Significance
Persisting proliferative capacity and regeneration in the adult brain are confined to a minority of regions. In the murine brain, the olfactory system is supplied by thousands of newly born neuroblasts daily to support sensory plasticity. Here, we reveal a neuronal scaffold that resides within and alongside this migratory route [termed the “rostral migratory stream” (RMS)] to guide forward migration of newly born neuroblasts. These neurons can externalize the enzyme matrix metalloprotease-2 to loosen the extracellular matrix, thus producing permissive corridors for migrating neuroblasts. This mechanism is likely phylogenetically conserved because it exists in the RMS equivalent in human fetal brains. This inducible mechanism might be pharmacologically targeted for therapeutic benefit.
Keywords: human fetus, calcium-binding protein, cell motility, restorative strategy, olfactory system
Abstract
The rostral migratory stream (RMS) is viewed as a glia-enriched conduit of forward-migrating neuroblasts in which chemorepulsive signals control the pace of forward migration. Here we demonstrate the existence of a scaffold of neurons that receive synaptic inputs within the rat, mouse, and human fetal RMS equivalents. These neurons express secretagogin, a Ca2+-sensor protein, to execute an annexin V-dependent externalization of matrix metalloprotease-2 (MMP-2) for reconfiguring the extracellular matrix locally. Mouse genetics combined with pharmacological probing in vivo and in vitro demonstrate that MMP-2 externalization occurs on demand and that its loss slows neuroblast migration. Loss of function is particularly remarkable upon injury to the olfactory bulb. Cumulatively, we identify a signaling cascade that provokes structural remodeling of the RMS through recruitment of MMP-2 by a previously unrecognized neuronal constituent. Given the life-long presence of secretagogin-containing neurons in human, this mechanism might be exploited for therapeutic benefit in rescue strategies.
The subventricular zone of adult rodents and its human fetal equivalent (1) generate neuroblasts that migrate through the rostral migratory stream (RMS) to supply the olfactory bulb with new neurons (2). Tangential neuroblast migration in the RMS is regulated by various environmental factors including growth factors (3–10), ephrins (11), and cell adhesion-related cues (12–14). Many of these regulatory events are executed through direct intercellular interactions: Chain-migrating neuroblasts move in contact with each other (15), and neuron–glia communication shapes the journey of new-born cells toward the olfactory bulb (16, 17). In particular, astrocytes can vector neuroblast movement by secreting netrin (18) and VGEF (19, 20) and regulate the speed of migration by releasing GABA (21, 22) while actively reconfiguring their own network in response to neuroblast-derived signals through the Slit/Robo machinery (16).
Brain extracellular matrix is a juxtacellular scaffold that defines the microenvironmental arrangements surrounding each cell. Extracellular matrix components were earlier implicated in neuroblast migration: Tenascin-R can mediate activity-dependent neuroblast recruitment, (23) and hyaluronan and its receptor, Rhamm, driving hyaluronan-mediated motility, are selectively expressed in the adult RMS (24). Notably, members of the matrix metalloproteinase (MMP) family are recognized as critical for restructuring the extracellular matrix by cleaving all its components (25, 26). However, MMP activity and efficacy in neurogenic niches were studied chiefly in the early postnatal nervous system (12), during axonal growth, and upon cancer cell invasion (27–29) with pathologically increased proliferative capacity. In the RMS, MMP inhibitors reduce the rate of neuroblast migration in young mice, but chain migration remains unaffected in the adult (12). Here, MMP activity was shown to surround chain-migrating neuroblasts (12). However, the cellular source of any specific MMP family member, the molecular determinants of its liberation, and its evolutionary conservation between rodents and human remain unknown.
Ca2+ sensor proteins undergo conformational changes upon Ca2+ binding, which then triggers intermolecular interactions with downstream effectors to regulate activity-dependent release or signaling events (30, 31). Secretagogin is one such protein (32) with a phylogenetically conserved distribution in vertebrates (33, 34). Explored first in pancreatic beta cells, secretagogin was shown to interact with the SNARE machinery (32). Similarly, secretagogin’s proteome-wide interactions posit a role in neuropeptide release from hypothalamic neurons (35).
Here, we identify a population of neurons that express secretagogin within the adult murine RMS to control MMP2, but not MMP9, externalization through annexin V recruitment. This focal and “on-demand” MMP2 release affects only motile neuroblasts but is unlikely to mobilize the source cells themselves. Based on histochemical analysis advancing to the ultrastructural level in rodent and human fetal brains, we suggest that secretagogin-containing neurons are a cell population in the RMS that form a scaffold around migrating neuroblasts. By developing in situ zymography and combining this technique with mass spectrometry, we show that secretagogin-containing neurons are the foci of MMP-2 release in the rodent brain. We then generated secretagogin knockout mice and performed in vivo and in vitro gene silencing to dissect molecular interactions among secretagogin, annexin V, and MMP-2. Cumulatively, we identified a hitherto undescribed neuronal contingent and its selective MMP-2–externalization mechanism that controls neuroblast migration by restructuring the extracellular matrix, a mechanism conserved between rodents and human.
Results
Secretagogin-Expressing Neurons Form a Scaffold in the Rodent RMS.
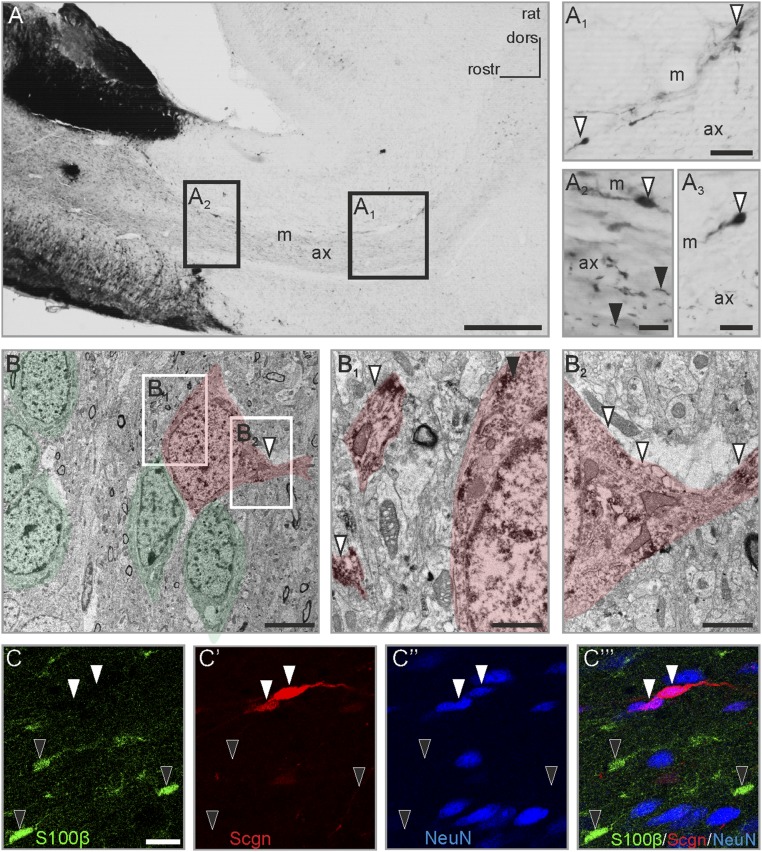
Cellular interactions in the RMS are mandatory for the coordinated chain migration of neuroblasts toward the olfactory bulb (3, 4, 6, 16). In rodents, adult-born neurons migrate in permissive corridors (“tunnels”) established by a complex network of astrocytes (2). Neuron–glia communication is pivotal in this process, together with the active participation of neuroblasts transforming the astrocytic tunnel (36). However, the mechanisms of communication between newborn neurons and a contingent of nonastroglial cells resident in the RMS remain unexplored. Here, we show that secretagogin labels a distinct subset of bipolar cells (Fig. 1 A1–A3) primarily populating the outer region of the adult rat RMS (Fig. 1 A1–A3) and significantly outnumbering the cells in its inner compartment (“axis”) (Fig. 1A2). In accord with recent human terminology (37), this localization pattern allows an onion skin-like cellular matrix suited to direct neuroblasts toward the olfactory bulb. Secretagogin immunoreactivity was detected in both somata (Fig. 1 A1–A3) and processes (Fig. 1A2). When the subcellular compartmentalization of secretagogin was analyzed at the ultrastructural level, particularly in cells contacting chain-migrating neuroblasts (Fig. 1B), secretagogin typically was detected in processes (Fig. 1B1 and Fig. S1E), many of which emanated perpendicular to the axis of the RMS (Fig. 1B2). Secretagogin immunoreactivity often was closely associated with the membrane of the endoplasmic reticulum (Fig. S1 A and A1) and mitochondria (Fig. S1B). In some cases, secretagogin+ focal accumulations were selectively seen in the RMS, which surrounded dendrite-like profiles (Fig. S1 F, F1, and G), suggesting focal signaling events through protein recruitment.
Fig. 1.
The presence and compartmentalization of secretagogin in rat RMS neurons. (A–A3) Secretagogin+ cells populate the rat RMS. In addition to somatic labeling (open arrowheads in A1–A3) a large number of individual processes were identified in the center of the middle domain of the stream (black arrowheads in A2; also Fig. S1E). Additionally, secretagogin+ cells often delineated the border of the RMS with their processes extended bipolarly (open arrowheads in A1 and A3). (B) Ultrastructural analysis identified secretagogin+ cells in close contact with chain-migrating neurons with secretagogin expression detectable both in the soma (black arrowhead in B1 and in processes arranged in parallel (open arrowheads in B1) or transversally (open arrowheads in B2). Chain-migrating neuronblasts and secretagogin+ cells are semitransparently color-coded in green and red, respectively. (C–C′′′) Secretagogin+ cells were invariably immunonegative for the gilal marker S100β (black arrowheads in C–C′′′) but were immunopositive for the neuron marker NeuN (white arrowheads in C′–C′′′). ax, axis of RMS; dors, dorsal; m, marginal zone of RMS; rostr, rostral; Scgn, secretagogin. (Scale bars: 500 µm in A, 30 µm in A1 and A2′; 20 µm in A3; 15 µm in C–C′′′; 10 µm in A2 and A4; 5 µm in B; 2 µm in B2; and 1 µm in B1.)
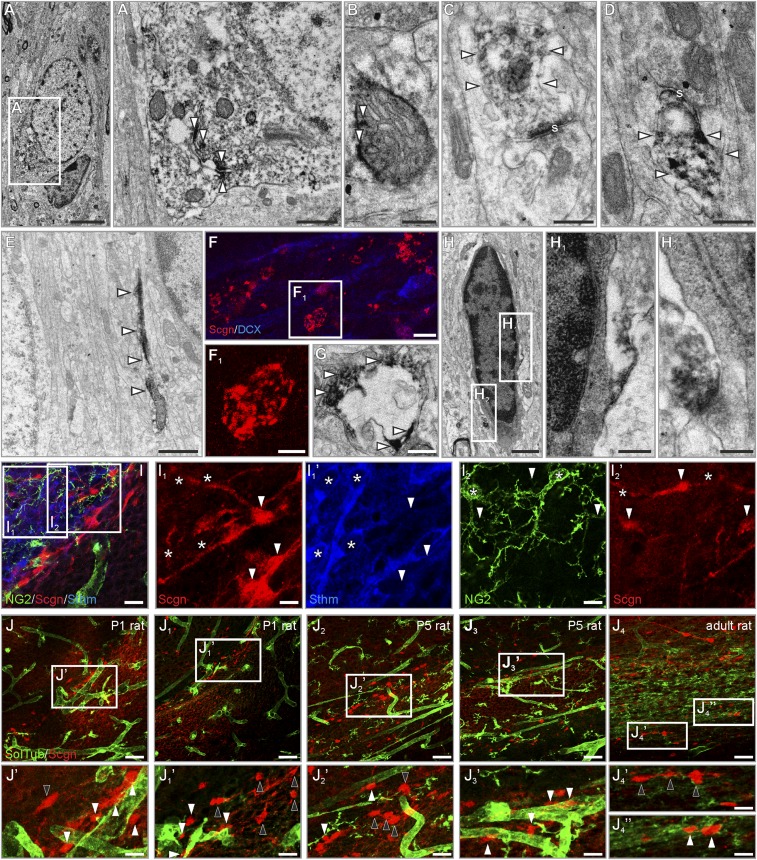
Fig. S1.
Presence and compartmentalization of secretagogin in rat RMS neurons. (A and B) Secretagogin was typically associated with intracellular membranes (open arrowheads in A1) in the cytoplasm or with mitochondria (open arrowheads in B). (C and D) In the neuropil, secretagogin was present in the postsynaptic compartment of immature synaptic-like structures (open arrowheads). (E) Secretagogin typically appeared in cellular processes. (F and G) Secretagogin+ round or oval pearl-like structures surrounded processes that probably were dendrites in the RMS. (H–H2) Dark, electron-dense glial cells remained immunonegative for secretagogin. (I–I2′) Secretagogin+ cells were invariably immunonegative for the glial marker NG2 (asterisks in I2 and I2′) and stathmin-2 (asterisks in I1 and I1′). (J–J4′′) Secretagogin+ neurons were randomly positioned with (white arrowheads) or without (black arrowheads) a close association with vessels in P1, P5, and adult rats. s, synapse; SolTub, Solanum Tuberosum agglutinin. (Scale bars: 50 m in J–J4; 10 µm in I–I2′ and J1′– J4′′; 3 µm in F; 2 µm in A; 1 µm in E; 300 nm in A1, B–D, F1, G, and H; and 200 nm in H1 and H2.)
Secretagogin+ cells were negative for neural/glial antigen 2 (NG2 proteoglycan) (Fig. S1 I, I2, and I2′), S100 Ca2+-binding protein B (S100B) (Fig. 1 C and C′ and Fig. S1 H–H2), and the microtubule destabilizing protein stathmin-2 (Fig. S1 I, I1, and I1′) typically present in motile neurite tips (38). In contrast, secretagogin+ cells expressed the neuronal nuclear antigen NeuN (Fig. 1 C–C′′), thus qualifying as bona fide differentiated neurons. Secretagogin+ neurons were positioned with no clear association to local microvessels in either early postnatal or adult rats (Fig. S1 J–J4′′). Recent studies show that mature neurons establish synapses in the RMS (37, 39), an observation corroborated by the presence of secretagogin in the postsynaptic compartment (Fig. S1 C and D). Taking these findings together, we suggest that mature neurons might reside in the murine RMS with a spatial configuration aimed at modulating chain migration.
Secretagogin+ Neurons in the Human Fetal Lateral Ventricle and Olfactory Tract.
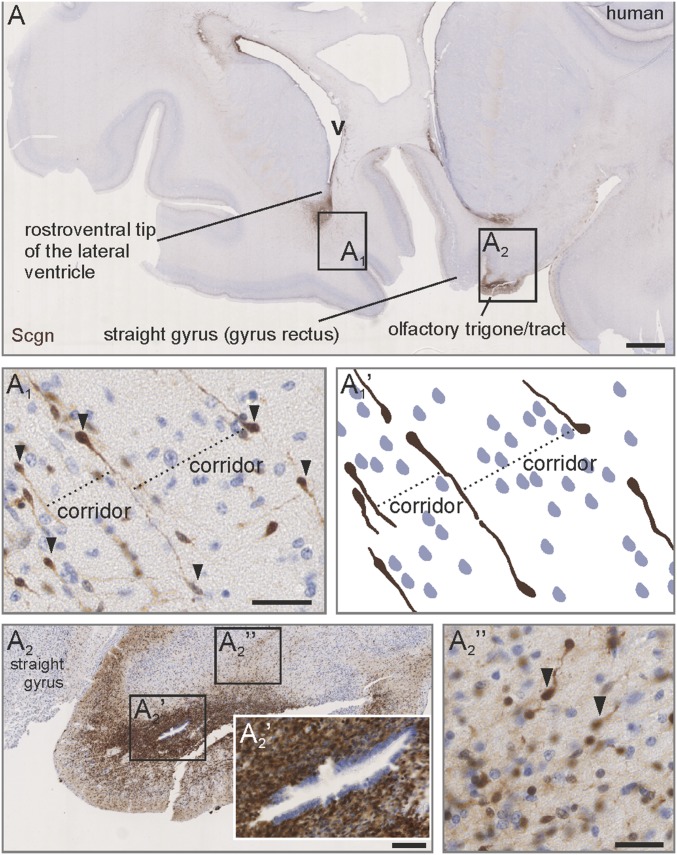
Many mammals retain the ability to generate neurons in adulthood (2, 15, 40–43). In humans, migratory cell streams leaving the subventricular zone cease to exist by 18 mo postpartum (1). Before this time, the infant human subventricular zone and RMS equivalent contain a pool of forward-migrating neuroblasts (1) with their uninterrupted stream connecting the ventral tip of the subventricular zone with the olfactory peduncle (1). By analyzing third-trimester human fetal brains, we find secretagogin+ bipolar cells at the rostroventral pole of the lateral ventricle (Fig. 2 A and A1). These cells establish a palisade-like parcellated structure that extends toward the base of the brain (Fig. 2 A1 and A1′). By exploiting oblique sectioning, which also includes the proximal domain of the contralateral olfactory tract (Fig. 2A), we showed dense secretagogin immunoreactivity (Fig. 2A2) with most cells concentrated around the olfactory ventricle (a ventricular space lined by ependymal cells) (Fig. 2A2′). A cap-shaped condensation of secretagogin+ neurons was seen dorsal to the olfactory trigone/tract (Fig. 2A2′′) that retained clear organization through parallel and equally spaced neurons (Fig. 2A2′′). Considering that all other cells, shown by nuclear staining resided between secretagogin+ “guide” neurons and along their processes (Fig. 2 A1 and A2′′), we hypothesize that this cell contingent might be functionally significant in organizing the human fetal RMS equivalent.
Fig. 2.
Secretagogin+ neurons outline a ventriculo-olfactory stream in human fetus. (A–A1′) Secretagogin+ neurons were concentrated at the rostroventral tip of the lateral ventricle (A1) and emanated in parallel strings ventrally forming corridors (A1 and A1′). The olfactory trigone harbored a high density of secretagogin+ neurons (A2), which typically were concentrated around a ventricle-like space surrounded by ependymal cells (A2′). Bipolar secretagogin+ neurons formed a cap-shaped region dorsal to the olfactory trigone which pointed toward the ventral pole of the lateral ventricle (A2′′). Images in A and A2 were acquired using the tile-and-stitch function. v, ventricle. (Scale bars: 1,000 µm in A; 200 µm in A2; 40 µm in A1′, A2′, and A2′′.)
Slow Turnover of Secretagogin+ Resident Neurons.
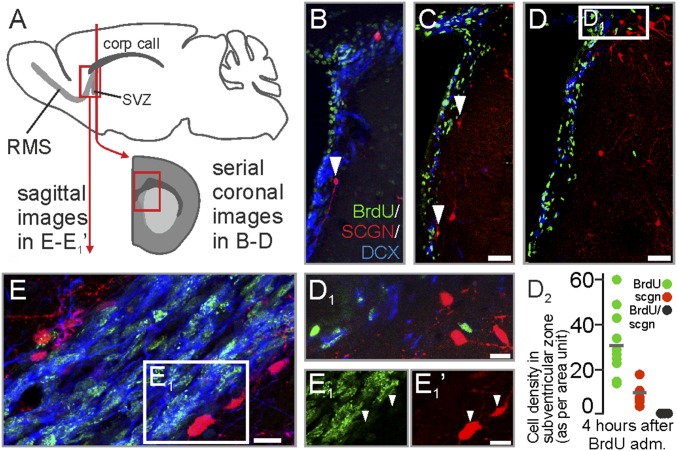
Chain-migrating neuroblasts arise from the ventricular wall of adult rat and mouse to enter the RMS (44). Comparative neuroanatomical studies in avians show that secretagogin+ neurons densely populate the rostroventral domain of the lateral ventricle (34). Using BrdU in rats, we labeled newly born neuroblasts at the ventricular wall (Fig. 3 A and B). Secretagogin+ neurons were BrdU− and surrounded the doublecortin (DCX)+/BrdU+ migrating neuroblasts (Fig. 3 B, C, and D2), thus producing a tunnel-like demarcation (Fig. 3 B–D1). This cellular arrangement extended into the adjacent, proximal part of the RMS (Fig. 3 E–E1′), reinforcing the notion that slow-turnover secretagogin+ neurons could be ideally positioned to direct the migration of neuroblasts.
Fig. 3.
Secretagogin+ cells in the subventricular zone region and on entry into the RMS. (A) Schema of tissue sampling. (B–D2) Coronal sections. BrdU+ neuroblasts in the subventricular zone were demarcated toward the neighboring striatum by secretagogin+/BrdU− neurons. (E–E1′) Saggital sections. BrdU+ neuroblasts in the most proximal part of the RMS are demarcated by secretagogin+ neurons (white arrowheads in E1 and E1′). med, medial. (Scale bars: 80 µm in C and D; 10 µm in D1, E, and E1′.)
RMS Injury Increases the Number of Secretagogin+ Neurons.
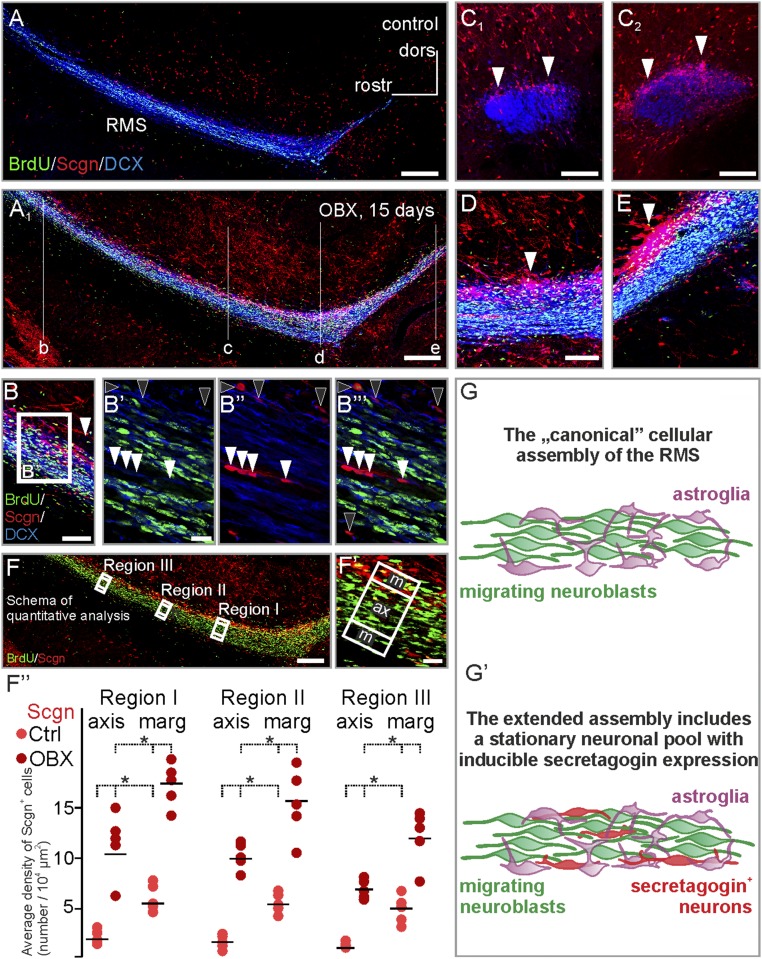
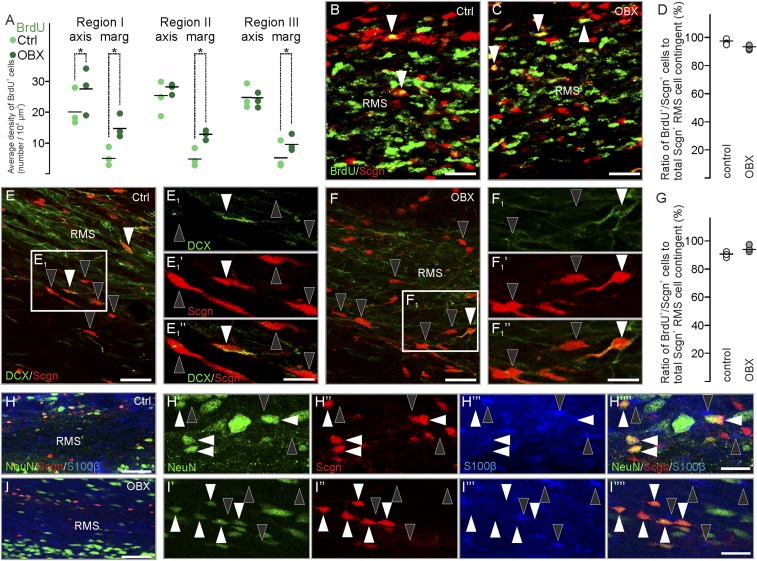
After unilateral surgical removal of the olfactory bulb, the subventricular zone and RMS persist and enlarge (43). We hypothesized that their increased size and cell number (43) could affect the number of secretagogin+ neurons, particularly if increased contingents of neuroblasts migrate toward the site of injury. Secretagogin expression significantly increased in the ipsilateral RMS 15 d after bulbectomy throughout the cranio-caudal extension of the RMS (Fig. 4 A–E). In addition to a primary increase in the outer regions of the RMS (Fig. 4 C2, D, and E), secretagogin+ neurons were seen with increasing frequency in the RMS axis (Fig. 4 B′–B′′′). This increased frequency was confirmed by quantitative immunohistochemistry (Fig. 4 F–F′′) showing that secretagogin+ neurons retained their native distribution irrespective of bulbectomy (Fig. 4F′′ and Table S1). Our domain- and region-specific analysis revealed an up to eightfold increase in secretagogin+ neurons in bulbectomized animals as compared with sham-lesioned controls (Fig. 4F′′). These data together with secretagogin cells remaining BrdU− even after injury (96.78 ± 1.15% in control animals vs. 92.81 ± 1.01% in bulbectomized animals) (Fig. S2 B–D) prompts the suggestion that secretagogin expression is inducible in resident neurons rather than introduced in a new contingent upon injury (Fig. 4 G and G′ and Fig. S2). This notion is further supported by secretagogin+ neurons being typically DCX− (Fig. S2 E–G) and retaining their NeuN+/S100β− identity (Fig. S2 H–I′′′′) upon bulbectomy.
Fig. 4.
Unilateral bulbectomy increases the number of secretagogin-expressing cells in the rostral migratory stream. (A and A1) Secretagogin+ cells not only appeared in the otherwise secretagogin-immunonegative axis of the stream but also typically accumulated in its shell region 15 d after olfactory bulbectomy. Saggital sections are shown in B, D, and E. Coronal sections at the rostrocaudal level indicated in A1 are demonstrated in C1 and C2. (B′–B′′′) Olfactory bulbectomy increased the number of secretagogin+ neurons both in the marginal zone (black arrowheads) and in the axis (white arrowheads) of the RMS. (F–F′′) Fifteen days after bulbectomy, the number of secretagogin-expressing cells increased robustly in the axis and shell domains of the RMS compared with the number of cells detected in sham-operated controls. Secretagogin+ neurons and BrdU+ neuroblasts were counted manually, and their density was calculated in the marginal and axial domains (F′) of the distal RMS. To exclude region-specific differences, we carried out identical measurements in three regions of the distal RMS (F). The density secretagogin+ neurons increased significantly in both the marginal and axial domains of the RMS. (G and G′) In addition to astroglia and migrating neuroblasts, the rodent RMS contains secretagogin+ neurons (P < 0.05, Student’s t test). Images in A, A1, and F were acquired using the tile-and-stitch function. (Scale bars: 300 μm in A and A1; 200 μm in F; 100 μm in B, C1, C2, and D; and 10 μm in B′ and F′.)
Table S1.
Two-factorial ANOVA and post hoc analysis of in vivo analysis in rats
| Effect | SS | Degree of freedom | MS | F | P | |||
| Distal 1 RMS | ||||||||
| Intercept | 15,620.13 | 1 | 15,620.13 | 1,360.608 | 0.000000 | |||
| Treatment | 1,794.45 | 1 | 1,794.45 | 156.308 | 0.000000 | |||
| Location | 451.78 | 1 | 451.78 | 39.353 | 0.000011 | |||
| Treatment*location | 12.05 | 1 | 12.05 | 1.050 | 0.320806 | |||
| Error | 183.68 | 16 | 11.48 | |||||
| Cell no. | Bonferroni test; variable sqrtr Scgn+ cell number | |||||||
| Probabilities for post hoc tests | ||||||||
| Error: between MS = 11.48, df = 16 | ||||||||
| Treatment | Location | [1] (24.003) | [2] (12.945) | [3] (41.395) | [4] (33.442) | |||
| 1 | Control | Marginal | 0.000569 | 0.000003 | 0.002658 | |||
| 2 | Control | Axis | 0.000569 | 0.000000 | 0.000000 | |||
| 3 | Bulbectomy | Marginal | 0.000003 | 0.000000 | 0.011379 | |||
| 4 | Bulbectomy | Axis | 0.002658 | 0.000000 | 0.011379 | |||
| Distal 2 RMS | ||||||||
| Intercept | 13,675.81 | 1 | 13,675.81 | 1,514.256 | 0.000000 | |||
| Treatment | 1,595.27 | 1 | 1,595.27 | 176.636 | 0.000000 | |||
| Location | 426.40 | 1 | 426.40 | 47.213 | 0.000004 | |||
| Treatment*location | 12.56 | 1 | 12.56 | 1.391 | 0.255439 | |||
| Error | 144.50 | 16 | 9.03 | |||||
| Cell no. | Bonferroni test; variable sqrtr Scgn+ cell number | |||||||
| Probabilities for post hoc tests | ||||||||
| Error: between MS = 9.03, df = 16 | ||||||||
| Treatment | Location | [1] (22.628) | [2] (11.808) | [3] (38.905) | [4] (31.256) | |||
| 1 | Control | Marginal | 0.000200 | 0.000001 | 0.002012 | |||
| 2 | Control | Axis | 0.000200 | 0.000000 | 0.000000 | |||
| 3 | Bulbectomy | Marginal | 0.000001 | 0.000000 | 0.005880 | |||
| 4 | Bulbectomy | Axis | 0.002012 | 0.000000 | 0.005880 | |||
| Distal 3 RMS | ||||||||
| Intercept | 10,765.17 | 1 | 10,765.17 | 1,254.685 | 0.000000 | |||
| Treatment | 985.11 | 1 | 985.11 | 114.815 | 0.000000 | |||
| Location | 446.42 | 1 | 446.42 | 52.031 | 0.000002 | |||
| Treatment*location | 7.82 | 1 | 7.82 | 0.911 | 0.353953 | |||
| Error | 137.28 | 16 | 8.58 | |||||
| Cell no. | Bonferroni test; variable sqrtr Scgn+ cell number | |||||||
| Probabilities for post hoc tests | ||||||||
| Error: between MS = 8.58, df = 16 | ||||||||
| Treatment | Location | [1] (21.532) | [2] (10.832) | [3] (34.318) | [4] (26.119) | |||
| 1 | Control | Marginal | 0.000170 | 0.000021 | 0.148985 | |||
| 2 | Control | Axis | 0.000170 | 0.000000 | 0.000002 | |||
| 3 | Bulbectomy | Marginal | 0.000021 | 0.000000 | 0.002545 | |||
| 4 | Bulbectomy | Axis | 0.148985 | 0.000002 | 0.002545 | |||
Quantitative analysis of secretagogin+ neurons in rats after bulbectomy in the axial and marginal regions of the RMS. Treatment: bulbectomy vs. control. Location: marginal vs. axial. MS, mean of squares; SS, sum of squares; variable sqrtr, variable under square root transformation.
Fig. S2.
Olfactory bulbectomy fails to recruit additional secretagogin+ cells in the RNA. (A) Bulbectomy increases the number of BrdU+ cells in the distal limb of the RMS (see Fig. 4 F and F′ for sampling). (B–D) Secretagogin+ neurons remain BrdU− following bulbectomy. (E–G) Secretagogin+ neurons do not express DCX in the RMS, irrespective of bulbectomy. (H–I′′′′) Removal of the olfactory bulb induces secretagogin expression in many NeuN+/S100β− neurons. (Scale bars: 30 µm in B, C, E, H, and J; 20 µm in F; 10 µm in E1′′, F1′′, H′′′′, and I′′′′.)
Secretagogin Loss of Function in Mice Reduces Neuroblast Migration.
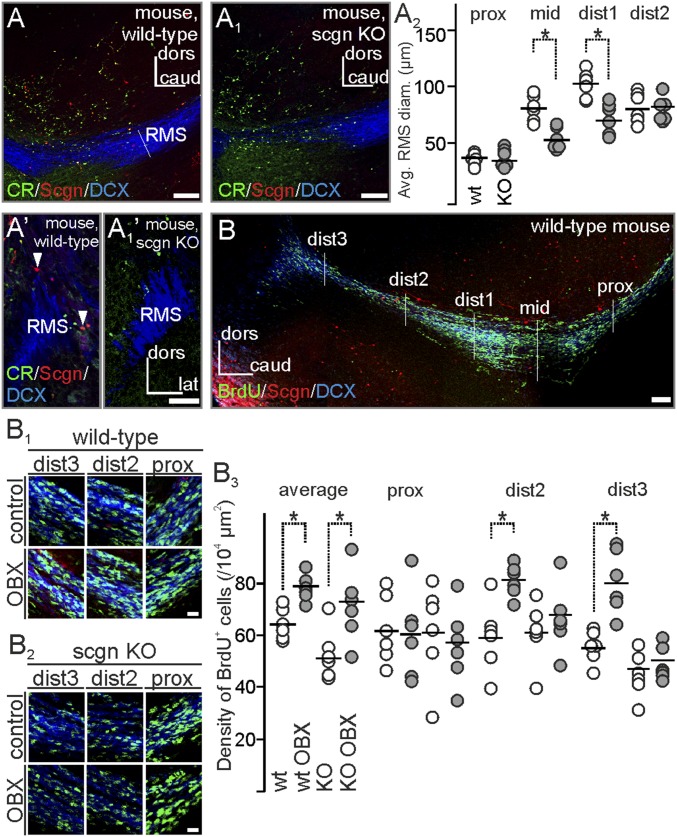
We used secretagogin−/− mice (Fig. S3) to show that the lack of secretagogin expression impacts neuroblast migration: Domain-specific analysis of the RMS revealed that the dorsoventral diameter of the RMS was reduced in secretagogin−/− animals (Fig. 5 A–A1′) despite the lack of any major phenotypic difference in morphological appearance, destination, or overall DCX content. To explore further if migration defects exist in secretagogin−/− mice, the density of BrdU+ cells in the proximal and distal RMS was analyzed in control animals and after olfactory bulbectomy (Fig. 5B). Bulbectomy increased the density of BrdU+ neuroblasts in both secretagogin−/− and wild-type mice (Fig. 5B1). Although the proximal RMS region harbored similar densities of newborn cells, irrespective of genotype or surgery (Fig. 5 B1–B3 and Table S2), the density of BrdU+ neuroblasts increased significantly only in the distal RMS domains of bulbectomized wild-type mice. In contrast, BrdU+ cell numbers were unchanged in secretagogin−/− mice following surgery (Fig. 5 B1–B3). This finding suggests that bulbectomy-induced mobilization of neuroblasts toward the injury site is slowed in the absence of secretagogin, implicating secretagogin in a mode of intercellular communication.
Fig. S3.
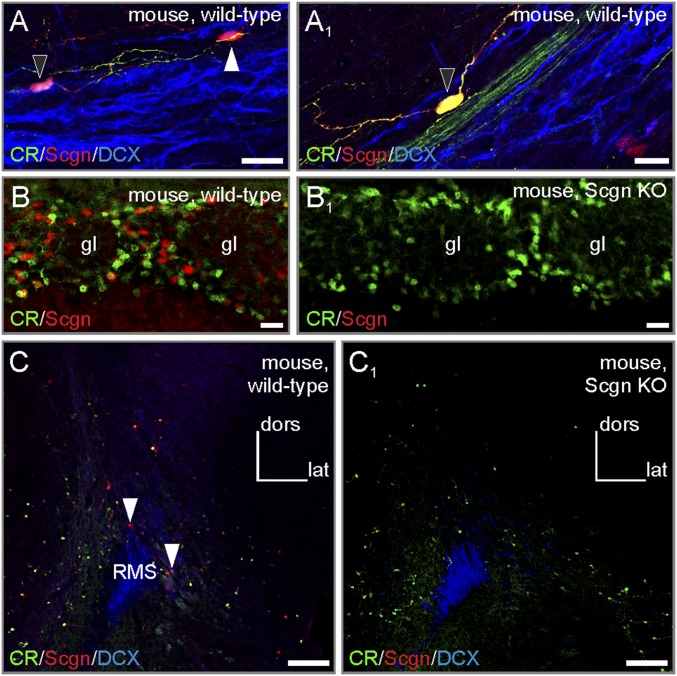
Secretagogin−/− mice demonstrate successful loss of function. (A and A1) Secretagogin+ neurons in wild-type RMSs typically expressed calretinin (black arrowheads). The white arrowhead points to a secretagogin+/calretinin− neuron). (B–C1) Secretagogin−/− mice lacked secretagogin expression in both the olfactory bulb (B and B1) and RMS (C and C1). (Scale bars: 100 µm in C and C1; 20 µm in B and B1; 10 µm in A and A1.)
Fig. 5.
Altered neuroblast morphology and migration in the RMS of secretagogin−/− mice. (A–A1′) Secretagogin−/− mice lacked any secretagogin expression in their RMS, validating this model. (A2) The dorsoventral diameter of the middle and distal domains of the RMS was narrower in secretagogin−/− mice. (B–B3) Olfactory bulbectomy increased the overall density of BrdU+ neurons in the RMS. No genotype or surgery effects were found in the proximal RMS domain, but olfactory bulbectomy increased the density of BrdU+ cells in the distal RMS regions of wild-type but not secretagogin−/− mice. P < 0.05, Student’s t test. Image in B was acquired using the tile-and-stitch function. caud, caudal; CR, calretinin; dist, distal; lat, lateral; prox, proximal. (Scale bars: 100 µm in A–A1′ and B; 10 µm for all captures in B1 and B2.)
Table S2.
Two-factorial ANOVA and post hoc analysis of in vivo analysis in mice
| Effect | SS | Degree of freedom | MS | F | P | |
| Average of all analyzed RMS regions | ||||||
| Intercept | 275,282.0 | 1 | 275,282.0 | 1,485.793 | 0.000000 | |
| Genotype | 1,334.7 | 1 | 1,334.7 | 7.204 | 0.009128 | |
| Treatment | 1,200.5 | 1 | 1,200.5 | 6.480 | 0.013185 | |
| Genotype*treatment | 800.0 | 1 | 800.0 | 4.318 | 0.041492 | |
| Error | 12,598.8 | 68 | 185.3 | |||
| Cell no. | Bonferroni test; variable proximal BrdU+ cell density | |||||
| Probabilities for post hoc tests | ||||||
| Error: between MS = 185.3, df = 68 | ||||||
| Genotype | Treatment | [1] (58.722) | [2[ (73.556) | [3] (56.778) | [4] (58.278) | |
| 1 | WT | Spared | 0.010162 | 1.000000 | 1.000000 | |
| 2 | WT | OBX | 0.010162 | 0.002618 | 0.007518 | |
| 3 | KO | Spared | 1.000000 | 0.002618 | 1.000000 | |
| 4 | KO | OBX | 1.000000 | 0.007518 | 1.000000 | |
| Proximal RMS | ||||||
| Intercept | 87,604.17 | 1 | 87,604.17 | 337.6317 | 0.000000 | |
| Genotype | 28.17 | 1 | 28.17 | 0.1086 | 0.745220 | |
| Treatment | 88.17 | 1 | 88.17 | 0.3398 | 0.566460 | |
| Genotype*treatment | 28.17 | 1 | 28.17 | 0.1086 | 0.745220 | |
| Error | 5,189.33 | 20 | 259.47 | |||
| Cell no. | Bonferroni test; variable proximal BrdU+ cell density | |||||
| Probabilities for post hoc tests | ||||||
| Error: between MS = 259.47, df = 20 | ||||||
| Genotype | Treatment | [1] (62.333) | [2] (60.667) | [3] (62.333) | [4] (56.333) | |
| 1 | WT | Spared | 1.000000 | 1.000000 | 1.000000 | |
| 2 | WT | OBX | 1.000000 | 1.000000 | 1.000000 | |
| 3 | KO | spared | 1.000000 | 1.000000 | 1.000000 | |
| 4 | KO | OBX | 1.000000 | 1.000000 | 1.000000 | |
| Distal 2 RMS | ||||||
| Intercept | 10,8676.0 | 1 | 10,8676.0 | 908.0931 | 0.000000 | |
| Genotype | 145.0 | 1 | 145.0 | 1.2120 | 0.284018 | |
| Treatment | 1,218.4 | 1 | 1,218.4 | 10.1807 | 0.,004592 | |
| Genotype*treatment | 442., | 1 | 442.0 | 3.6937 | 0.068983 | |
| Error | 2,393.5 | 20 | 119.7 | |||
| Cell no. | Bonferroni test; variable distal 2 BrdU+ cell density | |||||
| Probabilities for post hoc tests | ||||||
| Error: between MS = 119.7, df = 20 | ||||||
| Genotype | Treatment | [1] (58.333) | [2] (81.167) | [3] (62.000) | [4] (67.667) | |
| 1 | WT | Spared | 0.010359 | 1.000000 | 0.930331 | |
| 2 | WT | OBX | 0.010359 | 0.039268 | 0.270589 | |
| 3 | KO | Spared | 1.000000 | 0.039268 | 1.000000 | |
| 4 | KO | OBX | 0.930331 | 0.270589 | 1.000000 | |
| Distal 3 RMS | ||||||
| Intercept | 80,157.04 | 1 | 80,157.04 | 1,118.600 | 0.000000 | |
| Genotype | 2,109.38 | 1 | 2,109.38 | 29.437 | 0.000026 | |
| Treatment | 1,190.04 | 1 | 1,190.04 | 16.607 | 0.000590 | |
| Genotype*treatment | 513.38 | 1 | 513.38 | 7.164 | 0.014501 | |
| Error | 1,433.17 | 20 | 71.66 | |||
| Cell no. | Bonferroni test; variable distal 3 BrdU+ cell density | |||||
| Probabilities for post hoc tests | ||||||
| Error: between MS = 71.66, df = 20 | ||||||
| Genotype | Treatment | [1] (55.500) | [2] (78.833) | [3] (46.000) | [4] (50.833) | |
| 1 | WT | Spared | 0.000694 | 0.396748 | 1.000000 | |
| 2 | WT | OBX | 0.000694 | 0.000009 | 0.000079 | |
| 3 | KO | Spared | 0.396748 | 0.000009 | 1.000000 | |
| 4 | KO | OBX | 1.000000 | 0.000079 | 1.000000 | |
Quantitative analysis of BrdU+ neurons in wild-type and Scgn-KO mice after bulbectomy. Genotype: Scgn-KO vs. wild-type. Treatment: bulbectomy vs. control. MS, mean of squares; SS, sum of squares.
Secretagogin Gene Silencing Reduces DCX Expression and Slows Neuroblast Migration.
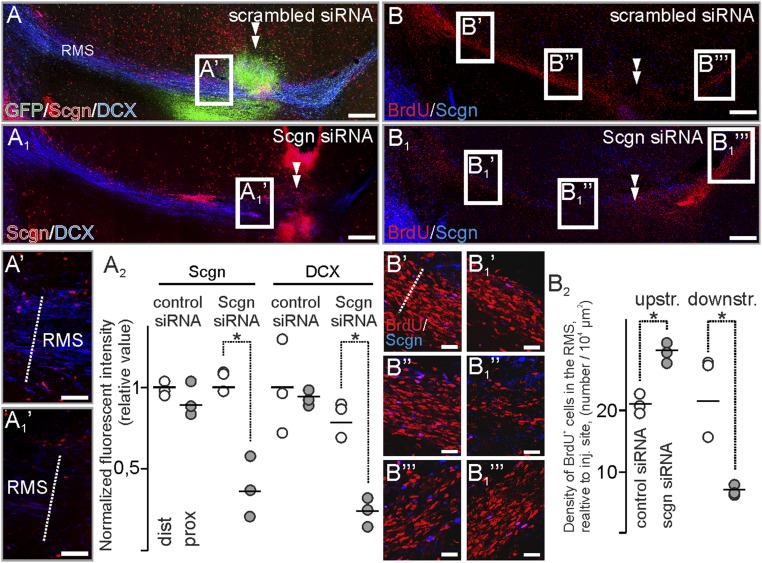
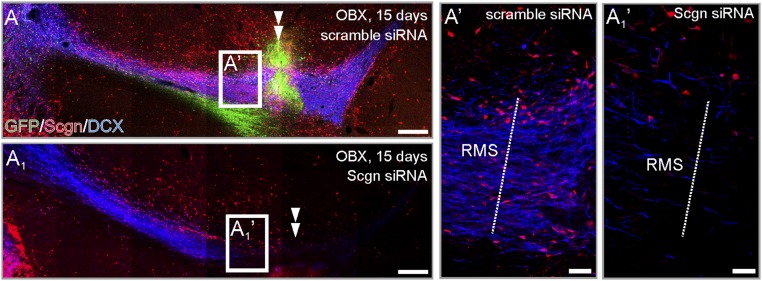
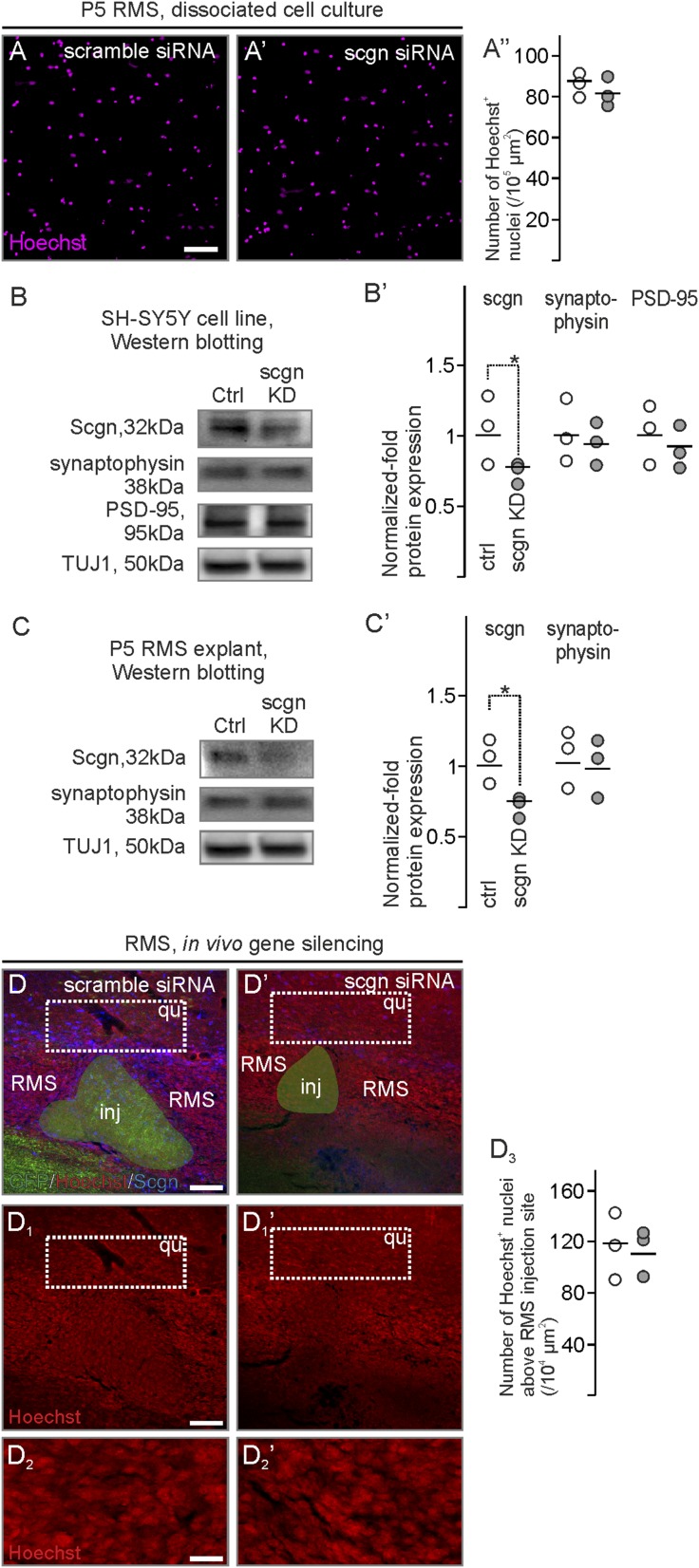
To extend our genetic data on secretagogin’s role in modulating neuroblast motility, we injected secretagogin siRNA in close proximity to the “knee region” of the RMS (Fig. 6 A–B1). Secretagogin immunoreactivity was significantly reduced in and around the injection site, compared with the most distal secretagogin+ somata (Fig. 6A2). In parallel, secretagogin silencing abolished immunoreactivity for DCX, an intermediate filament protein involved in neuronal migration (45), around the administration site (Fig. 6 A1 and A1′). DCX loss could not be rescued by olfactory bulbectomy (Fig. S4). The loss of DCX immunoreactivity prompted us to determine the density of BrdU+ cells upstream (toward the ventricle) and downstream (toward the olfactory bulb) from the gene-silencing site (Fig. 6 B and B1). We observed a decreased density of BrdU+ cells toward the olfactory bulb (Fig. 6 B′, B1′, and B2), particularly in the tissue bordering the lesion site (Fig. 6 B′′ and B1′′). This decrease was paralleled by an increased density of BrdU+ cells upstream (Fig. 6 B′′′ and B1′′′).
Fig. 6.
In vivo secretagogin silencing decreases DCX expression and slows neuroblast migration in the RMS. (A–A2) Secretagogin (A1 and A1′) but not scrambled siRNA (A and A′) reduced DCX expression locally (arrowheads in A and A1). This effect is independent of olfactory bulbectomy (Fig. S4). Note the reduced secretagogin expression upon using specific (A1′) but not scrambled (A′) siRNA and compared with distal RMS free of silencing effect (A2). (B–B2) The density of BrdU+ cells decreased rostral but increased caudal to the silencing site (B2). P < 0.05, Student’s t test. Images in A, A1, B, and B1 were acquired using the tile-and-stitch function. (Scale bars: 200 µm in A, A1, B, and B1; 25 µm in A′ and A1′; 15 µm in B′–B1′′′.)
Fig. S4.
Secretagogin expression in bulbectomized rats following in vivo gene silencing. Secretagogin (A1 and A1′) but not scrambled siRNA (A and A′) reduced DCX expression locally (double arrowheads in A and A′) in bulbectomized rats. Images in A and A′ were acquired using the tile-and-stitch function. (Scale bars: 200 µm in A and A1; 25 µm in A′ and A1′.)
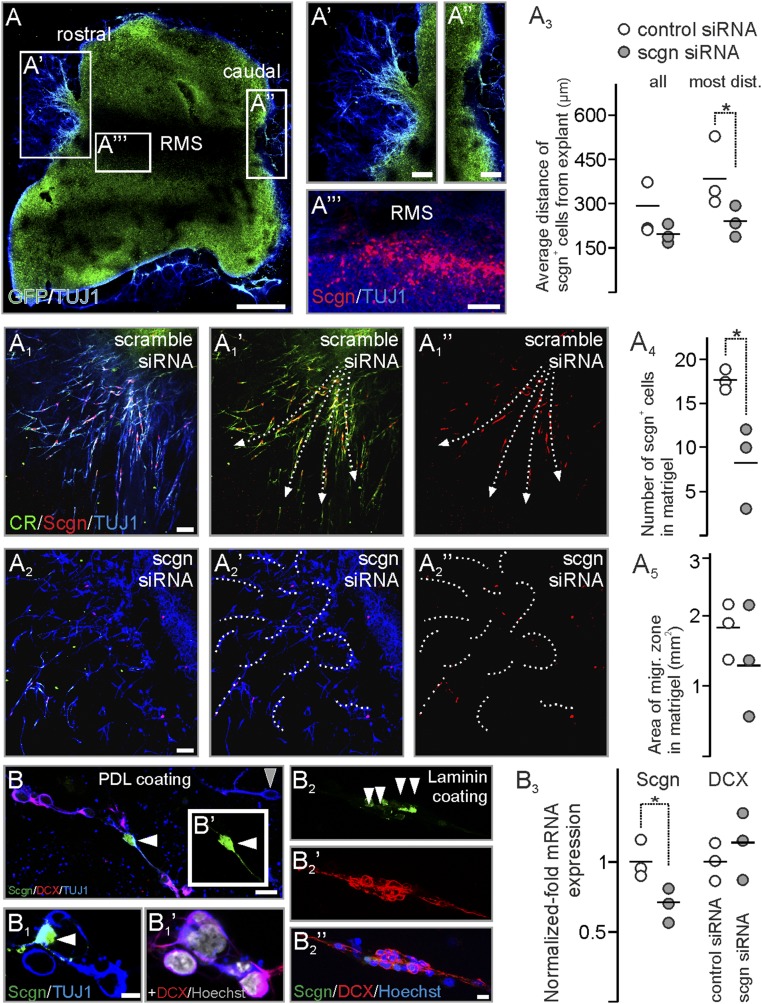
We next extended these in vivo results in vitro using RMS explants from postnatal day (P)5 rats by determining the rate of RMS-derived neuroblast migration into the surrounding embedding medium (Fig. 7A). Explants preserved their polarity so that neurons exited the explants exclusively at their rostral (Fig. 7A′) but not caudal (Fig. 7A′′) pole. Secretagogin+ neurons typically accumulated at the margins of the RMS (Fig. 7A′′′), recapitulating the typical arrangement in vivo (Fig. 1A). In control experiments, β-III-tubulin (TUJ1)+ neurons invaded the Matrigel along well-defined streams outlined by secretagogin+ neurons (Fig. 7 A1–A1′′), recapitulating in vivo arrangements in both rodents and humans. In contrast, secretagogin knockdown (Fig. 7A4) disrupted the migratory pattern of TUJ1+ neurons (Fig. 7 A2–A2′′). Notably, the distance covered by migratory secretagogin+ cells decreased significantly (Fig. 7A) with a marked reduction in the rate of migrating cells borne at the rostral tip of the explants (Fig. 7 A3 and A5).
Fig. 7.
Secretagogin silencing disrupts neuroblast migration in vitro. (A–A′′′) RMS explants from P5 rat pups preserve polarity with migrating cells at the rostral (A′) but not caudal (A′′) pole with secretagogin+ neurons retaining their positions within explants (A′′′). (A1–A2′′) In controls neuroblasts migrate from the rostral pole of the RMS explants into Matrigel along corridors (arrows in A1′ and A1′′), which are shaped by secretagogin+ neurons. Secretagogin silencing disrupts cell migration (dashed curves in A2′ and A2′′). (A3–A5) Although the total area in the Matrigel occupied by migratory neurons decreased only nonsignificantly upon gene silencing, both the maximal and the average distance of secretagogin+ cells was significantly reduced from RMS exit. (B–B1′) In primary culture from P5 RMS plated on PDL, neuroblasts contacting secretagogin+ neurons (white arrowheads in B and B1) typically expressed DCX. (The gray arrowhead in B points to a DCX− neuron.) (B2–B2′′) On laminin-coated surfaces, secretagogin+ neurons (white arrowheads in B2) formed scaffolds for DCX+ neuroblasts. (B3) Secretagogin silencing decreased secretagogin mRNA levels but left DCX mRNA expression unaltered. P < 0.05, Student’s t test. (Scale bars: 500 µm in A; 70 µm in A′–A′′′; 25 µm in A1–A1′′; 10 µm in B; and 3 µm in B2′′.)
To support the notion that secretagogin+ neurons are critical for neighboring neurons’ DCX expression, implying the determination of their motility, we have dissociated the RMS of P5 rats. Neuroblasts were identified by their TUJ1 immunoreactivity (Fig. 7B). DCX immunoreactivity in TUJ1+ neurons was typically confined to neurons with in contact with secretagogin+ cells (Fig. 7 B–B1′). In contrast to this even, nonpolarized distribution of cells seeded on poly-d-lysine (PDL) coating, DCX+ RMS neurons on laminin appeared in a string-like arrangement concentrated around spindle-shaped secretagogin+ cells (Fig. 7 B2–B2′′). Knockdown of secretagogin expression left overall DCX expression unaltered at the transcriptional level as shown by quantitative PCR (qPCR) (Fig. 7B3). This finding suggests that DCX expression is not directly regulated by secretagogin but instead that secretagogin expression regulates cell motility through an extracellular mechanism.
Secretagogin Regulates MMP-2 Release to Reorganize the Extracellular Matrix.
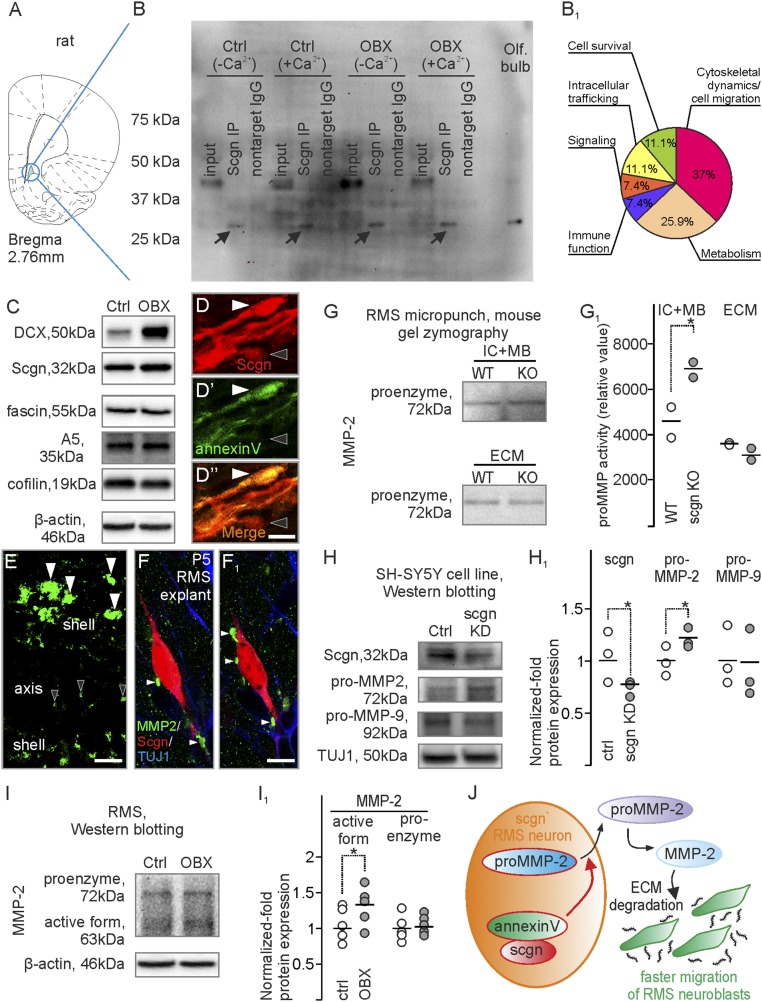
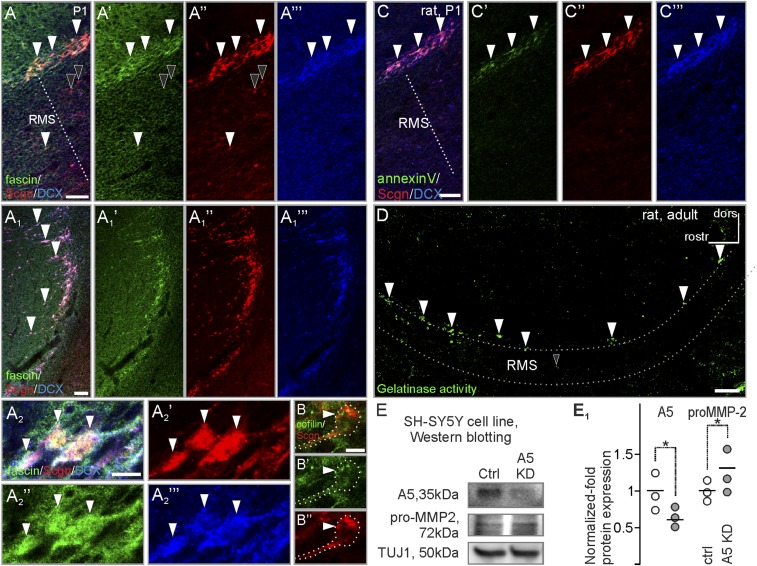
Migrating neurons traverse the extracellular matrix, which not only forms a mechanical barrier but also serves to anchor interacting guidance cues (46–48). We hypothesized that secretagogin+ neurons actively shape the surrounding matrix to impact neuroblast migration. To explore this notion, we looked for interacting partners of secretagogin in RMS micropunches (Fig. 8A) of control and bulbectomized rats. Secretagogin-interacting proteins were identified in both Ca2+-enriched and Ca2+-free conditions by mass spectrometry (Fig. 8 B and B1) (35). Among the 264 identified proteins, 64 were considered to be secretagogin-specific in at least one of four conditions (Table S3). Among these, we identified 44 proteins forming secretagogin’s Ca2+-dependent interactome, of which 18 were detected in samples from bulbectomized rats. In addition to the actin-bundling protein fascin (49) and the actin dynamic regulator cofilin (50), which reinforce the likely ability of secretagogin+ neurons to undergo morphological changes, we identified annexin V, a protein involved in the release of MMPs, particularly MMP-2 (51). Post hoc Western blotting confirmed the presence of fascin, cofilin, and annexin V in the RMS (Fig. 8C), and immunohistochemistry showed these proteins in secretagogin+ cells (Fig. 8 D–D1′′ and Fig. S5).
Fig. 8.
Secretagogin regulates MMP-2 release. (A and B) RMS micropunch samples from bulbectomized and control rats were immunoprecipitated with secretagogin in Ca2+-containing or Ca2+-free isolation buffers and were subjected to mass spectrometry. (B1) Functional distribution of secretagogin-interacting proteins recruited from samples of bulbectomized animals and homogenized in Ca2+-containing isolation buffer. (C) Western blotting verified DCX, secretagogin, fascin, annexin V, and cofilin expression in the RMS. (D–D′′) Post hoc immunohistochemistry resolved annexin V immunoreactivity in secretagogin+ neurons in the RMS (white arrowheads). The black arrowhead points to an annexin V−/secretagogin+ neuron. (E) In situ zymography revealed gelatinase activity in the RMS that was reminiscent of the distribution of secretagogin+ neurons. White and black arrowheads indicate profiles in the shell and axial domains of the RMS, respectively. (F and F1) MMP2+ profiles on the extracellular surface of secretagogin+ neurons in RMS explants (serial reconstruction, 700-nm thin optical slices, consecutive z-stack images). (G and G1) Gel zymography from RMS micropunches showed increased proMMP-2 levels in both intracellular and membrane-bound fractions of secretagogin−/− mice, but proMMP-2 levels decreased in the extracellular fraction. (H and H1) Secretagogin silencing increased the expression of the proenzyme form of MMP-2 but not of MMP-9 in SH-SY5Y neuroblastoma cells in vitro. (I and I1) Olfactory bulbectomy increased the level of the active but not of the proenzyme form of MMP-2 in rat RMS. (J) Schematic overview of the secretagogin-regulated mechanism in the RMS. Secretagogin regulates the externalization of proMMP-2, thereby limiting the amount of the active form of MMP-2. MMP-2 degrades the extracellular matrix to promote neuroblast migration. P < 0.05, Student’s t test. A5, annexin V; Ctrl, control; OBX, bulbectomized. (Scale bars; 40 µm in D; 10 µm in D′′ and E; and 5 µm in F1.)
Table S3.
Possible secretagogin-interacting molecules
| Identified proteins, n = 64 | Accession no. | Massin kDA | No. NTS IgG, minimum twofold less than the no. of NTS Scgn | Quantitative value (normalized total spectra) | Exclusive unique peptide counts | Protein identification probability, % | Percent coverage | ||||||||||||||||||||||||||||||||
| Description | Gene name | Ctrl, no Ca | Ctrl, Ca | OBX, no Ca | OBX, Ca | Ctrl, no Ca | Ctrl, Ca | OBX, no Ca | OBX, Ca | Ctrl, no Ca | Ctrl, Ca | OBX, no Ca | OBX, Ca | Ctrl, no Ca | Ctrl, Ca | OBX, no Ca | OBX, Ca | Ctrl, no Ca | Ctrl, Ca | OBX, no Ca | OBX, Ca | ||||||||||||||||||
| IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | IgG | Scgn | ||||||||
| 14-3-3 protein zeta/delta | Ywhaz | A0A0G2JV65_RAT | 29 | * | 6 | 2 | 7 | 14 | 12 | 11 | 10 | 10 | 4 | 2 | 4 | 5 | 5 | 4 | 3 | 3 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 19 | 9 | 19 | 26 | 26 | 16 | 13 | 13 | |||
| 2,4-dienoyl-CoA reductase, mitochondrial | Decr1 | DECR_RAT (+1) | 36 | * | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | |||
| 60S ribosomal protein L23 | Rpl23 | RL23_RAT | 15 | * | 1 | 2 | 0 | 0 | 2 | 3 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 2 | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 0 | 0 | 14 | 25 | 0 | 0 | 21 | 25 | 0 | 0 | |||
| Actn1 protein | Actn1 | Q6GMN8_RAT | 103 | * | 28 | 6 | 2 | 6 | 28 | 7 | 5 | 10 | 15 | 2 | 1 | 4 | 16 | 4 | 3 | 8 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 21 | 3 | 2 | 5 | 22 | 6 | 4 | 11 | |||
| ADP/ATP translocase 1 | Slc25a4 | ADT1_RAT | 33 | * | 1 | 2 | 1 | 2 | 6 | 6 | 2 | 4 | 1 | 2 | 1 | 2 | 4 | 3 | 2 | 2 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 4 | 7 | 3 | 8 | 14 | 11 | 8 | 7 | |||
| Annexin A5 | Anxa5 | ANXA5_RAT (+1) | 36 | * | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 98 | 0 | 0 | 5 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | |||
| ATP synthase subunit alpha, mitochondrial | Atp5a1 | ATPA_RAT | 60 | * | 0 | 0 | 4 | 8 | 0 | 1 | 1 | 4 | 0 | 0 | 2 | 3 | 0 | 1 | 1 | 3 | 0 | 0 | 100 | 100 | 0 | 100 | 100 | 100 | 0 | 0 | 5 | 6 | 0 | 2 | 3 | 7 | |||
| ATP synthase subunit O, mitochondrial | Atp5o | ATPO_RAT | 23 | * | 7 | 2 | 4 | 4 | 8 | 7 | 1 | 4 | 5 | 1 | 3 | 3 | 5 | 5 | 1 | 2 | 100 | 100 | 100 | 100 | 100 | 100 | 98 | 100 | 33 | 10 | 16 | 19 | 34 | 34 | 8 | 17 | |||
| ATP-dependent 6-phosphofructokinase | Pfkm | A0A0G2KBC7_RAT | 94 | * | 3 | 0 | 10 | 6 | 0 | 0 | 4 | 9 | 2 | 0 | 6 | 4 | 0 | 0 | 2 | 6 | 100 | 83 | 100 | 100 | 0 | 100 | 100 | 100 | 3 | 0 | 8 | 5 | 0 | 0 | 3 | 9 | |||
| ATP-dependent 6-phosphofructokinase, platelet type | Pfkp | PFKAP_RAT | 86 | * | 1 | 0 | 8 | 9 | 0 | 3 | 6 | 9 | 1 | 0 | 3 | 5 | 0 | 2 | 4 | 6 | 100 | 0 | 100 | 100 | 38 | 100 | 100 | 100 | 2 | 0 | 6 | 10 | 0 | 4 | 9 | 11 | |||
| ATP-dependent RNA helicase DDX1 | Ddx1 | DDX1_RAT | 82 | * | 7 | 2 | 1 | 0 | 1 | 2 | 2 | 1 | 3 | 2 | 1 | 0 | 1 | 2 | 1 | 1 | 100 | 100 | 100 | 98 | 100 | 100 | 99 | 98 | 5 | 3 | 2 | 0 | 2 | 3 | 2 | 2 | |||
| Beta-adducin | Add2 | F8WFS9_RAT | 81 | * | 8 | 1 | 4 | 1 | 7 | 5 | 2 | 9 | 3 | 1 | 3 | 1 | 4 | 3 | 2 | 4 | 100 | 90 | 100 | 100 | 100 | 100 | 100 | 100 | 6 | 1 | 6 | 1 | 7 | 6 | 4 | 7 | |||
| Calcium/calmodulin-dependent protein kinase type II subunit gamma | Camk2g | F1LMC3_RAT (+1) | 59 | * | 60 | 33 | 0 | 43 | 35 | 33 | 64 | 52 | 3 | 1 | 0 | 2 | 3 | 3 | 3 | 2 | 100 | 91 | 0 | 100 | 100 | 100 | 100 | 100 | 18 | 14 | 0 | 16 | 18 | 18 | 20 | 16 | |||
| Cofilin-1 | Cfl1 | COF1_RAT | 19 | * | 1 | 0 | 1 | 1 | 5 | 3 | 1 | 3 | 1 | 0 | 1 | 1 | 4 | 2 | 1 | 2 | 100 | 0 | 98 | 98 | 100 | 100 | 100 | 100 | 12 | 0 | 7 | 7 | 30 | 17 | 7 | 19 | |||
| Complement C1q subcomponent subunit C | C1qc | C1QC_RAT | 26 | * | * | * | 0 | 5 | 0 | 4 | 0 | 3 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 2 | 12 | 100 | 98 | 100 | 46 | 100 | 99 | 100 | 0 | 8 | 0 | 8 | 0 | 4 | 0 | 8 | |
| Complement C3 | C3 | M0RBF1_RAT | 186 | * | * | * | * | 12 | 61 | 10 | 54 | 7 | 45 | 6 | 29 | 5 | 8 | 3 | 8 | 4 | 8 | 2 | 7 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 3 | 5 | 2 | 5 | 3 | 5 | 1 | 4 |
| Coronin | Coro1b | G3V940_RAT | 54 | * | 1 | 0 | 0 | 2 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 100 | 48 | 0 | 100 | 100 | 100 | 99 | 100 | 2 | 0 | 0 | 4 | 2 | 2 | 2 | 2 | |||
| Coronin | Coro2a | A0A0G2QC56_RAT | 62 | * | * | 1 | 2 | 0 | 1 | 6 | 2 | 1 | 7 | 1 | 2 | 0 | 1 | 4 | 2 | 1 | 4 | 99 | 100 | 12 | 100 | 100 | 100 | 100 | 100 | 2 | 4 | 0 | 2 | 10 | 4 | 2 | 8 | ||
| Coronin | Coro2b | F1LMV9_RAT | 55 | * | * | 5 | 2 | 1 | 4 | 9 | 7 | 3 | 10 | 3 | 2 | 1 | 2 | 4 | 3 | 1 | 2 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 8 | 5 | 3 | 5 | 11 | 7 | 3 | 5 | ||
| Coronin-1A | Coro1a | COR1A_RAT | 51 | * | 0 | 0 | 0 | 3 | 2 | 1 | 3 | 4 | 0 | 0 | 0 | 2 | 2 | 1 | 2 | 3 | 54 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 0 | 5 | 4 | 2 | 4 | 6 | |||
| Cortactin-binding protein 2 | Cttnbp2 | sp|Q2IBD4|CTTB2_RAT | 179 | * | 6 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 100 | 67 | 0 | 94 | 100 | 100 | 0 | 96 | 4 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |||
| Creatine kinase B-type | Ckb | KCRB_RAT | 43 | * | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 99 | 100 | 0 | 6 | 0 | 0 | 0 | 0 | 3 | 6 | 0 | 0 | 0 | 0 | |||
| Creatine kinase, mitochondrial 1, ubiquitous | Ckmt1b | Q5BJT9_RAT | 47 | * | * | 14 | 11 | 0 | 2 | 6 | 12 | 0 | 1 | 4 | 3 | 0 | 2 | 3 | 4 | 0 | 1 | 100 | 100 | 68 | 100 | 100 | 100 | 8 | 99 | 13 | 11 | 0 | 9 | 11 | 13 | 0 | 5 | ||
| Elongation factor 1-alpha 1 | Eef1a1 | EF1A1_RAT | 50 | * | 8 | 10 | 4 | 3 | 10 | 12 | 2 | 6 | 4 | 3 | 3 | 1 | 4 | 5 | 2 | 4 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 13 | 7 | 6 | 2 | 13 | 11 | 4 | 9 | |||
| F-actin-capping protein subunit beta | Capzb | CAPZB_RAT | 31 | * | 14 | 2 | 3 | 8 | 19 | 12 | 9 | 11 | 6 | 2 | 2 | 4 | 7 | 7 | 3 | 5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 17 | 9 | 9 | 19 | 25 | 28 | 14 | 22 | |||
| Fascin | Fscn1 | FSCN1_RAT | 54 | * | * | 13 | 11 | 0 | 3 | 8 | 10 | 3 | 10 | 8 | 5 | 0 | 2 | 7 | 6 | 2 | 6 | 100 | 100 | 10 | 100 | 100 | 100 | 100 | 100 | 18 | 10 | 0 | 4 | 16 | 14 | 5 | 14 | ||
| Fc of IgG, low affinity IIIa, receptor | Fcgr3a | A0A0B4J2J1_RAT (+1) | 28 | * | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 27 | 0 | 10 | 0 | 97 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Flightless I homolog (Drosophila) | Flii | Q5RKI5_RAT | 145 | * | 16 | 5 | 3 | 5 | 22 | 15 | 1 | 11 | 9 | 2 | 2 | 5 | 9 | 7 | 1 | 6 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 9 | 2 | 2 | 4 | 9 | 7 | 1 | 6 | |||
| Fructose-bisphosphate aldolase A | Aldoa | ALDOA_RAT | 39 | * | 4 | 1 | 4 | 1 | 0 | 2 | 0 | 0 | 3 | 1 | 2 | 1 | 0 | 2 | 0 | 0 | 100 | 91 | 100 | 97 | 6 | 100 | 0 | 0 | 13 | 2 | 6 | 4 | 0 | 7 | 0 | 0 | |||
| Glutamate dehydrogenase 1, mitochondrial | Glud1 | DHE3_RAT | 61 | * | * | 9 | 6 | 0 | 4 | 11 | 25 | 0 | 2 | 5 | 3 | 0 | 2 | 8 | 14 | 0 | 1 | 100 | 100 | 23 | 100 | 100 | 100 | 16 | 100 | 9 | 5 | 0 | 3 | 17 | 31 | 0 | 1 | ||
| Hemoglobin alpha, adult chain 2 | Hba1 | B1H216_RAT (+1) | 15 | * | * | 2 | 6 | 5 | 8 | 2 | 3 | 2 | 6 | 2 | 3 | 4 | 5 | 2 | 3 | 2 | 3 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 22 | 26 | 46 | 49 | 20 | 33 | 15 | 26 | ||
| Hemoglobin subunit beta-1 | Hbb | HBB1_RAT | 16 | * | * | 7 | 9 | 7 | 6 | 3 | 7 | 1 | 9 | 5 | 5 | 4 | 4 | 2 | 4 | 1 | 4 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 41 | 44 | 30 | 28 | 14 | 35 | 8 | 28 | ||
| Histone H2A | Hist1h2af | Q6I8Q6_RAT | 14 | * | 3 | 0 | 0 | 2 | 0 | 0 | 1 | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 100 | 0 | 0 | 100 | 31 | 16 | 100 | 97 | 22 | 0 | 0 | 22 | 0 | 0 | 7 | 15 | |||
| Histone H4 | Hist1h4b | sp|P62804|H4_RAT | 11 | * | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 39 | 0 | 0 | 0 | 0 | |||
| Ig gamma-2B chain C region | Igh-1a | IGG2B_RAT | 36 | * | 2 | 4 | 0 | 2 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 0 | 1 | 100 | 100 | 0 | 100 | 100 | 100 | 13 | 100 | 3 | 3 | 0 | 3 | 3 | 9 | 0 | 3 | |||
| Ig kappa chain C region, A allele | N/A | KACA_RAT | 12 | * | * | 0 | 12 | 0 | 2 | 2 | 6 | 0 | 0 | 0 | 3 | 0 | 1 | 1 | 3 | 0 | 0 | 0 | 100 | 0 | 99 | 95 | 100 | 0 | 94 | 0 | 36 | 0 | 12 | 12 | 36 | 0 | 0 | ||
| Junction plakoglobin | Jup | PLAK_RAT | 82 | * | 1 | 0 | 0 | 7 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 5 | 1 | 0 | 0 | 0 | 98 | 57 | 0 | 100 | 95 | 0 | 0 | 0 | 2 | 0 | 0 | 9 | 2 | 0 | 0 | 0 | |||
| Microtubule-associated protein | Mapt | D3ZKD9_RAT | 39 | * | 6 | 19 | 0 | 0 | 7 | 10 | 0 | 0 | 3 | 6 | 0 | 0 | 3 | 4 | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 0 | 0 | 10 | 20 | 0 | 0 | 10 | 13 | 0 | 0 | |||
| Myelin basic protein | Mbp | sp|P02688|MBP_RAT | 22 | * | 73 | 134 | 1 | 7 | 70 | 71 | 7 | 4 | 7 | 8 | 1 | 5 | 7 | 7 | 5 | 3 | 100 | 100 | 99 | 100 | 100 | 100 | 100 | 100 | 38 | 41 | 5 | 28 | 38 | 38 | 28 | 17 | |||
| Nonspecific serine/threonine protein kinase | Cdc42bpb | A0A0G2KB58_RAT | 194 | * | 1 | 1 | 0 | 1 | 5 | 6 | 1 | 11 | 1 | 1 | 0 | 1 | 5 | 6 | 1 | 8 | 100 | 100 | 21 | 100 | 100 | 100 | 100 | 100 | 1 | 1 | 0 | 1 | 4 | 4 | 1 | 6 | |||
| O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase), isoform CRA_a | Ogt | A0A0G2K3V4_RAT | 117 | * | * | 7 | 25 | 1 | 0 | 6 | 14 | 0 | 0 | 5 | 13 | 1 | 0 | 4 | 10 | 0 | 0 | 100 | 100 | 99 | 0 | 100 | 100 | 0 | 0 | 6 | 15 | 1 | 0 | 5 | 12 | 0 | 0 | ||
| Peroxisomal multifunctional enzyme type 2 | Hsd17b4 | DHB4_RAT (+1) | 79 | * | 4 | 4 | 0 | 0 | 1 | 3 | 0 | 3 | 2 | 3 | 0 | 0 | 1 | 2 | 0 | 1 | 100 | 100 | 57 | 0 | 100 | 100 | 0 | 100 | 4 | 5 | 0 | 0 | 2 | 3 | 0 | 2 | |||
| Phosphorylase kinase, beta | Phkb | Q5RKH5_RAT | 126 | * | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 43 | 0 | 12 | 100 | 0 | 0 | 0 | 46 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | |||
| Protein Diras2 | Diras2 | D3ZHX3_RAT | 22 | * | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 100 | 0 | 96 | 0 | 100 | 0 | 97 | 0 | 6 | 0 | 6 | 0 | 12 | 0 | 6 | |||
| Protein Dmxl2 | Dmxl2 | F1M3W5_RAT | 341 | * | 3 | 0 | 3 | 4 | 1 | 1 | 1 | 5 | 2 | 0 | 3 | 2 | 1 | 1 | 1 | 4 | 100 | 12 | 100 | 100 | 100 | 100 | 100 | 100 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 2 | |||
| Protein Golga3 | Golga3 | D3ZLD5_RAT | 163 | * | * | 0 | 0 | 1 | 9 | 0 | 0 | 0 | 7 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 5 | 6 | 0 | 97 | 100 | 0 | 0 | 0 | 100 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 5 | ||
| Protein Hcfc1 | Hcfc1 | D3ZN95_RAT | 209 | * | * | 2 | 21 | 0 | 0 | 1 | 4 | 0 | 0 | 2 | 9 | 0 | 0 | 1 | 4 | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 0 | 0 | 1 | 5 | 0 | 0 | 1 | 3 | 0 | 0 | ||
| Protein LOC100362298 | LOC100362298 | D3ZM33_RAT | 18 | * | 3 | 7 | 0 | 0 | 5 | 5 | 0 | 0 | 3 | 4 | 0 | 0 | 5 | 4 | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 0 | 0 | 19 | 24 | 0 | 0 | 30 | 26 | 0 | 0 | |||
| Protein rogdi homolog | Rogdi | ROGDI_RAT | 32 | * | 0 | 0 | 1 | 1 | 0 | 2 | 0 | 4 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 3 | 41 | 61 | 99 | 100 | 32 | 100 | 30 | 100 | 0 | 0 | 5 | 7 | 0 | 7 | 0 | 12 | |||
| Protein Sacs | Sacs | D4A1D3_RAT | 521 | * | 3 | 1 | 1 | 2 | 6 | 4 | 2 | 14 | 2 | 1 | 1 | 2 | 4 | 4 | 2 | 8 | 100 | 99 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 2 | |||
| Protein Slc25a12 | Slc25a12 | F1LX07_RAT | 72 | * | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 95 | 26 | 0 | 0 | 21 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | |||
| Protein Vps35 | Vps35 | G3V8A5_RAT | 92 | * | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 3 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 100 | 0 | 100 | 100 | 87 | 0 | 99 | 100 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 3 | |||
| RAB10, member RAS oncogene family | Rab10 | Q5RKJ9_RAT | 23 | * | 0 | 0 | 2 | 3 | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 12 | 10 | 69 | 100 | 22 | 0 | 0 | 100 | 0 | 0 | 6 | 11 | 6 | 0 | 0 | 17 | |||
| Rho guanine nucleotide exchange factor 11 | Arhgef11 | A0A0G2JZC6_RAT | 174 | * | 2 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 100 | 26 | 17 | 0 | 100 | 100 | 11 | 86 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | |||
| Secretagogin | Scgn | F1LN07_RAT | 32 | * | * | * | * | 0 | 47 | 0 | 31 | 0 | 54 | 0 | 55 | 0 | 12 | 0 | 10 | 0 | 15 | 0 | 11 | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 56 | 0 | 51 | 0 | 60 | 0 | 50 |
| Septin-7 | Sept7 | D4A0F5_RAT | 51 | * | 27 | 20 | 0 | 0 | 13 | 11 | 0 | 2 | 6 | 6 | 0 | 0 | 4 | 4 | 0 | 2 | 100 | 100 | 0 | 10 | 100 | 100 | 74 | 100 | 19 | 19 | 0 | 0 | 11 | 11 | 0 | 4 | |||
| Stress-70 protein, mitochondrial | Hspa9 | GRP75_RAT | 74 k | * | 1 | 2 | 3 | 3 | 6 | 1 | 4 | 6 | 1 | 2 | 3 | 3 | 5 | 1 | 4 | 5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 2 | 4 | 6 | 6 | 9 | 2 | 7 | 10 | |||
| Synapsin II, isoform CRA_a | Syn2 | G3V733_RAT (+1) | 63 | * | * | 37 | 81 | 1 | 0 | 24 | 48 | 5 | 2 | 4 | 6 | 1 | 0 | 1 | 5 | 1 | 1 | 100 | 100 | 100 | 25 | 100 | 100 | 100 | 100 | 14 | 15 | 3 | 0 | 8 | 17 | 6 | 3 | ||
| Tenascin-R | Tnr | F1LQ63_RAT | 150 | * | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 100 | 97 | 0 | 0 | 97 | 100 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 2 | |||
| Transgelin-3 | Tagln3 | TAGL3_RAT | 23 | * | 30 | 22 | 0 | 6 | 46 | 57 | 8 | 12 | 11 | 8 | 0 | 3 | 12 | 12 | 6 | 8 | 100 | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 63 | 41 | 0 | 17 | 71 | 71 | 32 | 41 | |||
| Triosephosphate isomerase | Tpi1 | TPIS_RAT | 27 | * | * | 1 | 0 | 0 | 3 | 2 | 1 | 2 | 5 | 1 | 0 | 0 | 2 | 1 | 1 | 2 | 5 | 100 | 62 | 31 | 100 | 100 | 100 | 100 | 100 | 6 | 0 | 0 | 10 | 5 | 5 | 10 | 28 | ||
| Tropomodulin-2 | Tmod2 | TMOD2_RAT | 39 | * | 22 | 17 | 11 | 23 | 22 | 24 | 22 | 21 | 5 | 4 | 4 | 5 | 7 | 4 | 4 | 5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 17 | 14 | 14 | 17 | 29 | 14 | 14 | 16 | |||
| Tropomyosin alpha-3 chain | Tpm3 | sp|Q63610|TPM3_RAT | 29 | * | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 43 | 0 | 0 | 100 | 0 | 0 | 88 | 51 | 0 | 0 | 0 | 10 | 0 | 0 | 4 | 0 | |||
| Unconventional myosin-Ib | Myo1b | D4A2X2_RAT (+1) | 132 | * | 8 | 0 | 7 | 7 | 11 | 4 | 4 | 10 | 6 | 0 | 5 | 5 | 7 | 3 | 3 | 6 | 100 | 10 | 100 | 100 | 100 | 100 | 100 | 100 | 6 | 0 | 6 | 5 | 8 | 4 | 3 | 7 | |||
Asterisks indicate proteins identified with a minimum 99% probability, with a mininimum of two exclusive unique peptide counts, and with at least twofold higher normalized total spectra following immunoprecipitation. Taxonomic names for all: Rattus norvegicus. NTS, normalized total spectra.
Fig. S5.
Secretagogin is coexpressed with the motility proteins fascin and cofilin. (A–A2′′′) Fascin+/secretagogin+ neurons were identified not only in the marginal region of the RMS in P1 rats (white arrowheads in A–A′′′; black arrowheads point to fascin−/secretagogin+ neurons) but also in groups leaving the RMS from its proximal limb transversally (white arrowheads in A1–A1′′′). (B–B′′) Cofilin colocalized with secretagogin. The dotted line surrounds a cofilin+/secretagogin+ cell. (C–C′′′) Secretagogin+ cells coexpress annexin V in the dorsal margin of the RMS. (D) In situ zymography revealed gelatinase activity in the RMS, which was reminiscent of the distribution of secretagogin+ neurons. (E and E1) Knockdown of annexin V expression increased proMMP-2 expression. (Scale bars: 40 µm in A, A1, B, and C; 200 μm in D; and; 10 µm in A2.)
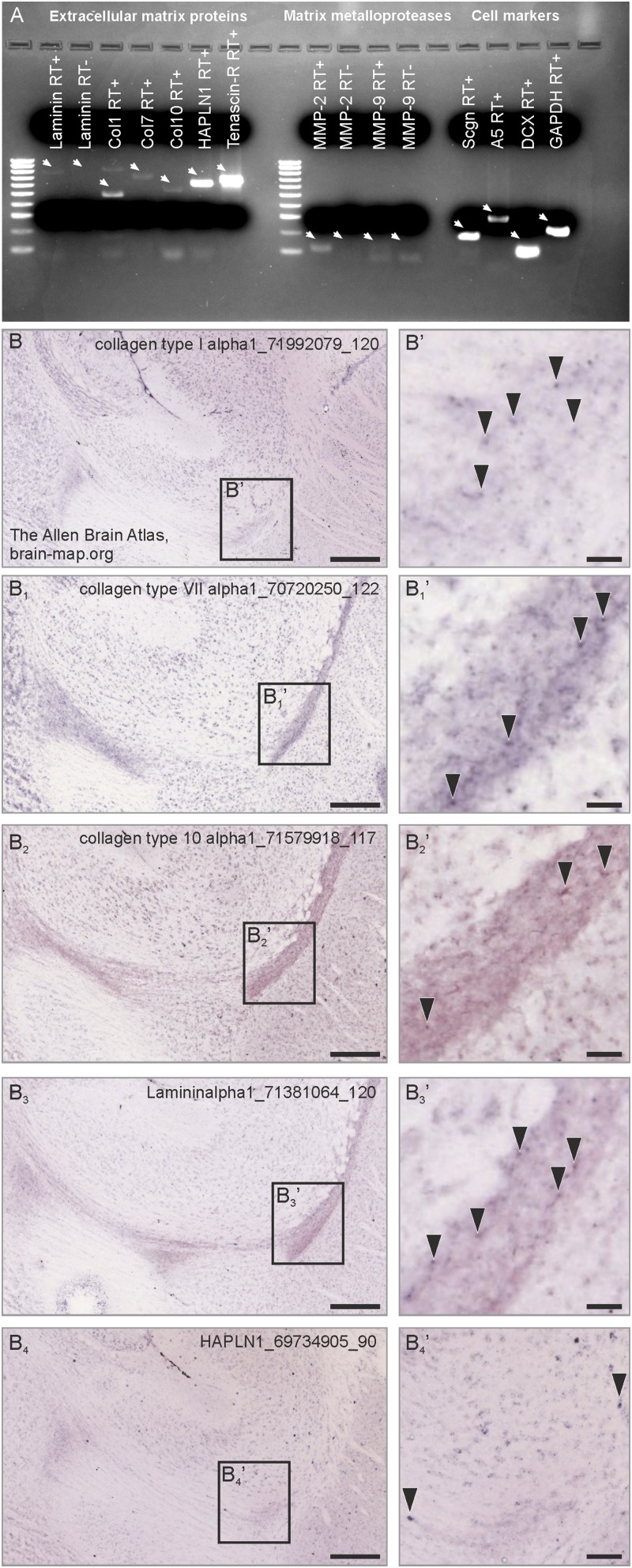
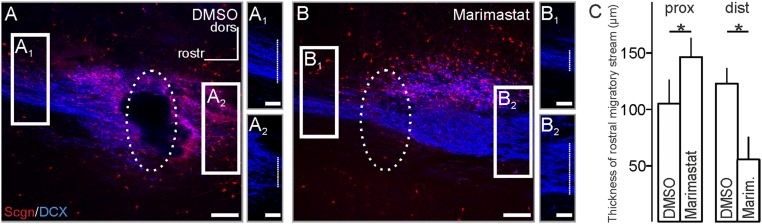
Annexin V’s presence in secretagogin+ neurons led us to hypothesize that secretagogin could trigger a secretory pathway to degrade the RMS extracellular matrix (Fig. S6) via regulated MMP-2 release. We first showed that MMP2/9 activity in the RMS is critical for migrating neurons because focal RMS application of Marimastat, a potent and selective MMP-2/MMP-9 inhibitor (52), increased the diameter of the proximal but decreased the diameter of the distal limbs of the RMS (Fig. S7), suggesting migration arrest. Notably, a large number of secretagogin+ neurons accumulated upstream from the drug injection site, especially in the dorsal RMS (Fig. S7B). To distinguish MMP-active foci in the RMS, we developed in situ zymography (Fig. S5D) and showed gelatinase activity in both axial and marginal regions (Fig. S5D and Fig. 8E), which harbor the bulk of secretagogin+ neurons (see also Fig. 1A). High-resolution orthogonal confocal imaging revealed that MMP-2 foci typically were anchored at the outside surface of secretagogin+ neurons in P5 RMS explants, as is compatible with MMP-2 secretion (Fig. 8 F and F1). Further support came through gel-zymography in secretagogin−/− mice: The proenzyme form of MMP-2 (proMMP-2) increased in the fraction containing intracellular and membrane-bound enzymes but decreased in the extracellular fraction prepared from RMS micropunches (Fig. 8 G and G1). We extended this finding by showing that secretagogin silencing increases the level of proMMP-2, but not proMMP-9, in the cytosolic fraction of SH-SY5Y neuroblastoma cells (Fig. 8 H and H1). Impaired externalization of proMMP-2 likely involved annexin V because annexin V knockdown led to a similar increase of intracellular proMMP-2 (Fig. S5 E and E1).
Fig. S6.
The presence of extracellular matrix components and matrix metalloproteases in the RMS. (A) PCR amplicons from RMS micropunches indicate the presence of laminin, collagen types 1, 7, and 10, the link protein HAPLN-1, and tenascin-R. In contrast to MMP-2, MMP-9 mRNA expression was at the detection limit. White arrows indicate cDNA bands (or the lack of, in case of the RT lanes). (B–B4′) Corresponding in situ hybridization images from the open source database of the Allen Brain Atlas (experiment numbers are indicated in the figure images) demonstrate the presence of collagen types 1, 7, and 10, laminin, and and HAPLN-1 (black arrowheads) in the adult mouse RMS. Col, collagen; HPLN-1, hyaluronan and proteoglycan link protein 1. (Scale bars: 300 μm for B, B1, B2, B3, and B4; 20 μm for B′, B1′, B2′, B3′, and B4′.)
Fig. S7.
Marimastat blocks forward neuroblast migration. (A–C) Focal administration of the dual MMP-2/MMP-9 inhibitor Marimastat (1 µ M/0.1 µL) (Tocris) increased the diameter of the proximal RMS (B2) but decreased the diameter of the distal RMS (B1), compared with control (DMSO) application (A2 and A1, respectively) with secretagogin+ cells accumulating upstream the application site. The effect of Marimmastat is quantified in C. (Scale bars: 100 µm in A and B; 20 µm in A1, A2, B1, and B2.) *P < 0.05.
Decreased levels of externalized proMMP-2 suggest reduced active MMP-2 extracelluarly. We have measured the protein level of active MMP-2 by Western blotting in RMS micropunches of control and bulbectomized rats. ProMMP-2 and active MMP-2 isoforms were separated by their molecular weights (53). Olfactory bulbectomy significantly increased the level of active MMP-2 but not overall proMMP-2 (Fig. 8 I and I1) in the RMS. Taken together, our data identify differentiated neurons in the mammalian RMS that express secretagogin to initiate a molecular cascade through annexin V to increase MMP-2 release, thus remodelling the extracellular matrix to aid neuroblast migration (Fig. 8J) (15, 16).
Discussion
Researchers have extensively investigated the mechanisms that regulate neuroblast migration in the fetal brain and have identified principal guidance rules in immature tissues. Nevertheless, the RMS of the adult rodent (2) and primate (40) brain is a differently structured, mature anatomical domain that relies on alternative migration cues. Much of our existing knowledge about how newborn cells move in this predominantly glial tube includes glia–neuroblast interactions. Overall, disruption of the glial tunnel as shown in neural cell adhesion molecule- (54) or β1-integrin– (55) deficient mice leads to neuroblast migration defects. In addition to factors related to nonsoluble cell-adhesion (56), astrocytes can use and release soluble signals to orchestrate the movement of RMS neuroblasts (18). At the same time, neuroblasts are proactive in shaping their own journey by relying on physiologically conserved mechanisms, such as Slit/Robo signaling, to clear astrocytic processes (16).
Ca2+-binding proteins are widely used to distinguish and characterize neuronal subsets in the mammalian brain. In addition to parvalbumin, calbindin, and calretinin, secretagogin is a useful marker for identifying forebrain neurons (57). Here we show that the rodent RMS harbors a secretagogin+ neuronal contingent that is non–self-renewing, morphologically differentiated, and may be wired into synaptically connected neuronal circuits.
Secretagogin+ neurons integrated along the total RMS. Thus, secretagogin+ neurons also occurred in the axial RMS domain sent perpendicularly oriented processes to reach the space between neuroblasts and to surround neurites. Secretagogin expression of RMS neurons could be induced by olfactory bulbectomy, suggesting its injury-induced activation to allow for enhanced forward neuroblast migration. Therefore we suggest that a latent neuronal cohort resides in the RMS that will up-regulate its secretagogin content only when pathophysiologically primed to partake in the MMP-2–mediated reorganization of the RMS.
Both insoluble and soluble signals are critically reliant upon the composition and dynamic regulation of the extracellular matrix. Innate components of the matrix can themselves operate as migration signals; for instance, hyaluronan acts on Rhamm receptors expressed on neuroblasts (24). However, the importance of the extracellular domain extends far beyond direct actions between matrix molecules and cell-surface receptors; instead, it offers a critical interface for a wide array of factors and signaling molecules that regulate migration (3–7, 25, 26, 58). Dynamic restructuring of the extracellular matrix is critical in RMS neuroblast migration. Therefore, a coordinated release of endogenous matrix-lytic enzymes, MMPs, can effectively and focally impact intercellular signaling and migration in select pericellular microdomains. We hypothesized that, instead of secreting specific ligands or factors, secretagogin+ neurons regulate neuroblast migration by controlling the reassembly of the extracellular matrix.
In addition to characterizing a hitherto unexplored cell cohort in the RMS, we describe an enzyme externalization mechanism which (i) works on an on-demand principle, (ii) was known only in tumor biology but not in focal pericellular mechanisms in the healthy adult, and (iii) promotes the migration of the neighboring cells but not the enzyme-releasing cell itself. More than 20 members of the MMP family cleave matrix components. In the developing central nervous system, MMP-2 and MMP-9 attracted much attention when rat postmitotic cerebellar granule neuron precursors (59) and human neuroepithelial stem cells (60) were shown to express MMP-2 and MMP-9–deficient mice showed granule cell migration defects (61). The expression profile of the different MMP members in the postnatal RMS showed the presence of MMP-2, but not MMP-9 (Fig. S6A) (12), and migrating neuroblasts were found to express membrane-type MMP 5 (MT5-MMP) (12). Using in situ zymography and immunohistochemistry, we now have identified secretagogin+ neurons as MMP-2–expressing foci. We argue that secretagogin regulates MMP-2 release because (i) mass spectrometry identified annexin V, a protein involved in MMP-2 release (51), in secretagogin’s interactome; (ii) secretagogin−/− RMS showed reduced proMMP-2 release; (iii) knockdown of secretagogin or annexin V expression in SH-SY5Y neuroblastoma cells selectively increased the level of intracellular proMMP-2, but not proMMP-9; and (iv) active MMP-2 increased in RMS micropunches after bulbectomy. Thus, secretagogin+ neurons can dissemble and rearrange the extracellular matrix to promote the migration of neighboring molecules. Blocking neuroblast migration by MMP inhibition using Marimastat led to increased secretagogin expression in marginal RMS cells upstream of the inhibition site, possibly reflecting a reactive and up-regulated secretagogin-mediated release of MMP activity in these cells to help neuroblast migration. These interactions enable secretagogin-containing neurons to control their own mobility via direct regulation of cytoskeletal proteins. Here, we found that knockdown of secretagogin expression led to reduced mobility of secretagogin+ neurons in RMS explants.
The migration of immature neuronal progeny from the ventricle to the olfactory bulb is a phylogenetically conserved mechanism in mammals (2, 62), including the human infant (1), with a largely similar guidance machinery and cellular expression profiles. Thus, the identification of new cellular components and intercellular signaling mechanisms is paramount to understand the development of the human brain as well as its malfunctions. The adult human subventricular zone principally lacks neural stem cells which could produce tangentially migrating neuroblasts for the olfactory system (1, 63, 64). During infancy, however, corridors of migrating neurons connect the anteroventral tip of the lateral ventricle to the olfactory tract. This pediatric RMS contains immature neurons that are organized in chains and express DCX (1). Here, we show a continuum of secretagogin+ cells that form corridors from the ventral tip of the lateral ventricle toward and reaching the olfactory trigone. Although neuroblast migration ceases in humans at the early postnatal age (1), we have previously demonstrated that secretagogin+ cells remain present throughout the olfactory tract even in the elderly (37). We suggest that secretagogin+ neurons are critical members of olfactory neuroblast migration in the human fetus, that their dysfunction could play a role in the development of neurological diseases when the olfactory system is compromised, and that, once remobilized, they might be used to for rescue strategies thus providing a tangible starting point for as yet unexplored avenues for redirecting neuroblasts to injury sites.
Materials and Methods
Fetal Tissue.
Two male fetal brains with normal development (between gestational weeks 31–33) were selected from the Brain Bank of the Institute of Neurology, Medical University of Vienna, Austria. Fetal brain tissue was obtained from spontaneous or medically induced abortions. Tissues were obtained and used in compliance with the Declaration of Helsinki and following institutional guidelines. The study was performed in the course of a study approved by the Ethical Committee of the Medical University of Vienna (No.104/2009).
Animals, Surgery, and Ethical Approval of Experimental Studies.
A total of 65 male rats (Wistar) and 24 male mice were used. Secretagogin-null(−/−) mice were custom-generated at the Mutant Mouse Resource and Research Center (University of California, Davis, Mouse Biology Program) using the “two-in-one” targeting strategy (65), which generates full knockouts by expressing a termination signal after exon 3 of the secretagogin gene. The ensuing truncated protein therefore terminates before the first EF-hand domain, excluding Ca2+-binding activity. Food and water were available ad libitum, and animals were kept under standard housing conditions using a 12/12 light/dark cycle. Experimental procedures, including stereotaxic injections and transcardial perfusion, were approved by the Ethical Review Boards of the Semmelweis University and the Medical University of Vienna and conformed to the European Convention for the Protection of Vertebrate Animals used for experimental and other scientific purposes (Protocols: ETS No. 170, ETS No.123, Tierversuchgesetz 2012, BGBI, Nr. 114/2012).
For all rat (n = 33) and mouse (n = 12) surgeries, 6-wk-old animals were used. During surgeries and transcardial perfusions animals were anesthetized i.m. or i.p. with a mixture of ketamine (50 mg/kg body weight) and xylazine (4 mg/kg body weight). After surgery, brains were perfusion-fixed by transcardially applying 4% (wt/vol) paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB). BrdU administration, olfactory bulbectomy, in vivo gene silencing, and in vivo inhibition of matrix MMP activity in rats or transgenic mice are described in SI Materials and Methods.
Immunohistochemistry and Imaging.
Chromogenic or multiple immunofluorescence histochemistry with select combinations of primary antibodies (Table S4) (1, 34, 35, 49, 51, 66–74) was performed according to published protocols (67). Results of chromogenic histochemistry were captured on an Olympus BX-51 microscope. Sections processed for multiple immunofluorescence histochemistry were inspected and images were acquired on a 780LSM confocal laser-scanning microscope (Zeiss). Procedural details of imaging described in SI Materials and Methods.
Table S4.
Markers used for immunolabeling
| Marker | Source | Host | IHC dilution | WB dilution | Ref. |
| Annexin V | Sigma | Mouse* | 1:1,000 | 1:1,000 | (51) |
| 5-Bromo-2'-deoxyuridine | Abcam | Rat* | 1:1,000 | N.a. | (66) |
| β-Actin | Sigma | Mouse* | n.a | 1:10,000 | (67) |
| β-III-Tubulin (TUJ1) | Sigma | Mouse* | 1:2,000 | N.a. | (74) |
| Calretinin | Synaptic Systems | Guinea pig† | 1:1,000 | N.a. | (68) |
| Cofilin | Sigma | Rabbit† | 1:5,000 | 1:5,000 | (69) |
| Doublecortin (DCX) | Santa Cruz | Goat† | 1:300 | 1:200 | (1) |
| Fascin | Dako | Mouse* | 1:50 | 1:100 | (49) |
| Matrix-metalloproteinase 2 (MMP-2) | R&D Systems | Mouse* | 1:50 | 1:400 | (70) |
| Neuronal nuclear antigen (NeuN) | Merck/Millipore | Mouse* | 1:1,000 | N.a. | (71) |
| Neural/glial antigen 2 (NG2 proteoglycan) | Merck/Millipore | Rabbit† | 1:200 | N.a. | (72) |
| S100 calcium-binding protein B (S100B) | Merck/Millipore | Rabbit† | 1:1,000 | N.a. | (73) |
| Secretagogin | L.Wagner | Rabbit† | 1:12,000 | 1:12,000 | (35) |
| Secretagogin | R&D Systems | Goat† | 1:100 | 1:1,000 | (34) |
| Solanum tuberosum | Vector | N.a. | 1:500 | (35) | |
| Stathmin | NeuroMab | Mouse* | 1:1,000 | N.a. | (74) |
IHC, immunohistochemistry; N.a., not applicable.
Monoclonal antibody.
Polyclonal antibody.
Electron Microscopy.
Sections processed for secretagogin immunohistochemistry (chromogenic detection) were postfixed in buffered 1% OsO4 at 22–24 °C for 1 h, and were flat-embedded in Durcupan ACM (Fluka). The RMS region was selected and embedded for ultrasectioning. Ultrathin sections (100 nm) were collected on single-slot nickel grids coated with Formvar and were studied on a Jeol 1200 EMX microscope. Primary magnifications ranged from 12,000× to 80,000×, as indicated.
Shotgun Proteomic Analysis.
Eluted fractions from secretagogin coimmunoprecipitates were separated on gradient gels (3–12%) and were in-gel digested. Their LC-MS/MS analysis was performed using an an Ultimate 3000 nanoRSLC (Thermo Scientific) coupled to a high-capacity ion trap mass spectrometer (AmaZon ETD; Bruker Daltonics) via a CaptiveSpray ion source (Bruker) (SI Materials and Methods) (75–78).
In Situ and Gelatin Zymography.
MMP gelatinase activity on sections was detected by in situ zymography (79, 80). Intracellular/cell membrane-bound and extracellular matrix-bound MMP activity was measured using gelatin zymography as previously published (SI Materials and Methods) (81, 82).
Cell Migration from RMS Explants.
P4/5 rat brains (n = 6) were sectioned sagittally on a vibratome in ice-cold DMEM containing penicillin (100 U/mL) and streptomycin (100 µg/mL) (all from Invitrogen). The RMS was excised and submerged in Matrigel (Corning) in complete culture medium (2 mM Glutamax, B27 supplement in Neurobasal medium) in a 3:1 ratio, which also contained secretagogin or scrambled siRNA (250 pmol/40 µL Matrigel mix) (GE Dharmacon). The explant–Matrigel mix was spread over the surface of Geltrex-coated (Invitrogen) coverslips and incubated in culture medium for 3 d. Coverslips were subsequently immersion-fixed and analyzed by immunocytochemistry (SI Materials and Methods).
SH-SY5Y Neuroblastoma and P5 Rat Primary Dissociated RMS Cultures.
SH-SY5Y neuroblastoma cells were cultured in DMEM:GlutaMAX containing 5% (vol/vol) FBS, penicillin (100 U/mL) and streptomycin (100 μg/mL) (all from Invitrogen) on PDL-coated coverslips. Using sagittal brain slices, the RMS region was dissected, cells were enzymatically dissociated and plated at a density of 50,000 cells per well on PDL- or laminin-coated coverslips in 24-well plates for morphometry or at a density of 200,000 cells per well in six-well plates for real-time qPCR. Primary neurons and neuroblasts were maintained in DMEM/F12 (1:1) containing B27 supplement [2% (vol/vol)], l-glutamine (2 mM), penicillin (100 U/mL), and streptomycin (100 μg/mL) (all from Invitrogen). In both cultures, secretagogin gene silencing was through the application of secretagogin siRNA (250 pmol/500 µL, diluted in culture medium) (GE Dharmacon) for 2 d. SH-SH-SY5Y cells and primary RMS cultures were subsequently lysed and collected for Western blotting or qPCR, respectively.
Western Blotting.
Protein samples from bulbectomized or sham-operated rat (n = 3 per group) RMS were prepared, and their concentrations were determined (83). The protein samples were analyzed under denaturing conditions. Western blotting was performed with the primary antibodies listed in Table S4. Blots were scanned using a Bio-Rad XRS+ imaging system and were quantified with Image Lab 3.01 (Bio-Rad Laboratories). Protein concentrations were normalized to β-actin (1:10.000) (Sigma).
Real-Time qPCR.
qPCR was performed on a Bio-Rad CFX 96 thermal cycler using primer sets listed in Table S5. Samples were run in triplicate to avoid processing-related deviations. Expression levels were normalized to the housekeeping gene encoding GAPDH.
Table S5.
Sequences of primers used
| Gene | Primer/probe | Sequence |
| Scgn | Primer (forward) | 5′–CCC AGA AGT GGA TGG ATT TG–3′ |
| Primer (reverse) | 5′–GTT GGG GAT CAG GGG TTT AT–3′ | |
| DCX | Primer (forward) | 5′–CTC CTA TCT CTA CAC CCA CAA GCC–3′ |
| Primer (reverse) | 5′–GAA TCG CCA AGT GAA TCA GAG TC–3′ | |
| GAPDH | Primer (forward) | 5′–AAC TTT GGC ATT GTG GAA GG–3′ |
| Primer (reverse) | 5′–ACA CAT TGG GGG TAG GAA CA–3′ |
Statistical Analysis.
Data were analyzed using Statistica Software Package version 13.2 (StatSoft, Inc.). Pairwise group comparisons in the in vitro histochemical and Western blotting experiments were evaluated using the Student’s t test (on independent samples). Data were normalized to controls as indicated. Data were expressed as means ± SEM. A P value of 0.05 was considered statistically significant. In the in vivo experiments, densities of secretagogin+ or BrdU+ cells in the RMS were analyzed by two-factorial ANOVA with genotype (transgenic vs. wild-type) and treatment (bulbectomized vs. control) as between-subjects factors in mice or with treatment (bulbectomized vs. control) and location of cells (marginal vs. axial) as between-subjects factors in rats. Two-factorial ANOVAs in rats were made using square root transformation of data. Post hoc analyses of between-subject effects were performed by Bonferroni’s t test. Homogeneity of variance was analyzed by Levene’s test. The normality of the sample was examined by Shapiro–Wilk W test. A P value of 0.05 was considered statistically significant.
SI Materials and Methods
Immunohistochemistry.
Free-floating sections (30 μm) were rinsed in PB (pH 7.4) and pretreated with 0.3% Triton X-100 (in PB) for 1 h at 22–24 °C to enhance the penetration of antibodies (1, 34, 35, 49, 51, 66–74). Nonspecific immunoreactivity was suppressed by incubating our specimens in a mixture of 5% (wt/vol) normal donkey serum (NDS) (Jackson), 2% (wt/vol) BSA (Sigma), and 0.3% Triton X-100 (Sigma) in PB for 1 h at 22–24 °C. Sections were exposed for 16–72 h at 4 °C to select combinations of primary antibodies (Table S3) diluted in PB to which 0.1% NDS and 0.3% Triton X-100 had been added. After extensive rinsing in PB, sections were processed by using chromogenic or immunofluorescence detection. In single-labeling experiments, hippocampal sections were exposed to biotinylated anti-rabbit IgG raised in donkey (1:1,000 dilution) (Jackson) for 2 h at 22–24 °C followed by preformed avidin–biotin complexes also incorporating HRP for 1 h at 22–24 °C. Immunosignals were visualized by 3,3′-diaminobenzidine (0.025%) (Sigma) as chromogen intensified with Ni-ammonium sulfate (0.05%) (Merck) in the presence of 0.001% H2O2 as substrate (dissolved in 0.05 M Tris buffer, pH 8.0). In multiple immunofluorescence labeling experiments, immunoreactivities were revealed by carbocyanine (Cy) 2-, 3-, or 5-tagged secondary antibodies raised in donkey (1:200) (Jackson) for 2 h at 22–24 °C. Glass-mounted sections were coverslipped with glycerol/gelatin (GG-1; Sigma).
Results of chromogenic stainings were captured on an Olympus BX-51 microscope at 2×, 10×, and 20× primary magnification. Sections processed for multiple immunofluorescence histochemistry were inspected, and images were acquired on a 780LSM confocal laser-scanning microscope (Zeiss) with optical zoom ranging from 1–3× at a primary magnification of 63× (Plan-Apochromat 63×/1.40) and pinhole settings limiting signal detection to 0.5–0.7 μm “optical thickness.” Emission spectra for each dye were limited as follows: Cy2, 505–530 nm; Cy3, 560–610 nm; and Cy5, 650–720 nm. Multipanel figures were assembled in CorelDraw X5 (Corel Corp.).
Animal Surgery.
BrdU administration and its combination with olfactory bulbectomy and in vivo gene silencing.
Rats (n = 5) were injected i.p.with BrdU (50 mg/kg body weight). After 4 h, animals were perfused transcardially, and their brains were removed, sectioned, and used for immunohistochemistry. In subsequent experiments, anesthetized rats (n = 12) were mounted in a stereotaxic frame, the skull was burr-holed, and the right olfactory bulb was aspirated using a vacuum pump. Sham-operated controls (equal number of rats) underwent an identical procedure except for the removal of their olfactory bulbs. Six bulbectomized and six sham-operated rats received BrdU (dose and administration as above) daily for a total of 4 or 15 d (n = 3 per time point). In an additional six bulbectomized and six sham-operated rats that had received identical BrdU treatment for 15 d, secretagogin or GFP-conjugated scrambled siRNA (500 µM, 0.2 µL) (GE Dharmacon) was administered with a Hamilton syringe into the RMS 11 d after bulbectomy at coordinates 1.2 mm lateral and 3.0 mm rostral from bregma, 6.0 mm ventral from the dural surface. One day after the last BrdU treatment, rats were transcardially perfused, and their brains were processed for immunohistochemistry.
BrdU administration combined with olfactory bulbectomy in secretagogin−/− mice.
Olfactory bulbs were removed from 12-wk-old mice (n = 3 wild-type, n = 3 secretagogin−/−), and an equal number of animals of the same genotypes were sham-operated as above. Eleven days after olfactory bulbectomy or sham surgery, mice received BrdU i.p. (50 mg/kg body weight) twice daily for three consecutive days. Animals were perfused for immunohistochemistry on day 15.
In vivo cell tracing combined with BrdU administration.
Rats (n = 3) received BrdU i.p. (50 mg/kg body weight) for a total of 15 d and then were killed. On day 11, 0.2 μL 10% (wt/vol) biotinylated dextran amine (BDA) (10,000 Da) (Molecular Probes) was injected into the RMS (coordinates: 1.2 mm lateral and 3.0 mm rostral from bregma, 6.0 mm ventral from the dural surface).
In vivo inhibition of matrix MMP activity.
Rats (n = 6) received Marimastat (1 µM/0.1 µL) (Tocris), a potent broad-spectrum MMP inhibitor with anti–MMP-2/MMP-9 activity (29), or DMSO (0.1/µL) (Sigma) in the distal limb of the RMS (at coordinates 1.2 mm lateral and 3.0 mm rostral from bregma, 6.0 mm ventral from the dural surface). Two days after surgery the animals were transcardially perfused, and their brains were processed for immunohistochemistry.
Human Fetal Tissue Preparation and Histochemistry.
Two male fetal brains with normal development (between gestational weeks 31–33) were selected from the Brain Bank of the Institute of Neurology, Medical University of Vienna, Austria. Fetal brain tissue was obtained from spontaneous or medically induced abortions. Only cases without genetic disorders, head injury, or neurological diseases were included. These cases showed no chromosome aberrations or postmortem autolysis. Neuropathological examination excluded major central nervous system malformations, severe hypoxic/ischemic encephalopathy, intraventricular hemorrhages, severe hydrocephalus, and meningitis or ventriculitis. Tissues were obtained and used in compliance with the Declaration of Helsinki and following institutional guidelines. The study was performed in the course of an study approved by the Ethical Committee of the Medical University of Vienna (No.104/2009).
Three-micrometer-thick tissue sections of formalin-fixed, paraffin-embedded tissue blocks containing the corpus callosum and the hippocampal formation were mounted on precoated glass slides (Star Frost). Shortly after deparaffinization and rehydration, the sections were pretreated in low-pH EnVision FLEX antigen retrieval solution at 98 °C for 20 min (PTLink; Dako) and subsequently were incubated with rabbit secretagogin antibody (gift of L.W.). The DAKO EnVision detection kit, peroxidase/3,3′-diaminobenzidine-tetrahydrochloride (DAB), and rabbit secondary antibody (K5007, ready-to-use; Dako) were used to visualize antibody binding. Sections were counterstained with hematoxylin, dehydrated in ascending concentrations of ethanol, cleared with xylene, and covered with Consil-Mount (Shandon; Thermo Scientific). Representative images containing the area of interest were exported from scanned sections (NanoZoomer 2.0; Hamamatsu).
Quantitative Analysis.
Images from serial sections (n = 4 per animal) were captured using a Zeiss 780 confocal laser-scanning microscope. BrdU+ and secretagogin+ cells were labeled, and their density was calculated in the demarcated area of interest using ZEN software (Carl Zeiss). Regions of interest were as follows: (i) the peri- and subventricular regions in rats; (ii) three regions (cranial, middle, and caudal) of the distal RMS limb; and (iii) the proximal limb, the knee region, and the distal limb of the RMS in secretagogin-null mice and rats after olfactory bulbectomy and in vivo gene silencing. Immunohistochemical sections were coded to avoid bias.
Fluorescence intensity was analyzed in gene-silencing experiments to quantify secretagogin and DCX expression. For secretagogin immunoreactivity, the fluorescence intensity of 50 secretagogin+ cell bodies >1,000 μm distal to the injection site but along the RMS was measured, and their average was compared with the average intensity of 50 secretagogin+ perikarya within 100 μm of the administration site in each animal. To explore changes in DCX immunoreactivity, fluorescence intensities in measurement frames were quantified. Frames were positioned around or >1,000 μm distal to the injection site within the RMS. Fluorescence intensities were measured using ZEN Image Analysis Software.
Immunoprecipitation and Shotgun Proteomic Analysis.
RMS regions were isolated from bulbectomized and sham-operated 6-wk-old rats (n = 8 animals in total with an equal number in each condition) and were incubated in artificial CSF (ACSF) (control stimulation) or in ACSF in which 52.5 mM NaCl was replaced by KCl to depolarize neurons by inducing Ca2+ influx (treatment condition). After 10 min of incubation, samples were collected in lysis buffer containing 50 mM NaCl, 20 mM Hepes, 10 μM CaCl2, 1 mM EDTA, 0.2% Triton X-100, and a mixture of protease inhibitors (Roche); pH was adjusted to 7.4. Meanwhile, samples treated by depolarizing solution were transferred into the lysis buffer supplemented with 10 μM CaCl2 without EDTA. Tissues were homogenized and centrifuged at 18,000 × g for 30 min, and supernatants were used in subsequent experiments. After preclearance, samples were incubated with rabbit anti-secretagogin primary antibody (1:2,000; provided by L.W.) overnight at 4 °C. An aliquot of each sample was exposed to rabbit IgG (Santa Cruz Biotechnology) to control nonspecific binding. Subsequently, samples were incubated with Dynabeads Protein G (Novex; Life Technologies) for 90 min. After repeated rinses, proteins were eluted with Laemmli buffer and separated on 10% (vol/vol) SDS/PAGE under denaturing conditions.
Eluted fractions from secretagogin and IgG control coimmunoprecipitations were loaded onto a 4–15% (vol/vol) gradient gel and slightly separated (within 2 cm) via SDS/PAGE in denaturing conditions. Sample lanes were visualized by colloidal Coomassie blue staining and were excised into 10 equal bands before LC-MS/MS analysis. In-gel digestion of sample bands using trypsin was carried out according to previously published protocols (75) with minor modifications. Following peptide extraction, final sample volumes were equalized to the extent that allowed two injections of each, which served as technical replicates.
LC-MS/MS analysis was performed using an Ultimate 3000 nanoRSLC system (Thermo Scientific) coupled to a high-capacity ion trap mass spectrometer (AmaZon ETD; Bruker Daltonics) via a CaptiveSpray ion source. Analysis settings and instrumental parameters were similar to those described elsewhere (76). Raw data files corresponding to one biological sample were merged into one search, and all MS/MS spectra were analyzed using Mascot version 2.5.1 (Matrix Science) against a UniProt rat (Rattus norvegicus) reference proteome database (as of November 2015; 31,457 sequences) assuming the digestion enzyme trypsin (allowing a maximum of two missed cleavages). The fragment ion and the parent ion mass tolerances were 0.40 Da and 0.35 Da, respectively. Carbamidomethylation of cysteine residues and the oxidation of methionine were specified as fixed and variable modifications, respectively. MS/MS-based peptide and protein identifications were validated using Scaffold version 4.5.3 (Proteomic Software Inc.). Peptide identifications were accepted if they were found with more than 95.0% probability by the Peptide Prophet algorithm (77). Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (78). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. To compare biological samples, normalized total spectra were calculated. A protein was considered specific for secretagogin if it was identified with a minimum 99% probability, with a minimum of two exclusive unique peptide counts, and with at least twofold higher normalized total spectra in the target immunoprecipitation (using an antibody against secretagogin) than the nonspecific IgG control in a given condition (i.e., control without Ca, control with Ca, bulbectomy without Ca, and bulbectomy with Ca).
In situ zymography.
Anesthetized rats (n = 3) were transcardially perfused with Tris buffer (40 mM Tris, 20 mM HCI, 152 mM NaCI) to remove red blood cells from the brain and to optimize enzymatic activity (80). Brains were removed, embedded in cryostat embedding medium (OCT; Bio-Optica), and frozen on dry ice. Sections (20 µm thick) were cut on a Leica cryostat, mounted onto SuperFrost Ultra Plus glasses, and incubated in a humidity chamber in the dark with the reaction buffer kept at 37 °C (0.05 M Tris⋅HCl, 0.15 M NaCl, 5 mM CaCl2, and 0.2 mM NaN3, pH 7.6) containing 20 µg/mL DQ gelatin (Molecular Probes). After 48 h, sections were rinsed with bidestillated water and fixed [4% (wt/vol) PFA, 0.1 M PB] for 30 min, washed in 0.1 M PB, and coverslipped with Surgipath Micromount medium. Gelatinase activity was inspected and images were acquired on a 780LSM confocal laser-scanning microscope (Zeiss).
Gelatin zymography.
Sample preparation and in-gel zymography were performed as published (81). Briefly, RMS samples were homogenized, and Triton X-100–soluble and –insoluble fractions (containing intracellular/cell membrane-bound and extracellular matrix-bound MMPs, respectively) were separated. After ethanol precipitation, samples were prepared for SDS/PAGE under nonreducing conditions. Proteins were separated on 7.5% (vol/vol) polyacrylamide gels copolymerized with 0.1% FITC-gelatin. Then in-gel renaturation and activation of the enzymes were carried out, leading to gelatinolysis at positions where gelatinases (MMP-2 and MMP-9) accumulated during SDS/PAGE. This gelatinolytic activity was detected as dark bands against a bright fluorescent background upon exposure to UV light using a ChemiDOC Imaging system (Bio-Rad Laboratories) (82). As a positive control (and to differentiate MMP-2 and MMP-9 gelatinolytic activity), human recombinant MMP-2 enzyme (Merck Millipore) was loaded onto the gels, also giving an estimate of the molecular weight of both proMMP2 and active MMP2 isoforms.
qPCR.
P4/5 rat brains were microdissected on ice to isolate the RMS and were snap frozen in liquid N2 until processing. RNA was extracted using the RNeasy Mini Kit (Qiagen) with a DNase I step performed to eliminate traces of genomic DNA30 and were reverse transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Reactions were performed after an initial denaturation at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, denaturation, annealing, and extension at calculated temperatures (60 s), and a dissociation stage from 60–95 °C with 0.5 °C steps for 10 s each (CFX96; Bio-Rad) with primer pairs amplifying short fragments for each gene (Table S4). Samples without template or reverse transcriptase served as negative controls. Expression levels obtained for every sample in parallel assays were normalized to the housekeeping gene encoding GAPDH.
Western Blotting.
P4/5 rat RMS micropunches or SH-SY5Y neuroblastoma cells were homogenized in TNE buffer containing 0.5% Triton X-100 (Sigma), 1% octyl-β-d-glucopyranoside (Calbiochem), 5 mM NaF, 100 μM Na3VO4, and a mixture of protease inhibitors (CompleteTM; Roche) by using a sonicator. Cell debris and nuclei were pelleted by centrifugation (800 × g for 10 min at 4 °C). Protein concentrations were determined by Bradford’s colorimetric method (83). Samples were diluted to a final protein concentration of 2 μg/μL, denatured in 5× Laemmli buffer, and analyzed by SDS/PAGE on 8% or 10% (vol/vol) resolving gels (Fig. S8). After transfer onto Immobilon-FL PVDF membranes (Millipore), membrane-bound protein samples were blocked for 1.5 h in 3% (wt/vol) BSA and 0.5% Tween-20 diluted in TBS and subsequently were exposed to primary antibodies (Table S3) overnight at 4 °C. Appropriate combinations of HRP-conjugated secondary antibodies from goat, rabbit, or mouse hosts (Jackson) (1:10,000; 2 h) were used for signal detection. Image acquisition and analysis were performed on a Bio-Rad XRS+ imaging platform.
Fig. S8.
Full lanes of Western blots and zymography gels. (A) Western blotting. (B) Gel zymography.
Secretagogin Silencing and Specificity.
We knocked down secretagogin mRNA expression by using specific siRNA in both in vivo and in vitro (explants and dissociated cell culture) experiments. To control specificity, we used scramble siRNA in parallel experiments. Application of secretagogin siRNA did not impact the density of living cells as estimated by the equal number of Hoechst+ nuclei in vitro (Fig. S9 A–A′′) or in vivo (Fig. S9 D–D3). Secretagogin silencing did not affect the quantity of either synaptophysin or postsynaptic density protein 95 (PSD-95) in SH-SY5Y neuroblastoma cells (Fig. S9B) or in P5 RMS explants (Fig. S9C).
Fig. S9.
In vitro, ex vivo, and in vivo secretagogin silencing (siRNA) does not affect cell survival or the expression of synaptic markers. (A–A′′) Control experiments for secretagogin siRNA selectivity. The density of live cells as determined by counting Hoechst+ nuclei was identical after the application of scrambled or secretagogin-specific siRNA in P5 dissociated RMS cultures. (B and B′) Secretagogin silencing in SH-SY5Y neuroblastoma cells left the expression of the pre- and postsynaptic markers synaptophysin and PSD-95, respectively, unaltered. (C and C′) Synaptophysin expression did change after secretagogin silencing in P5 RMS explants. (D–D3) In vivo gene silencing did not affect the density of Hoechst+ cells at the application site immediately dorsal to the RMS (areas within the dashed boxes). High-power images are shown in D2 and D2′. inj, injection site; qu, quantification. (Scale bars: 20 μm in A, D, and D1; 8 μm in D2.)
Acknowledgments
The excellent technical help of Andrea Németh is gratefully acknowledged. This work was supported by National Brain Research Program of Hungary Grant MTA-SE NAP B, KTIA_NAP_13-2014-0013 (to A.A.); the Swedish Research Council (Vetenskapsrådet) (T.H. and T.G.M.H.); the NovoNordisk Foundation (T.H. and T.G.M.H.); Hjärnfonden (T.H.), European Research Council Grant SECRET-CELLS, 2015-AdG-695136 (to T.H.); and Hungarian Scientific Research Fund Grant NKFI K115422 (to Z.E.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700662114/-/DCSupplemental.
References
- 1.Sanai N, et al. Corridors of migrating neurons in the human brain and their decline during infancy. Nature. 2011;478(7369):382–386. doi: 10.1038/nature10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264(5162):1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- 3.Chiaramello S, et al. BDNF/ TrkB interaction regulates migration of SVZ precursor cells via PI3-K and MAP-K signalling pathways. Eur J Neurosci. 2007;26(7):1780–1790. doi: 10.1111/j.1460-9568.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- 4.Hurtado-Chong A, et al. IGF-I promotes neuronal migration and positioning in the olfactory bulb and the exit of neuroblasts from the subventricular zone. Eur J Neurosci. 2009;30(5):742–755. doi: 10.1111/j.1460-9568.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 5.Garzotto D, Giacobini P, Crepaldi T, Fasolo A, De Marchis S. Hepatocyte growth factor regulates migration of olfactory interneuron precursors in the rostral migratory stream through Met-Grb2 coupling. J Neurosci. 2008;28(23):5901–5909. doi: 10.1523/JNEUROSCI.1083-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paratcha G, Ibáñez CF, Ledda F. GDNF is a chemoattractant factor for neuronal precursor cells in the rostral migratory stream. Mol Cell Neurosci. 2006;31(3):505–514. doi: 10.1016/j.mcn.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, et al. Regional effects of endocannabinoid, BDNF and FGF receptor signalling on neuroblast motility and guidance along the rostral migratory stream. Mol Cell Neurosci. 2015;64:32–43. doi: 10.1016/j.mcn.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagley JA, Belluscio L. Dynamic imaging reveals that brain-derived neurotrophic factor can independently regulate motility and direction of neuroblasts within the rostral migratory stream. Neuroscience. 2010;169(3):1449–1461. doi: 10.1016/j.neuroscience.2010.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marks C, Belluscio L, Ibáñez CF. Critical role of GFRα1 in the development and function of the main olfactory system. J Neurosci. 2012;32(48):17306–17320. doi: 10.1523/JNEUROSCI.1522-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindberg OR, Persson A, Brederlau A, Shabro A, Kuhn HG. EGF-induced expansion of migratory cells in the rostral migratory stream. PLoS One. 2012;7(9):e46380. doi: 10.1371/journal.pone.0046380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conover JC, et al. Disruption of Eph/ephrin signaling affects migration and proliferation in the adult subventricular zone. Nat Neurosci. 2000;3(11):1091–1097. doi: 10.1038/80606. [DOI] [PubMed] [Google Scholar]
- 12.Bovetti S, Bovolin P, Perroteau I, Puche AC. Subventricular zone-derived neuroblast migration to the olfactory bulb is modulated by matrix remodelling. Eur J Neurosci. 2007;25(7):2021–2033. doi: 10.1111/j.1460-9568.2007.05441.x. [DOI] [PubMed] [Google Scholar]
- 13.Murase S, Cho C, White JM, Horwitz AF. ADAM2 promotes migration of neuroblasts in the rostral migratory stream to the olfactory bulb. Eur J Neurosci. 2008;27(7):1585–1595. doi: 10.1111/j.1460-9568.2008.06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono K, Tomasiewicz H, Magnuson T, Rutishauser U. N-CAM mutation inhibits tangential neuronal migration and is phenocopied by enzymatic removal of polysialic acid. Neuron. 1994;13(3):595–609. doi: 10.1016/0896-6273(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 15.Doetsch F, Alvarez-Buylla A. Network of tangential pathways for neuronal migration in adult mammalian brain. Proc Natl Acad Sci USA. 1996;93(25):14895–14900. doi: 10.1073/pnas.93.25.14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneko N, et al. New neurons clear the path of astrocytic processes for their rapid migration in the adult brain. Neuron. 2010;67(2):213–223. doi: 10.1016/j.neuron.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peretto P, Merighi A, Fasolo A, Bonfanti L. Glial tubes in the rostral migratory stream of the adult rat. Brain Res Bull. 1997;42(1):9–21. doi: 10.1016/s0361-9230(96)00116-5. [DOI] [PubMed] [Google Scholar]
- 18.Mason HA, Ito S, Corfas G. Extracellular signals that regulate the tangential migration of olfactory bulb neuronal precursors: Inducers, inhibitors, and repellents. J Neurosci. 2001;21(19):7654–7663. doi: 10.1523/JNEUROSCI.21-19-07654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balenci L, et al. IQGAP1 regulates adult neural progenitors in vivo and vascular endothelial growth factor-triggered neural progenitor migration in vitro. J Neurosci. 2007;27(17):4716–4724. doi: 10.1523/JNEUROSCI.0830-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bozoyan L, Khlghatyan J, Saghatelyan A. Astrocytes control the development of the migration-promoting vasculature scaffold in the postnatal brain via VEGF signaling. J Neurosci. 2012;32(5):1687–1704. doi: 10.1523/JNEUROSCI.5531-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platel JC, Dave KA, Bordey A. Control of neuroblast production and migration by converging GABA and glutamate signals in the postnatal forebrain. J Physiol. 2008;586(16):3739–3743. doi: 10.1113/jphysiol.2008.155325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolteus AJ, Bordey A. GABA release and uptake regulate neuronal precursor migration in the postnatal subventricular zone. J Neurosci. 2004;24(35):7623–7631. doi: 10.1523/JNEUROSCI.1999-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saghatelyan A, de Chevigny A, Schachner M, Lledo PM. Tenascin-R mediates activity-dependent recruitment of neuroblasts in the adult mouse forebrain. Nat Neurosci. 2004;7(4):347–356. doi: 10.1038/nn1211. [DOI] [PubMed] [Google Scholar]
- 24.Lindwall C, Olsson M, Osman AM, Kuhn HG, Curtis MA. Selective expression of hyaluronan and receptor for hyaluronan mediated motility (Rhamm) in the adult mouse subventricular zone and rostral migratory stream and in ischemic cortex. Brain Res. 2013;1503:62–77. doi: 10.1016/j.brainres.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 25.Barkho BZ, et al. Endogenous matrix metalloproteinase (MMP)-3 and MMP-9 promote the differentiation and migration of adult neural progenitor cells in response to chemokines. Stem Cells. 2008;26(12):3139–3149. doi: 10.1634/stemcells.2008-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wojcik-Stanaszek L, Gregor A, Zalewska T. Regulation of neurogenesis by extracellular matrix and integrins. Acta Neurobiol Exp (Warsz) 2011;71(1):103–112. doi: 10.55782/ane-2011-1827. [DOI] [PubMed] [Google Scholar]
- 27.Yong VW. Metalloproteinases: Mediators of pathology and regeneration in the CNS. Nat Rev Neurosci. 2005;6(12):931–944. doi: 10.1038/nrn1807. [DOI] [PubMed] [Google Scholar]
- 28.Björklund M, Koivunen E. Gelatinase-mediated migration and invasion of cancer cells. Biochim Biophys Acta. 2005;1755(1):37–69. doi: 10.1016/j.bbcan.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Gabelloni P, et al. Inhibition of metalloproteinases derived from tumours: New insights in the treatment of human glioblastoma. Neuroscience. 2010;168(2):514–522. doi: 10.1016/j.neuroscience.2010.03.064. [DOI] [PubMed] [Google Scholar]
- 30.Brose N, Petrenko AG, Südhof TC, Jahn R. Synaptotagmin: A calcium sensor on the synaptic vesicle surface. Science. 1992;256(5059):1021–1025. doi: 10.1126/science.1589771. [DOI] [PubMed] [Google Scholar]
- 31.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323(5913):474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner L, et al. Cloning and expression of secretagogin, a novel neuroendocrine- and pancreatic islet of Langerhans-specific Ca2+-binding protein. J Biol Chem. 2000;275(32):24740–24751. doi: 10.1074/jbc.M001974200. [DOI] [PubMed] [Google Scholar]
- 33.Mulder J, et al. Secretagogin is a Ca2+-binding protein specifying subpopulations of telencephalic neurons. Proc Natl Acad Sci USA. 2009;106(52):22492–22497. doi: 10.1073/pnas.0912484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gáti G, Lendvai D, Hökfelt T, Harkany T, Alpár A. Revival of calcium-binding proteins for neuromorphology: Secretagogin typifies distinct cell populations in the avian brain. Brain Behav Evol. 2014;83(2):82–92. doi: 10.1159/000357834. [DOI] [PubMed] [Google Scholar]
- 35.Romanov RA, et al. A secretagogin locus of the mammalian hypothalamus controls stress hormone release. EMBO J. 2015;34(1):36–54. doi: 10.15252/embj.201488977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen-Ba-Charvet KT, et al. Multiple roles for slits in the control of cell migration in the rostral migratory stream. J Neurosci. 2004;24(6):1497–1506. doi: 10.1523/JNEUROSCI.4729-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Attems J, et al. Clusters of secretagogin-expressing neurons in the aged human olfactory tract lack terminal differentiation. Proc Natl Acad Sci USA. 2012;109(16):6259–6264. doi: 10.1073/pnas.1203843109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin K, et al. Proteomic and immunochemical characterization of a role for stathmin in adult neurogenesis. FASEB J. 2004;18(2):287–299. doi: 10.1096/fj.03-0973com. [DOI] [PubMed] [Google Scholar]
- 39.Blasko J, Fabianova K, Martoncikova M, Sopkova D, Racekova E. Immunohistochemical evidence for the presence of synaptic connections of nitrergic neurons in the rat rostral migratory stream. Cell Mol Neurobiol. 2013;33(6):753–757. doi: 10.1007/s10571-013-9956-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornack DR, Rakic P. The generation, migration, and differentiation of olfactory neurons in the adult primate brain. Proc Natl Acad Sci USA. 2001;98(8):4752–4757. doi: 10.1073/pnas.081074998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakic P. Principles of neural cell migration. Experientia. 1990;46(9):882–891. doi: 10.1007/BF01939380. [DOI] [PubMed] [Google Scholar]
- 42.Rakic P. Neurogenesis in adult primates. Prog Brain Res. 2002;138:3–14. doi: 10.1016/S0079-6123(02)38067-1. [DOI] [PubMed] [Google Scholar]
- 43.Kirschenbaum B, Doetsch F, Lois C, Alvarez-Buylla A. Adult subventricular zone neuronal precursors continue to proliferate and migrate in the absence of the olfactory bulb. J Neurosci. 1999;19(6):2171–2180. doi: 10.1523/JNEUROSCI.19-06-02171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doetsch F. A niche for adult neural stem cells. Curr Opin Genet Dev. 2003;13(5):543–550. doi: 10.1016/j.gde.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 45.des Portes V, et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92(1):51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 46.de Chevigny A, et al. Delayed onset of odor detection in neonatal mice lacking tenascin-C. Mol Cell Neurosci. 2006;32(1-2):174–186. doi: 10.1016/j.mcn.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Mobley AK, McCarty JH. β8 integrin is essential for neuroblast migration in the rostral migratory stream. Glia. 2011;59(11):1579–1587. doi: 10.1002/glia.21199. [DOI] [PubMed] [Google Scholar]
- 48.Curtis MA, et al. Human neuroblasts migrate to the olfactory bulb via a lateral ventricular extension. Science. 2007;315(5816):1243–1249. doi: 10.1126/science.1136281. [DOI] [PubMed] [Google Scholar]
- 49.Sonego M, et al. Fascin regulates the migration of subventricular zone-derived neuroblasts in the postnatal brain. J Neurosci. 2013;33(30):12171–12185. doi: 10.1523/JNEUROSCI.0653-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alberti S, et al. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc Natl Acad Sci USA. 2005;102(17):6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu L, et al. Annexin A5 promotes invasion and chemoresistance to temozolomide in glioblastoma multiforme cells. Tumour Biol. 2014;35(12):12327–12337. doi: 10.1007/s13277-014-2545-1. [DOI] [PubMed] [Google Scholar]
- 52.Drummond AH, et al. Preclinical and clinical studies of MMP inhibitors in cancer. Ann N Y Acad Sci. 1999;878:228–235. doi: 10.1111/j.1749-6632.1999.tb07688.x. [DOI] [PubMed] [Google Scholar]
- 53.Shimokawa Ki K, et al. Matrix metalloproteinase (MMP)-2 and MMP-9 activities in human seminal plasma. Mol Hum Reprod. 2002;8(1):32–36. doi: 10.1093/molehr/8.1.32. [DOI] [PubMed] [Google Scholar]
- 54.Chazal G, Durbec P, Jankovski A, Rougon G, Cremer H. Consequences of neural cell adhesion molecule deficiency on cell migration in the rostral migratory stream of the mouse. J Neurosci. 2000;20(4):1446–1457. doi: 10.1523/JNEUROSCI.20-04-01446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belvindrah R, Hankel S, Walker J, Patton BL, Müller U. Beta1 integrins control the formation of cell chains in the adult rostral migratory stream. J Neurosci. 2007;27(10):2704–2717. doi: 10.1523/JNEUROSCI.2991-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García-Marqués J, De Carlos JA, Greer CA, López-Mascaraque L. Different astroglia permissivity controls the migration of olfactory bulb interneuron precursors. Glia. 2010;58(2):218–230. doi: 10.1002/glia.20918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alpár A, Attems J, Mulder J, Hökfelt T, Harkany T. The renaissance of Ca2+-binding proteins in the nervous system: Secretagogin takes center stage. Cell Signal. 2012;24(2):378–387. doi: 10.1016/j.cellsig.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oudin MJ, et al. Endocannabinoids regulate the migration of subventricular zone-derived neuroblasts in the postnatal brain. J Neurosci. 2011;31(11):4000–4011. doi: 10.1523/JNEUROSCI.5483-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szklarczyk A, Lapinska J, Rylski M, McKay RD, Kaczmarek L. Matrix metalloproteinase-9 undergoes expression and activation during dendritic remodeling in adult hippocampus. J Neurosci. 2002;22(3):920–930. doi: 10.1523/JNEUROSCI.22-03-00920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frölichsthal-Schoeller P, et al. Expression and modulation of matrix metalloproteinase-2 and tissue inhibitors of metalloproteinases in human embryonic CNS stem cells. Neuroreport. 1999;10(2):345–351. doi: 10.1097/00001756-199902050-00025. [DOI] [PubMed] [Google Scholar]
- 61.Vaillant C, Didier-Bazès M, Hutter A, Belin MF, Thomasset N. Spatiotemporal expression patterns of metalloproteinases and their inhibitors in the postnatal developing rat cerebellum. J Neurosci. 1999;19(12):4994–5004. doi: 10.1523/JNEUROSCI.19-12-04994.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paredes MF, Sorrells SF, Garcia-Verdugo JM, Alvarez-Buylla A. Brain size and limits to adult neurogenesis. J Comp Neurol. 2016;524(3):646–664. doi: 10.1002/cne.23896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arellano JI, Rakic P. Neuroscience: Gone with the wean. Nature. 2011;478(7369):333–334. doi: 10.1038/478333a. [DOI] [PubMed] [Google Scholar]
- 64.Bergmann O, et al. The age of olfactory bulb neurons in humans. Neuron. 2012;74(4):634–639. doi: 10.1016/j.neuron.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 65.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474(7351):337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alpár A, et al. Slow age-dependent decline of doublecortin expression and BrdU labeling in the forebrain from lesser hedgehog tenrecs. Brain Res. 2010;1330:9–19. doi: 10.1016/j.brainres.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 67.Lendvai D, et al. Neurochemical mapping of the human hippocampus reveals perisynaptic matrix around functional synapses in Alzheimer’s disease. Acta Neuropathol. 2013;125(2):215–229. doi: 10.1007/s00401-012-1042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwaller B, Buchwald P, Blümcke I, Celio MR, Hunziker W. Characterization of a polyclonal antiserum against the purified human recombinant calcium binding protein calretinin. Cell Calcium. 1993;14(9):639–648. doi: 10.1016/0143-4160(93)90089-o. [DOI] [PubMed] [Google Scholar]
- 69.Moriyama K, Yahara I. Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are affected differentially by mutations around the actin-binding helix. EMBO J. 1999;18(23):6752–6761. doi: 10.1093/emboj/18.23.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nico B, et al. Increased matrix-metalloproteinase-2 and matrix-metalloproteinase-9 expression in the brain of dystrophic mdx mouse. Neuroscience. 2006;140(3):835–848. doi: 10.1016/j.neuroscience.2006.02.077. [DOI] [PubMed] [Google Scholar]
- 71.Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116(1):201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 72.Levine JM, Card JP. Light and electron microscopic localization of a cell surface antigen (NG2) in the rat cerebellum: Association with smooth protoplasmic astrocytes. J Neurosci. 1987;7(9):2711–2720. doi: 10.1523/JNEUROSCI.07-09-02711.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shashoua VE, Hesse GW, Moore BW. Proteins of the brain extracellular fluid: Evidence for release of S-100 protein. J Neurochem. 1984;42(6):1536–1541. doi: 10.1111/j.1471-4159.1984.tb12739.x. [DOI] [PubMed] [Google Scholar]
- 74.Tortoriello G, et al. Miswiring the brain: Δ9-tetrahydrocannabinol disrupts cortical development by inducing an SCG10/stathmin-2 degradation pathway. EMBO J. 2014;33(7):668–685. doi: 10.1002/embj.201386035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ghafari M, et al. Formation of GABA receptor complexes containing alpha1 and alpha5 subunits is paralleling a multiple T-maze learning task in mice. Brain Struct Funct. 2017;222(1):549–561. doi: 10.1007/s00429-016-1233-x. [DOI] [PubMed] [Google Scholar]
- 76.Zhang MD, et al. Comparative anatomical distribution of neuronal calcium-binding protein (NECAB) 1 and -2 in rodent and human spinal cord. Brain Struct Funct. 2016;221(7):3803–23. doi: 10.1007/s00429-016-1191-3. [DOI] [PubMed] [Google Scholar]
- 77.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 78.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 79.Oh LY, et al. Matrix metalloproteinase-9/gelatinase B is required for process outgrowth by oligodendrocytes. J Neurosci. 1999;19(19):8464–8475. doi: 10.1523/JNEUROSCI.19-19-08464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okabe M, Nyakas C, Buwalda B, Luiten PG. In situ blotting: A novel method for direct transfer of native proteins from sectioned tissue to blotting membrane: Procedure and some applications. J Histochem Cytochem. 1993;41(6):927–934. doi: 10.1177/41.6.8315283. [DOI] [PubMed] [Google Scholar]
- 81.Papp AM, et al. Visible light induces matrix metalloproteinase-9 expression in rat eye. J Neurochem. 2007;103(6):2224–2233. doi: 10.1111/j.1471-4159.2007.04917.x. [DOI] [PubMed] [Google Scholar]
- 82.Hattori S, Fujisaki H, Kiriyama T, Yokoyama T, Irie S. Real-time zymography and reverse zymography: A method for detecting activities of matrix metalloproteinases and their inhibitors using FITC-labeled collagen and casein as substrates. Anal Biochem. 2002;301(1):27–34. doi: 10.1006/abio.2001.5479. [DOI] [PubMed] [Google Scholar]
- 83.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]