Significance
A potential avenue to control the spread of mosquito disease vectors lies in reproductive events that follow a blood meal. A key component is the massive production of yolk proteins in the fat body tissue, governed by regulatory networks triggered by the available nutrients. MicroRNAs play a critical role in mosquito egg maturation, and deciphering their dynamics and targets is necessary to fully realize these regulatory processes. We carried out a tissue-specific and time-resolved characterization of microRNA expression in the Aedes aegypti fat body and integrated these results with transcriptome-wide determination of their mRNA targets, followed by validation. This extensive analysis lays the groundwork for a systemic understanding of the gene regulation that underpins reproductive events in the female mosquito.
Keywords: Aedes aegypti, fat body, microRNA, microRNA targets, Argonaute 1
Abstract
The mosquito Aedes aegypti is a major vector of numerous viral diseases, because it requires a blood meal to facilitate egg development. The fat body, a counterpart of mammalian liver and adipose tissues, is the metabolic center, playing a key role in reproduction. Therefore, understanding of regulatory networks controlling its functions is critical, and the role of microRNAs (miRNAs) in the process is largely unknown. We aimed to explore miRNA expression and potential targets in the female fat body of Ae. aegypti, as well as their changes postblood meal (PBM). Small RNA library analysis revealed five unique miRNA patterns sequentially expressed at five sampled time points, likely responding to, and affecting, waves of upstream hormonal signals and gene expression in the same period. To link miRNA identities with downstream targets, transcriptome-wide mRNA 3′ UTR interaction sites were experimentally determined at 72 h posteclosion and 24 h PBM through Argonaute 1 cross-linking and immunoprecipitation followed by high-throughput sequencing. Several target sites were validated by means of in vitro luciferase assays with wild-type and mutated 3′ UTRs for six miRNA families. With established transgenic lines, consistent results were observed with spatiotemporal knockdown of miR-8 and luciferase assays. We further investigated miRNAs potentially regulating various physiological processes based on Clusters of Orthologous Groups functional categories. Hence, the present work comprehensively elucidated miRNA expression and target dynamics in the female mosquito fat body, providing a solid foundation for future functional studies of miRNA regulation during the gonadotrophic cycle.
Mosquitoes transmit numerous pathogens of dangerous human diseases because of their obligatory blood feeding (1). The Aedes aegypti mosquito is the major vector of arboviral diseases, such as dengue fever, yellow fever, chikungunya, and Zika virus (2–4). With the exception of yellow fever, there are no specific drugs or vaccines against these illnesses. Therefore, integrative vector population control must be implemented as part of their prevention. Mosquitoes have evolved a plethora of adaptations for hematophagy, making them exceptionally efficient disease carriers. Studies of mosquito-specific processes could provide valuable information for designing novel control approaches using genetic manipulation.
In hematophagous mosquitoes, a blood meal activates numerous processes and genes essential for digestion of the blood in the gut, massive synthesis of yolk protein precursors (YPPs) in the fat body (FB), accumulation of yolk proteins in developing oocytes, and eventual egg production. These processes are governed by complex regulatory networks, which have been extensively studied (5–9). Recent studies have also implicated microRNAs (miRNAs) in these regulatory networks in mosquitoes (10, 11).
miRNAs are endogenous small noncoding RNAs of 21–24 nt that are generated from either the 5′ (-5p) or 3′ (-3p) side of longer double-stranded hairpin precursors. Mature miRNAs complex with Argonaute (AGO) proteins and target them to specific sites in mRNA 3′ UTRs through complementarity in the seed region of the miRNA (nucleotides 2–8). Once bound, AGO and associated factors posttranscriptionally modulate the expression of a large proportion of transcripts in both animals and plants (12). For many miRNAs, this regulatory activity has been demonstrated to carry functional importance in both developmental transitions and homeostasis. For example, the lin-4 and let-7 miRNAs control developmental timing in the nematode Caenorhabditis elegans (13, 14); miR-430 is essential for gastrulation and brain formation in the zebrafish Danio rerio (15); and a number of miRNAs impact the development of hematopoietic, muscle, and other tissues in mammals (16–19). Similarly, several miRNAs have been phenotypically characterized in the model insect Drosophila melanogaster, starting with control of growth and development by the bantam miRNA (20). Current transcriptome-wide profiling techniques serve as valuable tools to characterize the important tissue- and time-specific expression patterns of miRNAs and their targeting repertoires, adding to a comprehensive molecular understanding of animal development (21, 22).
Previously, the application of 454 pyrosequencing has revealed the first set of miRNA and precursor sequences from midgut samples of female Ae. aegypti mosquitoes, either nonblood fed (NBF) or blood fed at 24 h postblood meal (PBM) (23). Another recent study uncovered miRNA expression profiles in ovaries of Ae. aegypti at several time points before and after blood feeding (24). Observed differences in miRNA levels in NBF and PBM female mosquitoes suggest miRNA regulatory functions. Using high-throughput sequencing, several studies have conducted comparative investigations of miRNA levels in NBF and PBM Ae. aegypti and Anopheles gambiae mosquitoes (25, 26).
The mosquito FB, the counterpart of the vertebrate liver and adipose tissue, is the center of metabolism and protein synthesis (7). In female mosquitoes, a blood meal triggers a chain of regulatory signals that results in activation of mRNA expression and synthesis of YPPs in the FB. The amino acid (aa)-induced target of rapamycin (TOR) signaling pathway and 20-hydroxyecdysone (20E) are the main factors controlling YPP gene expression (5, 6, 27). The vitellogenin (Vg) gene, widely taken as an indicator of FB vitellogenic status, reaches its maximal expression at 24 h PBM and declines dramatically thereafter at around 36–40 h PBM (28). Later, autophagy is initiated and reprograms the FB, setting the stage for a successive gonadotrophic cycle (28). A recent study found four major expression patterns of genes throughout the 72 h PBM time span in Ae. aegypti FBs that are differentially regulated by 20E, JH, and nutritional amino acids (7). Previous studies illustrated that miR-275, miR-1174, miR-8, and miR-1890 are essential for the normal progression of blood meal digestion and egg development of Ae. aegypti (29–32). However, our knowledge of the role of miRNAs in posttranscriptional regulation of the FB genes affecting these tissue functions is limited.
Here, we have used two complementary approaches—small RNA sequencing and Argonaute 1 cross-linking and immunoprecipitation followed by high-throughput sequencing (AGO1 CLIP-seq)—to elucidate miRNA dynamics and their targets throughout the gonadotrophic cycle in Ae. aegypti FBs. Five major miRNA expression peaks were observed in the FB within the 48-h PBM period. AGO1 CLIP-seq has provided insight into their downstream gene targets. Therefore, the present work yields a systematic view of miRNA expression and targets in Ae. aegypti FBs, offering a solid foundation for future functional studies.
Results
MiRNA Expression Dynamics in the Fat Body of Female Ae. aegypti Mosquitoes.
To examine miRNA expression profiles in the FB during the gonadotrophic cycle, we collected this tissue at 72 h posteclosion (PE) and 6, 24, 36, and 48 h PBM. Small RNA libraries were constructed with three biological replicates at each of the time points and sequenced to the total depth of 299 million reads, 96.76% of which were mapped to the genome (SI Appendix, Table S1). Read counts for 124 Ae. aegypti mature miRNAs in miRBase v21 were normalized to numbers of genome-mapped reads in each library, expressed as reads per million (RPM) (Dataset S1). Principle component analysis (PCA) was performed on raw read counts (SI Appendix, Fig. S1). Based on PC1 separation, all three biological replicates correlated well at three time points—72 h PE, 6 h PBM, and 24 h PBM. For 36 h and 48 h PBM, two replicates were separated from the other time points. Averages of three replicates were taken to represent expression levels at various time points.
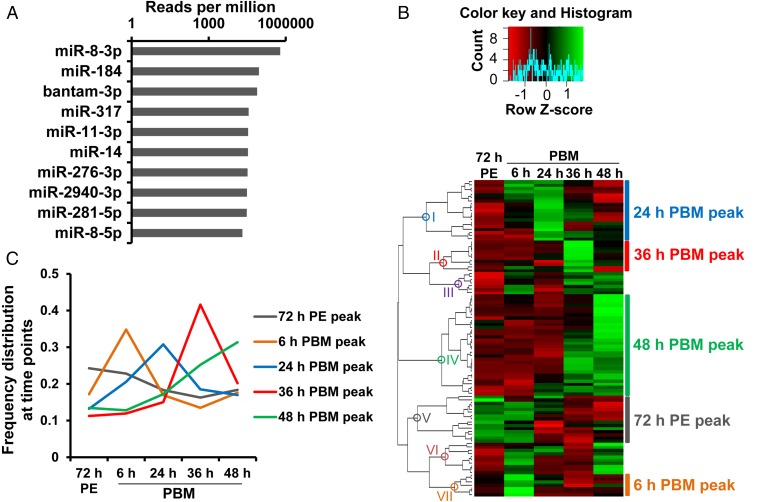
First, we examined the most abundant FB miRNAs throughout the time course indicated above. Averaging the expression value at the five time points, the top 10 miRNAs are presented in Fig. 1A. Notably, even the less abundant -5p arm of miR-8 was in the top 10, consistent with the idea that it is the most highly expressed FB miRNA precursor. In agreement with these data, miRNAs miR-8, miR-184, and bantam were also among the most abundant miRNAs in sugar-fed and blood-fed females of An. gambiae (25), suggesting function conservation of these miRNAs between these two mosquito species. Furthermore, miR-8–3p and miR-275–3p (ranked 11th; Dataset S1) have been shown previously to be critical in the normal progression of Ae. aegypti egg development PBM (29, 31).
Fig. 1.
A time course of miRNA expression in the Ae. aegypti fat body following a blood meal. (A) Top 10 abundant miRNAs over the time course (note the log10-based horizontal axis). (B) Hierarchical clustering (Pearson correlation and complete linkage) of top 100 mature miRNAs over the time course; expression levels are represented by averages of normalized expression from three biological replicates. (C) Expression trend lines of five expression patterns peaking at 72 h PE (clade V), 6 h (clade VII), 24 h (clade I), 36 h (clade II), and 48 h PBM (clade IV). For each miRNA, expression profiles were converted to represent percentages of normalized reads at each time point. Averages were taken at each time point within clusters to display trend lines. Cluster colors are consistent with those in B.
To examine changes in miRNA expression patterns during the time course, we analyzed mature miRNAs with a total normalized value over 10 RPM. Unsupervised hierarchical clustering revealed seven major clades representing distinct expression patterns (Fig. 1B and SI Appendix, Table S2). Five of these clades, encompassing 83 of 100 mature miRNAs, displayed expression peaks at a single time point—6 h, 24 h, and 36 h PBM (VII, I, and II, respectively, Fig. 1C), or two consecutive time points—designated as 48 h PBM and 72 h PE peaks (IV and V, respectively, Fig. 1C). Clades III and VI contained miRNAs with less defined expression peaks (Fig. 1B and SI Appendix, Table S2). Altogether, the observed expression patterns indicate substantial differential regulation of FB miRNAs during the 48 h PBM.
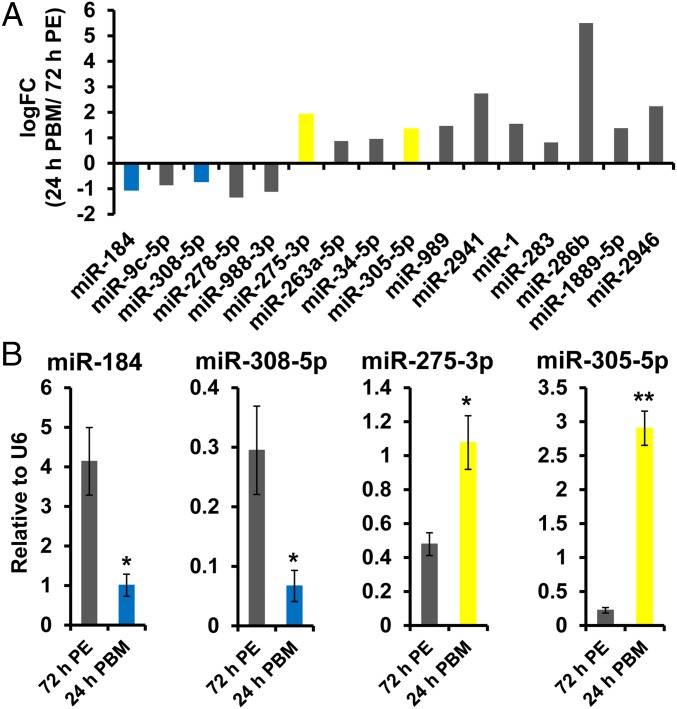
To define the miRNAs that may participate in the establishment of the vitellogenic transition in the FB more rigorously, we used the differential expression analysis package edgeR (33) to quantify the differences between the replicates at 72 h PE and 24 h PBM, where expression of Vg, an indicator of vitellogenic status, exhibits a peak. Sixteen miRNAs passed filtering criteria of statistical significance and abundance, with 5 down-regulated and 11 up-regulated (Fig. 2A). A total of 2 up-regulated miRNAs from the 24-h PBM expression peak (miR-275–3p and miR-305–5p) and 2 down-regulated species from the 72-h PE peak (miR-184 and miR-308–5p) were selected for validation using quantitative PCR (qPCR) relative to U6 snRNA. Levels of all four miRNAs in independently collected FB samples were significantly modulated at 24 h PBM, in agreement with analysis of the small RNA libraries (Fig. 2B). The four miRNAs ranked 2nd, 9th, 24th, and 46th, based on edgeR-computed abundance, confirming the broad dynamic range of our small RNA libraries in quantifying miRNA expression.
Fig. 2.
Differential expression of miRNAs at 72 h PE and 24 h PBM. (A) Differentially expressed miRNAs at 72 h PE and 24 h PBM (edgeR package). qPCR validation is highlighted by blue for down-regulation and yellow for up-regulation. (B) qPCR validation for four miRNAs, normalized to U6 snRNA. Three independent biological replicates were performed and statistical significance was calculated using unpaired t tests. Expression levels are represented as mean ± SEM (*P value < 0.05; **P value < 0.01).
Deep sequencing of small RNA libraries, particularly from specific tissues and developmental time points, also allows for novel miRNA identification in animals and plants (34). Using the default parameters of miRDeep2, we identified 206 new candidate miRNA loci in our FB libraries (Dataset S2). Eighteen of the loci, producing 15 unique miRNAs, agreed with two recent studies that applied additional screening to define genuine new miRNAs. Of 8 new miRNAs identified in small RNA libraries constructed at sequential developmental stages (35), 6 matched our candidate loci (aae-miR-new5, -9, -12, -17, -18, and -19; Dataset S2). Additionally, 9 out of another 30 new miRNAs identified in the Aag2 cell line with or without dengue virus 2 infection (36) agreed with our data (aae-miR-11900, -11903b, -11905, -11907, -11909, -11911, -11918, -11921, and -11926; Dataset S2). Further efforts are essential to confirm these candidate loci as genuine miRNAs and to reveal their biological functions.
Transcriptome-Wide Identification of miRNA Targets in the Fat Body Using AGO1 CLIP-Seq.
To understand miRNA-guided regulation, it is essential to uncover the downstream mRNA targets in addition to profiling miRNA identities. Cross-linking and immunoprecipitation of the core miRNA binding partner, Argonaute, has been used successfully to identify miRNA target sites and their changes during cellular transitions (37). In D. melanogaster, a single Argonaute protein (AGO1) mediates miRNA-guided repression (38), and its Ae. aegypti ortholog shares 86% amino acid identity (a second, truncated copy is also present) (39). Because immunoprecipitation is a critical step in CLIP-seq library preparation, we confirmed the efficiency and specificity of AGO1 pulldown using a GenScript aae-AGO1 antibody (order ID 218004–6; lot no. A313070458; SI Appendix, Fig. S2 and Experimental Procedures).
To identify miRNA target sites and capture their dynamics PBM, CLIP-seq libraries were constructed from three biological replicates of female FB tissues at 72 h PE and 24 h PBM using established protocols (40). Of the total ∼157 million sequenced reads, 105 million contained inserts longer than 15 nt, 53.62% of which mapped to the Ae. aegypti genome (AaegL3) (SI Appendix, Table S3). Reads were counted by the number of unique 4-nt random barcodes in the 5′-adapter (to avoid PCR amplification bias) and annotated based on the AaegL3.3 gene set and miRBase v21 coordinates (SI Appendix, Fig. S3A). Counts of mature miRNAs (normalized to total mapped counts) showed substantially higher Pearson correlation coefficients within, as opposed to between, the two conditions (SI Appendix, Fig. S3B), implying distinct physiological processes in the FB at 72 h PE and 24 h PBM, as well as high reproducibility of CLIP-seq library preparations. The proportions of annotated categories were consistent among the three replicates, and the high fraction of coding DNA sequence (CDS)-, UTR-, and miRNA-annotated reads confirmed the AGO1 specificity of the CLIP-seq (SI Appendix, Fig. S3A). The lower proportion of 3′ UTR vs. CDS reads (compared with mammalian CLIP libraries) is likely explained by the relatively short 3′ UTRs in Ae. aegypti, which take up only 15.83% of annotated transcript length (SI Appendix, Table S4).
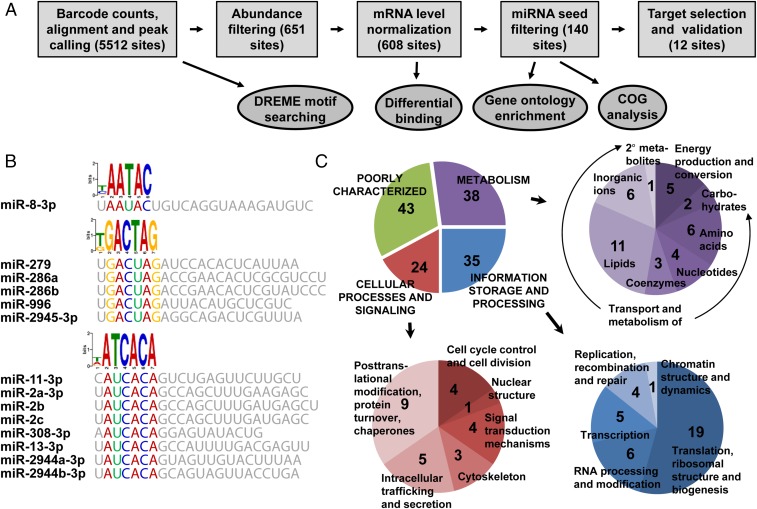
The CLIP-seq analysis pipeline is outlined in Fig. 3A. Because productive miRNA targeting occurs primarily in the 3′ UTR through perfect complementarity to the seed (nucleotides 2–7 or 2–8) of the miRNA (41), we focused on characterizing AGO1 interactions in these regions. Overlapping or adjoining reads were collected into well-resolved peaks representing individual AGO1 binding sites, 5,512 of which occurred in 3′ UTRs (Dataset S3). To assess the recovery of expected miRNA seed signatures, sequences underneath the 3′ UTR peaks were subjected to de novo motif discovery using DREME (42). Three out of 10 identified motifs matched in reverse complement to seeds of abundant miRNAs—miR-8–3p, -11–3p, and -279/286 (GTATTV, TGTGATH, and CTAGTCM, respectively; Fig. 3B). This finding provided independent confirmation that our CLIP-seq libraries captured sites of bona fide AGO1 association with mRNA 3′ UTRs through canonical miRNA targeting. To further narrow a set of reliable miRNA–mRNA targeting interactions, the AGO1 CLIP peaks were filtered for abundance: considering only those with a total sum of more than six read counts in the six libraries yielded 651 3′ UTR sites. To gauge the regulatory potential of these sites, we scored for the presence of 6mer (nucleotides 2–7) or the more efficacious 7mer (nucleotides 2–8) (41) seed complements to the top 60 miRNAs in their sequences. Confirming and extending the de novo motif discovery, 346 of 651 filtered 3′ UTR peaks harbored 6mers, and 147 contained 7mer matches. Consistent with the three significant motifs discovered by DREME (Fig. 3B), the most frequently encountered 6mers belonged to the miR-11–3p, miR-8–3p, and miR-279/286 families with 48, 46, and 22 instances, respectively.
Fig. 3.
Identification of miRNA targets in the fat body by CLIP-seq. (A) Summary of the analysis pipeline. (B) Characteristic motif logos of 3′ UTR peaks. Characteristic reverse complement logos identified by DREME analysis of 3′ UTR peaks are shown with corresponding miRNA seed regions. Peak calling was performed from six AGO1 CLIP-seq libraries, and 3′ UTR peaks were used as input for DREME with a minimal core width as 6 nt. (C) Clusters of Orthologous Groups functional assignments for the 140 high-confidence targets. In the detailed illustration of cellular processes and signaling, two instances were each counted twice, because both were assigned to two different functional groups.
We sought to define a set of high-confidence miRNA target candidates and to quantify the strength of these AGO1-target interactions, and their dynamics, before and after a blood meal. Because the observed CLIP signal for each peak depends on the abundance of the underlying mRNA, normalizing it to transcript abundance would be more reflective of the inherent regulatory power of the interaction. Furthermore, such normalization is particularly appropriate when comparing AGO1 binding between conditions, as it would take into account changes in mRNA levels. To this end, we intersected the filtered AGO1 peak data with previously measured corresponding transcript abundances in the female FB at 72 h PE and 24 h PBM (7), yielding 608 sites. Using the mRNA levels as peak-specific normalization factors for differential expression analysis of the CLIP-seq peak count data in DESeq2 (43), we identified 18 interactions with an adjusted P value of <0.05 (Dataset S4). Because the AGO1 targeting of these mRNAs is changing, these events may contribute to the reprogramming of expression patterns during the vitellogenic period. Two of the differentially targeted loci are potentially explained by interaction with a miRNA that is changing in the same direction (Fig. 2A and Dataset S4): makorin (AAEL007476) is targeted by miR-275–3p; and AAEL002512 is targeted by miR-184. Conversely, among the four miRNAs that themselves were shown to be differentially expressed between 72 h PE and 24 h PBM (Fig. 2B), we identified five, six, and three potential targets for miR-275–3p, -184, and -305–5p, respectively. Although the extent of AGO1 targeting may not be changing for some of these sites, their constitutive regulation may be necessary in the FB during vitellogenesis.
The resulting set of peaks was further filtered for sites containing 7mer matches of the top 60 miRNAs, producing 140 high-confidence 3′ UTR targeting sites, their normalized CLIP strength, and the normalized change in targeting efficiency before and after blood meal (Dataset S4). This set represents the likely miRNA-regulatory interactions that are important in the pre- or postblood meal state. It is worth noting that with the current analysis, the miRNAs that most frequently targeted these sites, with the exception of miR-316, were themselves highly expressed (SI Appendix, Table S5), reinforcing the expectation that abundant miRNAs regulate wider sets of transcripts. A Clusters of Orthologous Groups (COG) analysis of this gene set (Fig. 3C) indicated that the broad categories of metabolism, information storage and processing, and cellular processes and signaling, are well represented, with metabolic genes occupying the largest portion. Furthermore, we performed gene ontology (GO) enrichment analysis on the set of 140 high-confidence 3′ UTR miRNA-targeting sites against the background of 651 filtered 3′ UTR peaks. Again, our analysis revealed that miRNA-targeted mRNAs encoded proteins enriched in amino acid, lipid, and sugar metabolism and transport functions, consistent with the metabolic and secretory role of the fat body in vitellogenesis and the regulatory role of the miRNA machinery in orchestrating these functions (SI Appendix, Table S6).
In Vitro Validation of CLIP-Seq Targets.
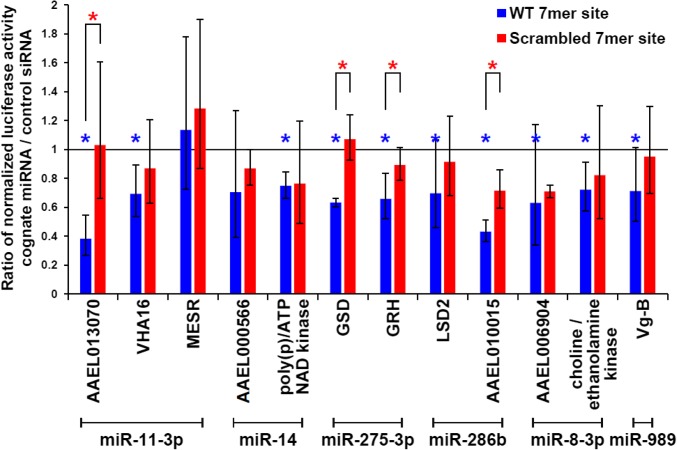
To assess the regulatory potential of the identified interactions, candidates for targeting by miR-8–3p, -275–3p, -279/286, -14, and -11–3p families were selected based on possessing relatively high normalized AGO1 binding site signal. The miR-989 target candidate Vitellogenin B (AAEL006138; Dataset S4) also passed these selection criteria and was added to the list due to its functional connection to the biological transition. Candidates were also filtered to exclude cases where the identified peak is among many comparable peaks on the 3′ UTR, suggesting unusual and potentially complicating regulation (SI Appendix, Fig. S4). DNA corresponding to 3′ UTRs of 12 candidate targets was successfully cloned into a luciferase reporter vector (Fig. 4). To measure reporter response to the targeting miRNAs, we took advantage of 293TRex cells, which do not endogenously express high levels of those miRNAs, but carry the conserved protein machinery to repress insect targets with exogenously provided miRNAs (44).
Fig. 4.
In vitro validation of miRNA targets. For each 3′ UTR, five and three biological replicates were measured for the wild-type (blue) and mutated (red) reporters, respectively. Statistical significance was calculated using unpaired one-tailed Student’s t tests. Data are represented as mean ± SD (*P value <0.05). Blue asterisks represent statistical significance in comparing the effect of miRNA mimic and negative control siRNA on wild-type reporters; red asterisks represent statistical significance between wild-type and mutated reporters with transfected miRNA mimic. Gene list is as follows: VHA16, AAEL000291, V-type proton ATPase 16-kDa proteolipid subunit; MESR, AAEL001659, misexpression suppressor of ras; poly(p)/ATP NAD kinase, AAEL000278; GSD, AAEL006834, glutamate semialdehyde dehydrogenase; GRH, AAEL000577, ortholog of D. melanogaster grainy head; LSD2, AAEL006820, lipid storage droplets surface binding protein 2; choline/ethanolamine kinase, AAEL008853; and Vg-B, AAEL006138, Vitellogenin isoform B.
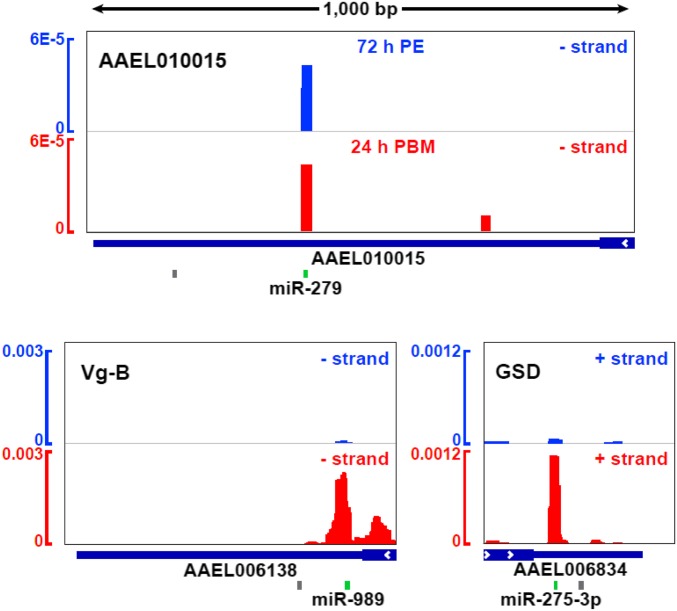
Normalized luciferase activity was measured in cells transfected with the reporter and cognate miRNA mimic (compared with a nontargeting control siRNA). Remarkably, 10 of the 12 candidates demonstrated statistically significant repression of luciferase activity through the 3′ UTR in the presence of the miRNA (Fig. 4). The strongest down-regulation was observed for miR-11–3p and AAEL013070 and miR-286b and AAEL010015. Distinct 3′ UTR CLIP signals (Fig. 5) and luciferase assay results suggest that those are authentic targets of corresponding miRNAs. In addition, to assess whether the miRNA binding sites within the CLIP-identified peaks are responsible for the 3′ UTR regulation, we mutated their seed complementarity regions in the reporters. Luciferase measurements showed a derepression for the majority of mutated constructs, reaching statistical significance in four of the targets. Thus, we have validated that most of the tested targets identified by biochemical association with AGO1 are repressed by their cognate miRNAs, and mutation of a single binding site leads to a significant loss of regulation in several cases.
Fig. 5.
Examples of distinct 3′ UTR binding sites of AGO1 on AAEL010015, Vg-B (AAEL006138, Vitellogenin isoform B), and GSD (AAEL006834, glutamate semialdehyde dehydrogenase). Read counts (vertical axis) are depicted as a fraction of all mapped counts. Seed sites for the microRNAs of interest are depicted in green; others are shown in gray.
Spatiotemporal Knockdown of miR-8 Confirms CLIP-Seq Targets in the Female Fat Body.
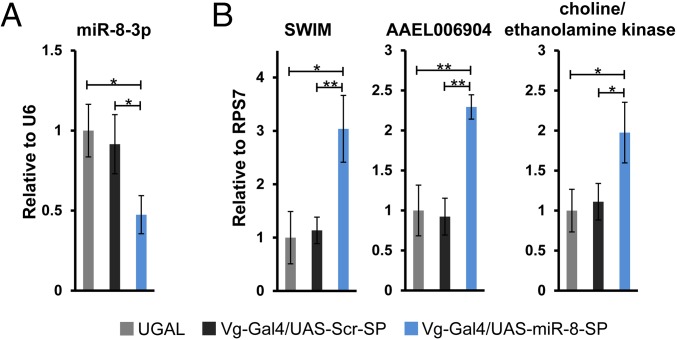
To validate the identified miRNA-target relationships in vivo, we turned to a transgenic mosquito system where a miR-8 sponge cassette has been previously used to achieve tissue-specific knockdown of miR-8 in the female fat body (31). Two responder lines containing an upstream activating sequence (UAS)–scramble–sponge (UAS–Scr–SP) and UAS–miR-8–sponge (UAS–miR-8–SP) were crossed with a driver line encoding an Aedes Vitellogenin (Vg) promoter-linked yeast transcriptional activator Gal4 protein (Vg–Gal4), and progeny expressing both were selected. Fat body samples were collected at 24 h PBM from Vg–Gal4/UAS–Scr–SP and Vg–Gal4/UAS–miR-8–SP female adult mosquitoes. Simultaneously, fat body tissues were collected from wild-type female mosquitoes (UGAL) at 24 h PBM. In agreement with previous observations (31), only Vg–Gal4/UAS–miR-8–SP hybrids exhibited reduced follicle size at 24 h PBM. We further confirmed miR-8–3p inhibition and the corresponding derepression of its previously identified target secreted Wg-interacting molecule (SWIM) in the fat body at 24 h PBM in Vg–Gal4/UAS–miR-8–SP mosquitoes (Fig. 6 A and B). To examine the specificity of miR-8–3p knockdown, we quantified the levels of miR-275–3p and one of its validated targets glutamate semialdehyde dehydrogenase (GSD) in luciferase assays (Fig. 4). As expected, neither showed significant changes in Vg–Gal4/UAS–miR-8–SP animals (SI Appendix, Fig. S5 A and B). We next tested two mRNAs, AAEL006904 and choline/ethanolamine kinase, whose 3′ UTRs showed significant repression with added miR-8–3p mimics in luciferase assays (Fig. 4). Consistently, expression levels of these two genes were significantly up-regulated in Vg–Gal4/UAS–miR-8–SP mosquitoes, indicating derepression upon reduction of miR-8–3p levels (Fig. 6B). Thus, the application and validation of AGO1 CLIP-seq expands our understanding of miRNA–mRNA interactions in the female fat body tissue.
Fig. 6.
Spatiotemporal knockdown of miR-8–3p using the Gal4/UAS system. At 24 h PBM, levels of miR-8–3p (A), and SWIM, AAEL006904 and choline/ethanolamine kinase (AAEL008853) (B), were measured by qPCR in the female fat body tissues from UGAL wild type, Vg–Gal4/UAS–Scr–SP, and Vg–Gal4/UAS–miR-8–SP mosquitoes. Expression levels were normalized to that of U6 snRNA for miR-8–3p and to RPS7 for protein coding genes. Three independent biological replicates were performed and statistical significance was calculated using unpaired t tests. For each gene, average of relative expression was taken as one from UGAL wild-type mosquitoes. Expression levels are represented as mean ± SD (*P < 0.05; **P < 0.01).
Discussion
Our study has revealed miRNA levels and their broad dynamics concomitant with the physiological changes that occur in the Ae. aegypti FB after a blood meal (Fig. 1C). We have identified five major expression patterns of miRNAs, with their respective peaks at 72 h PE, 6, 24, 36, and 48 h PBM. Previous work has found four major clusters of protein-coding genes in the FB with their peaks occurring at the same time points PBM as miRNA peaks detected in this work. The four protein-coding gene clusters, designated as early, early-mid, late-mid, and late genes, are under differential activation or inhibition effects of 20E, juvenile hormone (JH), and amino acid signaling pathways, as well as of hormone receptor 3 (HR3) (7). Expression patterns of miRNAs potentially provide relevant clues to their specific upstream regulation. Determination of miRNA patterns, the timing of which coincides with those of protein-coding genes, strongly suggests that they are under the control of similar factors.
Among the abundant and differentially expressed miRNAs, miR-275–3p, previously shown to be essential for blood digestion and ovary development (29), exhibited an expression peak at 24 h PBM (SI Appendix, Table S2). Interestingly, mRNAs that are up-regulated in the FB between 12 h and 36 h PBM respond to synergistic action of amino acids and 20E (7). In a similar manner, miR-275–3p is induced in the FB by a combined treatment of amino acids and 20E (29). Precursors of mir-305 and mir-275 are closely clustered in the genome (∼9 kb apart). Both miR-275–3p and miR-305–5p are up-regulated at 24 h PBM (Fig. 2B and SI Appendix, Table S2), suggesting common upstream transcriptional controls. In Drosophila, miR-8 and miR-14 are repressed by 20E and affect the insulin pathway (45). However, our small RNA library analysis revealed that miR-8–3p displayed a similar expression pattern to that of miR-275–3p, suggesting its activation by 20E (SI Appendix, Table S2). miR-8–3p has been previously shown to directly inhibit the expression of SWIM in the long-range Wingless signaling pathway and control the secretion of YPPs from FB (31). Whereas bantam mediates ecdysone production under the regulation of insulin signaling in the prothoracic gland (46), this miRNA peaked at 6 h PBM in the mosquito FB (SI Appendix, Table S2), indicating possible inhibition by 20E and activation by amino acids (7). In the cockroach Blattella germanica, combined treatment of 20E and JH significantly reduced the level of miR-276–3p (47), whereas miR-276–3p displayed an expression peaking at 24 h PBM in the mosquito FB (SI Appendix, Table S2), implying potential upstream regulation of 20E and amino acids. Because JH, 20E, amino acids, and HR3 signaling pathways modulate gene expression in Ae. aegypti (7), it is intriguing to further examine the regulatory functions of these miRNAs in the FB. Future in vitro fat body culture assays, similar to that of miR-275–3p (29), would shed light on how these signaling pathways affect miRNA levels.
Integration of miRNA (this study) and mRNA (7) abundance profiles with the targeting information obtained by CLIP-seq allows us to narrow a set of potential regulatory miRNA-target relationships that may play a role in several critical physiological processes. Importantly, the experimental identification by CLIP-seq can significantly reduce the set of bioinformatically predicted miRNA targets, which often number in the several hundred for a single miRNA. For the 140 high-confidence 3′ UTR sites, assignment of COG functional categories indicated that miRNAs extensively regulate metabolic processes, especially lipid transport and metabolism (Fig. 3C), consistent with the major physiological role of the FB. This observation was further corroborated by GO enrichment analysis (SI Appendix, Table S6). The largest proportion of miRNA targets in the “cellular processes and signaling” category is occupied by genes involved in folding, posttranslational modification, and turnover of proteins, as well as intracellular trafficking and secretion and molecular processes that are also crucial to yolk protein production and deposition. Among proteins involved in information storage and processing, miRNAs target a large number of transcripts responsible for translation, ribosomal structure, and biogenesis (Fig. 3C). This fraction included interaction sites on ribosomal proteins revealed through AGO1 CLIP-seq, which may be caused by high levels of the mRNAs themselves. However, this functional group also included interaction sites on eukaryotic translation initiation factor 4E binding protein (4EBP, AAEL001864) and eukaryotic translation elongation factor (AAEL004500), which are critical for the mRNA translation process. Finally, a large proportion of the identified miRNA targets (43 instances) lacked functional annotation, exemplified by AAEL006904, a validated target of miR-8–3p in vitro and in vivo (Figs. 4 and 6B), underscoring the need for further bioinformatics and experimental efforts to annotate the mosquito transcriptome. Interactions of individual miRNAs with specific functional groups are summarized in SI Appendix, Table S7, which provides clues into how a particular miRNA contributes to the intricate homeostasis within the female mosquito FB tissue.
Several individual miRNA-target relationships in the high-confidence set illustrate the potential relevance to the developmental and metabolic processes in the FB (SI Appendix, Fig. S7). miR-275–3p, which is essential to egg production (29), targets GSD (AAEL006834, Fig. 5). This enzyme is critical in metabolizing the abundant nitrogen from blood into proline through glutamate soon after a blood meal (48). The slow increase of GSD mRNA levels (peaking at 36 h PBM) (7) may be reversed toward the end of the digestion process by the observed up-regulation of miR-275–3p at 24 h PBM (SI Appendix, Table S2) and the corresponding increase in AGO1 targeting (Fig. 5). Supporting this view, the GSD 3′ UTR is shown to be regulated by miR-275–3p in luciferase assays (Fig. 4). Additionally, miR-275–3p is validated to target a Drosophila grh/elf-1 transcription factor ortholog (AAEL000577) (Fig. 4) and interacts with the makorin mRNA (AAEL007476) (SI Appendix, Fig. S6A), a protein known to control the juvenile-to-adult transition in C. elegans (49). Another protein, GS1 (AAEL001887, glutamine synthetase 1), involved in ammonia detoxification, is critical in both FB and midgut (50) and was found to be potentially targeted by miR-2940–5p (Dataset S4). Thus, it will be interesting to further examine the effects of miR-275–3p and miR-2940–5p on nitrogen metabolism. Furthermore, miR-8–3p, previously shown to affect Vg and lipid secretion (31), also targets choline/ethanolamine kinase (Figs. 4 and 6B), a phospholipid biosynthesis enzyme. Further efforts are required to elucidate the relationship between miR-8–3p and lipid metabolism.
The observed targeting by other abundant miRNAs in the FB is also likely to be involved in blood digestion and egg production. For example, the interaction between miR-11–3p and AAEL013070 results in robust repression in luciferase assays (Fig. 4). This mRNA encodes a membrane-bound acid phosphatase that is expected to play a role in removing protein phosphate groups in lysosomes during digestion. Similarly, the miR-279/286 family strongly down-regulates AAEL010015, a DNA-binding homeodomain protein (Fig. 4). Furthermore, the FOXL transcription factor (AAEL005741) was identified as a potential target for miR-184–3p (SI Appendix, Fig. S6B), and its knockdown significantly inhibits amino acid-induced expression of vitellogenin in the FB and reduces egg laying numbers (51). Accordingly, decreased levels of miR-184 at 24 h PBM (Fig. 2B) and a corresponding reduction in AGO1 targeting of FOXL (SI Appendix, Fig. S6B) may be necessary for homeostasis of FOXL and sustained egg development. Similarly, six potential targets were suggested for the bantam miRNA by 3′ UTR peak analysis (Dataset S4), including a recently duplicated acetyl-CoA synthetase (AcCoAS, AAEL015010, and AAEL000321) and a histone deacetylase (HDAC) RPD3 ortholog (AAEL004586). Both targeting events showed statistically significant changes between 72 h PE and 24 h PBM (adjusted P < 0.01). In D. melanogaster, knockdown of either AcCoAS or HDAC RPD3 results in extended life span (52, 53). Because acetyl-CoA is a critical metabolic intermediate and acetyl donor for epigenetic modifications, it would be intriguing to explore the regulatory functions of bantam in repressing these two targets. Future efforts, especially modulation of miRNA levels in vivo, would clarify their specific regulatory functions.
Overall, our study has provided a comprehensive analysis of miRNA changes that occur in the Ae. aegypti FB throughout the gonadotrophic cycle, coupled with quantitative measurements of the AGO1–mRNA targeting repertoire guided by the miRNAs. In it, we have delineated the dynamics of several abundant miRNAs and their possible connections to upstream signaling. Furthermore, we have identified a large number of downstream regulatory targets that provide a valuable foundation for future functional studies.
Experimental Procedures
Animals and Tissues.
Ae. aegypti mosquitoes were maintained under laboratory conditions as described previously (54). Three days PE, female abdominal walls with adhered FB tissue were collected as 72 h PE. Mosquitoes within the same cage were subsequently provided a blood meal from white leghorn chickens. Only fully engorged mosquitoes were dissected for FB collections at 6 h, 24 h, 36 h, and 48 h PBM. Biological materials were collected on three separate occasions for each experiment.
Small RNA Library Preparation and Sequencing.
Total RNA was extracted from tissues stored in TRIzol (Invitrogen). Small RNA libraries were prepared using Illumina TruSeq Small RNA Library Preparation Kits. For each time point, 1 µg of total RNA was used. Equal volumes of library PCR products were pooled and the 145- to 160-bp range was gel purified on a Novex TBE gel (Invitrogen). Samples were sequenced on an Illumina HiSeq2500 platform. Three independent biological replicates were prepared.
Computational miRNA Profiling of Small RNA Libraries.
Basic read processing was performed on the next-generation sequencing (NGS) files where individual FASTQ files were separated based on indexes. The systemPipeR pipeline was applied with modifications to obtain absolute read counts (https://github.com/tgirke/systemPipeR). Briefly, 3′ adapter sequences (TGGAATTCTCGGGTGCCAAGG) were trimmed, and reads were kept with lengths between 18 and 32 nt. The software Bowtie2 (version 2.2.3) was used to map reads onto the most recent genome assembly of Ae. aegypti (AaegL3, VectorBase: https://www.vectorbase.org) (55). Sequences of Ae. aegypti miRNAs and precursors were downloaded from miRBase (v21, https://mirbase.org). Genomic coordinate file was created in the format of GFF3 using BLAT on AaegL3. A similar rule was applied to extend profiling coordinates as that of miRDeep2: 2 nt upstream and 5 nt downstream (56). Raw read counts were obtained for miRNAs using the function of “summarizeOverlaps” based on profiling coordinates under the mode of IntersectionStrict (57). They were normalized to total number of reads mapped to the genome in each library and presented as RPM. A total of 124 mature miRNAs were subject to differential expression analysis using edgeR at 72 h PE and 24 h PBM. miRNAs with a false discovery rate (FDR) lower than 0.05 were considered statistically significant in differential expression, and subsequent abundance filtering included only the predominant (-3p or -5p) arms of the top 60 miRNAs based on edgeR-computed abundance. Default parameters were used with miRDeep2 for identification of novel miRNA candidates (56). Candidates with a minimal miRDeep2 score of 0 were listed.
Real-Time PCR.
qPCR for miRNA or mRNA expression was performed similarly to previous studies (29). Briefly, the enzyme DNase I (Invitrogen) was used to remove genomic DNA contamination and then inactivated by EDTA (Invitrogen). Reverse transcription was performed using 5× miScript HiSpec Buffer of miScript II RT kit (Qiagen) for small RNAs and using SupersScript III Reverse Transcriptase with oligo(dT)12–18 for protein coding mRNAs, whereas qPCR was conducted using QuantiFast SYBR Green PCR kit (Qiagen). Primer sequences are included in Dataset S5. Data were normalized to that of U6 snRNA or RPS7 using the method of 2-ΔCt (58).
Western Blotting and AGO1 Immunoprecipitation.
For Western blotting, cleared FB tissue lysates were loaded with LDS 4× NuPage sample buffer (Invitrogen) and 10× sample reducing reagent (Invitrogen) on Novex 10% Tris-glycine gels (Invitrogen) and transferred to PVDF membranes in 20% (vol/vol) methanol in 1× NuPage transfer buffer (Invitrogen). Membranes were blocked in StartingBlock (PBS) (Thermo Fisher), incubated in 1:1,000 primary antibody dilution, and in 1:2,000 dilution of anti-rabbit-HRP (Roche) secondary antibody after stringent washes. Antibody signals were detected using SuperSignal west pico chemiluminescent substrate (Thermo Fisher) and exposed to X-ray films.
Immunoprecipitation tests were done using the GenScript Inc AGO1 antibody (lot no. A313070458) and the protein A immunoprecipitation kit (Roche). Centrifuged FB tissue lysate was precleared with protein A agarose overnight at 4 °C, incubated with 5 µg of Ae. aegypti AGO1 antibody for 1 h at 4 °C, precipitated with protein A agarose for 3 h at 4 °C, and washed with wash buffers 1, 2, and 3.
The Drosophila Abcam ab5070 antibody detects AGO1 at ∼98 kDa in the Ae. aegypti Aag2 cell line and whole mosquito lysates (59), and we confirmed its specificity in FB protein extracts at 72 h PE with an observed ∼100 kDa band (SI Appendix, Fig. S2A). Next, immunoprecipitation from FB tissue lysates at 72 h PE was performed using a GenScript custom AGO1 antibody, probed with either the Abcam ab5070 antibody or the GenScript custom antibody in Western blot. A specific band at the same position (∼100 kDa) indicated efficient AGO1 pulldown (SI Appendix, Fig. S2B). Meanwhile, during CLIP-seq library preparation, AGO1–RNA complexes migrated above 100 kDa after UV crosslinking, and the “high” portion was excised for further analyses (SI Appendix, Fig. S2C). Those results suggested that the GenScript custom antibody provides sufficient AGO1 specificity.
CLIP-Seq Library Preparation.
CLIP-seq libraries were prepared as previously described (40). Briefly, 60 FBs were collected at 72 h PE and 24 h PBM each, finely sliced, and irradiated to 900 mJ/cm2. Following tissue lysis, lysates were treated with RQ1 DNase (Promega) and 1:1,000 RNase A (Ambion) dilution. GenScript custom AGO1 antibody (lot no. A313070458) was bound to Protein A Dynabeads (Invitrogen) and immunoprecipitation was performed overnight at 4 °C. Beads were further digested with 1:1,000 RNase A dilution, dephosphorylated with calf intestinal alkaline phosphatase (CIP) (NEB) and ligated to 32P-labeled 3′ RNA linker (RL3-OH) using T4 RNA ligase (Thermo Fisher Scientific). Nonradiolabeled 3′ RNA linker (5′-phosphorylated, RL3-P) and T4 RNA ligase were subsequently added for overnight incubation to ensure sufficient ligation. Upon treatment with protein loading buffer, AGO1–RNA complexes were separated on NuPAGE 4–12% Bis-Tris gels (Invitrogen) with Mops running buffer (Invitrogen) and then transferred to nitrocellulose membranes. Assisted with autoradiography, nitrocellulose membranes were excised separately corresponding to 110 kDa (“low”) and 130 kDa (“high”) regions (SI Appendix, Fig. S2C). RNAs were extracted from chopped membranes, 5′ phosphorylated with T4 PNK (NEB), ligated to 5′ RNA linker (RL5D) with T4 RNA ligase, and treated with RQ1 DNase. A degenerate 4-nt barcode was included in the 5′ RNA linker to increase the complexity of unique sequences and avoid artifacts of preferential PCR duplication. Recovered RNAs underwent reverse transcription with DP3 primer, amplification with DP5 and DP3 primers, and separation on 10% polyacrylamide gels. Gels were excised, recovered, and further amplified with PCR2–DP5 primer and corresponding index primers (PCR2–DP3, index X). After the second PCR, the anticipated length with miRNA inserts was around 168 nt. Regions between 150 and 175 nt and regions above 175 nt were taken as the low and high portions, respectively. PCR products were separated with agarose gels and recovered. Libraries of the high portion were pooled in equal amounts. Samples were sequenced on an Illumina HiSeq2500 platform for single-end data collection. Three independent biological replicates were performed for each time point. DNA and RNA oligonucleotides are included in Dataset S5.
Analysis of CLIP-Seq Libraries.
The 3′ adapters (GTGTCAGTCACTTCCAGCGG) were trimmed and reads were kept if RNA inserts were above 15 nt in length. Degenerate 4-nt barcode regions were introduced in the CLIP-seq protocol to differentiate individual ligations during library preparation. Each unique barcode was counted as one read count for each RNA insert to avoid preferential amplification bias. Barcode sequences were trimmed after this initial read collapse. Reads were mapped to VectorBase genome (AaegL3, https://www.vectorbase.org) using Bowtie2 and TopHat (55, 60). Reads with the same genomic coordinates were further collapsed and summed to represent read counts. Annotations were performed following base features of VectorBase AaegL3.3 gene set and miRNA coordinates from miRBase v21 (mirbase.org/). Browser extensible data (BED) files were generated for each library and visualized in Integrative Genomics Viewer (IGV) (61). To identify peaks, collapsed reads were pooled from six libraries and used as input for Findpeaks v4 (62). Peaks were separated if valleys occurred with peak heights below 50% and peak shoulders were trimmed if they were lower than 10% of peak heights. Based on peak genomic coordinates, peak read counts were then extracted from each library. Identified peaks were annotated similarly as described above. Because miRNAs mostly interact with 3′ UTRs (41), peaks falling into 3′ UTR regions were used for latter analyses. For DREME, motifs with an E-value lower than 0.05 and minimal widths of 6 nt were identified. As an abundance filtering step, peaks were retained if summed read counts across all six replicates were higher than 6. Transcript abundances were obtained for 72 h PE and 24 h PBM based on a microarray study of Ae. aegypti fat body (7) and appended to the peak count table. Raw peak counts and mRNA abundances were used in DESeq2 to perform differential analysis (43). Potential binding sites were scored for the presence of 6mer or 7mer seed site complements of top 60 miRNAs at 72 h PE and 24 h PBM identified from small RNA libraries. GO mappings of protein coding genes were obtained from BioMart on Vectorbase and GO enrichment analysis was performed with the package GOstats (63). COG functional groups were assigned according to inNOG for insects (higher priority) and NOG for all organisms with EggNOG database v4.5 (64). Genes with no mappings in inNOG or NOG were designated into functional group [S] Function unknown.
In Vitro Validation of CLIP-Seq Targets.
PCR products containing full-length Ae. aegypti 3′ UTRs (based on AaegL3.3 annotations) were prepared from genomic DNA (gDNA) or FB-specific cDNA templates. gDNA was extracted from whole-body female adults as previously described (65). TRIzol-isolated total RNA samples from female FBs at 72 h PE and 24 h PBM were reverse transcribed with SuperScript II (Invitrogen). The 3′ UTRs were amplified with primers containing BsmBI or BbsI sites and cloned downstream of Renilla luciferase in modified pRL-TK vectors (Promega) using Golden Gate Assembly (NEB): 4 µL H2O, 0.75 µL BsmBI or BbsI, 1 µL Tango Buffer, 1 µL 10 nM DTT, 0.25 µL T7 ligase, 1 µL pRL-TK plasmid, 1 µL 10 nM ATP, and 1 µL PCR product, with 10 cycles of 37 °C for 10 min and 20 °C for 10 min, followed by Plasmid-Safe DNase treatment (Epicentre Biotechnologies) to remove linear DNAs. For selected UTRs, 7mer miRNA seed complements in the AGO1 binding sites were mutated using a Q5 Site-Directed Mutagenesis kit (NEB). Cloning and mutagenesis primers are shown in Dataset S5. Insert sequences were validated by DNA sequencing.
HEK T-REx-293 cells were cultured in 96-well plates in 10% FBS in DMEM and transfected at 50–70% confluency with Renilla reporter plasmid and pGL3 (firefly luciferase) transfection normalization plasmid at 100 ng each, and miRNA mimic or AllStars negative control siRNA (Qiagen) at 100 nM using Attractene (Qiagen). Cells were assayed 24 h posttransfection using the Dual-Luciferase Reporter Assay System (Promega). The effects of miRNA mimics are presented by means of ratios of (Renilla/Firefly)miRNA-mimics/(Renilla/Firefly)Allstar. Each experiment consisted of three technical replicates (wells), and three to five total biological replicates were performed. Statistical significance of the biological replicates was calculated using unpaired one-tailed Student’s t tests, because it is generally accepted that miRNAs mostly repress target gene expression through posttranscriptional modulation.
In Vivo Validation of miRNA Targets.
Ae. aegypti transgenic responder lines—M3-1, UAS–scramble–sponge; M3-2, UAS–miR-8–sponge—have been established previously (31). These responder lines were crossed with a Vg–Gal4 driver line, also developed before (66). Progeny was screened for concurrent presence of eye-specific selection markers, EGFP and dsRed. Female FBs of the obtained hybrid line were collected at 24 h PBM, and transcript and miRNA abundance was measured by qPCR.
Sequencing Data Availability.
The sequencing data files for the miRNA profiling and CLIP-seq experiments are deposited in the Gene Expression Omnibus with the accession no. GSE93345.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01 AI113729 (to A.S.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE93345).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701474114/-/DCSupplemental.
References
- 1.Attardo GM, Hansen IA, Raikhel AS. Nutritional regulation of vitellogenesis in mosquitoes: Implications for anautogeny. Insect Biochem Mol Biol. 2005;35(7):661–675. doi: 10.1016/j.ibmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Barrett AD, Higgs S. Yellow fever: A disease that has yet to be conquered. Annu Rev Entomol. 2007;52:209–229. doi: 10.1146/annurev.ento.52.110405.091454. [DOI] [PubMed] [Google Scholar]
- 3.Tsetsarkin KA, Chen R, Weaver SC. Interspecies transmission and chikungunya virus emergence. Curr Opin Virol. 2016;16:143–150. doi: 10.1016/j.coviro.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver SC, et al. Zika virus: History, emergence, biology, and prospects for control. Antiviral Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen IA, Attardo GM, Park JH, Peng Q, Raikhel AS. Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proc Natl Acad Sci USA. 2004;101(29):10626–10631. doi: 10.1073/pnas.0403460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raikhel AS, Brown M, Belles X. 2005. Endocrine control of reproductive processes in Comprehensive Molecular Insect Science, eds Gilbert L, Gill S, Iatrou K, (Elsevier Press, Cambridge, MA), pp 433–491.
- 7.Roy S, et al. Regulation of gene expression patterns in mosquito reproduction. PLoS Genet. 2015;11(8):e1005450. doi: 10.1371/journal.pgen.1005450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saha TT, et al. Hairy and Groucho mediate the action of juvenile hormone receptor Methoprene-tolerant in gene repression. Proc Natl Acad Sci USA. 2016;113(6):E735–E743. doi: 10.1073/pnas.1523838113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou Z, et al. Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc Natl Acad Sci USA. 2013;110(24):E2173–E2181. doi: 10.1073/pnas.1305293110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas KJ, Myles KM, Raikhel AS. Small RNAs: A new frontier in mosquito biology. Trends Parasitol. 2013;29(6):295–303. doi: 10.1016/j.pt.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucas KJ, Zhao B, Liu S, Raikhel AS. Regulation of physiological processes by microRNAs in insects. Curr Opin Insect Sci. 2015;11:1–7. doi: 10.1016/j.cois.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 14.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 15.Takacs CM, Giraldez AJ. miR-430 regulates oriented cell division during neural tube development in zebrafish. Dev Biol. 2016;409(2):442–450. doi: 10.1016/j.ydbio.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song R, et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 2014;510(7503):115–120. doi: 10.1038/nature13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, et al. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209(9):1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 21.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: Approaches and considerations. Nat Rev Genet. 2012;13(5):358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1(2):266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Mead EA, Liang S, Tu Z. Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. BMC Genomics. 2009;10:581. doi: 10.1186/1471-2164-10-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akbari OS, et al. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 2013;3(9):1493–1509. doi: 10.1534/g3.113.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biryukova I, Ye T, Levashina E. Transcriptome-wide analysis of microRNA expression in the malaria mosquito Anopheles gambiae. BMC Genomics. 2014;15:557. doi: 10.1186/1471-2164-15-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussain M, Walker T, O’Neill SL, Asgari S. Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem Mol Biol. 2013;43(2):146–152. doi: 10.1016/j.ibmb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Smykal V, Raikhel AS. Nutritional control of insect reproduction. Curr Opin Insect Sci. 2015;11:31–38. doi: 10.1016/j.cois.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryant B, Raikhel AS. Programmed autophagy in the fat body of Aedes aegypti is required to maintain egg maturation cycles. PLoS One. 2011;6(11):e25502. doi: 10.1371/journal.pone.0025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryant B, Macdonald W, Raikhel AS. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2010;107(52):22391–22398. doi: 10.1073/pnas.1016230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Lucas KJ, Roy S, Ha J, Raikhel AS. Mosquito-specific microRNA-1174 targets serine hydroxymethyltransferase to control key functions in the gut. Proc Natl Acad Sci USA. 2014;111(40):14460–14465. doi: 10.1073/pnas.1416278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucas KJ, et al. MicroRNA-8 targets the Wingless signaling pathway in the female mosquito fat body to regulate reproductive processes. Proc Natl Acad Sci USA. 2015;112(5):1440–1445. doi: 10.1073/pnas.1424408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas KJ, Zhao B, Roy S, Gervaise AL, Raikhel AS. Mosquito-specific microRNA-1890 targets the juvenile hormone-regulated serine protease JHA15 in the female mosquito gut. RNA Biol. 2015;12(12):1383–1390. doi: 10.1080/15476286.2015.1101525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue) D1:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu W, Criscione F, Liang S, Tu Z. MicroRNAs of two medically important mosquito species: Aedes aegypti and Anopheles stephensi. Insect Mol Biol. 2015;24(2):240–252. doi: 10.1111/imb.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miesen P, Ivens A, Buck AH, van Rij RP. Small RNA profiling in dengue virus 2-infected Aedes mosquito cells reveals viral piRNAs and novel host miRNAs. PLoS Negl Trop Dis. 2016;10(2):e0004452. doi: 10.1371/journal.pntd.0004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seok H, Ham J, Jang ES, Chi SW. MicroRNA target recognition: Insights from transcriptome-wide non-canonical interactions. Mol Cells. 2016;39(5):375–381. doi: 10.14348/molcells.2016.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18(14):1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell CL, Black WC, 4th, Hess AM, Foy BD. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics. 2008;9:425. doi: 10.1186/1471-2164-9-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karginov FV, Hannon GJ. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013;27(14):1624–1632. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey TL. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27(12):1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16(2):144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luhur A, Chawla G, Sokol NS. MicroRNAs as components of systemic signaling pathways in Drosophila melanogaster. Curr Top Dev Biol. 2013;105:97–123. doi: 10.1016/B978-0-12-396968-2.00004-X. [DOI] [PubMed] [Google Scholar]
- 46.Boulan L, Martín D, Milán M. bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr Biol. 2013;23(6):473–478. doi: 10.1016/j.cub.2013.01.072. [DOI] [PubMed] [Google Scholar]
- 47.Rubio M, de Horna A, Belles X. MicroRNAs in metamorphic and non-metamorphic transitions in hemimetabolan insect metamorphosis. BMC Genomics. 2012;13:386. doi: 10.1186/1471-2164-13-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldstrohm DA, Pennington JE, Wells MA. The role of hemolymph proline as a nitrogen sink during blood meal digestion by the mosquito Aedes aegypti. J Insect Physiol. 2003;49(2):115–121. doi: 10.1016/s0022-1910(02)00267-6. [DOI] [PubMed] [Google Scholar]
- 49.Herrera RA, Kiontke K, Fitch DH. Makorin ortholog LEP-2 regulates LIN-28 stability to promote the juvenile-to-adult transition in Caenorhabditis elegans. Development. 2016;143(5):799–809. doi: 10.1242/dev.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scaraffia PY, Zhang Q, Thorson K, Wysocki VH, Miesfeld RL. Differential ammonia metabolism in Aedes aegypti fat body and midgut tissues. J Insect Physiol. 2010;56(9):1040–1049. doi: 10.1016/j.jinsphys.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen IA, et al. Forkhead transcription factors regulate mosquito reproduction. Insect Biochem Mol Biol. 2007;37(9):985–997. doi: 10.1016/j.ibmb.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenberg T, et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014;19(3):431–444. doi: 10.1016/j.cmet.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298(5599):1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- 54.Hays AR, Raikhel AS. A novel protein produced by the vitellogenic fat-body and accumulated in mosquito oocytes. Rouxs Arch Dev Biol. 1990;199(2):114–121. doi: 10.1007/BF02029559. [DOI] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawrence M, et al. Software for computing and annotating genomic ranges. PLOS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 59.Hussain M, O’Neill SL, Asgari S. Wolbachia interferes with the intracellular distribution of Argonaute 1 in the dengue vector Aedes aegypti by manipulating the host microRNAs. RNA Biol. 2013;10(12):1868–1875. doi: 10.4161/rna.27392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fejes AP, et al. FindPeaks 3.1: A tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24(15):1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 64.Huerta-Cepas J, et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44(D1):D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambrook J, Russell DW. 2006. Preparation of genomic DNA from mouse tails and other small samples. CSH Protoc 2006(1): 10.1101/pdb.p.
- 66.Kokoza VA, Raikhel AS. Targeted gene expression in the transgenic Aedes aegypti using the binary Gal4-UAS system. Insect Biochem Mol Biol. 2011;41(8):637–644. doi: 10.1016/j.ibmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.