Abstract
Background/Aims
Cancer is known to be a disease by many factors. However, specific results of reprogramming by pluripotency-related transcription factors remain to be scarcely reported. Here, we verified potential effects of pluripotent-related genes in hepatocellular carcinoma cancer cells.
Methods
To better understand reprogramming of cancer cells in different genetic backgrounds, we used four liver cancer cell lines representing different states of p53 (HepG2, Hep3B, Huh7 and PLC). Retroviral-mediated introduction of reprogramming related genes (KLF4, Oct4, Sox2, and Myc) was used to induce the expression of proteins related to a pluripotent status in liver cancer cells.
Results
Hep3B cells (null p53) exhibited a higher efficiency of reprogramming in comparison to the other liver cancer cell lines. The reprogrammed Hep3B cells acquired similar characteristics to pluripotent stem cells. However, loss of stemness in Hep3B-iPCs was detected during continual passage.
Conclusions
We demonstrated that reprogramming was achieved in tumor cells through retroviral induction of genes associated with reprogramming. Interestingly, the reprogrammed pluripotent cancer cells (iPCs) were very different from original cancer cells in terms of colony shape and expressed markers. The induction of pluripotency of liver cancer cells correlated with the status of p53, suggesting that different expression level of p53 in cancer cells may affect their reprogramming.
Keywords: Liver neoplasms, Induced pluripotent stem cells, Reprogramming
INTRODUCTION
Cancer is regarded as a fatal disease with multifactorial origins possessing uncontrolled proliferative potential. Even though the initial concept was first adopted very recently, the idea that certain population of cancer cells originate from cancer stem cells (CSCs), which have ability of differentiation into multilineage cells and self-renewal enough to transform into cancer, has recently re-highlighted.1,2 CSCs are also regarded as the cancer-initiating cells capable of carcinogenesis and can form tumor cells which can recur later and be very resistant to therapy. Progress has been made in verifying and isolating CSCs in hematologic malignancy and several solid tumors, including brain, colon, liver, and lung cancers.3–7 These studies indicate that CSCs are quiescent, rare and showing self-renewing capacity like normal stem cells. The origins of CSCs remain incompletely understood.
Hepatocellular carcinoma (HCC) is very fatal disease and causing a lot of social problems including death. Its incidence presents the fifth most commonly detected cancers world-widely, and second in men in Korea.8,9 Previous innovative studies have demonstrated the evidence of CSCs in HCC cell lines and during the development of primary HCC. Various stem cell candidate markers including the side population,10 CD133+,11,12 CD44+,12 CD90+,13 and epithelial cell adhesion molecules14 were identified. Most of CSC studies have been done for the detection of cell surface markers to purify CSC populations by serial transplantation method in the immune deficient mice.
Over the past several decades, remarkable progress has been done in the verification of molecular mechanisms of pluripotent differentiation as a result of intensive investigations into pluripotency and the development of embryonic stem (ES) cells from blastomere stages.15–18 Regarding the regulatory mechanisms maintaining pluripotency, several kinds of transcription factors typically identified in multipotent stem cells can be applied to CSC theory as a result of epigenetic manipulation.15–18
Recently, there have been some reports of the possibility of generating liver CSCs by the induction of reprogramming-related factors such as Oct4 or Nanog.19,20 The cellular reprogramming mediated by these factors may also act as a core player in the occurrence and recurrence of HCC.21 Therefore, understanding the control of cellular reprogramming in liver cancer is necessary to advance clinical treatment of HCC.
Here, authors analyzed the results of induction of these transcription factors, cancer-promoting oncogenes and tumor suppressor genes which were previously used for induced pluripotent stem (iPS) cells generation. Our results presented that induction of hepatocellular cancer cell into HCC stem cells using transcription factor genes resulted in reprogramming of cancer cells. Such induced cells were in a pluripotent stage and very different from original cancer cells.
MATERIALS AND METHODS
1. Culture of cancer induced pluripotent cells
Four liver cancer cell lines with different mutant states of p53 (HepG2, wild-type; Hep3B, null; Huh7, mutant; and PLC, mutant) were cultured in Dulbecco’s modified Eagle medium (DMEM) including 10% heat inactivated fetal bovine serum (FBS) at 37°C under a 5% humidified CO2 atmosphere. Newborn human foreskin fibroblasts (human dermal fibroblasts, HDFs) and gp-293 cells were purchased from Globalstem Inc. (Gaithersburg, MD, USA) and Takara Bio USA Inc. (Mountain View, CA, USA), respectively, and maintained in DMEM containing 10% FBS. Four liver cancer cell lines used for reprogramming and HDFs at passage 11 were also used as normal cell controls of reprogramming.19 The reprogrammed cells were grown on a mouse embryonic fibroblast feeder layer with human embryonic stem cells growth medium containing DMEM/F12, 20% knockout serum, 2 mM GlutaMAX, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol, 50 U/mL penicillin, and 50 g/mL streptomycin. The plasmids used in this study were purchased from Addgene (pMXs-hklf4, pMXs-hoct4, pMXs-hsox2, pMXs-hmyc, and pCMV-VSV-G; Cambridge, MA, USA) and Cell Biolabs, Inc. (pMXs-IRES-GFP; Huntsville, AL, USA).
2. Retrovirus generation and induced pluripotent cell induction from liver cancer cell lines
Retroviral particles including reprogramming human four factors (hKlf4, hOct4, hSox2, or hMyc, respectively) were generated from gp-293 cells that stably expressed packaging plasmids. In brief, cells were seeded at high confluence (>80%) into 10-cm dishes and transfected with 12 μg of each retroviral vector (hKlf4, hOct4, hSox2, and hc-Myc) plus 4-μg VSV-G plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). For virus infection, liver cancer cells and HDF cells were seeded in 6-well plates at 5×104 cells per well 1 day before transduction. The culture medium was changed into virus-containing supernatant, incubated for 6 hours with 8-μg polybrene, and then further cultured with fresh culture media. At day 7, the transduced cells were seeded at 1×104 per well in 6-well plates containing feeder cells, and incubated until the appearance of ES-like cells by changing medium every day.
3. Flow cytometry analysis of the efficiency of retrovirus infection
The cells transduced by retrovirus-GFP were resuspended in 500 μL of FACS buffer (1% bovine serum albumin in phosphate buffer saline). The efficiency of cell infection was analyzed by FACS using FACSCalibur (BD Biosciences, San Jose, CA, USA) and the CellQuest program (BD Biosciences).
4. Immunofluorescence and live cell imaging
The cells fixed with 4% paraformaldehyde were treated by permeabilization buffer (1% BSA and 0.1% Triton X-100) for 1 hour at room temperature. Primary antibodies against human Nanog (1:200; Cosmo Bio, Carlsbad, CA, USA) were incubated at 4°C overnight, consecutively incubation with secondary antibody (goat anti-Rabbit Texas Red-conjugated antibody; Invitrogen) was carried out. Mounted cells were examined with a fluorescence microscope or a Zeiss 510 confocal laser scanning microscope (Carl Zeiss, Jena, Germany). Live staining of Tra1-81 and SSEA4 was performed using StainAlive antihuman TRA1-81, DyLight 488 and antihuman SSEA-4, DyLight 550 (Stemgent, Cambridge, MA, USA). Immunostaining for each of the three germ layers after in vitro direct differentiation from Hep3B induced pluripotent cell was performed using human three germ layer 3-color immunocytochemistry kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instruction.
5. Embryonic body formation assay for putative cancer stem cell lines
The differentiation potential of induced pluripotent cancer (iPC) were investigated by induction of embryonic body (EB) formation through a 3-dimensional culture system. In brief, iPC colonies (at passage 5) on feeder cells were treated with collagenase (1 mg/mL; Invitrogen), transferred to 60-mm culture dishes, and incubated for 30 minutes to eliminate contamination with feeder cells. Suspended cells were then cultured with human embryonic stem media for 5 days to form spheroids (EBs) on no coated petri dishes (Greiner Bio-One, Monroe, NC, USA). The media was changed per day. The shape of EBs was analyzed by using an inverted light microscope (Olympus Optical, Melville, NY, USA).
6. Real-time quantitative polymerase chain reaction
Total RNA was prepared from both cells (Hep3B and Hep3B-iPC cells) and EB from Hep3B-iPC cells using RNeasy mini kit (Qiagen, Valencia, CA). A total of 1 μg of RNA was reverse transcribed by AccuPower RT PreMix (Bioneer, Seoul, Korea). Real-time polymerase chain reaction (PCR) was performed using FastStart Essential DNA Green Master (Roche, Indianapolis, IN, USA). All reactions were performed in triplicate. mRNA expression levels were normalized to endogenous glyceraldehydes 3-phosphate dehydrogenase (GAPDH) and expressed relative to control cells. The primer sequences are listed in Table 1.
Table 1.
Primer Sequences Used in qPCR Analysis
| Gene | Primer sequence | Tm, °C |
|---|---|---|
| Oct4-F | 5′-GATGTGGTCCGAGTGTGGTT-3′ | 60 |
| Oct4-R | 5′-AGCCTGGGGTACCAAAATGG-3′ | |
| Nanog-F | 5′-AAGGCCTCAGCACCTACCTA-3′ | 60 |
| Nanog-R | 5′-TGCACCAGGTCTGAGTGTTC-3′ | |
| Sox2-F | 5′-GCCCTGCAGTACAACTCCAT-3′ | 60 |
| Sox2-R | 5′-GACTTGACCACCGAACCCAT-3′ | |
| Foxa2-F | 5′-ATTGCTGGTCGTTTGTTGTG-3′ | 60 |
| Foxa2-R | 5′-CCTCGGGCTCTGCATAGTAG-3′ | |
| Sox17-F | 5′-GGCGCAGCAGAATCCAGA-3′ | 60 |
| Sox17-R | 5′-CCACGACTTGCCCAGCAT-3′ | |
| Goosecoid-F | 5′-GAGGAGAAAGTGGAGGTCTGGTT-3′ | 60 |
| Goosecoid-R | 5′-CTCTGATGAGGACCGCTTCTG-3′ | |
| Msx1-F | 5′-CGAGAGGACCCCGTGGATGCAGAG-3′ | 60 |
| Msx1-R | 5′-GGCGGCCATCTTCAGCTTCTCCAG-3′ | |
| Sox1-F | 5′-GGAATGGGAGGACAGGATTT-3′ | 60 |
| Sox1-R | 5′-AACAGCCGGAGCAGAAGATA-3′ | |
| Map2-F | 5′-CAGGTGGCGGACGTGTGAAAATTGAGAGTG-3′ | 60 |
| Map2-R | 5′-CACGCTGGATCTGCCTGGGGACTGTG-3′ | |
| GAPDH-F | 5′-GGACTCATGACCACAGTCCATGCC-3′ | 60 |
| GAPDH-R | 5′-TCAGGGATGACCTTGCCCACAG-3′ | |
| Endo-Oct4-F | 5′-GACAGGGGGAGGGGAGGAGCTAGG-3′ | 60 |
| Endo-Oct4-R | 5′-CTTCCCTCCAACCAGTTGCCCCAAAC-3′ | |
| Endo-Sox2-F | 5′-GGGAAATGGGAGGGGTGCAAAAGAGG-3′ | 60 |
| Endo-Sox2-R | 5′-TTGCGTGAGTGTGGATGGGATTGGTG-3′ | |
| Endo-Klf4-F | 5′-ACGATCGTGGCCCCGGAAAAGGACC-3′ | 60 |
| Endo-Klf4-R | 5′-TGATTGTAGTGCTTTCTGGCTGGGCTCC-3′ | |
| Endo-Myc-F | 5′-GCGTCCTGGGAAGGGAGATCCGGAGC-3′ | 60 |
| Endo-Myc-R | 5′-TTGAGGGGCATCGTCGCCGGAGGCTG-3′ | |
| Exo-Oct4-F | 5′-CAACGAGAGGATTTTGAGGCT-3′ | 60 |
| Exo-Sox2-F | 5′-TGCAGTAGAACTCCATGACCA-3′ | |
| Exo-Klf4-F | 5′-TGCGGCAAAACCTACACAAAG-3′ | |
| Exo-Myc-F | 5′-CAACAACCGAAATGCACCAGCCCCAG-3′ | |
| pMX-Tg-R | 5′-TACAGGTGGGGTCTTTCATTC-3′ | |
| hGAPDH-F | 5′-AACAGCCTCAAGATCATCAGC-3′ | 60 |
| hGAPDH-R | 5′-TTGGCAGGTTTTTCTAGACGG-3′ |
qPCR, real-time polymerase chain reaction; Tm, annealing temperature; F, forward; R, reverse.
7. In vitro tri-lineage differentiation from Hep3B-iPC cells
The direct differentiation of Hep3B-iPC cells into three germ layers in vitro was induced using human pluripotent stem cell functional identification kit (R&D Systems). In brief, the 2×105 cells were seeded on matrigel-coated slide chamber and cultured in media containing 10 μM of ROCK inhibitor Y27632 (STEM-CELL Technologies Inc., Vancouver, Canada). Next day, culture media was changed as differentiation media and each of three germ layers were induced according to the manufacturer’s instruction.
RESULTS
1. Infection efficiency of various liver cancer cells using retrovirus-GFP
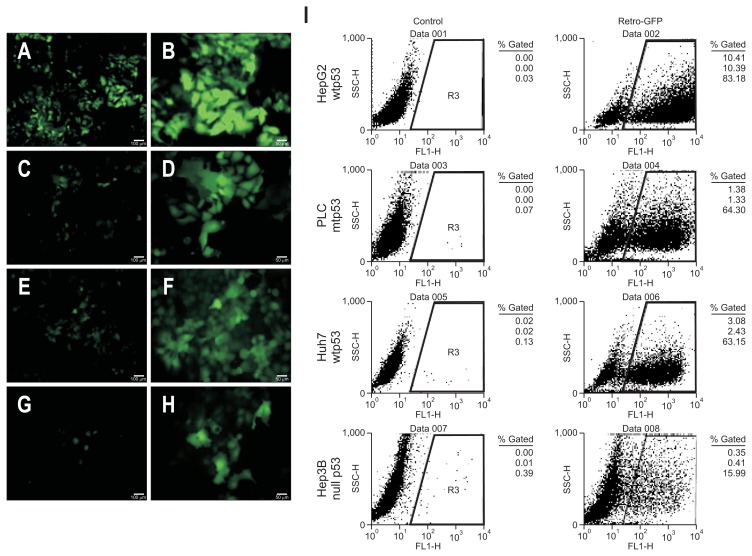
Induction of human cancer cells transduced with retroviral-GFP was performed for verification of the transduction efficiency of retrovirus. HepG2 (Fig. 1A and B), PLC (Fig. 1C and D), Huh7 (Fig. 1E and F), and Hep3B (Fig. 1G and H) cells were transduced with retroviral-GFP (Fig. 1A, C, E, and G, ×100; Fig. 1B, D, F, and H, ×200). Seven days after transduction we observed GFP-positive cells with various transduction efficiencies ranging from 83.18% to 15.99%, indicating different tumor cell characteristics according to p53 mutant status (Fig. 1I). HepG2 cells, which have wild-type p53, showed the highest transduction efficiency rate (83.18%) and Hep3B cells, which have null p53, had the lowest transduction rate (15.99%) (Fig. 1I).
Fig. 1.
Efficiency of infection in various liver cancer cells using retroviral-GFP. Induction of human cancer cells with retroviral-FP transduction. HepG2 (A and B), PLC (C and D), Huh7 (E and F), and Hep3B (G and H) were transduced with retroviral-GFP (A, C, E, and G, scale bar=100 μm; B, D, F, and H, scale bar=50 μm). We optimized the time course of the induction of human cancer cells. After seven days, we identified GFP-positive cells with various transduction efficiencies (I).
2. Characterization of iPS cells generated from Hep3B cancer cells
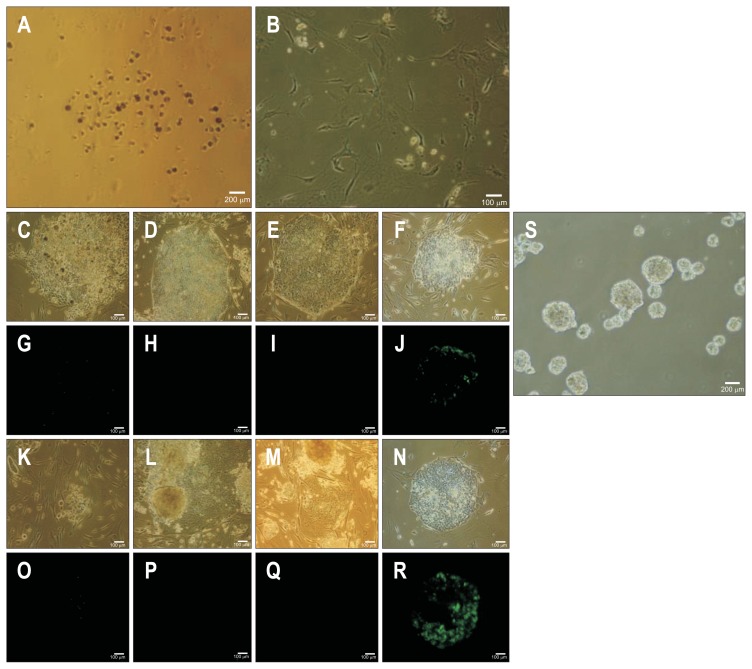
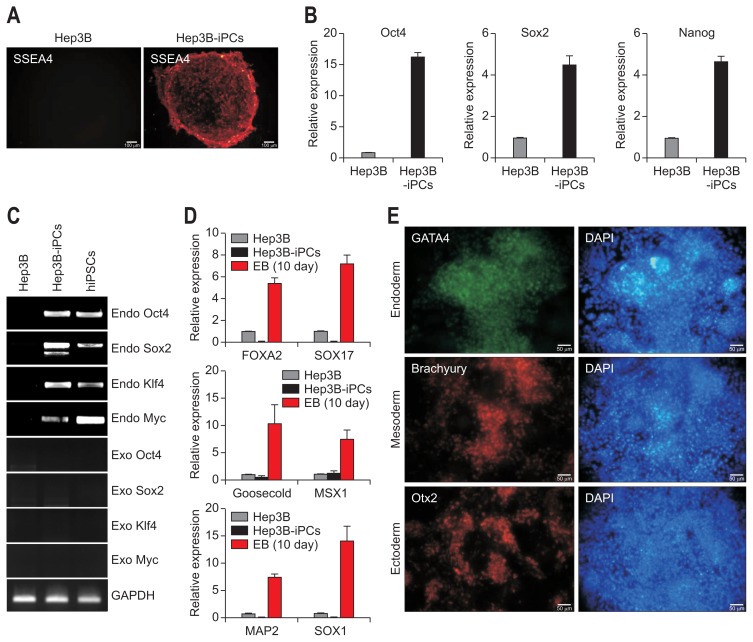
After 4 weeks of cultivation from transduction, typical types of cancer cell colonies which were very different from the negatively transduced original cells transfected with pMXs retroviral vector (Fig. 2A and B). After another 21 days, certain colonies have grown from the parental cancer cells after one passage (Fig. 2C–F). The numbers of human ES-like colony from all liver cancer cell lines was more than 100 during reprogramming, respectively (Table 2). However, we failed to identify Tra1-81–positive cells in the distinct colonies except for those from Hep3B cells (Fig. 2G–J). After two passages, distinct colonies were noticed in the Hep3B cells (Fig. 2K–N) and Tra1-81–positive cells were found among the Hep3B cells that had a staining pattern very similar to that of iPS cell colonies (Fig. 2O–R). This clone was examined to forming ability of EB, representative evidence of pluripotency in vitro (Fig. 2S), suggesting that Hep3B cells might be reprogrammed. We further characterized the established Hep3B-iPC clones by expression of the major pluripotent markers including SSEA4, Oct4, Sox2, and Nanog as measured by live-staining and real-time PCR (Fig. 3A and B). Also, we detected exogenous and endogenous expression level of four factors to address whether fully reprogramming (Fig. 3C). The pluripotency of Hep3B-iPC cells was confirmed by gene expression of the three germ layers (endoderm, mesoderm, and ectoderm) by real-time PCR and immunofluorescence staining (Fig. 3D and E) after culturing EB derived from Hep3B-iPC for 10 days and direct induction of tri-lineage differentiation using human pluripotent stem cell functional identification kit, respectively. In addition, in Hep3B cancer cells, we could not detect the expression of brachyury and Otx2, although GATA4 expression was weakly detected by immunostaining (data not shown). Therefore, these results indicated that Hep3B cells acquired induced pluripotent stem cell phenotype and those were truly reprogrammed.
Fig. 2.
Ability of embryonic body formation and staining of Tra1-81 in reprogrammed Hep3B cells by retrovirus-KOSM. HepG2 nontransduced cells were used as a negative control for KOSM transduction (A, scale bar=200 μm and B, scale bar=100 μm). HepG2 (C, G, K, and O), PLC (D, H, L, and P), Huh7 (E, I, M, and Q), and Hep3B (F, J, N, and R) were transduced with KOSM retroviral-vector (C–J, passage 1 and K–R, passage 2). Phase contrast (C–F and K–N) and Tra1-81 staining (G–J and O–R) are shown. Scale bar=100 μm. Embryonic body formation in Hep3B-iPC cells was demonstrated by culturing for 3 days (S, phase contrast; scale bar=200 μm).
Table 2.
Colony Numbers and Density of Reprogrammed Cells from Various Hepatocellular Cancer Cell Lines and Tra1-81 Staining after 3 to 4 Weeks on Mitotically Arrested-Mouse Embryonic Fibroblasts as Feeder Cells
| Numbers (ES-like colony) | Tra1-81 (live staining) | |
|---|---|---|
| HepG2 (wtp53) | >100 | X |
| PLC (mtp53) | >200 | X |
| Huh7 (mtp53) | 100–200 | X |
| Hep3B (null p53) | 100–200 | 2 |
ES, embryonic stem.
Fig. 3.
Analysis of gene expression related to the pluripotency and in vitro tri-lineage differentiation potential of Hep3B-iPC cells. Analysis of pluripotent marker expression in Hep3B and Hep3B-iPC cells at passage 4. (A) Live staining with antibody against stage-specific embryonic antigen-4 (SSEA4), which is a pluripotent marker. Scale bar=100 μm. (B) Quantitative real-time polymerase chain reaction (PCR) analysis with primers specific to pluripotent markers, including Oct4, Sox2, and Nanog. The mRNA expression values were normalized to GAPDH and are shown as the mean±SD of triplicate experiments, expressed relative to Hep3B. Evidence of the differentiation potential of Hep3B-iPCs into tri-lineage-type cells in vitro. (C) Reverse transcription polymerase chain reaction for exogenous and endogenous gene expression of the four reprogramming factors. (D) Quantitative real-time PCR analysis with primers specific to endoderm (Foxa2 and Sox17), mesoderm (Goosecoid and Msx1), and ectoderm (Map2 and Sox1). The embryonic body (EB) was formed from Hep3B-iPCs and cultured for 10 days. The mRNA expression values were normalized to glyceraldehydes 3-phosphate dehydrogenase (GAPDH) and are shown as the mean±SD of triplicate experiments, expressed relative to Hep3B. (E) Immunostaining with antibodies against the three germ layers, namely, antibodies to GATA4 (green, endoderm), Brachyury (red, mesoderm), and Otx2 (red, ectoderm). Nuclei were double-stained with DAPI (blue). Scale bar=50 μm. Tri-lineage differentiation of Hep3B-iPCs was performed as indicated in the materials and methods.
iPC, induced pluripotent cancer; iPSCs, induced pluripotent stem cell from fibroblasts.
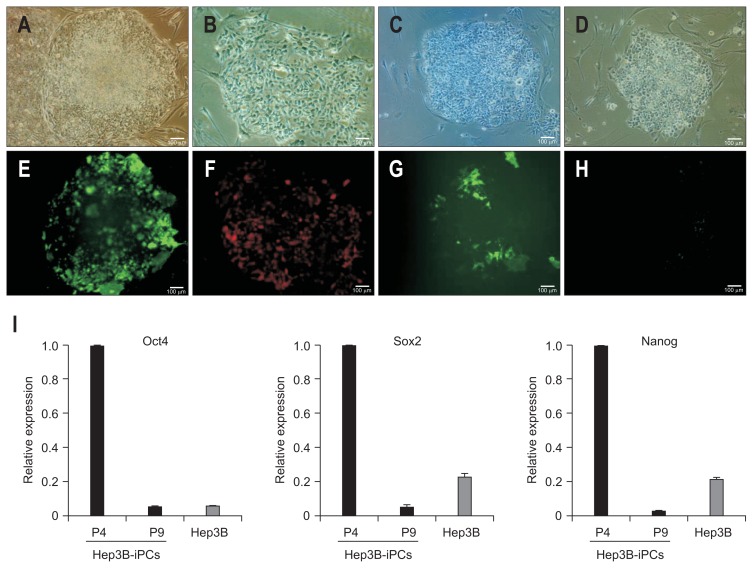
3. Diminished expression of Tra1-81 and pluripotent genes in Hep3B-iPCs after continual passage
Hep3B-iPC cells were stained with antibodies against Tra1-81 or Nanog, pluripotent stem cell markers and were analyzed in expression of major pluripotent genes during continual passage until 9 passages. Each antibody was used at passage 0, 5 or 3, 9. Data were shown in passage 0 (Fig. 4A and E), passage 3 (Fig. 4B and F), passage 5 (Fig. 4C and G), and passage 9 (Fig. 4D and H) of Hep3B-iPC cells under phase contrast (upper panel) and fluorescence (lower panel). Interestingly, expression of pluripotent markers was diminished upon continual passage of Hep3B-iPC as shown by immunofluorescence (Fig. 4H) and real-time PCR (Fig. 4I).
Fig. 4.
Staining of Tra1-81 and expression of pluripotent genes in Hep3B-iPCs after continual passage. Hep3B-iPCs were stained with Tra1-81 and Nanog and were analyzed regarding the expression of major pluripotent genes during continual passage until nine passages. Passage 0 (A and E), passage 3 (B and F), passage 5 (C and G), and passage 9 (D and H) are shown with phase contrast (A–D). Tra1-81 antibody staining was used for passage 0 and 5 Hep3B-iPCs (E and G). Nanog antibody staining was used for passage 3 and nine cells (F and H). Scale bar=100 μm. (I) Quantitative real-time polymerase chain reaction analysis with primers specific to pluripotent markers, including Oct4, Sox2, and Nanog. The mRNA expression values were normalized to glyceraldehydes 3-phosphate dehydrogenase (GAPDH) and are shown as the mean±SD of triplicate experiments, expressed relative to Hep3B-iPCs at passage 4 (P4).
iPCs, induced pluripotent cells.
DISCUSSION
It is obvious that stem cells play an important role not only in the various tissues differentiations and organ developments, but also in carcinogenesis. Here, we demonstrated that different genetic states in cancer cells might affect the maintenance and efficiency of cellular reprogramming of cancer cells. Using four HCC cell lines, we generated cancer iPCs through retroviral expression of reprogramming-related genes. Given the function of p53 as a blocker of reprogramming,21 Hep3B (null p53) cells showed better efficiency of reprogramming than the other liver cancer cell lines (Fig. 2). Induced cells, but not parental cells, exhibited the expression of known pluripotent marker genes and pluripotency, the ability to differentiate to cells derived from the three germ layers. However, continual passage induced the loss of stemness in Hep3B-iPCs.
It has been proposed that cells possessing stem cell characteristics are crucial for the generation of various types of human cancer.3–7 Complete elimination of CSCs of cancer tissue may be most important strategy of treatment of cancer. It can also achieve stable, lifelong remission, and sometimes a cure, especially for life-threatening cancers.3–7 Development in our understanding of the characteristics of stem cells enables us to make aiming and elimination of CSCs. Recent studies have shown that cellular reprogramming contributes to chemotherapy and/or radiotherapy resistance and the recurrence of cancers. Moreover, the aberrant expression of transcription factors, Oct4, Sox2, Nanog, Lin28, Klf4, and, c-Myc, is associated with unfavorable clinical outcomes in HCC.19 Therefore, understanding cellular reprogramming is necessary for its application in clinical HCC treatment. We introduced transcription factor genes into HCC cell lines, resulting in reprogramming of the cells to a pluripotent state. After 1 month, many colonies appeared in the transduced liver cancer cells. However, only Hep3B p53-null cells stained for Tra1-81 (one of the pluripotent stem cell markers) at very low efficiency (0.0002%), suggesting that p53 status may be associated with control of cellular reprogramming (Table 2, Fig. 2). Moreover, the efficiency of reprogramming was very low compared to HDFs, the commonly used donor cells for reprogramming.19,22 These differences may be due to cell type or different characteristics of cancers, as previously reported.20 We confirmed that the reprogrammed Hep3B cells acquired similar characteristics to pluripotent stem cells by observation of expression of SSEA4, Oct4, Sox2 and Nanog, major pluripotent markers (Fig. 3A and B). Furthermore, it is known that the ability to form EBs and direct differentiation into cells from three germ layers is representative evidence of pluripotency in vitro.23 Consistent with this, reprogrammed Hep3B cells showed the ability to form EBs (Fig. 2S) and potential of tri-lineage differentiation into cells derived from endoderm, mesoderm and ectoderm (Fig. 3D and E). In solid tumors, the CSC population, although it is regarded as a minor proportion of the total tumor volume, is closely connected to treatment failure due to resistance and causes of recurrence or spreading.1,2 There is also a lot of evidence supporting the presence of CSCs in HCC.7 Because specific surface markers of all CSCs which can isolate pure CSC population have not yet been verified, It is essential to understand the basic and oncological characteristics of these CSCs. Recently, some reports have shown that cancer development is related to not alone genetic but epigenetic changes of the tumor genome,19 but also to the induced reprogramming of tumor cells24 leading to tumor heterogeneity. Moreover, there have been several reports that definitely indicate the possibility of generating liver CSCs through the induction of reprogramming-related factors such as Oct4 or Nanog.23,24 Our study using transcription factor gene that were reported in previous study of iPS cells,19–21,25 show that cancer cells retaining stem cell characteristics of pluripotency could be achieved by cellular reprogramming following introduction of these transcription factor genes into liver cancer cells. Interestingly, continual passage reduced the stemness of these reprogrammed cancer cells (Fig. 4) suggesting that the properties of the cancer might affect the maintenance of pluripotency, although we could not exclude the possibility of partial reprogramming.
With this result, we raises the possible application of new cancer treatment using reprogramming approaches even in tumor cells which have very complex deranged genetic state. Various different HCC cell lines were induced with the same methods of iPS generation and successful iPC cells were made.20 For use as clinical therapeutic approaches, the heterogeneity and unstable state of reprogrammed cancer cells also remains to be elucidated with further studies.26
In conclusions, this study demonstrated that the generation of iPC cells by retroviral transfer of four factors will eventually allow us to accomplish some research and clinical goals in this field.
ACKNOWLEDGEMENTS
This study was supported by a grant of the Korea Institute of Radiological and Medical Sciences (KIRAMS), funded by Ministry of Science, ICT and Future Planning, Republic of Korea (No.1711031810/50586-2016).
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 3.Du L, Wang H, He L, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 4.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 5.Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 7.Ma S, Chan KW, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 8.McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis. 2011;15:223–243. doi: 10.1016/j.cld.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song IH, Kim KS. Current status of liver diseases in Korea: hepatocellular carcinoma. Korean J Hepatol. 2009;15(Suppl 6):S50–S59. doi: 10.3350/kjhep.2009.15.S6.S50. [DOI] [PubMed] [Google Scholar]
- 10.Haraguchi N, Utsunomiya T, Inoue H, et al. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24:506–513. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- 11.Ding W, Mouzaki M, You H, et al. CD133+ liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology. 2009;49:1277–1286. doi: 10.1002/hep.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, Hao X, Yan M, et al. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer. 2010;126:2067–2078. doi: 10.1002/ijc.24868. [DOI] [PubMed] [Google Scholar]
- 13.Yang ZF, Ho DW, Ng MN, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita T, Ji J, Budhu A, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 16.Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- 17.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 18.Wu Q, Chen X, Zhang J, et al. Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J Biol Chem. 2006;281:24090–24094. doi: 10.1074/jbc.C600122200. [DOI] [PubMed] [Google Scholar]
- 19.Zheng YW, Nie YZ, Taniguchi H. Cellular reprogramming and hepatocellular carcinoma development. World J Gastroenterol. 2013;19:8850–8860. doi: 10.3748/wjg.v19.i47.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun C, Liu YK. Induced pluripotent cancer cells: progress and application. J Cancer Res Clin Oncol. 2011;137:1–8. doi: 10.1007/s00432-010-0955-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong H, Takahashi K, Ichisaka T, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanabe K, Nakamura M, Narita M, Takahashi K, Yamanaka S. Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc Natl Acad Sci U S A. 2013;110:12172–12179. doi: 10.1073/pnas.1310291110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martí M, Mulero L, Pardo C, et al. Characterization of pluripotent stem cells. Nat Protoc. 2013;8:223–253. doi: 10.1038/nprot.2012.154. [DOI] [PubMed] [Google Scholar]
- 24.Welte Y, Adjaye J, Lehrach HR, Regenbrecht CR. Cancer stem cells in solid tumors: elusive or illusive? Cell Commun Signal. 2010;8:6. doi: 10.1186/1478-811X-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon YJ, Lee KG, Choi D. Clinical implications of advances in liver regeneration. Clin Mol Hepatol. 2015;21:7–13. doi: 10.3350/cmh.2015.21.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]