Scale bars, 20 μm.) The experiments were performed three times, and representative data are shown (means ± SD of three independent samples). *P < 0.05.
Fig. S5.
Cytoplasmic accumulation enhances anti-SINV activity of TIPARP. (A) Wild-type MEFs stably expressing TIPARP-FLAG and TIPARPNES-FLAG tagged protein were infected with SINV (MOI = 5) for 12 h. Fixed samples were subjected to immunocytochemistry analysis of FLAG-tagged protein and Hoechst 33342 staining of genomic DNA. (B and C) Wild-type MEFs stably expressing TIPARP-FLAG and TIPARPNES-FLAG-tagged protein were infected with SINV (MOI = 0.1) for the indicated durations (B) or VSV (MOI = 0.1) for 24 h (C). Viral titers in culture supernatants were determined by 50% tissue culture infectious dose assay. (D) Primary Mavs+/− or Mavs−/− MEFs stably expressing TIPARP-FLAG were infected with SINV (MOI = 5) for 12 h. Fixed samples were subjected to immunocytochemistry analysis of FLAG-tagged protein and Hoechst 33342 staining of genomic DNA. (E) Wild-type MEFs stably expressing TIPARP-FLAG were stimulated with IFN-β (10 ng/mL) or poly rI:rC (1 μg/mL) for 12 h. Fixed samples were subjected to immunocytochemistry analysis of FLAG-tagged protein and Hoechst 33342 staining of genomic DNA. (F) Wild-type MEFs were stably expressed with Nup62-specific and control shRNA. The levels of Nup62 mRNA were measured by quantitative reverse transcription-PCR. (G) Wild-type MEFs stably expressing TIPARP-FLAG with Nup62-specific shRNA or control shRNA. Fixed samples were subjected to immunocytochemistry analysis of TIPARP-FLAG and Hoechst 33342 staining of genomic DNA. (Scale bars, 20 μm.) Experiments were performed three times, and representative data are shown (means ± SD of three independent samples). *P < 0.05.
The RLR-IPS-1 signaling axis was dispensable for SINV-induced cytoplasmic accumulation of TIPARP (Fig. S5D). Neither dsRNA nor IFN-β induced TIPARP redistribution (Fig. S5E). Therefore, we examined whether organelle dysfunction, which often induces host defense responses to invading pathogens, is involved in TIPARP redistribution. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP, a mitochondria damage inducer) caused rapid TIPARP redistribution, whereas tunicamycin (an endoplasmic reticulum stress inducer), etoposide (a DNA damage inducer), and Leu-Leu methyl ester hydrobromide (a lysosome damage inducer) failed to do so at early times after treatment (Fig. 5B). These findings indicate that mitochondrial damage triggers TIPARP redistribution.
Mitochondrial Reactive Oxygen Species Trigger Cytoplasmic Accumulation of TIPARP.
Mitochondrial damage causes various cellular events such as reactive oxygen species (ROS) production and caspase-9 and voltage-dependent anion channel activation, and is often involved in innate immune responses (18). Butylated hydroxyanisole (BHA, a ROS scavenger), but not Z-VAD-fmk (a caspase inhibitor) or cyclosporin A (cyclophilin D inhibitor), partially inhibited the cytoplasmic localization of TIPARP induced by SINV (Fig. 5C). Hence, ROS generation was one cause for TIPARP redistribution induced by SINV. Consistently, both SINV and CCCP induced ROS generation by mitochondria (Fig. 5D).
These findings prompted us to examine the molecular mechanism of TIPARP redistribution by ROS. Oxidant stress is known to induce oxidization of the nucleoporin complex to affect nuclear-cytoplasmic cargo transport (19). TIPARP localized in the cytoplasms of cells lacking nucleoporin 62 (Nup62), a component of the nucleoporin complex (Fig. S5 F and G). SINV and CCCP induced oxidization of Nup62, which was prevented by the reducing agents BHA and DTT (Fig. 5E). Therefore, ROS produced by damaged mitochondria affect the function of the nucleoporin complex by oxidization, resulting in cytoplasmic accumulation of TIPARP.
BCL2 Family Members Regulate TIPARP Localization.
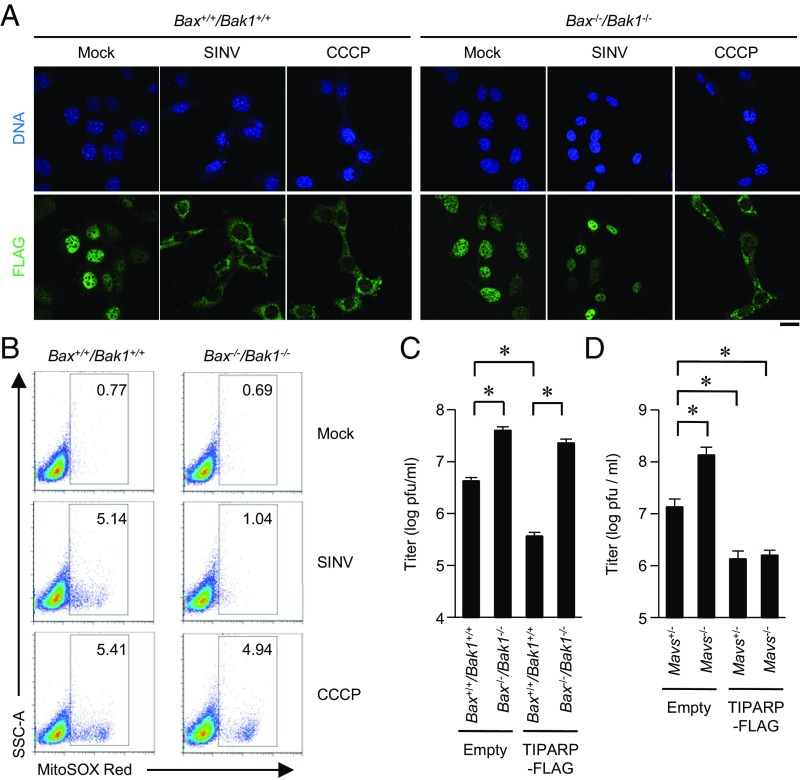
BCL2 family members such as BAX and BAK1 are regulators of mitochondrial damage. In Bax−/−/Bak1−/− MEFs, SINV failed to induce cytoplasmic accumulation of TIPARP (Fig. 6A). Consistently, in Bax−/−/Bak1−/− MEFs, SINV failed to induce ROS production by mitochondria (Fig. 6B). However, CCCP, a direct inhibitor of mitochondrial membrane potential, induced cytoplasmic accumulation of TIPARP and ROS production by mitochondria in Bax−/−/Bak1−/− MEFs. SINV replication was significantly enhanced in Bax−/−/Bak1−/− MEFs compared with that in Bax+/+/Bak1+/+ MEFs (Fig. 6C). Furthermore, enforced expression of TIPARP hardly inhibited the replication of SINV in Bax−/−/Bak1−/− MEFs (Fig. 6C). Enforced expression of TIPARP inhibited the replication of SINV in both Mavs+/− MEFs and in Mavs−/− MEFs (Fig. 6D). These findings indicate that, on SINV infection, BAX and BAK1 cause mitochondrial damage that induces a TIPARP-mediated antiviral response independent of the RLR-IPS-1 signaling axis.
Fig. 6.
BAX- and BAK1-dependent generation of mitochondrial reactive oxygen species causes cytoplasmic accumulation of TIPARP to limit SINV replication. (A) Bax+/+/Bak1+/+ and Bax−/−/Bak1−/− MEFs stably expressing TIPARP-FLAG were infected with SINV (MOI = 5) or stimulated with CCCP (10 μM) for 12 h. Fixed samples were subjected to immunocytochemistry analysis of TIPARP-FLAG and Hoechst 33342 staining of genomic DNA. (B) Bax+/+/Bak1+/+ and Bax−/−/Bak1−/− MEFs were infected with SINV (MOI = 5) or stimulated with CCCP (10 μM) for 12 h, and then stained with MitoSOX Red. Samples were subjected to flow cytometric analysis to measure levels of mitochondrial reactive oxygen species. (C and D) Bax+/+/Bak1+/+ and Bax−/−/Bak1−/− MEFs (C) or primary Mavs+/− and Mavs−/− MEFs (D) stably expressing TIPARP-FLAG were then infected with SINV (MOI = 5) for 24 h. Viral titers in culture supernatants were determined by 50% tissue culture infectious dose assay. (Scale bar, 20 μm.) Experiments were performed three times, and representative data are shown (means ± SD of three independent samples). *P < 0.05.
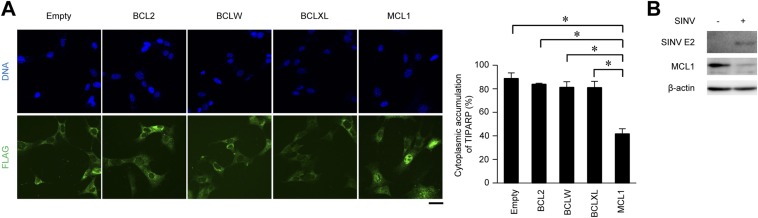
We next examined the involvement of anti-apoptotic BCL2 family members in the regulation of TIPARP localization. Enforced expression of myeloid cell leukemia sequence 1 (MCL1), but not of BCL2, BCLW, or BCLXL, disrupted cytoplasmic accumulation of TIPARP (Fig. S6A). Interestingly, the expression level of MCL1 was decreased by SINV infection (Fig. S6B). These findings suggest that a reduction in MCL1 promotes BAX- and BAK1-dependent mitochondrial damage, leading to cytoplasmic accumulation of TIPARP, which in turn induces an antiviral response.
Fig. S6.
MCL1 limits cytoplasmic accumulation of TIPARP. (A) Wild-type MEFs stably expressing TIPARP-FLAG with BCL2, BCLW, BCLXL, or MCL1 protein were infected with SINV (MOI = 5) for 12 h. Fixed samples were subjected to immunocytochemistry analysis of FLAG-tagged protein and Hoechst 33342 staining of genomic DNA. Frequencies of MEFs with cytoplasmic accumulation of TIPARP were determined. (B) Wild-type MEFs were infected with SINV (MOI = 0.5) for 12 h. Levels of viral protein, MCL1 protein, and β-actin in cell lysates were measured by immunoblotting analysis. (Scale bar, 20 μm.) Experiments were performed three times, and representative data are shown (means ± SD of three independent samples). *P < 0.05.
Discussion
We identified TIPARP as a viral RNA-sensing PRR that induces a host defense response to SINV and rubella virus. However, TIPARP failed to inhibit replication of VSV, CVB3, JEV, and IAV, suggesting that TIPARP senses RNA from specific RNA viruses for degradation. At present, the molecular mechanism underlying selective viral RNA recognition by TIPARP is unclear. Additional studies are needed to determine the sequence or secondary structure of viral RNA that is sensed by the CCCH-type zinc finger domain of TIPARP and to understand the target specificity of TIPARP.
Recent studies have shown that organelle stress as well as type I IFNs are driving forces in the induction of antiviral responses. Organelle stress acts as an alarm for viral infection to stimulate an innate immune response, leading to the establishment of an antiviral state. The NOD-like receptor (NLR) family pyrin domain containing 3-inflammasome, a component of the innate immune system, responds to damage of various organelles and induces an antiviral immune response by promoting the production of inflammatory cytokines (20, 21). We found that TIPARP is a unique PRR whose activation is regulated by mitochondrial damage and induces virus RNA degradation. Therefore, organelle stress regulates not only the production of immune modulatory factors but also direct virus elimination. It will be interesting to examine whether there are other components of the innate immune system that are activated by organelle stress, because TIPARP inhibits replication of RNA viruses within one family.
The CCCH-type zinc finger functions as an RNA-binding domain, and it is conserved from yeasts to mammals. To date, more than 50 members of the CCCH-type zinc finger domain-containing protein family have been identified and are known to regulate a variety of cellular events such as RNA degradation and protein translation (22). A number of studies have shown that TIPARP, ZAP, ZFP36, and regnase-1 regulate antiviral responses, suggesting these family members are representative viral RNA-sensing PRRs and effectors. However, the roles of the majority of these family members in antiviral responses are still unclear. Furthermore, crosstalk and synergistic effects between these family members have not been examined. In future studies, we will address these points for a comprehensive understanding of the role of these family members in host defenses against RNA viruses.
Materials and Methods
Mice and Cells.
Mice were maintained in our animal facility and treated in accordance with the guidelines of Osaka University. Research involving mice was approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University. Bax−/−Bak1−/− and Mavs−/− MEFs have been described previously (23, 24). Primary MEFs were prepared from pregnant female mice on embryonic day 13.5, as described previously (25). Spontaneously immortalized wild-type MEFs and Vero cells were described previously (26). U373 astrocytoma cells expressing CD14 were kindly donated by T. Fujita (Kyoto University). MEFs, U373-CD14 cells, and Vero cells were maintained in DMEM supplemented with 10% (vol/vol) FBS, penicillin (100 U/mL), and streptomycin (100 U/mL).
Reagents and Plasmids.
Anti-SINV E2 antibody was originally prepared by D. E. Griffin (Johns Hopkins University School of Medicine) and was obtained from Y. Yoshinaka (Tokyo Medical and Dental University). Anti-JEV core rabbit polyclonal antibody was prepared as described previously (27). The retroviral expression vector pMRX-ires-puro was kindly donated by S. Yamaoka (Tokyo Medical and Dental University). The expression vectors pcDNA3.1(+), pEGFP-N1, and pSUPER-retro-puro were described previously (26). Details are given in SI Materials and Methods.
Viruses.
SINV, VSV, JEV, and IAV (A/Puerto Rico/8/34, H1N1 strain) have been described previously (27, 28). HSV-1 was kindly donated by Y. Kawaguchi (University of Tokyo). Rubella virus (genotype 2B) was isolated from a throat swab of a rubella patient, using Vero cells. Human CVB3 was purchased from American Type Culture Collection. In a mouse model of SINV infection, 10-d-old pups were s.c. inoculated with 100 plaque-forming units SINV. Viral titers were determined with a 50% tissue culture infectious dose assay (26).
RNA Immunoprecipitation Assay.
Interaction between SINV RNA and TIPARP were detected using the Ribocluster Profiler/RIP-Assay kit (MBL), according to the manufacturer’s instructions.
Immunocytochemistry and RNA Fluorescence in Situ Hybridization.
Cells were fixed with paraformaldehyde. Immunocytochemistry was performed as described previously (25). Fluorescence in situ hybridization was performed using a QuantiGene ViewRNA ISH Cell Assay Kit (Veritas) according to the manufacturer’s instructions. A Cy5-labeled fluorescence in situ hybridization probe was designed to hybridize specifically to the region between nucleotide 6041 and nucleotide 6898 of SINV transcripts. The samples were examined under an LSM780 confocal laser-scanning microscope (Carl Zeiss) and a DMI6000B fluorescence microscope (Leica Microsystems).
Immunoblotting.
Total cell lysates were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with the indicated antibodies, as described previously (25).
Coimmunoprecipitation.
U373-CD14 cells were lysed in lysis buffer (150 mM NaCl, 20 mM Tris at pH 7.6, 1% TritonX-100, and a protease inhibitor [Roche]) on ice, and the lysates were clarified by centrifugation for 15 min at 4 °C at 12,000 rpm in a microcentrifuge (Centrifuge 5424R, Eppendorf). The supernatants were incubated with anti-FLAG antibody for 1 h at 4 °C, and protein G-Sepharose 4B Fast Flow beads (GE Healthcare) were added. After incubation for 2 h at 4 °C, the resins were washed three times with lysis buffer, and the bound proteins were detected by immunoblotting.
Flow Cytometric Analysis.
MEFs infected with SINV or stimulated with various ligands were incubated with MitoSOX Red (1 μM) for 60 min at 37 °C and washed three times. Fluorescent signals were detected by a FACS Canto II, and data were analyzed with FlowJo software.
Statistical Analyses.
The statistical significance of differences between two groups was determined by Student’s t test. The statistical significance of difference between more than two groups was determined by a one-way analysis of variance test, followed by a Bonferroni correction. A P value of < 0.05 was considered significant.
SI Materials and Methods
Mice and Cells.
The Tiparp gene was isolated from genomic DNA extracted from 129/sv-background wild-type embryonic stem cells by PCR. A targeting vector was constructed by replacing exon 2, including the zinc finger domain, with a neomycin resistance gene cassette and herpes simplex virus thymidine kinase driven by the phosphoglycerate kinase promoter. After the targeting vector was transfected into embryonic stem cells, G418 and gancyclovir doubly resistant colonies were selected and screened by PCR and further confirmed by Southern blotting. Homologous recombinants were microinjected into C57BL/6 female mice, and heterozygous F1 progenies were intercrossed to obtain Tiparp+/− mice. Tiparp+/− mice were then backcrossed to C57BL/6 mice for more than six generations before their use in this study. C57BL/6 mice were purchased from CLEA Japan, Inc. To prepare primary dendritic cells, mouse bone marrow cells were treated for 6 d with granulocyte macrophage colony-stimulating factor (10 ng/mL) or FMS-like tyrosine kinase 3 ligand (20 ng/mL) in RPMI medium 1640 supplemented with 10% (vol/vol) FBS, 50 mM 2-mercaptoethanol, penicillin (100 U/mL), and streptomycin (100 U/mL).
Reagents.
High-molecular-weight poly rI:rC, 5′PPP dsRNA, IFN stimulatory DNA, lipopolysaccharide, R848, ODN 1585, and ODN 1668 were purchased from InvivoGen. Recombinant mouse IFN-β was purchased from Prospec. Macrophage-activating lipopeptide-2 was purchased from Alexis. Cyclosporin A was purchased from Enzo Life Science. Tunicamycin was purchased from Nakalai Tesque. Z-VAD-fmk was purchased from Peptide Institute. Carbonyl cyanide 3-chlorophenylhydrazone, Leu-Leu methyl ester hydrobromide, etoposide, butylated hydroxyanisole, and mouse anti-FLAG antibody (F3165) for immunoblotting were purchased from Sigma. Rabbit anti-DYKDDDDK tag antibody (2368) for immunocytochemical staining and mouse IgG1 isotype control antibody (5415S) were purchased from Cell Signaling Technology. Anti-VSV-G (A190-131A) antibody was purchased from Bethyl Laboratory, anti-HSV1 (gC) antibody (11-332-C100) was purchased from Exbio, and anti-enterovirus antibody (M7064) was purchased from Dako. Anti-actin-HRP antibody (sc-1615 HRP) was purchased from Santa Cruz Biotechnology. Anti-IAV nucleoprotein antibody (ab20343) and anti-Mcl-1 antibody (ab32087) were purchased from Abcam. FITC anti-mouse CD3 antibody (100204) and APC anti-mouse/human CD45R/B220 antibody (103212) were purchased from BioLegend. PE Rat anti-mouse CD4 antibody (5537730), PE rat anti-mouse CD11b antibody (553311), PerCP-Cy5.5 rat anti-mouse Ly6G antibody (560602), APC-Cy7 hamster anti-mouse CD11c antibody (561241), and APC-rat anti-mouse CD8a antibody (561093) were purchased from BD Bioscience. The ELISA kits for IFN-α and IFN-β were purchased from PBL IFN Source. Recombinant mouse TNF and the ELISA kits to detect CXCL10, IL-6, and TNF were purchased from R&D Systems. MitoSOX Red, Hoechst 33342, Alexa dye-labeled secondary antibodies, and Lipofectamine 2000 were purchased from Invitrogen.
Plasmids.
The cDNA fragments encoding ZAP, TIPARP, TIPARP-N, and TIPARP-C were cloned into pMRX-FLAG-ires-puro, generating pMRX-ZAP-FLAG-ires-puro, pMRX-TIPARP-FLAG-ires-puro, pMRX-TIPARP-N-FLAG-ires-puro, and pMRX-TIPARP-C-FLAG-ires-puro, respectively. The cDNA fragment encoding the nuclear export signal (NES) was cloned into pMRX-TIPARP-FLAG-ires-puro, generating pMRX-TIPARP-NES-FLAG-ires-puro. The cDNA fragments were also cloned into pcDNA3-FLAG, generating pcDNA3-ZAP-FLAG, pcDNA3-TIPARP-FLAG, pcDNA3-TIPARP-N-FLAG, and pcDNA3-TIPARP-C-FLAG, respectively. The cDNA fragment encoding EXOSC5 was cloned into pMRX-Myc-ires-puro and pcDNA3-Myc, generating pMRX-EXOSC5-Myc-ires-puro and pcDNA3-EXOSC5-Myc, respectively. pCDH-CMV-MCS-EF1-puro was purchased from System Bioscience. The cDNA fragments encoding BCLW and MCL1 were cloned into pCDH-CMV-MCS-EF1-puro, generating pCDH-puro-BCLW and pCDH-puro-MCL1, respectively. pCDH-puro-Bcl-2 (#46971), pCDH-puro-Bcl-XL (#46972), pCMV-ΔR8.2 (#12263), and pMD2.G (#12259) were obtained from Addgene.
Viral Vectors.
Plat-E packaging cells were described previously (29) and kindly donated by T. Kitamura (University of Tokyo, Japan). 293T cells were described previously (30). Plat-E cells and 293T cells were maintained in DMEM supplemented with 10% (vol/vol) FBS, penicillin (100 U/mL), and streptomycin (100 U/mL). A retroviral vector was produced by Plat-E cells transfected with pMRX-ires-puro or pSUPER-retro-puro, as described previously (26). A lentiviral vector was produced by 293T cells transfected with pCDH-puro, pCMV-ΔR8.2, and pMD2.G, as described previously (30).
RNA-Mediated Interference.
U373-CD14 cells were transiently transfected with the indicated siRNAs (Ambion) using Lipofectamine RNAiMAX (Invitrogen). For stable knockdown of the target genes, short hairpin RNAs were expressed by retroviral vectors. The cDNA sequences inserted downstream of the H1 promoter of pSUPER-retro-puro were as follows (only the sense strand sequence is shown): Exosc5, 5′-GCAAAGAAATCTTCAACAA-3′; Nup62, 5′-GGAGCAAAGTGGCACCATA-3′. The unrelated control was as described previously (26).
Quantitative Reverse Transcription PCR.
Viral RNA was isolated from culture supernatants using a ZR Viral RNA kit (Zymo Research). Total RNA was isolated from cultured cells using a ZR RNA MicroPrep kit (Zymo Research). Total RNA was isolated from mouse brain tissue using an RNeasy Lipid Tissue Mini Kit (Qiagen). Reverse transcription (RT) was performed with ReverTra Ace reverse transcriptase (Toyobo). For the quantitative PCR, cDNA fragments were amplified from the RT products with Real-time PCR Master Mix (Toyobo). Fluorescence from the TaqMan probe was detected with a StepOnePlus Real-Time PCR System (Applied Biosystems). The levels of TIPARP, ZC3HAV1, and PARP12 mRNAs were normalized to that of the 18S rRNA. The levels of Tiparp, Zc3hav1, Parp12, Ifnb1, Cxcl10, Il1b, and Il6 mRNAs were normalized to that of Actb mRNA. The assays were performed in accordance with the manufacturer’s instructions.
Flow Cytometric Analysis.
Cell suspensions from spleens were prepared by sieving and gentle pipetting. Before immunostaining, Fc receptors were blocked for 10 min at 4 °C. Cell surface markers were stained with anti-CD3, anti-B220, anti-CD4, anti-CD8, anti-CD11b, anti-CD11c, and anti-Ly6G antibodies for 15 min at 4 °C and washed twice with FACS buffer (1% FBS and 0.01% EDTA in PBS). Fluorescence signals were detected by FACS Canto II, and data were analyzed with FlowJo software.
Acknowledgments
We thank T. Fujita for U373-CD14 cells, T. Kitamura for Plat-E cells, S. Yamaoka for pMRX-ires-puro, Y. Kawaguchi for HSV-1, D. E. Griffin and Y. Yoshinaka for anti-SINV E2 antibody, J. Wang for pCDH-puro-Bcl-2 (#46971) and pCDH-puro-Bcl-XL (#46972) plasmids, and D. Trono for pCMV-ΔR8.2 (#12263) and pMD2.G (#12259) plasmids. This work was partly supported by the Japan Society for the Promotion of Science KAKENHI (Grant numbers 15H01380, 26713005, 23659231, to T. Saitoh), a research grant from the Takeda Science Foundation (T. Saitoh), a research grant from the Japan Foundation for Pediatric Research (T. Saitoh), a research grant from the Tokyo Biochemical Research Foundation (T. Saitoh), and a National Institutes of Health (Grant PO1-AI070167 to S.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621508114/-/DCSupplemental.
References
- 1.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21(4):317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koshiba T. Mitochondrial-mediated antiviral immunity. Biochim Biophys Acta. 2013;1833(1):225–232. doi: 10.1016/j.bbamcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: A complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11(6):389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong S, Mu T, Wang G, Jiang X. Mitochondria-mediated apoptosis in mammals. Protein Cell. 2014;5(10):737–749. doi: 10.1007/s13238-014-0089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rongvaux A, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159(7):1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159(7):1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297(5587):1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, et al. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci USA. 2011;108(38):15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozaki T, et al. Role of zinc-finger anti-viral protein in host defense against Sindbis virus. Int Immunol. 2015;27(7):357–364. doi: 10.1093/intimm/dxv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol. 2004;78(23):12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA. 2007;104(1):151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda M, et al. Tristetraprolin inhibits HIV-1 production by binding to genomic RNA. Microbes Infect. 2006;8(11):2647–2656. doi: 10.1016/j.micinf.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Lin RJ, et al. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 2013;41(5):3314–3326. doi: 10.1093/nar/gkt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jose J, Snyder JE, Kuhn RJ. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 2009;4(7):837–856. doi: 10.2217/fmb.09.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryman KD, Klimstra WB. Host responses to alphavirus infection. Immunol Rev. 2008;225:27–45. doi: 10.1111/j.1600-065X.2008.00670.x. [DOI] [PubMed] [Google Scholar]
- 17.MacPherson L, et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 2013;41(3):1604–1621. doi: 10.1093/nar/gks1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tait SW, Green DR. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11(9):621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimura SH, Otsuka S, Kumeta M, Taga M, Takeyasu K. Intermolecular disulfide bonds between nucleoporins regulate karyopherin-dependent nuclear transport. J Cell Sci. 2013;126(Pt 14):3141–3150. doi: 10.1242/jcs.124172. [DOI] [PubMed] [Google Scholar]
- 20.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11(5):404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupfer C, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013;14(5):480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One. 2008;3(8):e2880. doi: 10.1371/journal.pone.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu S, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6(12):1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 24.Kumar H, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203(7):1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456(7219):264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 26.Saitoh T, et al. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7(6):598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 27.Moriishi K, Koura M, Matsuura Y. Induction of Bad-mediated apoptosis by Sindbis virus infection: Involvement of pro-survival members of the Bcl-2 family. Virology. 2002;292(2):258–271. doi: 10.1006/viro.2001.1206. [DOI] [PubMed] [Google Scholar]
- 28.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morita S, Kojima T, Kitamura T. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7(12):1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 30.Saitoh Y, et al. Overexpressed NF-kappaB-inducing kinase contributes to the tumorigenesis of adult T-cell leukemia and Hodgkin Reed-Sternberg cells. Blood. 2008;111(10):5118–5129. doi: 10.1182/blood-2007-09-110635. [DOI] [PubMed] [Google Scholar]