Abstract
The Mallard (Anas platyrhynchos) is an important reservoir species for influenza A viruses (IAV), and in this host, prevalence and virus diversity are high. Studies have demonstrated the presence of homosubtypic immunity, where individuals are unlikely to be reinfected with the same subtype within an autumn season. Further, evidence for heterosubtypic immunity exists, whereby immune responses specific for one subtype offer partial or complete protection against related HA subtypes. We utilized a natural experimental system to determine whether homo‐ or heterospecific immunity could be induced following experimental vaccination. Thirty Mallards were vaccinated with an inactivated H3, H6 or a sham vaccine and after seroconversion were exposed to naturally infected wild conspecifics. All ducks were infected within 2 days and had both primary and secondary infections. Overall, there was no observable difference between groups; all individuals were infected with H3 and H10 IAV. At the cessation of the experiment, most individuals had anti‐NP antibodies and neutralizing antibodies against H10. Not all individuals had H3 neutralizing antibodies. The isolated H3 IAVs revealed genetic dissimilarity to the H3 vaccine strain, specifically substitutions in the vicinity of the receptor‐binding site. There was no evidence of vaccine‐induced homosubtypic immunity to H3, a likely result of both a poor H3 immune response in the ducks and H3 immune escape. Likewise, there was no observed heterosubtypic protection related to H6 vaccination. This study highlights the need for experimental approaches to assess how exposure to pathogens and resulting immune processes translates to individual and population disease dynamics.
Keywords: Anas platyrhynchos, H3, homosubtypic immunity, humoral immunity, immunity, influenza A virus, Mallard
Introduction
Pathogenic microorganisms are a reality for all living organisms, from bacteriophages infecting bacteria in the ocean to economically relevant and devastating viruses in humans. However, not all encounters between hosts and pathogens result in infections, and not all infections are severe. Rather, the outcome will depend on intrinsic and acquired properties of the host, including the various compartments of the immune system, specific properties of the pathogen, such as variation in virulence and pathogenicity traits, and the interaction between pathogen and host mediated by the environment. In this context, the adaptive part of the immune system has received considerable interest, and how variation in immune genes translates into functional protection from infection. However, for most pathogens affecting wildlife, we still know very little on how previous exposure to a pathogen affects future infections and how these processes are influenced by variation in pathogen genes.
An important wildlife pathogen, which also has zoonotic potential, is the influenza A virus (IAV). An open question in IAV research relates to the maintenance of antigenic diversity. In contrast to the mammalian systems such as in humans, pigs and horses, numerous subtypes are maintained and circulated in wild birds, annually (Rambaut et al. 2008; Daly et al. 2011; Vincent et al. 2014). Wild birds are the reservoir for IAV in nature, and there are 16 hemagglutinin (HA) and nine neuraminidase (NA) subtypes identified in wild birds (Webster et al. 1992; Olsen et al. 2006). Thus far, 116 of 144 possible subtype combinations have been recovered in birds, globally (Olson et al. 2014). All subtypes are not recovered equally; some subtypes such as H3 in North America and H4 in Europe are recovered annually at high frequencies, whereas other HA subtypes and subtype combinations are infrequently detected (Wilcox et al. 2011; Bahl et al. 2013; Latorre‐Margalef et al. 2014). Seasonal patterns of subtype presence are somewhat predictable; it has been demonstrated that some subtypes are consistently isolated earlier or later in the autumn season (Latorre‐Margalef et al. 2014), or during fall or spring migrations (Ramey et al. 2014b).
Several hypotheses have been developed to explain these patterns, such as differences in general susceptibility (Hill et al. 2010; Daoust et al. 2012; Slusher et al. 2014), patterns of infection and virus shedding (Tolf et al. 2013) or response to infection (Vanderven et al. 2012), and these differ between individual, population and species levels. Host response to infection, particularly the immune response, may be an important driver of annual and interannual patterns, at both individual and population levels (Latorre‐Margalef et al. 2013, 2014; Tolf et al. 2013; Avril et al. 2016). While most studies have demonstrated an effect related to previous exposure, none have adequately explained the immune processes behind it.
Innate immune processes have been demonstrated in ducks (Vanderven et al. 2012), but both homosubtypic immunity and heterosubtypic immunity relate to acquired immunity. In contrast to experimental studies (Brown et al. 2012), most IAV infections in naturally infected Mallards are of short duration, in the order of 3.7–8.33 days (Latorre‐Margalef et al. 2014; Avril et al. 2016), suggesting that previous IAV infections affect IAV dynamics. In wild Mallards, across an autumn season, both homosubtypic immunity and heterosubtypic immunity have been demonstrated at the HA clade level, defined by phylogenetic and antigenic relatedness (Latorre‐Margalef et al. 2013). Based on patterns of infections in recaptured wild birds across an autumn season, it has been demonstrated that individuals infected with a particular subtype are unlikely to be reinfected with the same HA subtype later in the season (homosubtypic immunity). Further, there are fewer infections with phylogenetically closely related subtypes, suggesting partial or complete protection (heterosubtypic immunity) (Latorre‐Margalef et al. 2013, 2016). Homosubtypic immunity and heterosubtypic immunity have also been demonstrated in infection experiments, especially for accessing vaccine outcomes against highly pathogenic H5 and H7 viruses (Fereidouni et al. 2009, 2010; Costa et al. 2010, 2011).
In addition to infection probability, host population immunity also drives the antigenic divergence between and within HA types, and to a lesser degree, NA and NS. More specifically, the host immune response will result in phylogenetic branching resulting in discrete antigenic subtypes with little or no overlapping antigenic spaces (Gupta et al. 1998). Change of viruses into new antigenic spaces is best exemplified by escape from specific antibody‐mediated neutralization by accumulating mutations in the HA (antigenic drift), eloquently demonstrated in the human H3N2 viruses since 1968 (Koel et al. 2013). Antigenically unique viruses can emerge following coinfection and reassortment (antigenic shift), illustrated by the emergence of triple reassorted IAV affecting pigs and humans (Shinde et al. 2009), and of more concern the 2009 pH1N1 pandemic (Bokhari et al. 2012). While human IAV studies and experimental studies are invaluable to further our understanding of virus–immunity dynamics, wild ducks are exposed to numerous viral subtype combinations, resulting in unique and sometimes very complex infection histories (Tolf et al. 2013), and have to mitigate infection during severe physiological stress such as migration (van Gils et al. 2007; Altizer et al. 2011). Thus, the reality of antigenically distinct subtypes cocirculating in a host population creates a complex immunity landscape and high levels of individual variation in transmission probability.
We specifically wanted to test for the presence and effect of homo‐ and heterosubtypic immunity of Mallards, that is the role of IAV‐specific antibodies on the probability of reinfection against naturally circulating viruses in the dabbling duck reservoir. We used a natural challenge system, wherein Mallards were vaccinated with two divergent HA subtypes in addition to a sham‐inoculated control group. These individuals were naturally challenged by contact with infected wild conspecifics in a duck trap, which are annually infected with a diversity of different HA and NA subtypes. Given the outcome of the experiment, we further addressed an ad hoc hypothesis where the failure of homosubtypic protection was due to antigenic differences between the vaccine strains and cocirculating viruses. This is a unique study which utilizes an experimental approach, but is executed in a natural system, thus incorporating system‐level factors that cannot be addressed in classic experimental studies.
Methods
Ethics statement
Prior to the start of the experiment, ducks were raised at the Swedish National Veterinary Institute (SVA). Vaccination, sample collection, handling and euthanasia of ducks was undertaken by qualified personnel and under protocols approved by the Swedish Animal Research Ethics Board (Linköpings djurförsöksetiska nämnd, reference numbers 46–12, 37–13).
Experimental design
To determine whether ducks with pre‐exposure to certain HA lineages develop homo‐ and/or heterosubtypic immunity, we vaccinated ducks against two different HA subtypes, H3 and H6, and compared individuals and groups. Two treatment groups, each comprising of 10 individuals were included: H6 (representing Group 1, H1 Clade) and H3 (representing Group 2, H3 Clade) (Latorre‐Margalef et al. 2013), in addition to a control group of 10 individuals vaccinated with a sham. The goal was to include viruses from the different phylogenetic HA groups in the study design, therefore subtype choice was informed by presence and prevalence of subtypes that circulate annually at this study site (Tolf et al. 2013; Latorre‐Margalef et al. 2014). To allow for natural infections, the flock was introduced to a duck trap situated at Ottenby, a major stopover site for migratory waterfowl in the southern Baltic Sea (56°13′N, 16°27′E). This site is located in the European waterfowl flyway, and influenza surveillance has occurred here since 2002 (Latorre‐Margalef et al. 2014). Within the duck trap, primarily used to catch wild ducks, is a compartment used to house ‘lure’ ducks, separated from the wild ducks by only nylon mesh. As both compartments of the trap share water, birds placed in the experimental compartment are infected with IAV, introduced by their wild conspecifics through water transmission, aerosolization of viruses via water droplets and physical contact through the nylon mesh. All 30 experimental ducks shared the experimental compartment and were observed as a flock; they were not separated by treatment group. Infection patterns from these sentinel ducks have allowed us to analyse individual‐level responses of infection and immunity (Tolf et al. 2013), and thus allow for hypothesis testing in a natural experimental setting.
Birds were reared in an indoor facility at SVA. One week following vaccination, the experimental birds were transported to an outdoor enclosure near the study site to acclimatize to outdoor conditions. The flock was introduced to the duck trap on 16 September 2013 [27 days postvaccination; dpv] and euthanized 3 weeks later on 7 October 2013 [45 dpv, 21 days after introduction to the trap]. Blood was collected on four occasions: 9 weeks following hatching (5 August 2013), 14 dpv (3 September 2013), directly prior to introduction to the trap at 24 dpv (13 September 2013) and the cessation of the experiment (7 October 2013). Faecal samples were collected from the flock 9 weeks following hatching, and daily while the birds occupied the duck trap.
Vaccine
In order to immunologically prime birds, vaccines were developed in accordance with Fereidouni et al. (2010). Two viruses, A/Mallard/Sweden/101487/H3N8(2009) and A/Mallard/Sweden/99825/H6N2(2009), were used, both isolated from Mallards at Ottenby in 2009. Viruses were cultured in 10‐day‐old embryonated specific pathogen‐free (SPF) chicken eggs following inoculation by the allantoic route (WHO 2005). Eggs were incubated for 2 days prior to harvest, and fluid was tested for the presence of IAV by hemagglutination assay (HA). Viruses were subsequently inactivated using beta‐propiolactone (Sigma), and full inactivation was confirmed by the absence of replicating virus in two subsequent passages in SPF eggs. The hemagglutination titres of both antigens were 1:32 following inactivation. The administered vaccine consisted of 0.5 mL inactivated antigen and 0.5 mL Freud's complete adjuvant (Sigma‐Aldrich, St. Louis, USA), and the sham was comprised of 0.5 mL phosphate‐buffered saline (PBS) and 0.5 mL Freud's complete adjuvant (Sigma‐Aldrich). Approximately half of the vaccine was administered intramuscularly into the breast muscle and half subcutaneously at the base of the leg. No booster vaccine was administered as all individuals, but two, in the treatment groups, had raised anti‐NP‐ELISA values 14 days postvaccination.
Development of immunity: NP‐ELISA and microneutralization assay
Approximately 1.2 mL of blood was collected from the brachial vein. Sera were screened for anti‐NP antibodies using a commercially available ELISA kit (FlockCheck, Avian Influenza Virus Antibody Test Kit, IDEXX, Hoofddorp, the Netherlands). The results were interpreted following the manufacturers’ recommendations, where a sample‐to‐negative control ratio (S/N) value below 0.5 was considered positive.
Development of specific HA serum antibodies was tested using a virus microneutralization assay (VN) in Madin–Darby canine kidney cells (MDCK; ATCC, Manassas, Virginia, USA) as previously described (Ramey et al. 2014a), using antigens listed in Table S1 (Supporting information). A two‐step approach was used: (i) a screen for the presence of antibodies at a dilution 1:20, followed by (ii) titrations of positive sera 1:20 to 1:1280 dilutions. Plates were visually read after 48 h, and all tests were validated including controls and back titrations of the antigens.
IAV detection, isolation and characterization
IAV was detected using a rRT–PCR assay (Spackman et al. 2002; Tolf et al. 2013). Briefly, samples were diluted 1:4 in PBS and RNA was extracted using the MagNA Pure 96 robot and the Viral NA Large Volume Kit (Roche, Mannheim, Germany) according to the manufacturers’ specifications. RNA was assayed for a short fragment of the matrix gene segment by rRT–PCR using the Light Cycler480 (Roche) and the One Step Real‐Time PCR Kit (Qiagen, Hilden, Germany) (Spackman et al. 2002; Tolf et al. 2013). Default settings were used to determine Cq values, and a value of less than 40 was considered positive.
rRT–PCR‐positive samples were cultured in SPF eggs and agglutinating activity was assessed using previously described methods (Ellström et al. 2008). The HA subtype of virus isolates was characterized using the HAI test with both antisera raised in rabbits against HA subtypes (Latorre‐Margalef et al. 2014) and, to confirm, antisera raised in chickens against all HA subtypes. The NA subtype was determined using PCR assays (Wille et al. 2013). To assess our post hoc hypothesis, the HA, NA, NS and NP segments of nine randomly selected H3 viruses, in addition to the H3 vaccine strain, were amplified using previously published methods (Wille et al. 2013). The PCR products were cleaned using the Wizard® SV Gel and PCR Clean‐Up System (Promega, Madison, USA) and sequenced at Eurofins MWG Operon (Elmsberg, Germany). Sequences were assembled and subsequently aligned with the MAFFT algorithm (Katoh et al. 2009) within Geneious R7 (Biomatters, New Zealand). Phylogenetic models were determined and maximum‐likelihood trees were built and bootstrapped 1000 times. The amino acid positions that differ between the vaccine strain and circulating H3 viruses were plotted on the crystal structure of A/Aichi/2/1968(H3N2) (Protein Databank accession 5HMG) using Geneious R7 (Biomatters, New Zealand). All sequences generated have been deposited in GenBank, Accession nos. KT725399–KT725428.
Results
Prevaccination virological and serological status
Cloacal swabs and blood were collected from all individuals at SVA on 5 August 2013 (2 weeks prior to vaccination). All birds tested negative for IAV by rRT–PCR and were negative for anti‐NP antibodies tested by ELISA.
Immune status: anti‐NP antibodies and H3/H6 neutralizing antibodies prior to natural IAV challenge
Fourteen days postvaccination, all birds in the H6 group and 8 of 10 birds in the H3 group demonstrated serum anti‐NP antibodies. Birds 90A92759 and 90A92764 in the H3 group did seroconvert by the start of the experiment (24 dpv); however, two individuals (90A92766 and 90A92774) in the H6 group demonstrated a decrease in inverted S/N value to below the threshold. The sham‐vaccinated group was negative for serum anti‐NP antibodies prior to field challenge. On the final day of the experiment, almost all ducks tested antibody positive (Figs 1, S1, Supporting information).
Figure 1.
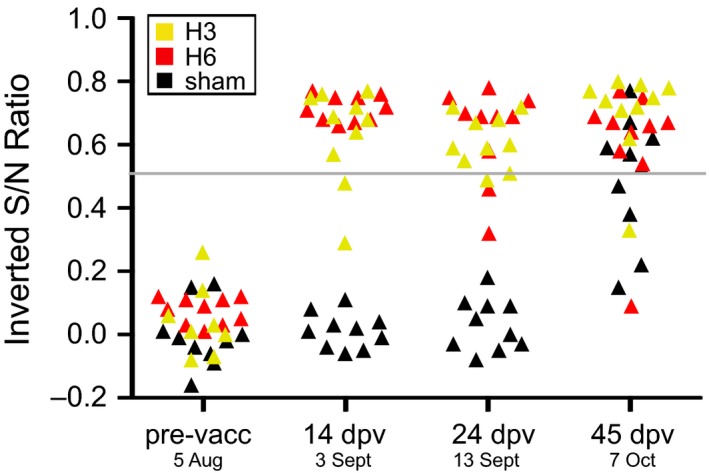
Temporal change of serum anti‐NP antibodies across the experiment. Time points are presented as both days postvaccination (dpv), and date in 2013. Ducks were placed in the duck trap on 16 September (27 dpv) and the final day in the duck trap was 7 October (45 dpv). Data are presented as the inverted sample‐to‐negative control ratio, so that values above 0.5 are positive. The positive to negative threshold (0.5) is a grey line across the plot. Individual changes are presented in Fig. S1 (Supporting information).
Serum antibodies to H6 were detected by microneutralization assays in 8 of 10 birds in the H6 group, and antibodies were detected within 14 dpv. These subtype‐specific antibodies were maintained to the start of the experiment, with a decrease in titres towards the end of the experiment (Fig. 2, Table S2, Supporting information). Specific serum H3 antibodies were detected in few individuals at lower titres 14 dpv, but 9 of 10 individuals had anti‐H3 antibodies against the H3 vaccine strain by 24 dpv (Fig. 2, Table S2, Supporting information). The sham vaccine group did not have detectable H3 or H6 antibodies until the end of the experiment (45 dpv), which corroborates the NP‐ELISA results (Fig. 2).
Figure 2.
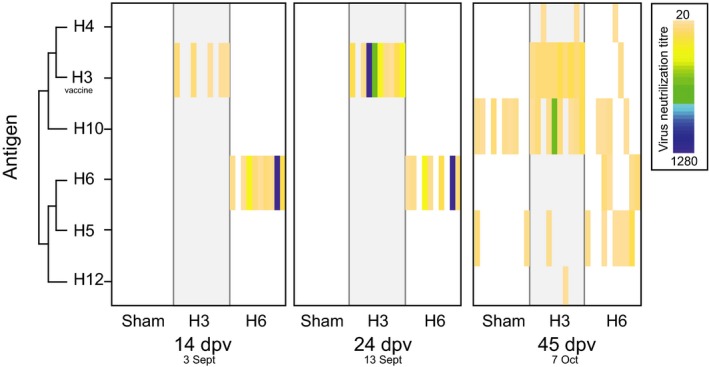
Development of specific serum antibodies following vaccination (14 dpv), prior to being placed in the duck trap (24 dpv), and following natural infection in the duck trap (45 dpv). Time points are presented as both days postvaccination (dpv), and date in 2013, where birds were placed in the duck trap on 16 September (27 dpv) and the final day in the duck trap was 7 October (45dpv). Each individual is presented as a column within each group. Virus neutralization titre ranges are presented as a heatmap, where for each individual (column) the associated titre is given by colour. Titres from 20 (light orange) to 1280 (dark blue), which is the highest dilution or titre at which the sera are still able to neutralize a fixed amount of the antigen. White spaces indicate no neutralization occurred. Results for H1, H2, H7–H9, H11, H14 are not plotted due to negative results in all individuals in all groups. HA subtypes are plotted by phylogenetic relatedness, illustrated by a cladogram. Results of H3 neutralizations are those using the vaccine strain as antigen. Full virus names for the antigens utilized are listed in Table S1 (Supporting information).
Due to encouraging anti‐NP results, wherein birds in the H3 and H6 groups had detectable anti‐NP antibodies, a boost was not administered. Furthermore, results from the VN assay indicate that these ducks had HA‐specific neutralizing serum antibodies prior to being placed in the duck trap.
Patterns of infection and specific immune response
Ducks were sampled daily for 21 days while in the duck trap (27–45dpv), and all birds were infected within 3 days of being introduced to the duck trap, regardless of treatment group. A total of 214 positive infection days were detected (34%), with the sham group having the longest duration of infection (mean infection days for: H3 = 5.8, H6 = 6.7, sham = 8.9). The duration of shedding during primary infections lasted <7 days, and secondary infections were present in a larger number of individuals in the sham group. There was no significant difference in daily Cq values or mean Cq values across the three groups in either the primary or secondary infections (Fig. S2, Supporting information).
Both H3 and H10 IAV infections were detected in all three groups, including the H3‐vaccinated group. Of the 164 isolates, 94 were H3 and 51 were H10. Furthermore, 62 of these H3 viruses were recovered from primary infections. We defined primary infections as the first infection detected by rRT–PCR, complimented with subtype information. The end of primary infection was defined by the first rRT–PCR‐negative day, which allowed for coinfections with multiple subtypes (Wille et al. 2013). H3 is the only subtype with which ducks were reinfected during secondary infections (11 individuals) excluding one individual that was infected with H10. This further demonstrates lack of homosubtypic protection from natural H3 primary infections. H3N8 was more commonly isolated in primary infections, compared with H3N6 during secondary infections; H3N8 occurred as secondary infections in individuals that were not infected with this subtype during primary infections. In some instances, individuals shed both H3N6 and H3N8 during an infection period. Similarly, both H10N6 and H10N8 were isolated. A few individuals were also infected with H4, H5 and H12 viruses; however, these appeared on 16 occasions across the entire data set. On 50 occasions, samples were PCR positive but culture negative (76.6% isolation efficiency), likely due to high Cq values or the detection of viral RNA following the abrogation of infection. As predicted, more viruses were isolated from the sham ducks in total (total number viruses: H3 = 49, H6 = 49, sham = 66). This pattern, whereby more viruses were isolated from sham ducks, is retained in the total number of H3 viruses, total number of H10 viruses and average number of H3 viruses across groups; however, the H3 group had more H10 isolates per Mallard compared with the H6 and sham groups (mean H10 viruses: H3 = 2.6, H6 = 1.8, sham = 1.8) (Fig. 3).
Figure 3.
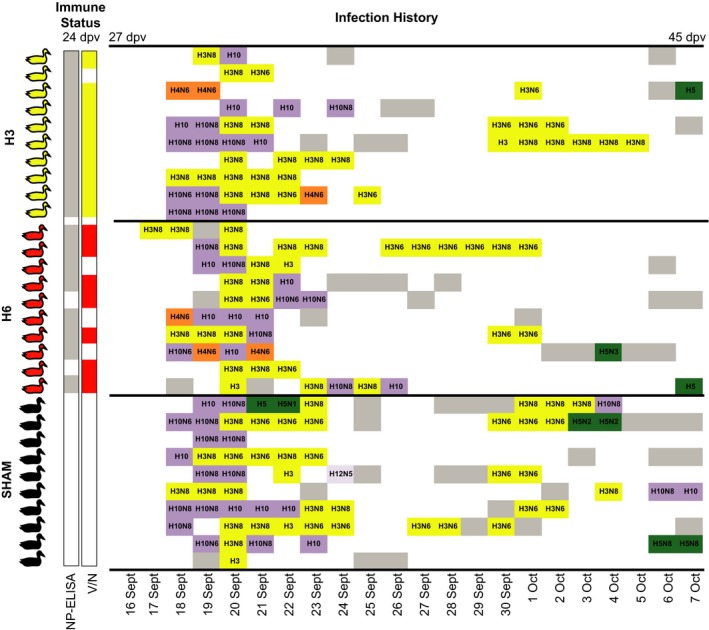
Immune status prior to being placed in the ducks trap, and infection histories of individuals. Individuals are ordered numerically by ring number within treatment group, vertically. Individuals with anti‐NP antibodies (S/N < 0.5 by NP‐ELISA) (13 September; 24 dpv) are shaded grey in the corresponding column, those with subtype‐specific antibodies (H3 antibodies – H3 group, H6 antibodies – H6 group by virus neutralization) (13 September, 24 dpv) are coloured in the V/N column. Infection histories are the daily infection status for each individual while in the duck trap [ranging from 16 September (27 dpv) to 7 October (45 dpv)]. White space indicates rRT–PCR‐negative samples. Isolates that were subtyped are coloured by HA subtype; samples that were rRT–PCR positive and not successfully isolated are in grey.
At the culmination of the experiment, almost all individuals had H10 neutralizing serum antibodies (number of individuals: H3 = 8, H6 = 4, sham = 6) (Fig. 2, Table S3, Supporting information). Not only did more individuals in the H3 vaccine group have H10 neutralizing antibodies, the H10 titres were also higher in this group (geometric mean H3 = 6.07, H6 = 4.57, sham = 5.61), with one individual reaching titres of 640 (Fig. 2, Table S3, Supporting information). Despite H3 infections in 25 of 30 ducks, only 13 individuals had H3 neutralizing serum antibodies at the end of the experiment, three of which were in the H6 treatment group (Fig. 4). Only individuals in the H6 vaccine group had neutralizing serum antibodies to H6 viruses; no H6‐specific neutralizing antibodies are detected in other groups due to the absence of H6 viruses circulating at Ottenby during the experiment (Fig. 2, Fig. 3). The number of positive individuals testing antibody positive to H5 (n = 6) was highest in the H6 vaccination group (geometric mean 4.99), but only two H5 viruses were isolated from this group. In contrast, five H5 viruses were isolated from the sham group and only two individuals had H5 neutralizing serum antibodies. No difference in number of H4 viruses isolated or the number of individuals with H4 neutralizing antibodies was detected between H3 and H6 vaccination groups, but H4 isolates and neutralizing serum antibodies were absent from the sham‐vaccinated group. Finally, as compared to the H3 and H6 groups, the sham group had few individuals with neutralizing serum antibodies; there were antibodies to only H5 and H10, and titres were lower (Figs 2 and 3, Table S3, Supporting information).
Figure 4.
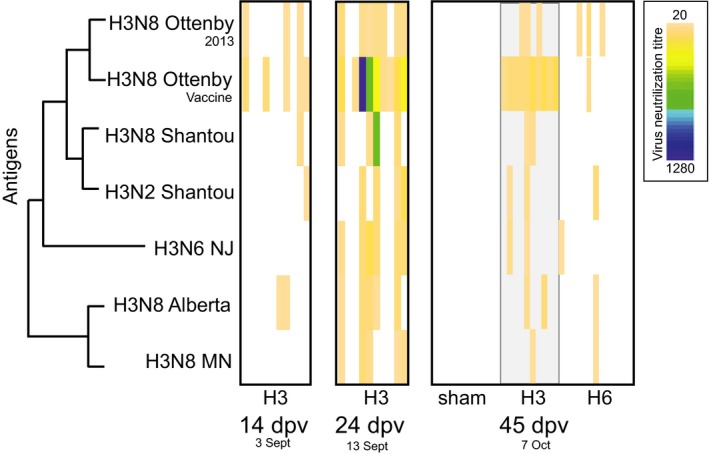
Comparison of H3 virus neutralization using different H3 antigens from North America, Asia and those utilized in this experiment. Time points are presented as both days postvaccination (dpv), and date in 2013. Ducks were placed in the duck trap 16 September (27 dpv) and the final day in the duck trap was 7 October (45 dpv). Each individual is presented as a column within each group. Antigens are presented on the y‐axis, and relatedness is demonstrated by a cladogram. The H3 strain isolated from the experimental ducks is indicated with ‘2013’. Full names for the virus antigens are listed in Table S1 (Supporting information). Virus neutralization titre ranges are presented as a heatmap, where for each individual (column) the associated titre is given by colour. Titres from 20 (light orange) to 1280 (dark blue), which is the highest dilution or titre at which the sera are still able to neutralize a fix amount of the antigen (100 TCID50/25 μL). White spaces indicate no neutralization occurred.
Genetic and antigenic comparison of H3 infections to H3 vaccination strain
The H3N8 strain used for vaccination was phylogenetically different from the H3 viruses isolated from the experimental ducks. For the HA, all the circulating viruses in 2013 were within 99% identity, but only 95.7% similar to the HA of the vaccine strain isolated in 2009 (Table 1, Fig. S3, Supporting information). Most of the substitutions were synonymous, resulting in only seven amino acid differences (amino acid position 22–545). Five of these amino acid substitutions occurred in HA1: T144A, N159S, A160T, D193N and V260K, while the remaining two were located in the HA2 peptide: I490M and S504G. Amino acids 159, 160 and 193 are located adjacent to the receptor‐binding site (Fig. S8, Supporting information).
Table 1.
Comparison of nine H3 viruses isolated during the experiment to the strain used in vaccine preparation
| Sample number | Date collected | Vaccine group | Subtype | HA (%)a | NA (%) | NP (%) | NS (%) |
|---|---|---|---|---|---|---|---|
| 140 901 | 17 September | H6 | H3N8 | 95.7 | 95.6 | 94.4 | 72.0 |
| 140 958 | 18 September | H3 | H3N8 | 95.7 | 95.6 | 94.4 | 72.0 |
| 140 959 | 18 September | H6 | H3N8 | 95.7 | 95.6 | 94.4 | 72.0 |
| 141 342 | 22 September | Sham | H3N6 | 95.7 | 53.9 | 93.9 | 71.8 |
| 141 462 | 25 September | H3 | H3N6 | 95.7 | 53.9 | 93.9 | 71.8 |
| 141 546 | 27 September | Sham | H3N6 | 95.7 | 53.9 | 93.9 | 96.8 |
| 141 675 | 30 September | H6 | H3N6 | 95.7 | 53.9 | 94.3 | 72.0 |
| 141 711 | 01 October | Sham | H3N8 | 95.7 | 95.6 | 97.3 | 72.0 |
| 141 811 | 03 October | H3 | H3N8 | 95.7 | 95.6 | 97.3 | 72.0 |
As with the HA, the N8 sequences from the circulating viruses were only within 95% identity to the vaccine strain, and all circulating N8 and N6s were within 99% identity of each other (Figs S4, S5, Supporting information). The NS subtype of the vaccination strain (Allele B) was genetically distinct from those detected in a majority (8/9) of circulating viruses (Allele A). Within these eight circulating viruses, there were two clusters within 97% identity (Table 1, Fig. S6, Supporting information). This pattern was maintained for the NP segment; however, two viruses (141 711 and 141 811) were 97% similar to the vaccine strain (Table 1, Fig S7, Supporting information). Combined, these results support the hypothesis that the circulating viruses are potentially antigenically different, fitting with some level of immune escape by putative antigenic drift and shift.
To assess H3 neutralizing antibody specificity, that is how well the H3 neutralizing antibodies recovered from the ducks in this study cross‐react with genetically (and presumably antigenically) different H3 viruses, the H3 VN was rerun utilizing an array of antigens representing viruses from North America, Asia and those isolated in this study (Table S1, Supporting information, Fig. 4). This array of H3 antigens revealed variations in immunological responses, whereby antigens from Eurasia detected H3‐specific antibodies viruses in a higher number of individuals, or by inducing higher neutralization titres as compared to North American virus antigens. In addition, specific antibody detection 14 and 24 dpv was higher using antigens from the vaccine strain as compared to North American antigens, as expected. Specific antibody detection following natural challenge in the duck trap was also best utilizing the vaccine strain for the H3‐vaccinated group, but the antigens from a virus isolated from these ducks in 2013 was better for detecting viruses circulating during the study such as those in the H6 group (three positives), suggesting poor or low‐level antibody development despite intense H3 infection (Fig. 4, Table S3, Supporting information). Indeed, H3 neutralization titres of recovered serum antibodies were different when comparing the virus used for vaccination and circulating viruses, supporting our hypothesis of escape. Despite utilizing a broader array of antigens, the H3 response following infection in the sham and H6 groups was undetectable or poor, respectively.
Discussion
No evidence of homosubtypic immunity in vaccinated ducks
This is the first experiment to assess vaccine‐induced homo‐ and/or heterosubtypic IAV immunity under conditions of natural challenge, whereby ducks are naturally infected by their conspecifics following vaccination. We predicted that if homosubtypic immunity was induced by vaccination, then the H6 vaccine group would be protected against H6 viral infections and potentially against related HA within the same clade, and the H3 vaccine groups would be protected against H3 viral infections, and these infections would be present in the other experimental group and the sham‐vaccinated group. Because ducks were naturally infected by wild conspecifics visiting the duck trap, we had no control related to challenge viruses. While H6 viruses are usually present at Ottenby (Tolf et al. 2013; Latorre‐Margalef et al. 2014), H6 was unfortunately not detected at the study site during the experiment. H3 was present and was isolated from all three experimental groups, demonstrating a failure of homosubtypic immunity despite the presence of specific H3 serum antibodies in the H3‐vaccinated group. Furthermore, most individuals were reinfected with H3 viruses as secondary infections in this study, demonstrating a failure of homosubtypic immunity following natural infection. If heterosubtypic immunity were present, we would predict partial or complete protection against closely related subtypes. The most closely related subtypes to H3 (Group 2, HA Clade 3) is H4 and H14, and in turn, the most closely related subtypes to H6 (Group 1, HA Clade 1) are H1, followed by H5 and H2 (Latorre‐Margalef et al. 2013). Subtypes other than H3 and H10 were rare during this experiment, and therefore, we were not able to confidently ascertain whether there was partial immunity against H4 or H5.
Immunity and protection against IAV has been assessed in experimental systems, but results to date are inconclusive. Experimental studies assessing homosubtypic immunity are largely limited to characterizing protective effects of low pathogenic strains to future highly pathogenic strains. In ducks challenged with low pathogenic H5, partial protection against highly pathogenic H5 has been reported (Fereidouni et al. 2009; Costa et al. 2011; Chaise et al. 2014). Most ducks shed virus following reinfection; however, a marked decrease in shedding intensity, duration and mortality as a result of infection has been demonstrated (Fereidouni et al. 2009; Costa et al. 2011; Chaise et al. 2014). In Wood Ducks (Aix sponsa), there is a difference in outcome depending on which low pathogenic H5 strain was used in initial infection (Costa et al. 2011). Studies have also assessed heterosubtypic immunity, and partial heterosubtypic immunity has been demonstrated. Ducks first exposed to low pathogenic H5 have reduced shedding (intensity, but not duration) of H7 (Chaise et al. 2014), and initial priming with H4 also has been reported to protect against highly pathogenic H5 (lower mortality rates) (Fereidouni et al. 2009). Additionally, infection with low pathogenic subtypes has resulted in decreased duration and intensity of shedding in heterosubtypic reinfections; specifically, H5 and H4 infection resulted in partial protection against H3 (H5 × H3, H4 × H3) (Costa et al. 2010; Pepin et al. 2012). Putative heterosubtypic protection has also been observed in naturally reinfected Mallards, where duration and intensity of infection are shorter in secondary and tertiary infections (Tolf et al. 2013; Avril et al. 2016). In wild birds, using infection and reinfection data, it has been demonstrated that there is a decreased probability for reinfection with the same subtype, and even closely related subtypes (Latorre‐Margalef et al. 2013). Such protection was not apparent in our results where reinfections with the vaccine subtype occurred in the H3 group as primary infections, but also secondary infections with the same subtype across all groups (H3). However, H3N6 was more common in secondary infections, in contrast to H3N8 in primary infections. Potentially, this subtype switch from N8 to N6 somehow contributes to maintaining infectivity, despite the fact that the H3 segment of circulating viruses was conserved between primary and secondary infections.
Following vaccination, anti‐NP antibodies were used as an indicator of immune development/response in the ducks; indeed, following vaccination, only ducks in the H3 and H6 groups had anti‐NP antibodies. Neither these antibodies, which are used for serological pan‐IAV detection and may not be neutralizing, nor the neutralizing HA antibodies conferred protection against influenza A infection, that is all ducks were rapidly infected after being placed in the duck trap. Measurements of viral RNA showed no differences in infection intensity (Cq values), total number of infection days or average number of infection days during the primary infections between groups. There is, however, an observable difference during secondary infections, wherein there were fewer infection‐positive days in the H3 and H6 groups compared with the sham group. This suggests that while there is no immediate protective effect, there was perhaps a delayed or combined effect, which would decrease the overall number of infections the ducks had over an autumn season. This lack of overall effect could, however, be due to the utility of vaccination. Application of a vaccine rather than actual infection in this study was due to ethical restrictions, whereby because we were using a natural system placing previously infected ducks into the environment is prohibited. Specifically, the potential limitation is that vaccination with an inactivated virus stimulates only the humoral response, whereas vaccination with a live attenuated vaccine, or a natural infection, elicits a cellular, mucosal and humoral response (Swayne & Kapczynski 2008). Given IAV is a gastrointestinal infection, mucosal immunity is important – the protective effect of serum antibodies on reinfection of influenza viruses in water birds is largely unknown (i.e. which titres correlate to protection). Regardless, it has been demonstrated that inactivated vaccines do elicit an IAV‐specific antibody response within 1 week of vaccination by both competitive ELISA (cELISA) and hemagglutination inhibition (HI), and Fereidouni et al. (2010) suggests that is due to the similarity of the outcome between vaccination and infection. In this study, both anti‐NP antibodies and specific HA neutralizing serum antibodies were detected prior to placement in the duck trap. This suggests that the administered vaccines had sufficient antigen to produce an immune response (potency). Administering a booster may have elicited a more efficacious response; we did not administer a booster due to encouraging NP‐ELISA results. Despite the presence of antibodies, the vaccines did not protect against the circulating field of viruses. Indeed, we do not know whether detectable immune response translates into actual protection against infection. Certainly, the maximum immune response following vaccination can only be as good as the overall immune response developed by the host to actual infection. Long‐term immune responses to influenza, as well as the level of protection induced by heterosubtypic immunity, are largely unknown in waterfowl species (Swayne & Kapczynski 2008; Magor 2011). Further, there is potential evidence that some subtypes might be less immunogenic; that is, the host develops a poor immune response to H3 compared with other HA subtypes (Costa et al. 2010; Globig et al. 2012; Pepin et al. 2012). For example, Globig et al. (2012) reported reinfections of sentinel ducks with H3 viruses. Furthermore, in experimental infections assessing heterosubtypic immunity, when challenged first with H4, ducks induced higher protection to H3 compared with the converse experimental set‐up utilizing H3 as the first challenge virus (Pepin et al. 2012).
Antigenic drift and shift within subtypes allow for long‐term maintenance
IAV has an extraordinary ability to evolve over time, making it hard to predict the (dominant) viruses that cause or shape the seasonal patterns (in humans) or the emergence of highly pathogenic strains (such as H5N1 and H7N9) in poultry. Only a few mutations (genetic drift) may result in substantial antigenic changes. For example, a small number of amino acid substitutions near the receptor‐binding site of HA in human H3N2 allows for immune escape (Koel et al. 2013). This is mirrored in the avian system, where a few amino acid substitutions near the receptor‐binding site of H5N1 allow for vaccine escape (Lee et al. 2004; Swayne & Kapczynski 2008). Indeed, we see amino acid substitutions differentiating the HA segment of the vaccine strain compared with circulating H3 viruses in this study, a number of which occur adjacent to the receptor‐binding site. Further, the specific positions of the observed amino acid changes are consistent with important antigenic changes in human H3 viruses (Koel et al. 2013). In addition to drift, IAV are able to escape immunity through antigenic shift following coinfection in the process of reassortment. All pandemic strains of IAV in humans are the result of a novel virus following reassortment (Scholtissek et al. 1978; Lindstrom et al. 2004; Taubenberger et al. 2005) and reassortment is an important feature in wild bird IAV dynamics (Dugan et al. 2008; Bahl et al. 2013; Wille et al. 2013). This feature is best exemplified by the interchangeability of HA‐NA combinations; within the H3 viruses circulating, despite identical HA segments, both N6 and N8 were present. Similarly, there was differentiation between the vaccine strain NP and the diversity of NP segments present in the circulating viruses; the role of anti‐NP antibodies in neutralizing avian IAV infection is unclear, and to date, no studies have assessed anti‐NP antibody as a function of infection frequency, or viral shedding (Kaminski & Lee 2011 ). It is hypothesized to play a role in protection, and anti‐NP antibody kinetics mimic anti‐HA antibodies (Marché et al. 2016). The presence of NS A in the circulating strains as compared to NS B is a potential contributing factor for the lack of homosubtypic immunity in this study. NS is important in evading the host immune system and augmenting viral replication; for example, NS1 is important in suppressing host interferon (Li et al. 2006; Dundon & Capua 2009). The presence of two different alleles suggests there is some avian‐induced selection pressure maintaining this diversity, with no apparent frequency differences, despite reported evidence of fitness advantages for the B allele in waterfowl (Adams et al. 2013). Due to the presence of both amino acid differences in the HA receptor‐binding site, the presence of two different NA subtypes and the shift in NS type, we hypothesize that the H3 viruses circulating during the experiment may be sufficiently different from the H3 vaccine strain to have escaped the vaccine strain selected. This is supported by mismatch in the virus neutralization assays, where H3 viruses other than the vaccine strain do not adequately cross‐react with H3 antibodies in the ducks.
Conclusions
Here, we present an innovative way to investigate host–pathogen interactions, specifically, the role of pre‐exposure of influenza on subsequent infection probability and dynamics. Following immunological priming, and utilizing a natural experimental set‐up, ducks were naturally infected by the diversity of viruses that circulate in the wild dabbling duck reservoir. This is in contrast to laboratory experimental infections, which while imperative do not incorporate system‐level factors. We aimed to assess presence and protection of vaccine‐induced homosubtypic (and/or heterosubtypic) immunity in ducks. Lack of protection may have been related to the combined effects of incomplete immunity related to vaccination, a poor vaccine response and an antigenic mismatch between vaccine strains and circulating viruses. While Mallards induced H3 neutralizing antibodies following vaccination, the vaccine was not protective even in individuals with neutralizing serum antibodies, and thus, an important follow‐up study would be to utilize a model of previous infection rather than vaccination; in this study, we specifically utilized vaccines due to ethical constraints of placing ducks previously exposed into the environment. Low titres of H3 neutralizing antibodies were not limited to vaccine response, and all ducks had low titres following H3 infections while in the duck trap. This suggests poor response to H3 infection, compared with the high titres of H10 neutralizing antibodies following infection. Steps towards more in‐depth characterization of immune dynamics are imperative if we are to better explain the patterns of diversification and maintenance of IAV in wild birds.
M.W., N.L.M., C.T., D.E.S. and J.W. conceived and designed the experiments and wrote the manuscript; M.W., N.L.M. and C.T. performed the experiments and analysed the data; D.E.S. and J.W. contributed reagents/materials/analysis tools.
Data accessibility
All sequences generated have been deposited in GenBank, Accession nos. KT725399–KT725428.
Supporting information
Fig. S1 Individual anti‐NP antibodies for (a) H6, (b) H3, and (c) PBS sham treatment groups.
Fig. S2 Infection intensity by rRT‐PCR during natural challenge. (A) rRT‐PCR results for each individual over the course of the experiment.
Fig. S3 Maximum Likelihood Tree of the HA, where the vaccination strain only has 95.7% identity to the H3 viruses circulating during the experiment.
Fig. S4 Maximum Likelihood Tree of the N8, where the vaccination strain only has 95.7% identity to the H3 viruses circulating during the experiment.
Fig. S5 Maximum Likelihood Tree of the N6, where all sequences are within 99% identity and similar to N6 sequences from viruses isolated in Sweden in 2010.
Fig. S6 Maximum Likelihood Tree of the NS segment, where all circulating viruses have the NS A subtype and the vaccine strain has the NS B subtype.
Fig. S7 Maximum Likelihood Tree of the NP.
Fig. S8 Amino acid differences between the vaccine strain and the circulating H3 viruses.
Table S1 Viruses used in neutralization assays
Table S2 Virus neutralization assays for H3 and H6 vaccination groups prior to being placed in the duck trap
Table S3 Results of virus neutralization assays on the final day of the experiment (7 Oct; 45 dpi and 21 days post release in the trap)
Acknowledgements
We are very grateful to all individuals who helped with the execution of this study; the staff at SVA animal house for care of the ducks prior to the experiment; J. Järhult, C. Atterby and A. Gillman for assisting in blood collection and vaccinations at SVA; H. Lundkvist for care of the ducks on Öland prior to release into the trap; J. Stedt for assisting in building the enclosure; and M. Olsson and M. Algotsson for being available for any veterinary concerns. Duck trappers M. Svensson and O. Persson were essential in daily sample collection. We are grateful to A. Jawad for assistance with egg isolation and C. Nordberg for NA typing and J. Chapman, A. Avril and D. Bengtsson for always providing a hand when one was needed. Finally, we are grateful to K. Subbarao and S. Krauss for sharing viruses used in the H3 VN comparison.
This study was supported by grants from the Swedish Research Councils VR (2010‐3067, 2010‐5399, 2011‐3568), FORMAS (2009‐1220) and the EC FP6 program NewFlubird. Work in DES laboratory (UGA) was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract HHSN272201400006C. MW was supported by a PGS‐D2 fellowship from the Natural Sciences and Engineering Research Council of Canada. NLM was supported by the Wenner‐Gren Foundations, Stockholm, and the Swedish Research Council VR. The funding agencies were not involved in the design, implementation or publishing of this study, and the research presented herein represents the opinions of the authors, but not necessarily the opinions of the funding agencies. This is publication number 301 from Ottenby Bird Observatory.
References
- Adams S, Xing Z, Li J et al (2013) The effect of avian influenza virus NS1 allele on virus replication and innate gene expression in avian cells. Molecular Immunology, 56, 358–368. [DOI] [PubMed] [Google Scholar]
- Altizer S, Bartel R, Han A (2011) Animal migration and infectious disease risk. Science, 331, 296–302. [DOI] [PubMed] [Google Scholar]
- Avril A, Grosbois V, Latorre‐Margalef N et al (2016) Capturing individual‐level parameters of influenza A virus dynamics in wild ducks using multistate models. Journal of Applied Ecology, 53, 1289–1297. [Google Scholar]
- Bahl J, Krauss S, Kuhnert D et al (2013) Influenza A virus migration and persistence in North American wild birds. PLoS Pathogens, 9, e1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhari SH, Pomeroy LW, Janies DA (2012) Reassortment Networks and the evolution of pandemic H1N1 swine‐origin influenza. IEEE/ACM Transactions on Computational Biology and Bioinformatics/IEEE, 9, 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Berghaus RD, Costa TP et al (2012) Intestinal excretion of a wild bird‐origin H3N8 low pathogenic avian influenza virus in mallards (Anas Platyrhynchos). Journal of Wildlife Diseases, 48, 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaise C, Lalmanach AC, Marty H et al (2014) Protection patterns in duck and chicken after homo‐ or hetero‐subtypic reinfections with H5 and H7 low pathogenicity avian influenza viruses: a comparative study. PLoS One, 9, e105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TP, Brown JD, Howerth EW, Stallknecht DE (2010) Effect of a prior exposure to a low pathogenic avian influenza virus in the outcome of a heterosubtypic low pathogenic avian influenza infection in Mallards (Anas platyrhynchos). Avian Diseases, 54, 1286–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TP, Brown JD, Howerth EW, Stallknecht DE, Swayne DE (2011) Homo‐ and heterosubtypic low pathogenic avian influenza exposure on H5N1 highly pathogenic avian influenza virus infection in wood ducks (Aix sponsa). PLoS One, 6, e15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly JM, MacRae S, Newton JR, Wattrang E, Elton DM (2011) Equine influenza: a review of an unpredictable virus. The Veterinary Journal, 189, 7–14. [DOI] [PubMed] [Google Scholar]
- Daoust P‐Y, van de Bildt MWG, van Riel D et al (2012) Replication of 2 subtypes of low pathogenicity avian influenza virus of duck and gull origins in experimentally infected Mallard ducks. Veterinary Pathology, 50, 548–559. [DOI] [PubMed] [Google Scholar]
- Dugan VG, Chen R, Spiro DJ et al (2008) The evolutionary genetics and emergence of avian influenza A viruses in wild birds. PLOS Pathogens, 4, e1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundon WG, Capua I (2009) A closer look at the NS1 of influenza virus. Viruses, 1, 1057–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellström P, Latorre‐Margalef N, Griekspoor P et al (2008) Sampling for low‐pathogenic avian influenza A virus in wild mallard ducks: oropharyngeal versus cloacal swabbing. Vaccine, 26, 4414–4416. [DOI] [PubMed] [Google Scholar]
- Fereidouni SR, Starick E, Beer M et al (2009) Highly pathogenic avian influenza virus infection of Mallards with homo‐ and heterosubtypic immunity induced by low pathogenic avian influenza viruses. PLoS One, 4, e6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fereidouni SR, Grund C, Häuslaigner R et al (2010) Dynamics of specific antibody responses induced in Mallards after infection by or immunization with low pathogenic avian influenza viruses. Avian Diseases, 54, 79–85. [DOI] [PubMed] [Google Scholar]
- van Gils JA, Munster VJ, Radersma R et al (2007) Hampered foraging and migratory performance in swans infected with low‐pathogenic avian influenza A virus. PLoS One, 2, e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globig A, Fereidouni SR, Harder TC et al (2012) Consecutive natural influenza A virus infections in sentinel Mallards in the evident absence of subtype‐specific hemagglutination inhibiting antibodies. Transboundary and Emerging Diseases, 60, 395–402. [DOI] [PubMed] [Google Scholar]
- Gupta S, Ferguson N, Anderson R (1998) Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science, 280, 912–915. [DOI] [PubMed] [Google Scholar]
- Hill NJ, Takekawa JY, Cardona CJ et al (2010) Waterfowl ecology and avian influenza in California: do host traits inform us about viral occurrence? Avian Diseases, 54, 426–432. [DOI] [PubMed] [Google Scholar]
- Kaminski DA, Lee FE‐H (2011) Antibodies against conserved antigens provide opportunities for reform in influenza vaccine design. Frontiers in Immunology, 2, 76. doi:10.3389/fimmu.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H (2009) Multiple alignment of DNA sequences with MAFFT. Methods in Molecular Biology, 537, 39–64. [DOI] [PubMed] [Google Scholar]
- Koel BF, Burke DF, Bestebroer TM et al (2013) Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science, 342, 976–979. [DOI] [PubMed] [Google Scholar]
- Latorre‐Margalef N, Grosbois V, Wahlgren J et al (2013) Heterosubtypic immunity to influenza A virus infections in Mallards may explain existence of multiple virus subtypes. PLOS Pathogens, 9, e1003443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre‐Margalef N, Tolf C, Grosbois V et al (2014) Long‐term variation in influenza A virus prevalence and subtype diversity in a migratory Mallards in Northern Europe. Proceedings of the Royal Society B, 281, 20140098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre‐Margalef N, Brown JD, Fojtik A et al (2016) Competition between IAV subtypes through heterosubtypic immunity modulates re‐infection and antibody dynamics in the mallard reservoir. bioRxiv. doi:10.1101/063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Senne DA, Suarez DL (2004) Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. Journal of Virology, 78, 8372–8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang Y, Jiao P et al (2006) The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. Journal of Virology, 80, 11115–11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom SE, Cox NJ, Klimov AI (2004) Genetic analysis of human H2N2 and early H3N2 influenza viruses, 1957‐1972: evidence for genetic divergence and multiple reassortment events. Virology, 328, 101–119. [DOI] [PubMed] [Google Scholar]
- Magor KE (2011) Immunoglobulin genetics and antibody responses to influenza in ducks. Developmental and Comparative Immunology, 35, 1008–1016. [DOI] [PubMed] [Google Scholar]
- Marché S, van den Berg T, Lambrecht B (2016) Evaluation of the kinetics of anti‐NP and anti‐HA antibody after infection of Pekin ducks with low pathogenic avian influenza virus. Veterinary Medicine and Science, 2, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B, Munster VJ, Wallensten A et al (2006) Global patterns of influenza A virus in wild birds. Science, 312, 384–388. [DOI] [PubMed] [Google Scholar]
- Olson SH, Parmley J, Soos C et al (2014) Sampling strategies and biodiversity of influenza A subtypes in wild birds. PLoS One, 9, e90826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin KM, VanDalen KK, Mooers NL et al (2012) Quantification of heterosubtypic immunity between avian influenza subtypes H3N8 and H4N6 in multiple avian host species. The Journal of General Virology, 93, 2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Pybus OG, Nelson MI et al (2008) The genomic and epidemiological dynamics of human influenza A virus. Nature, 453, 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey AM, Poulson RL, Gonzalez‐Reiche AS et al (2014a) Genomic characterization of H14 subtype Influenza A viruses in new world waterfowl and experimental infectivity in mallards (Anas platyrhynchos). PLoS One, 9, e95620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey AM, Poulson RL, Gonzalez‐Reiche AS et al (2014b) Evidence for seasonal patterns in the relative abundance of avian influenza virus subtypes in blue‐winged teal (Anas discors). Journal of Wildlife Diseases, 50, 916–922. [DOI] [PubMed] [Google Scholar]
- Scholtissek C, Rohde W, Von Hoyningen C, Rott R (1978) On the origin of human influenza virus subtypes H2N2 and H3N2. Virology, 87, 13–20. [DOI] [PubMed] [Google Scholar]
- Shinde V, Bridges CB, Uyeki TM et al (2009) Triple‐reassortant swine influenza A (H1) in humans in the United States, 2005‐2009. The New England Journal of Medicine, 360, 2616–2625. [DOI] [PubMed] [Google Scholar]
- Slusher MJ, Wilcox BR, Lutrell MP et al (2014) Are passerine birds reservoirs for influenza a viruses? Journal of Wildlife Diseases, 50, 792–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Senne DA, Myers TJ et al (2002) Development of a Real‐Time Reverse Transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. Journal of Clinical Microbiology, 40, 3256–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne DE, Kapczynski D (2008) Strategies and challenges for eliciting immunity against avian influenza virus in birds. Immunological Reviews, 225, 314–331. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM et al (2005) Characterization of the 1918 influenza virus polymerase genes. Nature, 437, 889–893. [DOI] [PubMed] [Google Scholar]
- Tolf C, Latorre‐Margalef N, Wille M et al (2013) Individual variation in influenza A virus infection histories and long‐term immune responses in mallards. PLoS One, 8, e61201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderven HA, Petkau K, Ryan‐Jean KE et al (2012) Avian influenza rapidly induces antiviral genes in duck lung and intestine. Molecular Immunology, 51, 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Awada L, Brown I et al (2014) Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health, 61, 4–17. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (1992) Evolution and ecology of influenza A viruses. Microbiological Reviews, 56, 152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2005) WHO manual on animal influenza diagnosis and surveillance. Available from: http://www.who.int/vaccine_research/diseases/influenza/WHO_manual_on_animal-diagnosis_and_surveillance_2002_5.pdf.
- Wilcox BR, Knutsen GA, Berdeen J et al (2011) Influenza A viruses in ducks in Northwestern Minnesota: fine scale spatial and temporal variation in prevalence and subtype diversity. PLoS One, 6, e24010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille M, Tolf C, Avril A et al (2013) Frequency and patterns of reassortment in natural influenza A virus infection in a reservoir host. Virology, 443, 150–160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Individual anti‐NP antibodies for (a) H6, (b) H3, and (c) PBS sham treatment groups.
Fig. S2 Infection intensity by rRT‐PCR during natural challenge. (A) rRT‐PCR results for each individual over the course of the experiment.
Fig. S3 Maximum Likelihood Tree of the HA, where the vaccination strain only has 95.7% identity to the H3 viruses circulating during the experiment.
Fig. S4 Maximum Likelihood Tree of the N8, where the vaccination strain only has 95.7% identity to the H3 viruses circulating during the experiment.
Fig. S5 Maximum Likelihood Tree of the N6, where all sequences are within 99% identity and similar to N6 sequences from viruses isolated in Sweden in 2010.
Fig. S6 Maximum Likelihood Tree of the NS segment, where all circulating viruses have the NS A subtype and the vaccine strain has the NS B subtype.
Fig. S7 Maximum Likelihood Tree of the NP.
Fig. S8 Amino acid differences between the vaccine strain and the circulating H3 viruses.
Table S1 Viruses used in neutralization assays
Table S2 Virus neutralization assays for H3 and H6 vaccination groups prior to being placed in the duck trap
Table S3 Results of virus neutralization assays on the final day of the experiment (7 Oct; 45 dpi and 21 days post release in the trap)
Data Availability Statement
All sequences generated have been deposited in GenBank, Accession nos. KT725399–KT725428.


