Significance
Damage to DNA bases due to oxidative stress is thought to be deleterious, leading to stalled replication forks and mutations. Similarly, folding of DNA strands into G-quadruplexes slows the progression of polymerases, requiring specialized helicases for unfolding before transcription. In the case of oxidative damage in a G-quadruplex–forming sequence of a promoter, we show that the presence of the DNA damage lesion 8-oxoguanine (OG) leads to an ∼300% increase in gene expression. This concept was demonstrated by chemical synthesis of a segment of the vascular endothelial growth factor (VEGF) or endonuclease III-like protein 1 (NTHL1) promoter with a site specifically incorporated lesion in a reporter plasmid. This observation is direct evidence that OG represents an epigenetic modification and G-quadruplex–forming sequences can serve as sensors of oxidative stress.
Keywords: oxidative damage, epigenetics, G-quadruplex, gene regulation, base excision repair
Abstract
Reactive oxygen species (ROS) have emerged as important cellular-signaling agents for cellular survival. Herein, we demonstrate that ROS-mediated oxidation of DNA to yield 8-oxo-7,8-dihydroguanine (OG) in gene promoters is a signaling agent for gene activation. Enhanced gene expression occurs when OG is formed in guanine-rich, potential G-quadruplex–forming sequences (PQS) in promoter-coding strands, initiating base excision repair (BER) by 8-oxoguanine DNA glycosylase (OGG1), yielding an abasic site (AP). The AP enables melting of the duplex to unmask the PQS, adopting a G-quadruplex fold in which apurinic/apyrimidinic endonuclease 1 (APE1) binds, but inefficiently cleaves, the AP for activation of vascular endothelial growth factor (VEGF) or endonuclease III-like protein 1 (NTHL1) genes. These details were mapped via synthesis of OG and AP analogs at single-nucleotide precision within the promoter of a luciferase reporter system. The reporters were analyzed in human and mouse cells while selectively knocking out or down critical BER proteins to identify the impact on luciferase expression. Identification of the oxidatively modified DNA base OG to guide BER activity in a gene promoter and impact cellular phenotype ascribes an epigenetic role to OG.
The conventional view of oxidatively induced DNA damage, such as 8-oxo-7,8-dihydroguanine (OG, Fig. 1A), is that it is detrimental to cellular processes. For example, when OG is located in recognition elements of nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB) (1), specificity protein 1 (SP1) (2), or CAMP responsive element binding protein 1 (CREB) (3) transcription factors, protein-binding affinity was significantly reduced. When OG was present in the template strand of protein-coding regions, modest stalling of RNA pol II occurred (4), whereas initiation of DNA repair at OG to yield an abasic site (AP) stopped RNA pol II, leading to truncated transcripts (5). These observations support a hypothesis of OG decreasing gene transcription. In contrast, livers of mice with infection-induced colitis exhibit increased levels of genomic OG in tandem with enhanced expression of many DNA repair, cell cycle, and stress response genes (6). Another notable example appears when rat pulmonary artery endothelial cells are subject to hypoxia; a strong positive correlation between OG in promoter regions and elevated expression of >100 genes was observed (7). One gene in particular is vascular endothelial growth factor (VEGF), for which OG was found in the G-rich potential G-quadruplex sequence (PQS) (7) demonstrated to be responsible for transcriptional regulation of the gene (8). Strong cellular evidence for enhancement of transcription via folding of promoter G-quadruplexes was recently demonstrated by the Balasubramanian laboratory (9). Therefore, we hypothesize formation of OG in the VEGF PQS under oxidative stress conditions functions as a signaling mark to unmask the G-quadruplex fold, thus leading to transcriptional activation. Experiments supporting this hypothesis are described herein.
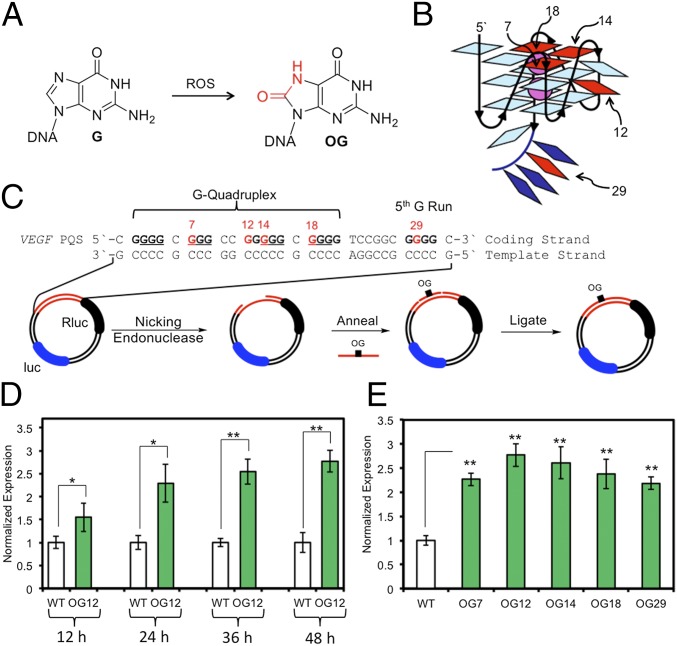
Fig. 1.
Oxidation of G in the VEGF PQS induces transcription. (A) G oxidation to OG. (B) VEGF G-quadruplex, labeled with positions studied. (C) VEGF sequence with Gs in the core underlined, reporter system design, and method for site-specific incorporation of DNA modifications. (D) Time-dependent and (E) position-dependent expression at 48 h posttransfection of OG-containing reporters in glioblastoma cells. WT refers to the plasmid containing the VEGF PQS with undamaged Gs. Error bars represent 95% CI on the basis of four or eight replicates. Significance values for each comparison were calculated by a Student’s t test. Significance at *P < 0.05 or **P < 0.01 is indicated.
The VEGF PQS possesses five G-tracks in which the four tracks required for folding to a parallel-stranded G-quadruplex are the four 5′-most (Fig. 1B) (10). Our previous work mapped the most reactive guanine (G) bases in VEGF toward oxidation, leading to OG and other secondary oxidation products (11). When DNA damage resides in the four G-tracks of the VEGF G-quadruplex (G4), DNA repair was not observed (12); to our surprise, addition of the fifth G-track allowed a structural transition to a competent G-quadruplex fold by extruding the damaged G run into a long loop, folding in the fifth track to reform the G4, and allowing faithful DNA repair of the looped out, damaged G (11). In the present work, a mechanism for gene induction driven by G oxidation to OG that induces a structural switch in the VEGF PQS promoter element is proposed and experimentally validated in human and mouse cells. Induction of transcription was found to require 8-oxoguanine DNA glycosylase (OGG1) and apurinic/apyrimidinic endoDNase 1 (APE1), also known as redox effector factor 1 (Ref-1), in the base excision repair (BER) pathway. The coupling of BER of OG and transcriptional activation observed in the present study leads to a hypothesis that G modification to OG may be epigenetic as a modification that regulates gene expression.
Results and Discussion
Demonstration that OG drives the VEGF PQS promoter element to induce transcription was accomplished using a luciferase reporter plasmid. Key features of the reporter system include the VEGF PQS promoter element with all five G runs regulating the Renilla luciferase gene (Rluc). The regulatory sequence has flanking nicking endonuclease sequences allowing replacement of the G-rich sequence with a synthetic oligomer containing a single, site-specific OG (Fig. 1C). Additionally, the plasmid possessed the firefly luciferase gene (luc) regulated by an unmodified promoter used as an internal standard (Fig. 1C and SI Appendix, Fig. S1). The OG positions selected are based on the VEGF G-quadruplex structure solved by nuclear magnetic resonance (NMR) (Fig. 1 B and C) (10).
Changes in gene expression as a function of OG position focused on the oxidation-prone VEGF PQS sites 7, 12, 14, and 18 (11). Position 12 is in a loop, whereas 7, 14, and 18 occupy core positions in the G4, providing contrasting views on DNA damage and structure (Fig. 1B). Also studied was position 29, residing in the fifth G-track 3′ to the principal G4 structure. First, the time-dependent expression of Rluc with OG incorporated at position 12 was evaluated upon transfection of the plasmid into glioblastoma cells (U-87 MG), and the reported expression was normalized against the expression of the internal standard luc. From 12 to 48 h posttransfection, the expression of Rluc significantly increased to nearly threefold when OG was present compared with the WT plasmid (i.e., the all-G–containing plasmid, Fig. 1D). Next, when OG was analyzed at other sites (i.e., 7, 14, 18, or 29), measurements made 48 h posttransfection found Rluc expression was enhanced by 2.2- to 3.0-fold (Fig. 1E). These results demonstrate that the presence of OG in the VEGF promoter PQS enhances the transcriptional output of the reporter gene; significantly, OG was not detrimental, but rather increased transcription. This observation is consistent with a previous study monitoring VEGF expression under hypoxic conditions that identified approximately threefold greater expression in tandem with the presence of OG in the PQS promoter element (7).
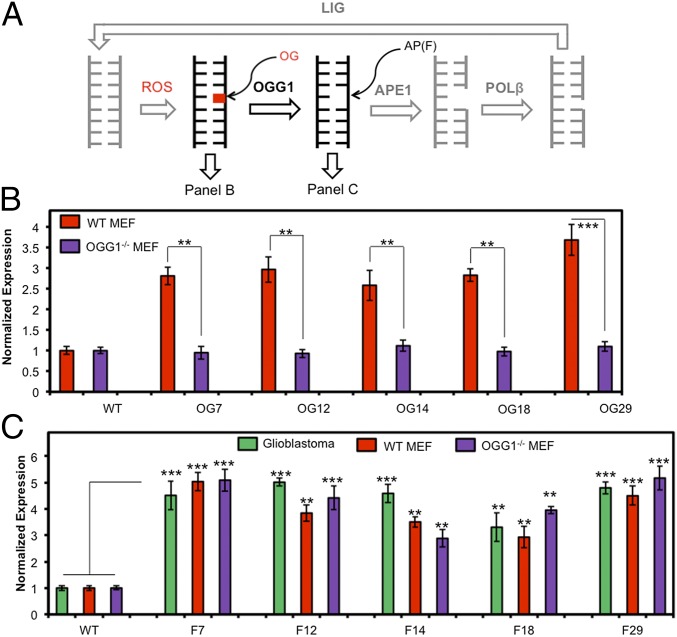
Experiments to reveal molecular details by which OG induced gene expression were conducted. In the mammalian genome, OG is bound and cleaved by OGG1 in the first step of BER (Fig. 2A). Whether OGG1 cleaves the phosphodiester backbone or APE1 catalyzes this step remains unanswered (Fig. 2A) (13). To establish whether OGG1 was involved in gene induction, we conducted comparative studies in WT and OGG1−/− mouse embryonic fibroblasts (MEFs) transfected with OG-containing plasmids. In WT MEFs, depending on the position of OG, Rluc expression increased by 2.5- to 3.9-fold, whereas OG-containing plasmids in the OGG1−/− MEFs yielded essentially no change in the amounts of Rluc expression compared with the WT plasmid (Fig. 2B). Because OG was in the coding strand of the promoter, it does not interfere with advancement of RNA pol II on the promoter to the transcription start site in the OGG1−/− MEFs, and, therefore, the same gene output was observed with and without OG.
Fig. 2.
BER initiates gene activation when OG is located in a promoter PQS. (A) The BER pathway. (B) Positional dependency in expression of OG-containing reporters in WT and OGG1−/− MEFs. (C) Positional and cell line dependency in expression of F-containing reporters, where F = the stable AP analog THF. Error bars represent 95% CI on the basis of four or eight replicates. Significance values for each comparison were calculated by a Student’s t test. Significance at **P < 0.01 or ***P < 0.001 is indicated.
The observation that OG does not increase transcription in the OGG1−/− MEFs confirms OGG1 was critical to expression enhancement; however, more questions remain. In WT cells, after removal of OG by OGG1, an AP is present in DNA, and we questioned whether an AP could also impact transcription. Consequently, plasmids with APs (THF analog, F) at the reactive positions were introduced to establish whether gene induction still occurs. Transfection of the AP-containing plasmids into all three cell lines yielded Rluc expression enhancement at levels of 2.5- to 6.0-fold relative to the WT plasmid (Fig. 2C). These results identify a stronger sequence and cell line dependency in transcriptional amplification than observed with OG. The AP analog F is more stable than a natural AP, likely resulting in the increased luciferase expression detected. The enhanced expression observed with an abasic site, especially in the OGG1−/− MEFs, identifies APE1 as the possible BER enzyme responsible for enhancing gene expression in this promoter sequence.
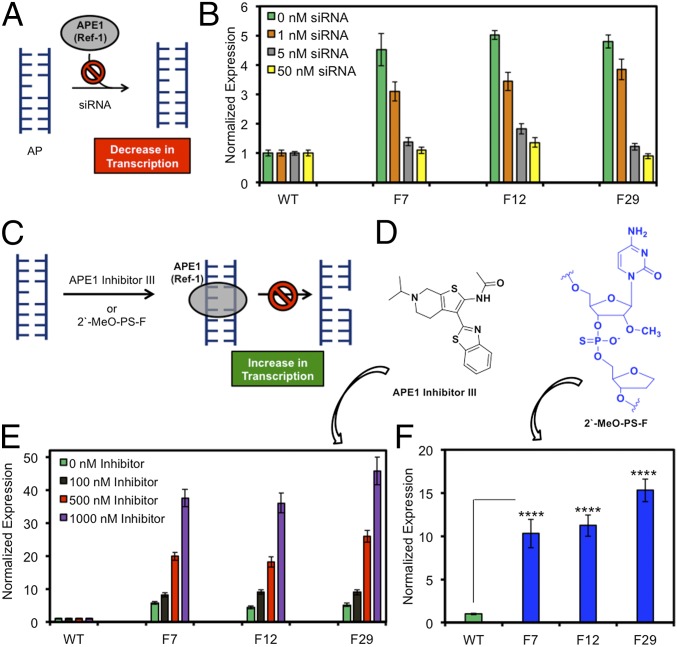
To further confirm that APE1 induces transcription, we treated glioblastoma cells with siRNA to knock down APE1 and prevent further processing of the AP (Fig. 3A). Glioblastoma cells titrated with APE1-specific siRNA in the range of 1–50 nM yielded a dose–response in the extent of Rluc expression when AP-containing plasmids were transfected (Fig. 3B). At the highest siRNA concentration studied, Rluc expression for each position studied was similar to the WT plasmid (Fig. 3B). This initial observation adds further support for the importance of APE1 in gene activation when an AP is present in the VEGF promoter PQS. Next, we wanted to understand if the ability of APE1 to bind the AP or cleave the site was responsible for gene activation (Fig. 3C). First, APE1 inhibitor III (Fig. 3D) was titrated at a concentration range of 100–1,000 nM into glioblastoma cells before transfection of the AP-containing plasmids. The inhibitor prevents cleavage of the backbone, whereas APE1 binding is not strongly impacted (Fig. 3C) (14). In these studies, Rluc expression increased up to >30-fold with a dose–response in the inhibitor concentration for the F-containing plasmid, whereas the WT plasmid remained the same throughout the titration (Fig. 3E).
Fig. 3.
Binding of APE1 (Ref-1) to AP in the VEGF PQS promoter element enhances gene transcription in glioblastoma cells. (A) Knockdown of APE1 and prevention of AP binding leads to decreased Rluc expression. (B) Expression levels measured when F-containing reporter plasmids were transfected into APE1 knockdown cells with 0–50 nM siRNA. (C) Mechanism for prevention of APE1 cleavage without impacting binding of an AP leading to increased gene expression. (D) Structures of APE1 inhibitor III and the 2′-MeO-PS-F–modified AP site. (E) Expression level measured when cells were treated with 100–1,000 nM APE1 inhibitor III. (F) Expression level measured when cells were transfected with the poorly cleavable 2′-MeO-PS-F analog. Error bars represent 95% CI on the basis of four or eight replicates. Significance values for each comparison were calculated by a Student’s t test. Significance at ****P < 0.0001 is indicated.
Further verification that APE1 binding to APs without causing a strand break leads to gene induction was achieved by studying APs poorly cleaved by APE1 (Fig. 3C). Previous experiments found APE1 binds normally but poorly cleaves oligomers containing a phosphorothioate and 2′-OMe nucleotide 5′ to the AP analog F (2′-MeO-PS-F, Fig. 3D) (15). Therefore, 2′-MeO-PS-F modified plasmids were transfected into glioblastoma cells, and >10-fold Rluc expression was observed (Fig. 3F). To rule out a strand break leads to the increased expression observed, an OG-containing plasmid was treated with Fpg and APE1 to yield a strand break before transfection. Transfection of this plasmid did not yield an observable Rluc expression. This result is likely a consequence of the poor efficiency of transfecting strand-broken plasmids into cells. The combined results of these studies support OG inducing transcription via OGG1 generation of an AP in the VEGF PQS followed by APE1 binding. More importantly, gene induction occurs while APE1 was bound (Fig. 3 C, E, and F), and it does not require the phosphodiesterase activity of APE1 to yield a strand break for gene activation.
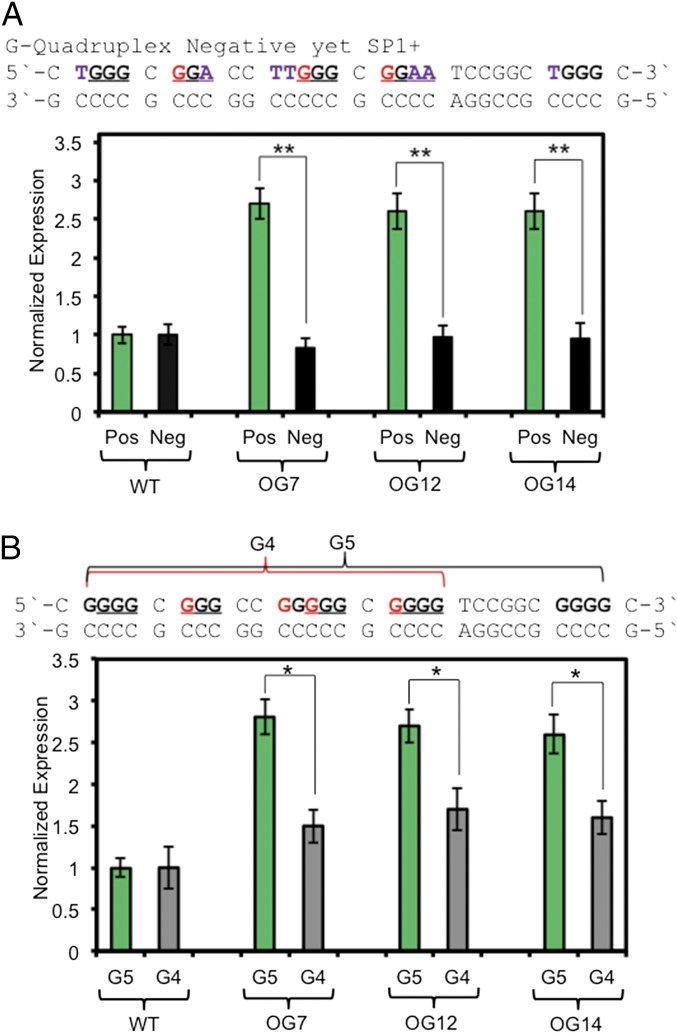
The next set of studies aimed to understand the possible involvement of G-quadruplex formation for induction of Rluc with the VEGF PQS promoter element. Previous experiments found the VEGF PQS to be bound by three equivalents of the SP1 transcription factor, and G4 formation is also involved in activation of this gene (8, 16). Decoupling of G-quadruplex formation from SP1 binding is complicated by the observation that this transcription factor binds both duplex and G-quadruplex folded DNA of the same sequence with similar affinity (17); additionally, OG in duplex DNA decreases SP1 binding by 70–90% depending on the position (2), and the impact in G-quadruplex DNA is not known. A sequence recognizable by SP1 but incapable of G-quadruplex formation (SI Appendix, Fig. S2) was designed for incorporation of OG (G-quadruplex negative SP1+; Fig. 4A). This new OG-containing plasmid was transfected into glioblastoma cells yielding a similar expression as observed in the WT plasmid with the same G-quadruplex negative sequence (Fig. 4A). This null result strongly supports the conclusion that the ability to adopt a G-quadruplex fold is important in gene activation when OG is present.
Fig. 4.
Gene induction with OG in the VEGF PQS requires the G-quadruplex fold. (A) Expression observed when OG is in a G-quadruplex positive or negative folding sequence context. (B) Requirement of the fifth G run for maximal expression when OG is present. Error bars represent 95% CI on the basis of four or eight replicates. Significance values for each comparison were calculated by a Student’s t test. Significance at *P < 0.05 or **P < 0.01 is indicated.
In another experiment, the VEGF sequence was judiciously mutated by removing a G run and converting one run to all T nucleotides to prevent G-quadruplex formation and SP1 binding. Transfection of this sequence with and without OG into glioblastoma cells also found OG did not impact transcription (SI Appendix, Fig. S3). These control experiments do not rule out all sequence context effects or the role of all protein activators and repressors but they do provide strong support for this sequence to adopt a G-quadruplex fold to induce the gene. The observation of transcriptional activation by G4 formation in the present study is consistent with a recent demonstration that these folds in promoters up-regulate transcription on the basis of G-quadruplex ChIP-Seq in tandem with RNA-Seq analysis (9).
We previously demonstrated the concept that the five-track VEGF G5 can switch structures to extrude damaged DNA bases to maintain the fold by replacing a damaged run with the fifth track (11). To verify if there is an essential role for the fifth G-track in gene activation, plasmids containing OG with only the four G runs required for folding (10) were transfected. The G4 samples gave significantly less gene expression than the G5 analogs, identifying the fifth G-track as essential to achieve the maximal amount of gene induction (Fig. 4B). Negative supercoiling in DNA was demonstrated by the Hurley laboratory to promote G4 formation (18); therefore, a comparison of Rluc expression in relaxed vs. supercoiled plasmids (plasmid state of all studies) with OG at position 12 of the VEGF G5 was made to find a similar result for both systems (SI Appendix, Fig. S4). This experiment supports the state of the plasmid leading to expression in the cell is similar for both supercoiled and relaxed plasmids. Furthermore, this observation hints that folding of VEGF to a G-quadruplex may be an essential step in the transcriptional activation process.
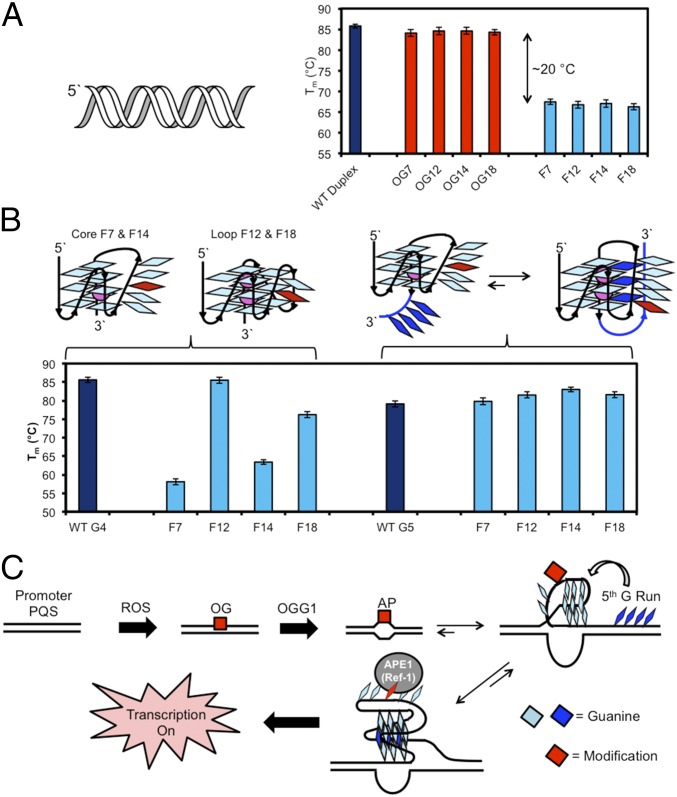
After establishing the essential role for a G-quadruplex in gene activation, we circled back to better understand how an AP site favors G-quadruplex formation and how APE1 functions in this context. When DNA is comprised of canonical Watson–Crick base pairs, the duplex context is thermodynamically favored, and even OG base paired with C does not significantly impact the duplex state (Fig. 5A); however, an AP site (i.e., F) significantly impacts the thermal stability of duplex DNA, leading to opening of the duplex, allowing the PQS to adopt a G-quadruplex fold. The ability of an AP to unmask the G-quadruplex was demonstrated by conducting a series of thermal melting experiments (Tm). In the VEGF duplex an AP site decreased the Tm by ∼20 °C at the positions studied (Fig. 5A). Evaluation of Tm values in the G4 context found loop APs decreased the Tm by >20 °C; in contrast, when the fifth domain was present in the G5 context the Tm values remained nearly the same as the nonmodified sequence while maintaining a G-quadruplex structure (Fig. 5B and SI Appendix, Fig. S5). We propose that maintaining the thermal stability results from extrusion of the damaged G run and replacing it with the fifth G-track (Fig. 5B). This observation is consistent with our previous studies that found extrusion of a damaged G-track by the fifth domain yields a more stable fold for highly helix-distorting modified DNA bases (11). Furthermore, recruitment of the fifth domain was most efficient for APs in core positions because they caused the greatest disruption to the G4-fold. This observation supports a mechanism by which the AP can facilitate G-quadruplex formation by shifting the duplex–quadruplex equilibrium.
Fig. 5.
Formation of an AP in the VEGF PQS shifts the duplex–quadruplex equilibrium. (A) Tm studies for the VEGF duplex derived from the G4 sequence provide comparisons between WT, OG-containing, and F-containing strands. (B) Tm studies for the VEGF G-quadruplex comparing positional dependency of an F residue in G4 vs. G5 sequences. (C) Proposed mechanism by which initiation of OG removal in the VEGF promoter PQS allows a structural switch to occur for binding of APE1 and activation of transcription.
The interaction between APE1 and an AP was previously studied in the G4 context by the C.J.B. laboratory and others, and it was found that APE1 binds APs in G4s, but the cleavage rate was highly attenuated (12, 19). The attenuated cleavage by APE1 on the promoter may provide the trigger to increase the rate of transcription observed by allowing APE1 to idle on the G4/G5 structural motif. The Ref-1 domain of APE1 is known to interact with protein factors (e.g., HIF1-α, STAT3, and CBP/p300) that bind promoter sequences such as the VEGF PQS and increase gene transcription (20); however, the detailed molecular interactions are not known at present. This proposed mechanism suggests any structure in DNA that prevents APE1 cleavage of an AP would lead to transcriptional activation. To test this hypothesis, we transfected plasmids into glioblastoma cells with noncleavable APs in both G-quadruplex negative sequences studied previously and found a significant increase of >1.5-fold in Rluc expression (SI Appendix, Fig. S6). The increase observed was less than that observed in the G-quadruplex context, suggesting the folded structure must play an additional role for induction of the gene that is not currently well understood. On the basis of these studies, gene induction by OG in the VEGF promoter PQS occurs when OGG1 binds and cleaves OG from the duplex to yield an AP. The AP shifts the duplex–quadruplex equilibrium to the G-quadruplex fold followed by APE1 binding and gene activation via the gene regulatory Ref-1 domain of APE1 (Fig. 5C).
Activation of the VEGF promoter PQS in the genomic context would require site-specific G oxidation to OG to initiate the BER process in a gene promoter. Oxidation can result from cellular oxidants (6), via long-range electron transfer (21), or active chromatin remodeling catalyzed by lysine demethylase 1A or 1B (LSD1 or LSD2) (22, 23). The latter pathway identified by Perillo et al. (22) occurs by the flavin-dependent LSD1 during demethylation of H3K9me2 to generate H2O2 near the DNA; LSD2 also catalyzes a similar reaction. The H2O2 so formed is most likely activated by the Fe(II)- or Cu(I)-mediated Fenton reaction to oxidize G to OG (and other base oxidation products) or an AP (24, 25). Perillo et al. demonstrated estrogen-induced activation of the BCL-2 gene occurs by this LSD1-mediated DNA oxidation mechanism in MCF7 cells, and they noted an essential role for OGG1 of BER in the activation process. Curiously, the region found to be oxidized in the BCL-2 promoter houses a PQS hypothesized to be responsible for gene regulation (18). The present results support a proposal that oxidation of G to OG in the BCL-2 PQS directs OGG1 to unmask the G4-fold to alter gene expression ultimately through APE1 (Fig. 5C). Thus, G oxidation in a regulatory PQS may serve as an epigenetic DNA modification to alter cellular phenotype.
A recent report by Boldogh and coworkers (26) found cellular oxidative stress oxidized G most likely to OG in a G-rich region adjacent to an NF-κB binding site in the TNF-α promoter of HEK293 cells. They propose OG was then bound by OGG1 that recruited NF-κB to its binding site and increased transcription. In a study by Antoniali et al., gene activation by OG was suspected in SIRT1 expression under oxidative stress conditions in HeLa cells (27); their work highlighted the importance of OGG1 and APE1 in the gene activation process. Together, these three genome-level studies support the concept that the actions of BER in gene promoters guided by oxidative modification of G can increase transcription; however, they could not definitively claim that OG was the oxidative modification responsible for the observed phenotypic change, or if the presence of OG was a consequence of the change in cellular state. Our studies reported here provide this evidence.
In the present work, we addressed these questions by synthetically installing OG with single-nucleotide precision in a reporter system that allowed unambiguous demonstration of OG enhancing transcription. Furthermore, we confirmed OGG1 is required to initiate BER on OG, leading to increased transcription in the VEGF PQS promoter. The VEGF PQS can adopt a G4/G5-fold with an AP that APE1 binds yet poorly cleaves to increase transcription (Fig. 5C). Because the action of APE1 on an AP results in increased transcription, the present experiments address how oxidations yielding APs, such as LSD1-generated H2O2 followed by Fenton chemistry, can change the transcriptional output of a gene without requiring OG. However, other G oxidation products from Fenton chemistry, such as the hydantoin products (24, 25), are substrates for the NEIL1 and 2 glycosylases and do not require APE1 (28, 29) and, therefore, would not impact transcription via the mechanism outlined unless additional factors are involved. Thus, further questions remain concerning how an indiscriminate oxidant like H2O2 liberated by LSD1 or LSD2 would have evolved to write OG at critical locations for gene induction. Regardless, the present studies in tandem with the earlier reports (22, 26, 27) identify a common thread in which OGG1 activity on OG in gene promoters can regulate transcription.
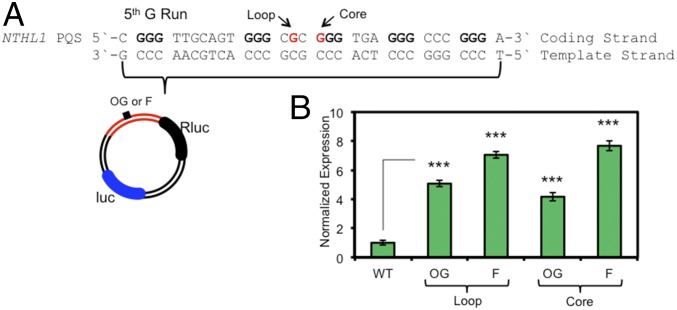
The sequence of events described provides an alternative mechanism for gene activation under oxidative stress. Is this mechanism specific to the VEGF PQS or can it be generalized to other genes? Because genomic OG concentrations are elevated in tandem with some DNA repair, cell cycle, and stress response genes during inflammation (6), we investigated DNA repair genes to identify another PQS with the potential to regulate transcription similarly to the VEGF PQS. The analysis found the endonuclease III-like protein 1 (NTHL1) gene harbors a coding strand PQS with five G runs (Fig. 6A). Additionally, the NTHL1 gene can be up-regulated under oxidative stress conditions (30). Structural studies (1H-NMR, CD, and Tm) found that the five-track NTHL1 PQS adopts a parallel-stranded G-quadruplex fold in vitro (SI Appendix, Fig. S7). Plasmids were prepared with the NTHL1 PQS with OG or an F located at a reactive loop or core position in the promoter of the Rluc reporter gene. Transfection of these plasmids into glioblastoma cells followed by a 48-h incubation found >fourfold increase in gene expression for all modifications studied (Fig. 6B). This second example provides some generality to the mechanism proposed in this work.
Fig. 6.
Gene activation is observed when OG or F is present in the NTHL1 PQS. (A) The NTHL1 PQS sequence and locations in which OG or F were synthesized. (B) Expression enhancement observed when OG or F is found in the NTHL1 PQS. Error bars represent 95% CI on the basis of four or eight replicates. Significance values for each comparison were calculated by a Student’s t test. Significance at ***P < 0.001 is indicated.
The alternative gene activation pathway identified requires base oxidation of G to OG in the PQS context to guide OGG1 for generation of an AP. The AP unmasks the G-quadruplex from the duplex state for prolonged binding by APE1 and associated factors (Fig. 5C). The function of oxidative base modifications in DNA to direct proteins to alter transcription ascribes an epigenetic role to OG. The protein readers and erasers are members of the BER pathway and, therefore, these proteins activate genes in addition to guarding the genome against insults such as oxidative stress. Coupling of DNA repair with transcription provides an efficient mechanism to complete two necessary cellular tasks during oxidative stress. An intertwining of these pathways is starting to emerge in other studies (22, 26, 27, 31). The present study demonstrates that cells can harness oxidized modifications of DNA bases for altering phenotype under oxidative stress and identifies a mechanism by which ROS are cellular-signaling agents, as previously hypothesized (32). Last, the observation that classically defined forms of DNA damage have an epigenetic role in the cell has surfaced with the recent demonstrations of 5-hydroxymethyluracil (33) and N6-methyladenine (34) also guiding cellular processes in higher eukaryotes. These studies extend the epigenetic landscape in DNA beyond methylation of cytosine and its oxidized derivatives (35–37).
Materials and Methods
Detailed materials and methods are described in SI Appendix, Materials and Methods. Characterization of the VEGF PQS with AP and structural characterization of the NTHL1 PQS can be found in the SI Appendix. The complete methods and data are located in the SI Appendix.
Plasmid Construction.
The plasmids were constructed from the psiCHECK2 plasmid (Promega) that contains genes for the Renilla luciferase (Rluc) and firefly luciferase (luc) proteins. The luc gene is regulated by the HSV-TK promoter and used as the internal standard, whereas the Rluc gene was originally regulated by the SV40 early enhancer/promoter that was modified to include the PQS of interest. Additionally, the PQSs of interest were flanked by Nt.BspQ1-nicking endonuclease recognition sequences. Insertion of the PQS and nicking endonuclease recognition sequences was achieved using restriction-free cloning, followed by transformation to competent Escherichia coli and isolation by miniprep kit (Qiagen), as described previously (38). The site-specific modifications were synthesized into short oligomers with the sequence between the two nicking endonuclease sites and they were inserted into the plasmid via literature methods (38, 39). Confirmation that the DNA modifications were introduced into the plasmid was achieved using a protocol established in the C.J.B. laboratory, in which the modification was removed by Fpg and APE1 to yield a ligatable gap (40). After ligation of the gap with T4-DNA ligase, Sanger sequencing provided a characteristic nucleotide loss at the modification site to confirm its presence (SI Appendix, Fig. S8).
Cell Culture Studies.
All cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with FBS, gentamicin, glutamax, and nonessential amino acids. The WT MEF and OGG1−/− MEF cells were previously developed and reported on in the literature (41), and the glioblastoma cells (U87 MG) were purchased from ATCC. Transfection experiments were conducted in white, 96-well plates using X-tremeGene HP DNA transfection agent (Roche) with 200–750 ng of plasmid following the manufacturer’s protocol. The Dual-Glo luciferase (Promega) assay was conducted following the manufacturer’s protocol to monitor Rluc and luc expression levels. Each experiment was conducted in four or eight replicates, as recommended by the Dual-Glo luciferase assay. The APE1 inhibitor III studies were conducted by titration of APE1 inhibitor III from 0 to 1,000 nM from a DMSO stock solution to the cell culture media during transfection. Controls in which only DMSO was added to the cells found no change in expression level of the WT plasmid, ensuring the data obtained resulted from the inhibitor and not the DMSO. The APE1 siRNA knockdown studies were conducted with FlexiTube siRNAs (Qiagen) following the manufacturer’s protocol. The siRNAs were transfected 12 h before addition of the reporter plasmid in a concentration range from 0 to 50 nM.
Data Analysis.
The data were analyzed by converting the luminescence measured into normalized relative response ratios (RRR), which is the luminescence of Rluc divided by the luminescence of luc (i.e., RRR = Rluc/luc). To obtain the normalized expression values reported, each RRR was divided by the RRR for the WT sequence in that data set. The error bars represent 95% confidence intervals obtained from the data. Statistical analysis was achieved using a two-tailed Student’s t test.
Supplementary Material
Acknowledgments
The MEF cell lines were provided by Dr. Tomas Lindahl (Imperial Cancer Research Fund, UK). We thank the University of Utah core facilities for synthesizing the OG- and F-containing oligomers, as well as conducting Sanger sequencing of the plasmids. This work was supported by a National Cancer Institute Grant R01 CA090689.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619809114/-/DCSupplemental.
References
- 1.Hailer-Morrison MK, Kotler JM, Martin BD, Sugden KD. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2′-deoxyguanosine lesions in the NF-kappaB promoter element. Biochemistry. 2003;42(32):9761–9770. doi: 10.1021/bi034546k. [DOI] [PubMed] [Google Scholar]
- 2.Ramon O, et al. Effects of 8-oxo-7,8-dihydro-2′-deoxyguanosine on the binding of the transcription factor Sp1 to its cognate target DNA sequence (GC box) Free Radic Res. 1999;31(3):217–229. doi: 10.1080/10715769900300781. [DOI] [PubMed] [Google Scholar]
- 3.Moore SP, Toomire KJ, Strauss PR. DNA modifications repaired by base excision repair are epigenetic. DNA Repair (Amst) 2013;12(12):1152–1158. doi: 10.1016/j.dnarep.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 2004;3(5):483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Allgayer J, Kitsera N, Bartelt S, Epe B, Khobta A. Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 2016;44(15):7267–7280. doi: 10.1093/nar/gkw473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangerich A, et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA. 2012;109(27):E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pastukh V, et al. An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am J Physiol Lung Cell Mol Physiol. 2015;309(11):L1367–L1375. doi: 10.1152/ajplung.00236.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D, et al. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol Cancer Ther. 2008;7(4):880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hänsel-Hertsch R, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48(10):1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal P, Hatzakis E, Guo K, Carver M, Yang D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: Insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013;41(22):10584–10592. doi: 10.1093/nar/gkt784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleming AM, Zhou J, Wallace SS, Burrows CJ. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent Sci. 2015;1(5):226–233. doi: 10.1021/acscentsci.5b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43(8):4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace SS. Base excision repair: A critical player in many games. DNA Repair (Amst) 2014;19(0):14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rai G, et al. Synthesis, biological evaluation, and structure-activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem. 2012;55(7):3101–3112. doi: 10.1021/jm201537d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freudenthal BD, Beard WA, Cuneo MJ, Dyrkheeva NS, Wilson SH. Capturing snapshots of APE1 processing DNA damage. Nat Struct Mol Biol. 2015;22(11):924–931. doi: 10.1038/nsmb.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schäfer G, et al. Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J Biol Chem. 2003;278(10):8190–8198. doi: 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- 17.Raiber EA, Kranaster R, Lam E, Nikan M, Balasubramanian S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012;40(4):1499–1508. doi: 10.1093/nar/gkr882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat Rev Drug Discov. 2011;10(4):261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broxson C, Hayner JN, Beckett J, Bloom LB, Tornaletti S. Human AP endonuclease inefficiently removes abasic sites within G4 structures compared to duplex DNA. Nucleic Acids Res. 2014;42(12):7708–7719. doi: 10.1093/nar/gku417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid Redox Signal. 2009;11(3):621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genereux JC, Barton JK. Mechanisms for DNA charge transport. Chem Rev. 2010;110(3):1642–1662. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perillo B, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319(5860):202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 23.van Essen D, Zhu Y, Saccani S. A feed-forward circuit controlling inducible NF-κB target gene activation by promoter histone demethylation. Mol Cell. 2010;39(5):750–760. doi: 10.1016/j.molcel.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Fleming AM, Muller JG, Ji I, Burrows CJ. Characterization of 2′-deoxyguanosine oxidation products observed in the Fenton-like system Cu(II)/H2O2/reductant in nucleoside and oligodeoxynucleotide contexts. Org Biomol Chem. 2011;9(9):3338–3348. doi: 10.1039/c1ob05112a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alshykhly OR, Fleming AM, Burrows CJ. 5-Carboxamido-5-formamido-2-iminohydantoin, in addition to 8-oxo-7,8-dihydroguanine, is the major product of the iron-Fenton or X-ray radiation-induced oxidation of guanine under aerobic reducing conditions in nucleoside and DNA contexts. J Org Chem. 2015;80(14):6996–7007. doi: 10.1021/acs.joc.5b00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan L, et al. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase1-mediated epigenetic regulation of nuclear factor kappaB-driven gene expression. J Biol Chem. 2016;1(49):25553–25566. doi: 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoniali G, et al. SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol Biol Cell. 2014;25(4):532–547. doi: 10.1091/mbc.E13-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47(27):7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alshykhly OR, Fleming AM, Burrows CJ. Guanine oxidation product 5-carboxamido-5-formamido-2-iminohydantoin induces mutations when bypassed by DNA polymerases and is a substrate for base excision repair. Chem Res Toxicol. 2015;28(9):1861–1871. doi: 10.1021/acs.chemrestox.5b00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hironaka K, Factor VM, Calvisi DF, Conner EA, Thorgeirsson SS. Dysregulation of DNA repair pathways in a transforming growth factor alpha/c-myc transgenic mouse model of accelerated hepatocarcinogenesis. Lab Invest. 2003;83(5):643–654. doi: 10.1097/01.lab.0000067483.89649.11. [DOI] [PubMed] [Google Scholar]
- 31.Fong YW, Cattoglio C, Tjian R. The intertwined roles of transcription and repair proteins. Mol Cell. 2013;52(3):291–302. doi: 10.1016/j.molcel.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trachootham D, Lu W, Ogasawara MA, Nilsa RD, Huang P. Redox regulation of cell survival. Antioxid Redox Signal. 2008;10(8):1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffeneder T, et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol. 2014;10(7):574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 34.Zhang G, et al. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161(4):893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 35.Chen K, Zhao BS, He C. Nucleic acid modifications in regulation of gene expression. Cell Chem Biol. 2016;23(1):74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Booth MJ, Raiber E-A, Balasubramanian S. Chemical methods for decoding cytosine modifications in DNA. Chem Rev. 2015;115(6):2240–2254. doi: 10.1021/cr5002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner M, et al. Age-dependent levels of 5-methyl-, 5-hydroxymethyl-, and 5-formylcytosine in human and mouse brain tissues. Angew Chem Int Ed Engl. 2015;54(42):12511–12514. doi: 10.1002/anie.201502722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riedl J, Ding Y, Fleming AM, Burrows CJ. Identification of DNA lesions using a third base pair for amplification and nanopore sequencing. Nat Commun. 2015;6:8807. doi: 10.1038/ncomms9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You C, et al. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat Chem Biol. 2012;8(10):817–822. doi: 10.1038/nchembio.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riedl J, Fleming AM, Burrows CJ. Sequencing of DNA lesions facilitated by site-specific excision via base excision repair DNA glycosylases yielding ligatable gaps. J Am Chem Soc. 2016;138(2):491–494. doi: 10.1021/jacs.5b11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klungland A, et al. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA. 1999;96(23):13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.