Significance
The Amazon is largely covered by contiguous rain forest. Nevertheless, previous studies have suggested that past geological and climatic events, as well as limited seed dispersal, may have restricted the movement of tree lineages across the Amazon. Using a phylogenetic approach, we show that dispersal into local communities and larger regions in the Amazon appears to not have been limited on evolutionary timescales, but instead, local communities have been assembled by lineages from across the Amazon. These results contrast with those from seasonally dry tropical forests, where closely related species are clustered in geographic space. Furthermore, our results suggest a role for dispersal as an initiator for geographic isolation that might lead to speciation in Amazonian trees.
Keywords: Amazonia, biogeography, community assembly, phylogenetic structure, tropical trees
Abstract
We investigate patterns of historical assembly of tree communities across Amazonia using a newly developed phylogeny for the species-rich neotropical tree genus Inga. We compare our results with those for three other ecologically important, diverse, and abundant Amazonian tree lineages, Swartzia, Protieae, and Guatteria. Our analyses using phylogenetic diversity metrics demonstrate a clear lack of geographic phylogenetic structure, and show that local communities of Inga and regional communities of all four lineages are assembled by dispersal across Amazonia. The importance of dispersal in the biogeography of Inga and other tree genera in Amazonian and Guianan rain forests suggests that speciation is not driven by vicariance, and that allopatric isolation following dispersal may be involved in the speciation process. A clear implication of these results is that over evolutionary timescales, the metacommunity for any local or regional tree community in the Amazon is the entire Amazon basin.
Amazonia is well known to have the most species-rich tree communities on the planet, with more than 300 species (≥10 cm diameter) found in a single hectare (1). These communities are assembled from the species pool of Amazonia, which is estimated to number 16,000 species (2). Although some species are widespread across the Amazon basin (3), the majority are more restricted geographically (2), which provides the basis for schemes dividing the Amazon into floristic regions, including distinguishing the flora of the Guianan Shield from that of the Brazilian Shield or the western Amazon basin (4, 5).
The pattern of diverse local Amazonian tree communities assembled from a species pool composed mostly of regionally restricted species raises the question of how the regional communities are assembled through time. Regional communities could result from extensive local in situ speciation (6–8) with little subsequent dispersal. This would predict a pattern of geographically structured phylogenies with closely related species found in the same region. However, an idea that has been little tested using phylogenies of Amazonian plant species (9) is that the assembly of regional rain forest tree communities has been heavily influenced by historical dispersal of species. This would predict a pattern for communities that lacked geographic phylogenetic structure in which species from a single genus found in a regional community would be phylogenetically scattered.
Biogeographic studies of tree families that form important components of Amazonian forest, such as legumes (10), Annonaceae (11), Burseraceae (12), Chrysobalanaceae (13), and Meliaceae (14), have demonstrated that dispersal has been important in developing the distributions of such components across continents and oceans (15, 16). The existence of long-distance, transoceanic dispersal at an intercontinental scale suggests that there should be little to hinder dispersal across the flat, continuously forested Amazon basin given its lack of present-day physical barriers. Although there is some debate about the role of potential historical dispersal barriers in the Amazon, such as forest fragmentation during Pleistocene climate changes (17–19) and a large wetland complex (Pebas) or marine incursions that occupied much of western Amazonia in the Miocene (20, 21), these are far less substantial impediments to plant dispersal than major oceans.
Once a species does successfully disperse to a new location, it still needs to establish a population. Establishment can be challenging, given that any immigrant seed is numerically swamped by locally produced seeds (22), but nonetheless large-scale resident mortality in rain forests may be sufficiently common, owing to drought mortality or landscape rearrangements from radical movement of river courses, to allow for establishment of immigrant species (20, 23). Thus, we suggest that there has been ample opportunity for historical immigration to play a key role in the assembly of Amazonian tree communities, as has been proposed by Lavin (24) and Pennington and Dick (25), and we tested this hypothesis in the present work.
To do so, we used a new phylogeny of Inga [Leguminosae (Fabaceae): Mimosoideae] that samples local and regional communities in Amazonia, including the Guiana Shield, plus the Inga community on Barro Colorado Island in central Panama, to investigate patterns of historical community assembly (Fig. 1). The neotropical tree genus Inga is species-rich (>300 species), is widely distributed, and has consistently high local abundance (2, 26) and species richness, with up to 43 species recorded in 25 Ha (27). Thus, it is an excellent exemplar for use in the study of community assembly in neotropical rain forests. Our phylogeny of Inga is notable in that it samples thoroughly across multiple, geographically dispersed, local Amazonian tree communities in the context of good phylogenetic coverage of an entire clade. We compare our results for Inga at a regional scale with those for three other tree lineages—Swartzia (Leguminosae: Papilionoideae), Protieae (Burseraceae) and Guatteria (Annonaceae)—which are also ecologically important, diverse, and abundant in Amazonia, to investigate whether patterns in Inga are general for Amazonian tree communities. Finally, we contrast the picture of community assembly that we uncovered for Amazonian rain forest communities with patterns in the seasonally dry tropical forest biome, which has greater physical barriers to dispersal and different ecological barriers to establishment.
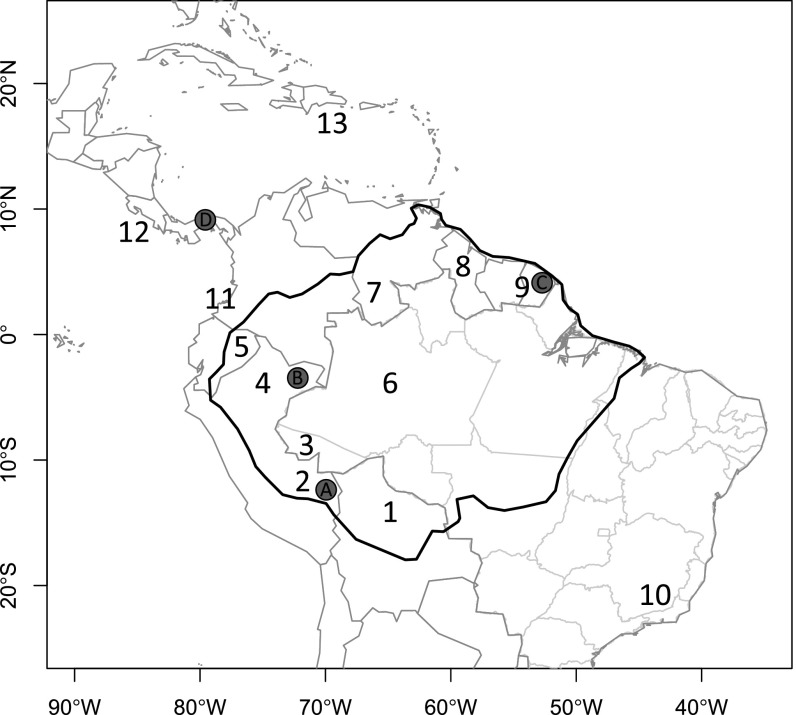
Fig. 1.
Map of the 13 neotropical regions used in the analyses of phylogenetic geographic structure for the four focal genera: 1, Amazonian Bolivia; 2, Madre de Dios, southern Peru; 3, Acre, Brazil; 4, Loreto, northern Peru; 5, Amazonian Ecuador; 6, Amazonas, Brazil; 7, Amazonas, Venezuela; 8, Guyana; 9, French Guiana; 10, Mata Atlantica (Atlantic rain forest); 11, Choco (trans-Andean) Colombia and Ecuador; 12, Central America, and 13, the Caribbean. Letters denote the location of the local communities of Inga (Leguminosae) that received in-depth sampling: A, Los Amigos Biological Station; B, Madreselva Biological Station; C, Nouragues Research Station; and D, Barro Colorado Island. The dark black line denotes our delimitation of Amazonia, which includes wet and moist forests across the Amazon basin and the Guianan Shield.
Results
Our phylogeny for Inga, which is based on eight molecular markers and includes 210 accessions of 124 species (Dataset S1), resolves relationships among major clades and shows that Inga communities in Peru, French Guiana, and Panama are composed of phylogenetically scattered species (Fig. 2 and Fig. S1). These findings, which show a clear lack of geographic structure in the phylogeny of Inga, are mirrored by the other tree lineages with numerous Amazonian species that we analyzed. We evaluated geographic phylogenetic structure by calculating phylogenetic diversity metrics for local communities and regions and comparing the observed values with a null expectation generated by randomly sampling species from the phylogenies. We used three phylogenetic diversity metrics (28, 29): (i) phylogenetic diversity sensu stricto (PDss), the total phylogenetic branch length present among species in a given community/region; (ii) mean pairwise distance (MPD), the mean of all pairwise phylogenetic distances among species in a given community/region; and (iii) mean nearest taxon distance (MNTD), the mean of the phylogenetic distance between each species and its closest relative in a given community/region. Communities showing significantly lower values than the null expectation indicate geographic phylogenetic structure or clustering, whereas significantly higher values than expected indicate phylogenetic overdispersion. None of the three local Amazonian communities showed phylogenetic clustering for any of the metrics evaluated (Dataset S2), whereas the Nouragues Research Station showed slight phylogenetic overdispersion. The Inga community on Barro Colorado Island, Panama, showed significant phylogenetic clustering, as evaluated by PDss and MPD.
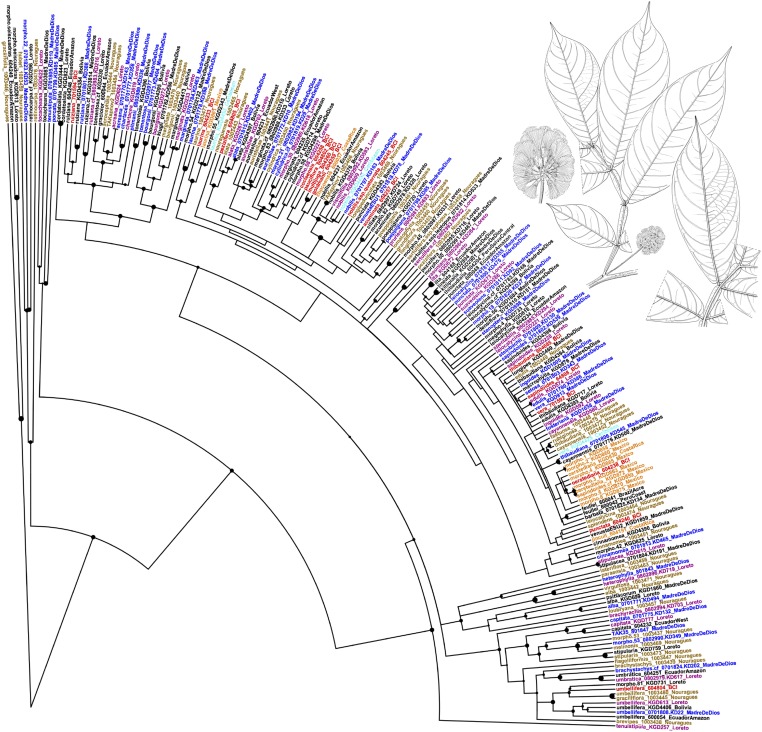
Fig. 2.
Phylogeny of 210 accessions representing 124 Inga (Leguminosae) species, with a maximum of one individual per species per region. Accessions from focal communities are color-coded as follows: blue, Los Amigos Biological Station; purple, Madreselva Biological Station; brown, Nouragues Research Station; red, Barro Colorado Island. Additional accessions are color-coded by biogeographic region: black, Amazon; orange, Central America; cyan, Caribbean. The size of the circles at nodes is proportional to bootstrap support. Details of tip labels and node support values are shown in Fig. S1. The line drawing at the top right is of I. pitmanii, a regionally restricted species, endemic to Madre de Dios, Peru. Reproduced with permission from ref. 74.
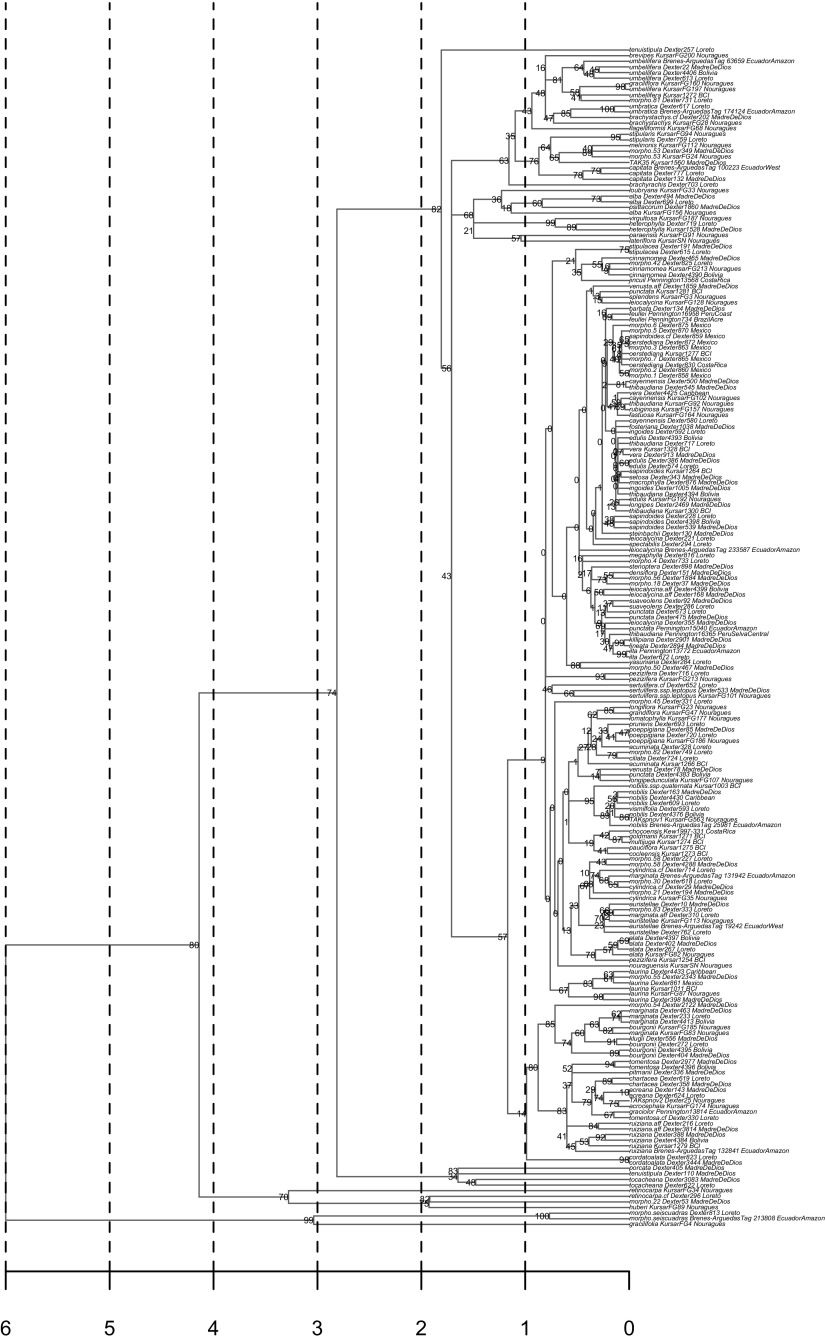
Fig. S1.
Maximum likelihood phylogeny for Inga (Leguminosae) after rate smoothing via penalized likelihood. The numbers to the left side of the nodes indicate the percentage of 1,000 maximum likelihood bootstrap replicates that support the relationship. Branch lengths are in terms of millions of years.
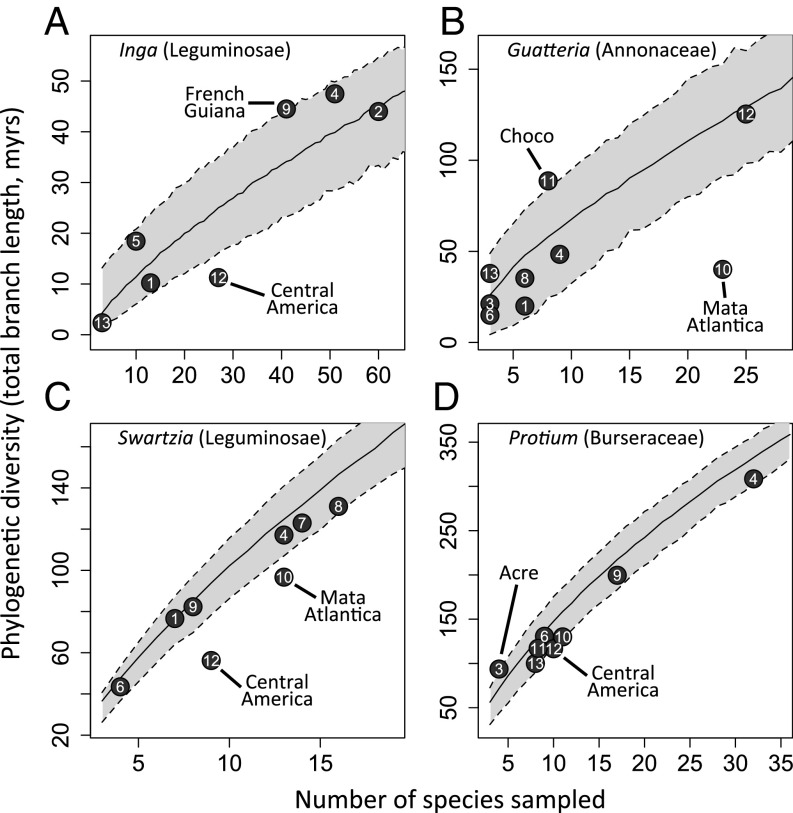
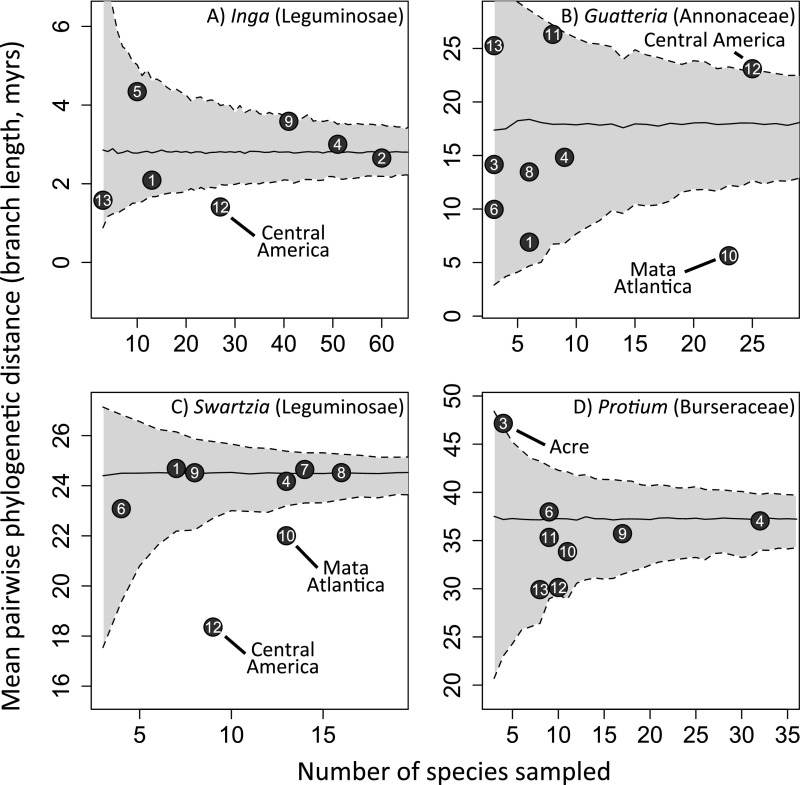
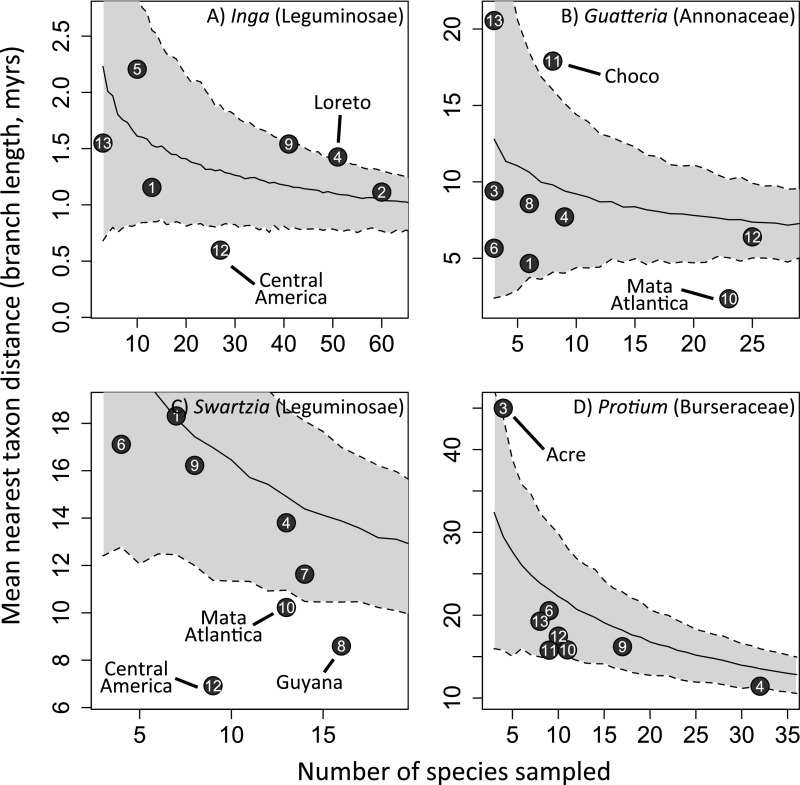
For Inga, we obtained sufficient samples from five Amazonian regions to test more broadly for geographic phylogenetic structure. As with local Amazonian Inga communities, no Amazonian region showed significant phylogenetic clustering by any metric (i.e., no points in Fig. 3 or in Figs. S2 and S3 below the gray area encompassing the 95% confidence interval; see also Dataset S2), whereas French Guiana showed slight phylogenetic overdispersion according to the PDss metric (Fig. 3; i.e., above the gray area encompassing the 95% confidence interval) and Loreto showed overdispersion using the MPD metric (Fig. S2). Meanwhile, Central America was the only region showing significant phylogenetic clustering for all three metrics (Fig. 3 and Figs. S2 and S3; i.e., in every case below the gray area encompassing the 95% confidence interval).
Fig. 3.
Relationship between number of taxa sampled and phylogenetic diversity in neotropical regions for four emblematic Amazonian tree genera. Phylogenetic diversity was evaluated as the sum of branch lengths in an ultrametric, temporally calibrated phylogeny including the taxa from a given region. Regions are numbered as in Fig. 1. The solid black line represents the mean null expectation for phylogenetic diversity given the number of taxa sampled, for 1,000 random draws of that number of taxa from the phylogenies. The shaded gray area denotes the 95% confidence intervals of the null expectation for the relationship. Regions that fall outside of the 95% confidence intervals are labeled.
Fig. S2.
Relationship between the number of taxa sampled and MPD in Neotropical regions for four emblematic Amazonian tree genera. Regions are numbered following Fig. 1. The solid black line represents the mean null expectation for MPD given the number of taxa sampled for 1,000 random draws of that number of taxa from the phylogenies. The shaded gray area denotes the 95% confidence intervals of the null expectation for that relationship. Regions that fall outside of the 95% confidence intervals are labeled.
Fig. S3.
Relationship between the number of taxa sampled and MNTD in Neotropical regions for four emblematic Amazonian tree genera. Regions are numbered following Fig. 1. The solid black line represents the mean null expectation for MNTD given the number of taxa sampled for 1,000 random draws of that number of taxa from the phylogenies. The shaded gray area denotes the 95% confidence intervals of the null expectation for that relationship. Regions that fall outside of the 95% confidence intervals are labeled.
This lack of geographic structure was duplicated in regional Amazonian communities of Swartzia, Protieae, and Guatteria, as measured by all metrics (Fig. 3, Figs. S2 and S3, and Dataset S2). All species in regional Amazonian communities represented a random draw from each phylogeny, as measured by all metrics, with the sole exception of the Swartzia community in Guyana as measured by MNTD (Fig. S3 and Dataset S2). The only cases in which species in regional communities were consistently more closely related than would be expected by chance were in Central America [Inga (all metrics), Swartzia (all metrics), Protieae (PDss)] and the Atlantic coastal rain forest of Brazil [Guatteria and Swartzia (all metrics)] (Fig. 3, Figs. S2 and S3, and Dataset S2). The level of sampling of different geographic regions varied widely (see the x-axes in Fig. 3 and Figs. S2 and S3), but well-sampled and poorly sampled Amazonian regions showed similar results. In general, neither departed significantly from null expectations for the phylogenetic diversity metrics.
Our results for geographic structure in Protieae differ slightly from those presented by Fine et al. (30), who calculated MTD and MNTD for major biogeographic regions in a global scale study of Protieae that included paleotropical species. First, the three Amazonian regions studied by Fine et al. (30)—eastern Amazonia, western Amazonia, and Guianas—are larger than those used here and thus not directly comparable. Furthermore, we analyzed only the neotropical clade of Protieae, given our focus on local and regional Amazonian communities, for which the neotropics alone may be a more appropriate wider metacommunity from which to draw random communities. Including paleotropical species, which form two clades basal to the neotropical species of Protieae, would have the effect of inflating values of phylogenetic diversity in the random communities, which also may have contributed to the greater evidence of phylogenetic clustering reported by Fine et al. (30) in the regional communities that they studied.
Discussion
Primacy of Historical Dispersal in the Assembly of Local and Regional Communities.
Our results demonstrate that tree communities at a local scale (for Inga) and a regional scale (for Inga, Swartzia, Protieae, and Guatteria) are assembled by dispersal across Amazonia. Species in all local Amazonian Inga communities and virtually all regional communities across all lineages are a random draw from the phylogeny in each of our exemplar taxa. This shared pattern is seen despite the different fruit morphologies of these lineages, reflecting a variety of vertebrate dispersers. Inga is dispersed primarily by primates; Protieae’s small endozoochorous fruits attract a wide variety of birds, bats, and terrestrial mammal species (31); Guatteria has been observed to be eaten by primates and birds (32); and Swartzia is dispersed by birds (33), primates (34), and, in one species, water (35).
The sole exception to this lack of phylogenetic geographic structuring is found outside of Amazonia in the rain forests of Atlantic coastal Brazil (in Swartzia and Guatteria) and Central America (Swartzia, Inga, Protieae). The phylogenetic clustering found in these areas may reflect their isolation from the Amazon by major physical barriers—the Andes mountains for Central America and a “dry diagonal” of seasonally dry vegetation formations across eastern Brazil for the Brazilian Atlantic coast (36, 37). In addition, the presence of physical barriers isolating these non-Amazonian areas has been suggested as an explanation for the greater phylogeographic structure found there among populations of Symphonia globulifera, a widespread tree species (38).
The lack of geographic phylogenetic structure demonstrated here implies that, on evolutionary timescales, the metacommunity for any regional or local tree community in the Amazon is the entire Amazon basin. This does not preclude a role for ecological filtering in the assembly of local communities. Previous work by us and others has demonstrated that Inga species in Madre de Dios have clear habitat preferences, and that environmental filtering affects species composition of Inga communities (39–41). Furthermore, our work has shown that Inga species that defend themselves against herbivores in distinct ways are more likely to co-occur, signifying filtering based on herbivore defense traits (42). Thus, ecological processes clearly can play a role in local community assembly. Nonetheless, the species that may populate any given region and provide species for local communities could have ancestry from anywhere in the Amazon and from any clade of the Inga phylogeny.
Interestingly, the average relatedness of co-occurring congeneric species differs markedly among the four genera that we have studied in this work (Fig. S2). For example, the average phylogenetic distance between co-occurring Inga species is 3 My (divergence time of 1.5 My), whereas that among Protieae species is 36 My. This difference could have significant implications for the level of ecological interaction among co-occurring Inga and Protieae species; for example, competition might be considered more intense among Inga species because of their recent divergence (43), which in turn could influence the composition of local and regional communities. However, our analyses tend to suggest that the average phylogenetic distance among co-occurring species of a given genus may depend simply on the age of the genus, although the exact phylogenetic distance estimates will depend on how well the genus has been sampled phylogenetically. Furthermore, the high degree of sympatric co-occurrence observed for the species-rich genera studied here suggests that there might not be strong constraints on the number of co-occurring congeneric species, especially if they differ in terms of herbivore defense traits (42, 44, 45). One of the key factors influencing the number of co-occurring species of a given genus in a given Amazonian tree community may simply be the total diversity of that genus in the Amazon, because dispersal into regions, which provide species for local communities, does not seem to be limited (46).
We emphasize that the generality of our results may apply only to larger trees, and that there are indications that patterns of geographic structure in phylogenies of shrubs, understory trees, and other tropical plant life forms may differ (47). For example, the phylogeny of the tropical rain forest herb genus Pilea is highly congruent with geography, which may reflect limited pollen dispersal and mechanical dispersal of seeds over very short distances of a few millimeters (48). Our results also contrast with studies published for large terrestrial birds (49) and primates (50), which show more geographically structured phylogenetic patterns.
Contrasting Patterns of Community Assembly in Different Biomes.
The pattern of assembly of regional tree communities reported for the neotropical seasonally dry forest biome (24, 51, 52) differs markedly from that discovered here for regional Amazonian communities. Phylogenies of several genera of woody plants characteristic of seasonally dry tropical forests in the neotropics (e.g., Coursetia, Poissonia, Cyathostegia, Amicia) demonstrate that clades of species are confined to single regions of dry forest, such as the Brazilian caatingas (53) or seasonally dry Andean valleys (52). These differences are not artifacts of the age of clades, because the crown clades of these dry forest genera are older than that of Inga; despite historical dispersal having had less time to operate in Inga, successful dispersal and establishment events are more prevalent.
The geographic phylogenetic structure seen in dry forest clades may reflect two factors (51). First, unlike the continuous Amazon rain forest, dry forest areas are scattered across the neotropics and are physically isolated by high mountains or areas of mesic vegetation, which may limit dispersal among them (51). Second, ecological factors operating over evolutionary timescales are different in dry forests, which may affect the likelihood of propagules establishing after dispersal (51, 54). For example, there may be more opportunities for immigration into rain forests, where drought can cause widespread tree mortality (23), and landscape evolution is known to be dynamic over evolutionary timescales in Amazonia, especially via radical movement of river courses (20, 55), which may be an additional source of environmental instability, creating opportunities for successful immigration.
Implications for Processes of Diversification in Amazonian Rain Forest Trees.
A key role for dispersal in Inga and other important tree genera has implications for understanding speciation histories in Amazonian rain forests. For Amazonian trees, the lack of geographic phylogenetic structure that we find in local and regional communities provides little support for large-scale reconfigurations of the landscape causing common vicariance of continuous populations of multiple species, a conclusion reached recently for Amazonian birds (56). Large-scale geological events that subdivide populations would lead to congruent geographic phylogenetic patterns across lineages, but there is little evidence suggesting common deep imprints of geological events in Amazonian tree phylogenies. For example, a geographic phylogenetic structure across the Miocene Pebas wetlands is not detected in the phylogenies of Inga, Swartzia, Guatteria, or Protieae; instead, geographic patterns are particular to lineages, reflecting a primacy for idiosyncratic historical dispersal in generating distributions (25, 53). The lack of congruent patterns suggests that allopatric speciation involving population vicariance caused by common geological factors is unlikely.
Rather than geological phenomena that isolate regions, our results for multiple Amazonian tree lineages are more consistent with the founding of isolated peripheral populations by dispersal, which could then lead to speciation. This model is also consistent with patterns of phylogenetic nesting of species within paraphyletic progenitor species seen in some Amazonian tree lineages (57). An alternative model would be more localized speciation followed by extensive dispersal, which also could result in the random phylogenetic composition of tree communities that we report here, as well as nesting of species within paraphyletic ancestors. Such local speciation could result from hybridization or adaptation to soil types (6, 8, 30, 58). The documented intersterility of sympatric Inga species (59) argues against a role for hybridization in speciation of that genus; however, our greatest challenge to understanding the mechanism of speciation is the possibility that rampant dispersal may overwrite the original signature of genetic divergence. To distinguish the relative importance of ecological divergence, breeding systems, and allopatric isolation in driving diversification of Amazonian trees, it would be fruitful to further characterize the variations in the functional ecology, biology, and underlying genetics of species of Inga and other diverse tree genera across their ranges.
Materials and Methods
Sampling.
In the Amazon basin and Guianas, together composing what we term Amazonia, we sampled 181 Inga individuals, representing 105 total species (including 20 unidentified morphospecies). Outside of the Amazon basin, we sampled two species in Ecuador west of the Andes, three species in the Caribbean, and 23 species in Central America. In total, our phylogenetic sampling for Inga included four local communities and seven regional communities and comprised 210 individuals from 124 species (Datasets S1 and S2). This represents many more accessions and more than double the species sampling in previously reported Inga phylogenies (sampled from 37–55 species) (39, 42, 60). Because our goal was to sample as many species as possible in individual local and regional communities, we sampled 44 of the total 124 species more than once, because these species were present in more than one region. We did not sample any species more than once within any one local or regional community.
Swartzia (Leguminosae: Papilionoideae) contains ∼200 neotropical species found from southern Mexico to southern Brazil, including the Caribbean Islands (61). Swartzia occurs in a variety of habitats but is especially typical of lowland rain forests, where 10 or more species can be found growing in sympatry (62). Phylogenetic data and the sampling locality for each accession of Swartzia come from Torke and Schaal (63), who sampled 76 species, including multiple exemplar species of each of the infrageneric groupings (64), covering the full geographic range of the genus.
The tribe Protieae (Burseraceae), comprising Protium together with Tetragastris and Crepidospermum nested within it, is an important tree lineage in terms of its diversity and abundance in neotropical and paleotropical rain forests (2, 30). The majority of Protieae species are found in the Amazon basin and the Guianas, but smaller numbers of species occur in other areas, including Central America, the Caribbean, and the Brazilian Atlantic forest. Phylogenetic data for Protieae come from Fine at al. (30), who sampled 102 species covering 75% of accepted species names and all pantropical areas of distribution.
Guatteria (Annonaceae) is an abundant and diverse component of lowland rain forests in the neotropics and is a member of the magnoliids, a basally divergent angiosperm lineage. The genus is hypothesized to have originated in Africa and to have colonized South America via North and then Central America during the late Miocene (65). Nevertheless, Guatteria is most diverse in lowland Amazonia (66, 67). The published phylogeny of the genus covers 97 of 265 named species from Central America to the Mata Atlantica, with 39 accessions covering 38 species sampled from Amazonia (Bolivia, Peru, Colombia, Brazil, and the Guianas), representing 40% of the species found in these areas (67).
Phylogenetic Reconstruction.
For Inga, we sequenced seven chloroplast regions (rpoCI, psbA-trnH, rps16, trnL-F, trnD-T, ndhF-rpl32, and rpl32-trnL; 5,916 aligned bp) and the nuclear ribosomal internal transcribed spacer regions (ITS 1 and 2; 572 aligned bp) (Dataset S1). PCR and sequencing protocols for chloroplast regions have been reported by Kursar et al. (42), and those for ITS have been published by Richardson et al. (60) and Dexter et al. (39). Sequences were initially aligned using MAFFT (68) and then adjusted manually, which was straightforward given the low sequence divergence. The phylogeny was estimated under a maximum likelihood framework using RAxML with separate partitions and models for ITS and cpDNA and 1,000 bootstrap replicates to estimate node support (69). The phylogeny was subsequently time-calibrated using penalized likelihood (70), with the crown age constrained to 6 My, following previous work (24, 60). The phylogeny is available as Dataset S3.
The Inga phylogeny resolves numerous clades with reasonable bootstrap support (Fig. 2 and Fig. S1) and is the best-resolved Inga phylogeny to date, although within major clades the relationships among closely related species are not always well resolved, reflecting the recent evolutionary radiation of the genus (60). The topology of our phylogeny is largely congruent with that presented by Nicholls et al. (71) based on 194 nuclear loci, which shows high support for all branches. There are only two strongly supported incongruencies between the two phylogenies, involving two species, Inga laurina and Inga ruiziana, and a formal statistical test (72) showed that the phylogenies are significantly congruent (Icong = 1.46; P = 0.0016). Although Nicholls et al. (71) sampled only 22 Inga species, the topological congruence gives confidence that our less well-supported phylogeny accurately reflects phylogenetic relationships.
For Swartzia, aligned sequences from Torke and Schaal (63) were downloaded from TreeBase and a phylogeny was estimated under a maximum likelihood optimality criterion as described for Inga using separate partitions and models for ITS, AAT1, and chloroplast DNA. This phylogeny was subsequently time-calibrated using penalized likelihood, with the crown age constrained to 13.6 My, following previous authors (73). For Protieae, the time-calibrated Bayesian phylogeny reported by Fine et al. (30) was downloaded from TreeBase. For Guatteria, sequences reported by Erkens et al. (66) were downloaded from GenBank, and a phylogeny was estimated under a maximum likelihood optimality criterion as described above for Inga with a single partition and model, because all loci reported are from the chloroplast genome. This phylogeny was subsequently time-calibrated using penalized likelihood, with the crown age constrained to 17.2 My following Erkens et al. (65).
Analyses of Geographic Phylogenetic Structure.
We analyzed geographic phylogenetic structure at two scales (Fig. 1): local communities (Inga only) and regions (across all groups). In the case of Inga, we were able to sample all or nearly all species in four local communities (see above) at Los Amigos and Madreselva Biological Stations (Peru), Nouragues Research Station (French Guiana), and Barro Colorado Island (Panama) (Fig. 1). The scale of the local communities varied from ∼6 km2 (Madreselva) to 15.6 km2 (Barro Colorado Island).
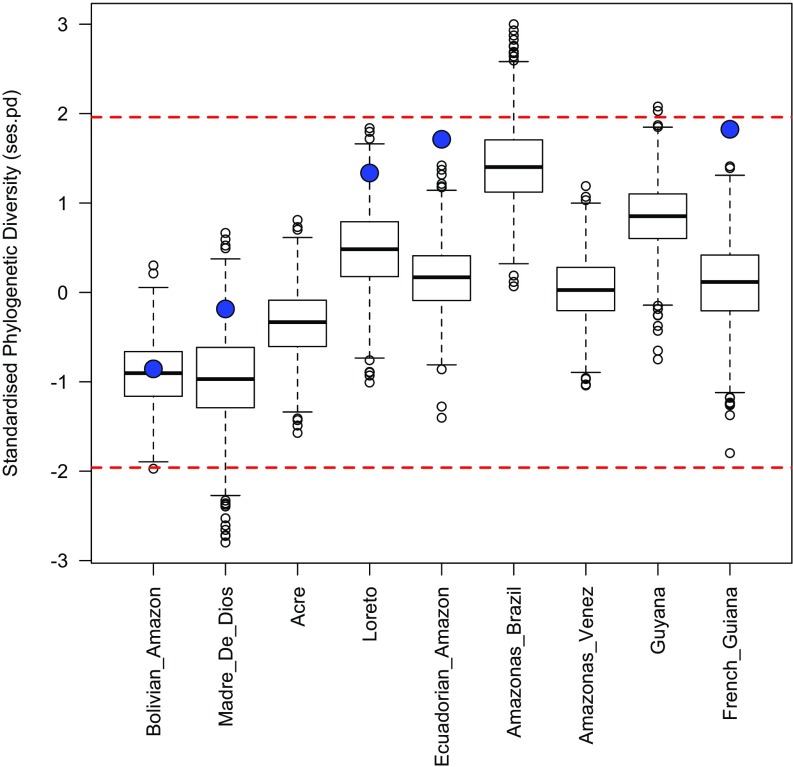
We defined 13 geographic regions with sufficient sampling (five or more species in nearly all cases) that could be analyzed across the different phylogenies (Fig. 1) using our knowledge of Inga and Swartzia and information provided by Fine et al. (30) for Protieae and by Erkens et al. (66) for Guatteria. In Amazonia and the Guianas, these are geographic political units of similar size, such as states in Brazil, departments in Peru, and countries such as Guyana. Beyond Amazonia and the Guianas, the defined regions were the Mata Atlantica (Atlantic coastal rain forest) of Brazil, the Choco of Colombia and Ecuador (i.e., South American rain forests on the Pacific coast west of the Andes), Central America (Panama north to Mexico), and the Caribbean. If an accession sampled in our phylogenies came from one of these regions, as indicated by its published locality (30, 63, 66), it was scored as present there. An alternative approach would be to assign a given species in the phylogeny to every region in which it is known to occur (30). This approach might be problematic if accessions are misidentified or not positively identified (i.e., morphospecies) or if species distributions are imperfectly known. For Inga, we conducted a series of sensitivity analyses to assess whether results were robust to our approach of assigning accessions only to the regions in which they were collected, and this revealed no effect on our results (SI Materials and Methods and Figs. S4–S6).
Fig. S4.
Distribution of ses.pd values for different Amazonian regions across 1,000 iterations of the sensitivity analyses. The values from the analyses presented in the main text are shown by the large blue circles. These are available only for Amazonian regions actually sampled in our phylogeny. Values <−1.96 indicate significant phylogenetic clustering, whereas values >1.96 indicate significant phylogenetic overdispersion. These threshold values are indicated by dashed red lines. Overall, these results demonstrate that most iterations of the sensitivity analyses do not result in significant phylogenetic clustering or overdispersion for Amazonian regions.
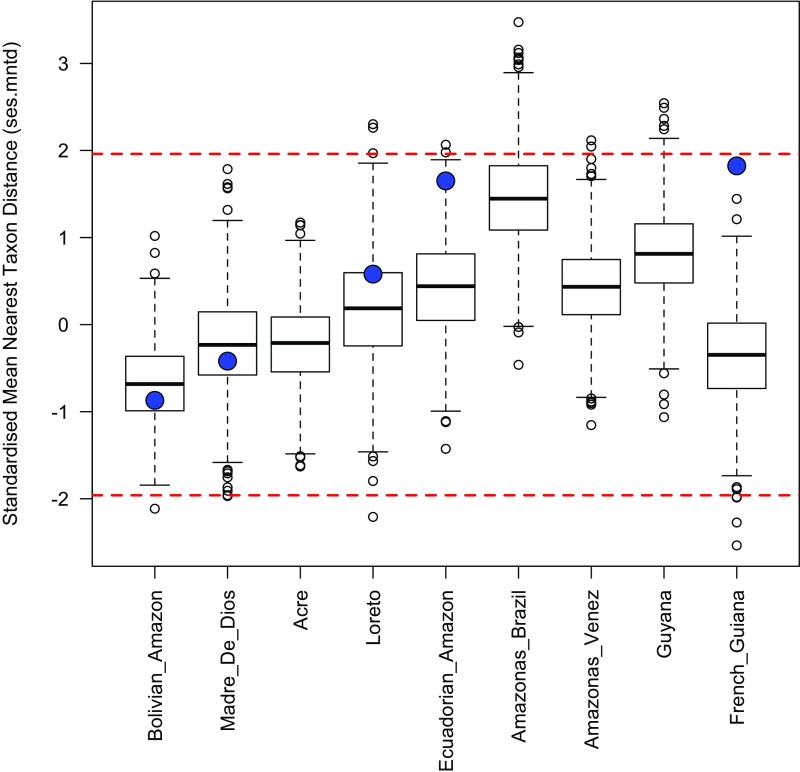
Fig. S6.
Distribution of ses.mntd values for different Amazonian regions across 1,000 iterations of the sensitivity analyses. The values from the analyses presented in the main text are shown by the large blue circles. These are available only for Amazonian regions actually sampled in our phylogeny. Values <−1.96 indicate significant phylogenetic clustering, whereas values >1.96 indicate significant phylogenetic overdispersion. These threshold values are indicated by dashed red lines. Overall, these results demonstrate that most iterations of the sensitivity analyses do not result in significant phylogenetic clustering or overdispersion for Amazonian regions.
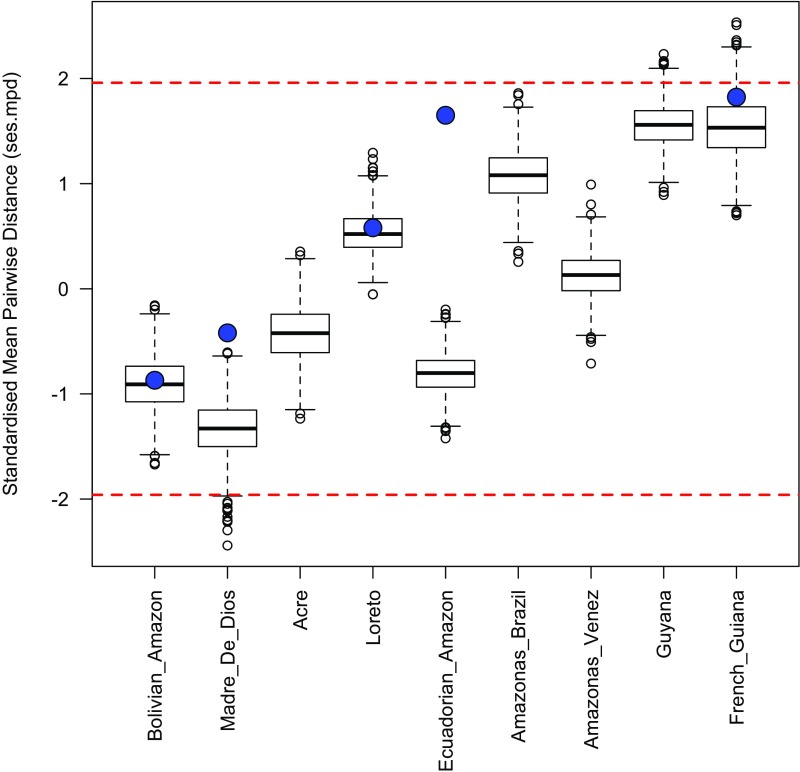
Fig. S5.
Distribution of ses.mpd values for different Amazonian regions across 1,000 iterations of the sensitivity analyses. The values from the analyses presented in the main text are shown by the large blue circles. These are available only for Amazonian regions actually sampled in our phylogeny. Values <−1.96 indicate significant phylogenetic clustering, whereas values >1.96 indicate significant phylogenetic overdispersion. These threshold values are indicated by dashed red lines. Overall, these results demonstrate that most iterations of the sensitivity analyses do not result in significant phylogenetic clustering or overdispersion for Amazonian regions.
If closely related species within a clade (in this case Inga, Swartzia, Protieae, or Guatteria) are found near each other in geographic space because they originated by local, in situ speciation with little subsequent dispersal, then we would expect the phylogenetic diversity represented by species in regions and local communities to be less than that if the same number of species were drawn randomly from across the phylogeny. Conversely, if distant dispersal is common over one or multiple generations, causing local and regional communities to be assembled stochastically from a wide geographic pool, then we would expect the phylogenetic diversity in communities and regions to be more commensurate with a random draw from the phylogeny. We evaluated phylogenetic diversity using three metrics described above. The null expectations for each of these metrics, and the uncertainty around them, were calculated by randomly drawing the same number of species as present in communities/regions from the phylogeny and repeating this process 999 times. Significant phylogenetic clustering for a given community/region was considered present when the observed phylogenetic diversity metric was less than the lower 2.5% quantile of the randomly generated distribution for that species richness, whereas significant overdispersion was indicated by a value exceeding the 97.5% quantile.
SI Materials and Methods
We conducted sensitivity analyses to assess the robustness of our results to uncertainty in the age of Inga clades, the topology of the Inga phylogeny, and the assignment of Inga species to different geographic regions. We ran a Bayesian analysis to calibrate the Inga phylogeny temporally while simultaneously estimating its topology, using BEAST v1.8.2 (75). Because there are no definitively identified fossils for Inga, we constrained the crown age of Inga in this phylogeny (using a lognormal prior with a mean of 6 My and an SD of 0.5) based on dates reported by Richardson et al. (60) and Lavin (24). For each iteration of the sensitivity analyses, we sampled one tree at random from the post–burn-in posterior distribution of trees from the BEAST analysis.
In our primary analyses presented in the main text, the species lists for a given geographic region comprise all species in a region that were sampled by accessions in the phylogeny. An alternative approach would be to include all species present in the phylogeny that are known to occur in the region based on their overall distribution (rather than just those sampled by accessions from the region in our phylogeny). Our primary approach has the advantages of not assuming monophyly of species (and not all Inga species are monophyletic; Fig. S1) and not assuming perfect taxonomy and knowledge of species distributions; however, this means that species lists for a given region might not include many species found in the region. As can be seen by examining the x-axes in Fig. 3 and Figs. S2 and S3, our level of sampling for different regions varied widely. Thus, we conducted additional analyses assigning Inga species to each region in which they are known to occur, based on distributions reported by Pennington (26) and our own field work. Because many species in the phylogeny are represented by multiple accessions, we selected a single accession for each species at random. This random selection introduces stochasticity into calculations, so we repeated this process 999 times. For each repetition, we started with a topology selected at random from the posterior distribution of trees (see above), which served to generate a range of results representing uncertainty in phylogenetic topology and ages.
For each iteration, we assessed whether a given Amazonian tree community showed more or less phylogenetic diversity than would be expected by chance by calculating the standardized effect size for each phylogenetic diversity metric (ses.pd, ses.mpd, and ses.mntd). Positive values indicate phylogenetic overdispersion, whereas negative values indicate phylogenetic clustering. Because these metrics are standardized (with an expected value of 0 and an SD of 1), values <−1.96 or >1.96 represent communities that show significant phylogenetic overdispersion or clustering, respectively. To assess how our results compare with those obtained using our primary approach, we assessed the value for each metric across the 1,000 iterations and compared these values with the values generated with the approach described in the main text (Figs. S4–S6). As can be seen, the median values from this alternative approach are slightly lower than those obtained in our analyses presented in the main text (on average); however, for the large majority of the iterations, none of the Amazonian communities show significant phylogenetic clustering (or overdispersion) by any metric. Thus, these sensitivity analyses demonstrate that Amazonian Inga communities represent a random draw from the Inga phylogeny, and that this result is robust to uncertainty in the age of Inga clades, the topology of the Inga phylogeny, and the method of assignment of Inga species to different geographic regions.
Supplementary Material
Acknowledgments
We thank Terry Pennington for help with Inga species identification, and Paul Fine and Hans ter Steege for helpful comments and discussions. R.T.P. thanks the Leverhulme Trust for Study Abroad Fellowship RF/2/2006/0142, which supported DNA sequencing and the development of ideas in this paper in the M.L. laboratory. K.G.D., P.D.C., and T.A.K. thank the Peruvian Ministry of Agriculture for permission to conduct research in Peru, and the Nouragues Research Station for grants to fund fieldwork in French Guiana (2009 Call for Research at Nouragues). K.G.D. and R.T.P. acknowledge the Natural Environment Research Council for Support via NE/I028122/1. Field research by P.D.C. and T.A.K. was supported by National Science Foundation Grants DEB-0640630 and Dimensions 1125733. A.D.T. is supported by Natural Environment Research Council Fellowship NE/L011336/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.C.D. is a Guest Editor invited by the Editorial Board.
Data deposition: The novel DNA sequences generated for this publication have been deposited in GenBank (accession nos. KY592383–KY593119). Dataset S1 details the sequences associated with each sequenced accession.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613655114/-/DCSupplemental.
References
- 1.Valencia R, Balslev H, Paz Y, Miño CG. High tree alpha-diversity in Amazonian Ecuador. Biodivers Conserv. 1994;3:21–28. [Google Scholar]
- 2.ter Steege H, et al. Hyperdominance in the Amazonian tree flora. Science. 2013;342(6156):1243092. doi: 10.1126/science.1243092. [DOI] [PubMed] [Google Scholar]
- 3.Pitman NCA, et al. Dominance and distribution of tree species in upper Amazonian terra firme forests. Ecology. 2001;82:2101–2117. [Google Scholar]
- 4.Prance GT. The phytogeographic subdivisions of Amazonia and their influence on the selection of biological reserves. In: Prance GT, Elias TS, editors. Extinction is Forever. New York Botanical Garden Press; Bronx, NY: 1977. pp. 195–213. [Google Scholar]
- 5.ter Steege H, et al. Continental-scale patterns of canopy tree composition and function across Amazonia. Nature. 2006;443(7110):444–447. doi: 10.1038/nature05134. [DOI] [PubMed] [Google Scholar]
- 6.Gentry AH. Distributional patterns and an additional species of the Passiflora vitifolia complex: Amazonian species diversity due to edaphically differentiated communities. Plant Syst Evol. 1981;137:95–105. [Google Scholar]
- 7.Moritz C, Patton JL, Schneider CJ, Smith TB. Diversification of rainforest faunas: An integrated molecular approach. Annu Rev Ecol Syst. 2000;31:533–563. [Google Scholar]
- 8.Fine PVA, Daly DC, Villa Muñoz G, Mesones I, Cameron KM. The contribution of edaphic heterogeneity to the evolution and diversity of Burseraceae trees in the western Amazon. Evolution. 2005;59(7):1464–1478. [PubMed] [Google Scholar]
- 9.Fiaschi P, Pirani JR. Review of plant biogeographic studies in Brazil. J Syst Evol. 2009;47:477–496. [Google Scholar]
- 10.Lavin M. Metacommunity process rather than continental tectonic history better explains geographically structured phylogenies in legumes. Philos Trans R Soc Lond B Biol Sci. 2004;359:1509–1522. doi: 10.1098/rstb.2004.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson JE, Chatrou LW, Mols JB, Erkens RHJ, Pirie MD. Historical biogeography of two cosmopolitan families of flowering plants: Annonaceae and Rhamnaceae. Philos Trans R Soc Lond B Biol Sci. 2004;359(1450):1495–1508. doi: 10.1098/rstb.2004.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weeks A, Daly DC, Simpson BB. The phylogenetic history and biogeography of the frankincense and myrrh family (Burseraceae) based on nuclear and chloroplast sequence data. Mol Phylogenet Evol. 2005;35(1):85–101. doi: 10.1016/j.ympev.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 13.Bardon L, et al. Origin and evolution of Chrysobalanaceae: Insights into the evolution of plants in the Neotropics. Bot J Linn Soc. 2013;171:19–37. [Google Scholar]
- 14.Muellner AN, Savolainen V, Samuel R, Chase MW. The mahogany family “out-of-Africa”: Divergence time estimation, global biogeographic patterns inferred from plastid rbcL DNA sequences, extant, and fossil distribution of diversity. Mol Phylogenet Evol. 2006;40(1):236–250. doi: 10.1016/j.ympev.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Renner SS. Tropical trans-Atlantic disjunctions, sea surface currents, and wind patterns. Int J Plant Sci. 2004;165:S23–S33. [Google Scholar]
- 16.Pennington RT, Dick CW. The role of immigrants in the assembly of the South American rainforest tree flora. Philos Trans R Soc Lond B Biol Sci. 2004;359(1450):1611–1622. doi: 10.1098/rstb.2004.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haffer J. Speciation in Amazonian forest birds. Science. 1969;165(3889):131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- 18.Colinvaux PA, DeOliveira PE, Moreno JE, Miller MC, Bush MB. A long pollen record from lowland Amazonia: Forest and cooling in glacial times. Science. 1996;274:85–88. [Google Scholar]
- 19.Bueno ML, et al. Effects of quaternary climatic fluctuations on the distribution of Neotropical savanna species. Ecography. 2016 doi: 10.1111/ecog.01860. [DOI] [Google Scholar]
- 20.Hoorn C, et al. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science. 2010;330(6006):927–931. doi: 10.1126/science.1194585. [DOI] [PubMed] [Google Scholar]
- 21.Latrubesse EM, et al. The Late Miocene paleogeography of the Amazon Basin and the evolution of the Amazon River system. Earth Sci Rev. 2010;99:99–124. [Google Scholar]
- 22.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton Univ Press; Princeton, NJ: 2001. [DOI] [PubMed] [Google Scholar]
- 23.Rowland L, et al. Death from drought in tropical forests is triggered by hydraulics not carbon starvation. Nature. 2015;528(7580):119–122. doi: 10.1038/nature15539. [DOI] [PubMed] [Google Scholar]
- 24.Lavin M. Floristic and geographic stability of discontinuous seasonally dry tropical forests explains patterns of plant phylogeny and endemism. In: Pennington RT, Ratter JA, Lewis GP, editors. Neotropical Savannas and Seasonally Dry Forests: Plant Biodiversity, Biogeography and Conservation. CRC Press; Boca Raton, FL: 2006. pp. 433–447. [Google Scholar]
- 25.Pennington RT, Dick CW. Diversification of the Amazonian flora and its relation to key geological and environmental events: a molecular perspective. In: Hoorn C, Vonhof H, Wesselingh F, editors. Amazonia, Landscape and Species Evolution: A Look into the Past. Wiley-Blackwell; Hoboken, NJ: 2010. pp. 373–385. [Google Scholar]
- 26.Pennington TD. The Genus Inga: Botany. Royal Botanic Gardens; Kew, UK: 1997. [Google Scholar]
- 27.Valencia R, et al. Tree species distributions and local habitat variation in the Amazon: Large forest plot in eastern Ecuador. J Ecol. 2004;92:214–229. [Google Scholar]
- 28.Webb CO. Exploring the phylogenetic structure of ecological communities: An example for rain forest trees. Am Nat. 2000;156(2):145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 29.Honorio-Coronado EN, et al. Phylogenetic diversity of Amazonian tree communities. Divers Distrib. 2015;21:1295–1307. [Google Scholar]
- 30.Fine PVA, Zapata F, Daly DC. Investigating processes of neotropical rain forest tree diversification by examining the evolution and historical biogeography of the Protieae (Burseraceae) Evolution. 2014;68(7):1988–2004. doi: 10.1111/evo.12414. [DOI] [PubMed] [Google Scholar]
- 31.Daly DC. 1987. A taxonomic revision of Protium Burm. F. (Burseraceae) in Eastern Amazonia and the Guianas. PhD dissertation, City University of New York, New York.
- 32.Van Roosmalen MGM. A Guide to the Fruits of the Guianan Flora. University of Utrecht Press/Veenman; Wageningen, The Netherlands: 1985. [Google Scholar]
- 33.Hamrick JL, Murawski DA, Nason JD. The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. In: Fleming TH, Estrada A, editors. Frugivory and Seed Dispersal: Ecological and Evolutionary Aspects. Springer; Netherlands: 1993. pp. 281–297. [Google Scholar]
- 34.Chapman CA. Primate seed dispersal: The fate of dispersed seeds. Biotropica. 1989;21:148–154. [Google Scholar]
- 35.Williamson GB, Costa F, Vera CVM. Dispersal of Amazonian trees: Hydrochory in Swartzia polyphylla. Biotropica. 1999;31:460–465. [Google Scholar]
- 36.Pennington RT, Prado DE, Pendry CA. Neotropical seasonally dry forests and Quaternary vegetation changes. J Biogeogr. 2000;27:261–273. [Google Scholar]
- 37.Neves DMR, Dexter KG, Pennington RT, Bueno ML, Oliveira-Filho AT. Environmental and historical controls of floristic composition across the South American dry diagonal. J Biogeogr. 2015;42:1566–1576. [Google Scholar]
- 38.Dick CW, Heuertz M. The complex biogeographic history of a widespread tropical tree species. Evolution. 2008;62(11):2760–2774. doi: 10.1111/j.1558-5646.2008.00506.x. [DOI] [PubMed] [Google Scholar]
- 39.Dexter KG, Pennington TD, Cunningham CW. Using DNA to assess errors in tropical tree identifications: How often are ecologists wrong and when does it matter? Ecol Monogr. 2010;80:267–286. [Google Scholar]
- 40.Dexter KG, Terborgh JW, Cunningham CW. Historical effects on beta diversity and community assembly in Amazonian trees. Proc Natl Acad Sci USA. 2012;109(20):7787–7792. doi: 10.1073/pnas.1203523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Endara MJ, Jaramillo J. The influence of microtopography and soil properties on the distribution of the speciose genus of trees, Inga (Fabaceae: Mimosoideae), in Ecuadorian Amazonia. Biotropica. 2011;43:157–164. [Google Scholar]
- 42.Kursar TA, et al. The evolution of antiherbivore defenses and their contribution to species coexistence in the tropical tree genus Inga. Proc Natl Acad Sci USA. 2009;106(43):18073–18078. doi: 10.1073/pnas.0904786106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerhold P, Cahill JF, Winter M, Bartish IV, Prinzing A. Phylogenetic patterns are not proxies of community assembly mechanisms (they are far better) Funct Ecol. 2015;29:600–614. [Google Scholar]
- 44.Sedio BE, Wright SJ, Dick CW. Trait evolution and the coexistence of a species swarm in the tropical forest understorey. J Ecol. 2012;100:1183–1193. [Google Scholar]
- 45.Salazar D, Jaramillo MA, Marquis RJ. Chemical similarity and local community assembly in the species rich tropical genus Piper. Ecology. 2016;97(11):3176–3183. doi: 10.1002/ecy.1536. [DOI] [PubMed] [Google Scholar]
- 46.Ricklefs RE. Community diversity: Relative roles of local and regional processes. Science. 1987;235(4785):167–171. doi: 10.1126/science.235.4785.167. [DOI] [PubMed] [Google Scholar]
- 47.Givnish TJ. On the causes of gradients in tropical tree diversity. J Ecol. 1999;87:193–210. [Google Scholar]
- 48.Monro AK. The revision of species-rich genera: A phylogenetic framework for the strategic revision of Pilea (Urticaceae) based on cpDNA, nrDNA, and morphology. Am J Bot. 2006;93(3):426–441. doi: 10.3732/ajb.93.3.426. [DOI] [PubMed] [Google Scholar]
- 49.Ribas CC, Aleixo A, Nogueira ACR, Miyaki CY, Cracraft J. A palaeobiogeographic model for biotic diversification within Amazonia over the past three million years. Proc Biol Sci. 2012;279(1729):681–689. doi: 10.1098/rspb.2011.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morales-Jimenez AL, Disotell T, Di Fiore A. Revisiting the phylogenetic relationships, biogeography, and taxonomy of spider monkeys (genus Ateles) in light of new molecular data. Mol Phylogenet Evol. 2015;82(Pt B):467–483. doi: 10.1016/j.ympev.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 51.Pennington RT, Lavin M, Oliveira-Filho A. Woody plant diversity, evolution, and ecology in the tropics: Perspectives from seasonally dry tropical forests. Annu Rev Ecol Evol Syst. 2009;40:437–457. [Google Scholar]
- 52.Särkinen T, Pennington RT, Lavin M, Simon MF, Hughes CE. Evolutionary islands in the Andes: Persistence and isolation explain high endemism in Andean dry tropical forests. J Biogeogr. 2012;39:884–900. [Google Scholar]
- 53.Hughes CE, Pennington RT, Antonelli A. Neotropical plant evolution: Assembling the big picture. Bot J Linn Soc. 2013;171:1–18. [Google Scholar]
- 54.Pennington RT, et al. Contrasting plant diversification histories within the Andean biodiversity hotspot. Proc Natl Acad Sci USA. 2010;107(31):13783–13787. doi: 10.1073/pnas.1001317107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkinson JM, Marshall LG, Lundberg JG, Kreslavsky MH. Megafan environments in northern South America and their impact on Amazon Neogene aquatic ecosystems. In: Hoorn C, Vonhof H, Wesselingh F, editors. Amazonia, Landscape and Species Evolution: A Look into the Past. Wiley-Blackwell; Hoboken, NJ: 2010. pp. 162–184. [Google Scholar]
- 56.Smith BT, et al. The drivers of tropical speciation. Nature. 2014;515(7527):406–409. doi: 10.1038/nature13687. [DOI] [PubMed] [Google Scholar]
- 57.Pennington RT, Lavin M. The contrasting nature of woody plant species in different neotropical forest biomes reflects differences in ecological stability. New Phytol. 2016;210(1):25–37. doi: 10.1111/nph.13724. [DOI] [PubMed] [Google Scholar]
- 58.Misiewicz TM, Fine PVA. Evidence for ecological divergence across a mosaic of soil types in an Amazonian tropical tree: Protium subserratum (Burseraceae) Mol Ecol. 2014;23(10):2543–2558. doi: 10.1111/mec.12746. [DOI] [PubMed] [Google Scholar]
- 59.Koptur S. Outcrossing and pollinator limitation of fruit set: Breeding systems of neotropical Inga trees (Fabaceae: Mimosoideae) Evolution. 1984;38:1130–1143. doi: 10.1111/j.1558-5646.1984.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 60.Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science. 2001;293(5538):2242–2245. doi: 10.1126/science.1061421. [DOI] [PubMed] [Google Scholar]
- 61.Torke BM, Perez AJ. Notes on the genus Swartzia (Leguminosae) in Ecuador, with descriptions of two new species. Phytotaxa. 2013;147:13–25. [Google Scholar]
- 62.de Oliveira AA, Mori SA. A central Amazonian terra firme forest, I: High tree species richness on poor soils. Biodiv Cons. 1999;8:1245–1259. [Google Scholar]
- 63.Torke BM, Schaal BA. Molecular phylogenetics of the species-rich neotropical genus Swartzia (Leguminosae, Papilionoideae) and related genera of the swartzioid clade. Am J Bot. 2008;95(2):215–228. doi: 10.3732/ajb.95.2.215. [DOI] [PubMed] [Google Scholar]
- 64.Torke BM, Mansano V. A phylogenetically based sectional classification of Swartzia (Leguminosae, Papilionoideae) Taxon. 2009;58:913–924. [Google Scholar]
- 65.Erkens RHJ, Maas JW, Couvreur TLP. From Africa via Europe to South America: Migrational route of a species-rich genus of Neotropical lowland rain forest trees (Guatteria, Annonaceae) J Biogeogr. 2009;36:2338–2352. [Google Scholar]
- 66.Erkens RHJ, Chatrou LW, Maas JW, van der Niet T, Savolainen V. A rapid diversification of rainforest trees (Guatteria; Annonaceae) following dispersal from Central into South America. Mol Phylogenet Evol. 2007;44(1):399–411. doi: 10.1016/j.ympev.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 67.Maas PJM, et al. Confronting a morphological nightmare: Revision of the Neotropical genus Guatteria (Annonaceae) Blumea. 2015;60:1–219. [Google Scholar]
- 68.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 70.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19(1):101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 71.Nicholls JA, et al. Using targeted enrichment of nuclear genes to increase phylogenetic resolution in the neotropical rain forest genus Inga (Leguminosae: Mimosoideae) Front Plant Sci. 2015;6:710. doi: 10.3389/fpls.2015.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Vienne DM, Giraud T, Martin OC. A congruence index for testing topological similarity between trees. Bioinformatics. 2007;23(23):3119–3124. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- 73.Simon MF, et al. Recent assembly of the Cerrado, a neotropical plant diversity hotspot, by in situ evolution of adaptations to fire. Proc Natl Acad Sci USA. 2009;106(48):20359–20364. doi: 10.1073/pnas.0903410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dexter KG, Pennington TD. Inga pitmanii (Fabaceae), a new species from Madre de Dios, Peru. Novon. 2011;21:322–325. [Google Scholar]
- 75.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.