Summary
Proteins that contain iron–sulfur (Fe–S) clusters play pivotal roles in various metabolic processes such as photosynthesis and redox metabolism. Among the proteins involved in the biosynthesis of Fe–S clusters in plants, the SUFB subunit of the SUFBCD complex appears to be unique because SUFB has been reported to be involved in chlorophyll metabolism and phytochrome‐mediated signaling. To gain insights into the function of the SUFB protein, we analyzed the phenotypes of two SUFB mutants, laf6 and hmc1, and RNA interference (RNAi) lines with reduced SUFB expression. When grown in the light, the laf6 and hmc1 mutants and the SUFB RNAi lines accumulated higher levels of the chlorophyll biosynthesis intermediate Mg‐protoporphyrin IX monomethylester (Mg‐proto MME), consistent with the impairment of Mg‐proto MME cyclase activity. Both SUFC‐ and SUFD‐deficient RNAi lines accumulated the same intermediate, suggesting that inhibition of Fe‐S cluster synthesis is the primary cause of this impairment. Dark‐grown laf6 seedlings also showed an increase in protoporphyrin IX (Proto IX), Mg‐proto, Mg‐proto MME and 3,8‐divinyl protochlorophyllide a (DV‐Pchlide) levels, but this was not observed in hmc1 or the SUFB RNAi lines, nor was it complemented by SUFB overexpression. In addition, the long hypocotyl in far‐red light phenotype of the laf6 mutant could not be rescued by SUFB overexpression and segregated from the pale‐green SUFB‐deficient phenotype, indicating it is not caused by mutation at the SUFB locus. These results demonstrate that biosynthesis of Fe–S clusters is important for chlorophyll biosynthesis, but that the laf6 phenotype is not due to a SUFB mutation.
Keywords: chloroplast, Fe–S cluster, chlorophyll biosynthesis, Mg‐protoporphyrin monomethyl ester cyclase, far‐red light, Arabidopsis thaliana
Significance Statement
We demonstrate that Fe‐S biosynthesis is required for Mg‐protoporphyrin monomethylester cyclase activity, an essential step in the synthesis of the chlorophyll precursor protochlorophyllide. However, we show that the long hypocotyl phenotype reported for the laf6 mutant is not due to a deficiency in the Fe‐S biosynthesis protein SUFB, but instead is the result of a second site mutation.
Introduction
Iron–sulfur (Fe–S) cluster cofactors play pivotal roles in various biological processes that are critical to life, such as photosynthesis and redox metabolism (Sheftel et al., 2010; Balk and Pilon, 2011). In plastids, Fe–S proteins are involved in photosynthetic electron transport and nitrogen and sulfur assimilation; therefore Fe–S cluster biogenesis is essential for these processes. Several proteins have been shown to take part in the biogenesis of Fe–S clusters in plastids, with the sulfur mobilization (SUF) system suggested to play a central role (Balk and Pilon, 2011). In an accompanying paper (Hu et al., 2017) we provided in planta evidence that demonstrates the importance of the SUF system in general biosynthesis of Fe–S clusters in plastids.
In the SUF system, the SUFB, SUFC and SUFD proteins form a complex, which is suggested to act as a scaffold to form Fe–S clusters. The Fe–S clusters are subsequently transferred to specific Fe–S carrier proteins before they are incorporated into Fe–S apoproteins (Balk and Pilon, 2011; Couturier et al., 2013; Balk and Schaedler, 2014). This model is based on the mechanism revealed for the bacterial SUF proteins. In Escherichia coli, one SUFB, two SUFC and one SUFD protein form a SUFBC2D complex that has been shown to function as a scaffold for Fe–S cluster assembly in vitro (Outten et al., 2003; Chahal et al., 2009; Wollers et al., 2010). The protein sequences of plant SUFBs exhibit high similarity to their prokaryotic counterparts, and Arabidopsis SUFB can complement SUFB deficiency in E. coli (Xu et al., 2005). In addition, SUFB has been shown to interact with the evolutionarily conserved plastidic SUFC protein (Xu et al., 2005), which in turn can interact with Arabidopsis plastidic SUFD (Xu and Møller, 2004). Recently, we demonstrated that Arabidopsis SUFB, SUFC and SUFD form a protein complex similar to their bacterial counterparts, indicating that the plant SUFBCD complex also functions as a scaffold for Fe–S cluster biosynthesis (Hu et al., 2017). For better understanding of the roles of Fe–S cluster biosynthesis in plastids, various mutant plants in which Fe–S synthesizing activities are compromised have been analyzed. These mutants typically show a reduction in photosystem I (PSI) subunits that bind several Fe–S proteins, as well as reduced levels of chlorophyll and a loss of accumulation of many photosynthetic proteins, including those without Fe–S clusters (Lezhneva et al., 2004; Touraine et al., 2004; Yabe et al., 2004; Van Hoewyk et al., 2007). However, the underlying reason for the reduced chlorophyll phenotypes is not currently established.
In higher plants, chlorophyll is synthesized from glutamate (Tanaka and Tanaka, 2007; Mochizuki et al., 2010; see Figure S1 in the Supporting Information). Three enzymatic steps synthesize 5‐aminolevulinic acid (ALA) from glutamate and eight molecules of ALA are subsequently converted into protoporphyrinogen IX. Protoporphyrinogen IX is then oxidized by protoporphyrinogen IX oxidase to protoporphyrin IX (Proto IX), which is converted into either Mg‐protoporphyrin IX (Mg‐proto) by Mg‐chelatase or protoheme by ferrochelatase. This is the main branching point of chlorophyll biosynthesis. Mg‐proto is further esterified to form Mg‐proto monomethyl ester (Mg‐proto MME). Conversion of Mg‐proto MME to divinyl protochlorophyllide a (DV‐Pchlide) by Mg‐proto MME cyclase follows, and DV‐Pchlide is then modified to form monovinyl protochlorophyllide a (MV‐Pchlide). Upon illumination, MV‐Pchlide is reduced to chlorophyllide a, which is then esterified to form chlorophyll.
A number of Arabidopsis SUF mutants have been characterized to date and all show a pale green phenotype. This includes the SUFB mutant allele, long after far‐red 6 (laf6), which harbors a transposon insertion in the upstream untranslated region of the SUFB gene resulting in reduced SUFB expression (Møller et al., 2001), and hmc1 that has a base substitution in its coding sequence resulting in the replacement of a conserved proline with a leucine (Nagane et al., 2010). Similarly, a SUFD T‐DNA insertion mutant of Arabidopsis also showed a pale green phenotype with a reduced chlorophyll content (Hjorth et al., 2005). These observations demonstrate that chlorophyll accumulation requires Fe–S cluster synthesis in Arabidopsis (Nagane et al., 2010; Balk and Schaedler, 2014). In addition to reduced chlorophyll accumulation, the two SUFB alleles mentioned above were both reported to accumulate intermediates of chlorophyll metabolism: Proto IX for laf6 (Møller et al., 2001) and 7‐hydroxymethyl‐chlorophyll a and pheophorbide a, both of which are intermediates of chlorophyll breakdown, for hmc1 (Nagane et al., 2010). Since there has been no report of mutants in other SUF proteins accumulating intermediates of chlorophyll metabolism, it was speculated that SUFB might play a specific role in the regulation of chlorophyll metabolism in addition to functioning in the biosynthesis of Fe–S clusters (Nagane et al., 2010). SUFB has also been proposed to be involved in phytochrome signaling. The laf6 mutant exhibited long hypocotyl when seedlings were grown in far‐red (FR) light, suggesting that the laf6 mutation affects phytochrome A signaling (Møller et al., 2001). However, long hypocotyl phenotype has not been reported for the other two SUF mutants and the role of SUFB in phytochrome signaling remains unclear.
Since it was reported that a knockout of SUFB was lethal at the embryonic stage (Nagane et al., 2010), we have investigated the physiological roles of SUFB and Fe–S cluster biosynthesis by constructing conditional SUFB silencing lines using RNA interference (RNAi) technology. In addition, we have expressed SUFB in the laf6 mutant background to evaluate the effects of SUFB overexpression on its hypocotyl elongation phenotype. Through in‐depth analysis of these lines, we have been able to identify potential target sites of Fe‐S cluster biosynthesis deficiency within the chlorophyll biosynthesis pathway and have clarified the hypotheses about the specific functions of SUFB in chlorophyll biosynthesis and phytochrome signaling.
Results
Depletion of SUFB by RNAi results in reduced chlorophyll levels
The two SUFB‐deficient mutants characterized to date, hmc1 and laf6, show rather different phenotypes with respect to chlorophyll biosynthesis and phytochrome responses. In the absence of available knock‐out lines due to embryo lethality, we constructed conditional SUFB‐silencing lines to further investigate the impact of SUFB depletion on these responses. In these lines, a dexamethasone (Dex)‐inducible promoter drives the expression of artificial sequences that are complementary to part of the Arabidopsis SUFB gene, leading to degradation of SUFB mRNA (see Experimental Procedures). It is possible that an RNAi construct suppresses some off‐target gene expression, and we therefore tested two independent RNAi constructs (SUFB‐RNAi‐1 and SUFB‐RNAi‐2) that were designed to interact with two different sequences within the SUFB gene. In agreement with previous SUFB‐deficient lines, both SUFB RNAi lines showed a pale green phenotype in 4‐week‐old plants that was dependent on Dex treatment (Figure 1a). The pale phenotype of the SUFB RNAi lines correlated with the Dex‐dependent loss of SUFB protein in these lines (Figure 1b). Analysis of chlorophyll levels showed that both RNAi lines were equally depleted in chlorophyll a and b and that the observed phenotype was stronger than that seen for the hmc1 mutant (Figure 1c), which shows a moderate reduction in chlorophyll levels in agreement with a previous report (Nagane et al., 2010). Identical results were obtained from 7‐day‐old mutant and SUFB RNAi seedlings grown on agar plates (Figure S2). Together, these results confirm that a reduction in SUFB leads to reduced chlorophyll accumulation.
Figure 1.
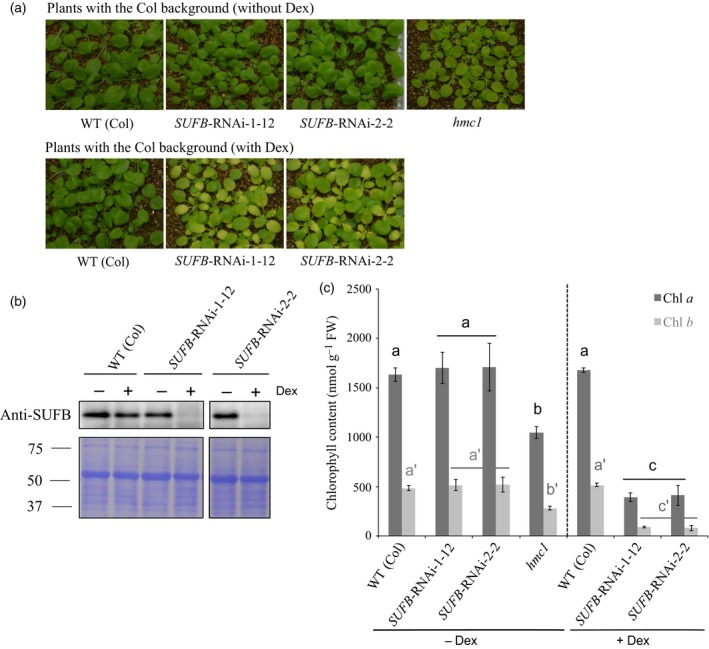
Phenotype of 4‐week‐old SUFB‐deficient plants grown on soil under long‐day conditions.
Conditional SUFB‐silenced lines (SUFB‐ RNAi‐1‐12 and 2‐2) were analysed with and without 10 μm dexamethasone (Dex) together with the SUFB‐deficient hmc1 mutant.
(a) Phenotype of SUFB‐deficient plants.
(b) SUFB protein levels in SUFB‐deficient mutant and transgenic lines. Total protein extracts (8 μg protein) of developing leaves were analyzed by immunoblotting using anti‐SUFB antiserum (upper row) and the membrane was subsequently stained by Coomassie Brilliant Blue (CBB) as a loading control (bottom row).
(c) Chlorophyll a and b content of developing leaves of SUFB‐deficient plants. Data points represent the mean ± SD of four biological replicates. Letters in black (chlorophyll a) or in grey (chlorophyll b) above each bar indicate significant differences (P < 0.05) by Tukey's multiple‐comparison test.
The SUFBC2D complex is required for chlorophyll synthesis
To determine how depletion of SUFB affects chlorophyll biosynthesis we measured the levels of key chlorophyll biosynthesis intermediates in the hmc1 mutant and SUFB RNAi lines using high‐performance liquid chromatography (Mochizuki et al., 2008). As shown in Figure 2, Mg‐proto MME accumulated in the hmc1 mutant and the SUFB RNAi lines treated with Dex to levels in excess of wild‐type (WT) lines. This was true both for mature plants (Figure 2a) and seedlings (Figure 2b). In contrast, all SUFB‐deficient lines had reduced levels of Proto IX and showed reductions in Mg‐proto (Figure S3). These results indicate that Mg‐proto MME cyclase activity is likely to be compromised in the hmc1 mutant and the conditional SUFB RNAi lines because Arabidopsis mutants that are defective in Mg‐proto MME cyclase activity accumulate this substrate (Tottey et al., 2003; Peter et al., 2009). As levels of Mg‐proto MME cyclase protein are unaltered in SUFB‐deficient plants (Figure 2c) we conclude that this effect is at the level of Mg‐proto MME cyclase activity. In addition, we interpret the lower levels of Proto IX and Mg‐proto to be due to feedback inhibition in the tetrapyrrole pathway. A blockage in the cyclase step could result in more porphyrin being directed towards heme, leading to inhibition of ALA synthesis (Cornah et al., 2003). It is also possible that oxidative stress effects are impairing tetrapyrrole biosynthesis via an alternative mechanism (Schlicke et al., 2014).
Figure 2.
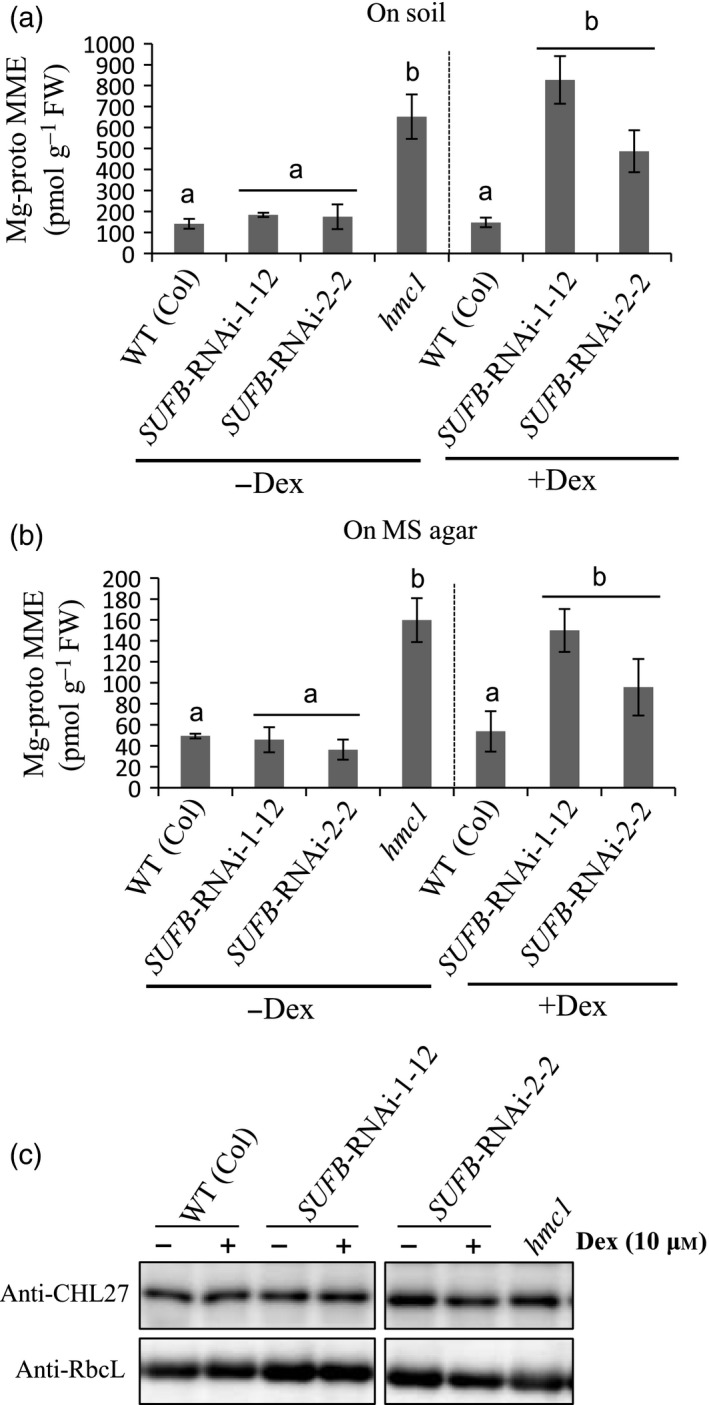
Analysis of chlorophyll biosynthetic intermediates in SUFB‐deficient plants.
Conditional SUFB‐silenced lines (SUFB‐ RNAi‐1‐12 and 2‐2) were analysed with and without 10 μm dexamethasone (Dex) together with the SUFB‐deficient hmc1 mutant.
Mg‐protoporphyrin IX monomethylester (Mg‐proto MME) content in (a) developing leaves of 4‐week‐old plants grown on soil under long‐day conditions or (b) in 7‐day‐old seedlings grown on 1/2 MS medium under long‐day conditions. Data points represent the mean ± SD of four biological replicates. Letters above each bar indicate significant differences (P < 0.05) by Tukey's multiple‐comparison test.
(c) The abundance of Mg‐proto MME cyclase (CHL27) in SUFB‐deficient mutant and transgenic lines. Total protein extracts (8 μg protein) of developing leaves of 4‐week‐old plants grown on soil under long‐day conditions were analysed by immunoblotting using anti‐CHL27 antisera. Anti‐Rubisco large subunit (RbcL) antisera were used to detect RbcL as the loading control.
The effect of SUFB deficiency could be due to an inhibition of Fe–S cluster synthesis or to an independent function of the SUFB protein. To test this we analyzed the phenotype of conditional RNAi‐silencing lines for SUFD and SUFC (Hu et al., 2017). Both SUFC and SUFD RNAi lines showed pale‐green leaves and were similar in appearance to SUFB lines (Figure 3a). In addition, in each case Mg‐proto MME accumulated to a similar extent to that observed for the SUFB RNAi line or hmc1 (Figure 3b). These results therefore indicate that accumulation of Mg‐proto MME is due to an impaired function of the SUFBC2D complex and not to a unique function of SUFB.
Figure 3.
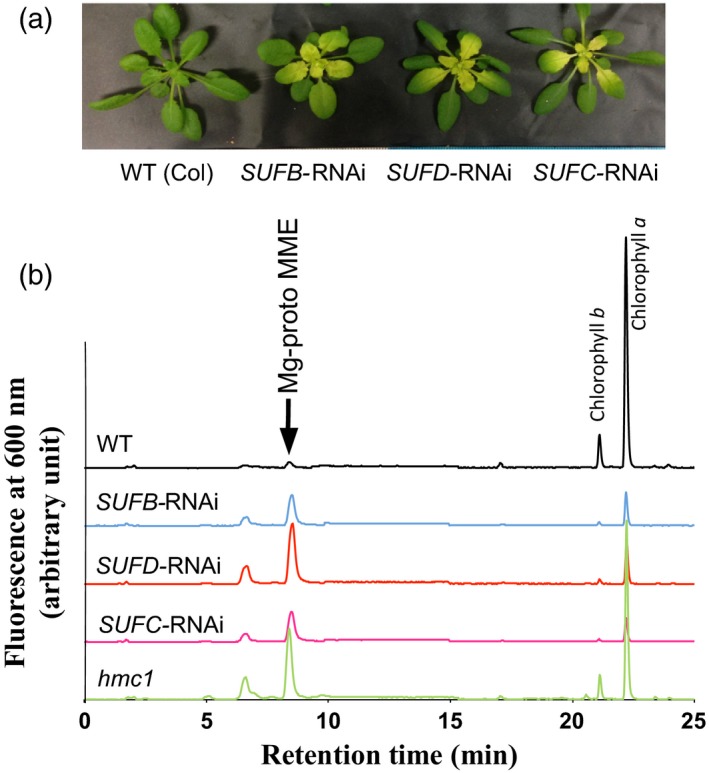
The phenotype of SUFB‐, SUFC‐ and SUFD‐deficient plants.
Conditional SUFB‐, SUFC‐ and SUFD‐silenced lines were analyzed after treatment with 10 μm dexamethasone (Dex) together with the SUFB‐deficient hmc1 mutant.
(a) Phenotype of 4‐week‐old wild‐type (WT) and transgenic plants 7 days after Dex treatment.
(b) The HPLC profiles of pigments extracted by acetone from young leaves of WT and transgenic plants after Dex treatment. The x‐axis indicates retention time. The y‐axis indicates fluorescence intensity at 600 nm which was excited at 415 nm in arbitrary units. Mg‐proto MME, Mg‐protoporphyrin IX monomethyl ester.
Overexpression of SUFB complements the white‐light‐grown phenotype of the laf6 mutant
To determine whether all the phenotypes observed in the laf6 mutant allele were due to SUFB deficiency, we generated several independent transgenic lines that expressed 35S::SUFB in the laf6 mutant, which is in the Landsberg erecta (Ler) background. Two overexpression lines (35S::SUFB/laf6‐4 and ‐7) were selected and used for further analysis (Figure 4a). The laf6 mutant shows a reduced level of the SUFB protein (to 15% of the WT (Ler) level), and this was increased in the two SUFB‐complemented lines, although the abundance of SUFB was still less than WT, at 64 and 70% of the WT level, respectively (Figure 4b). Nevertheless, both 35S::SUFB lines restored the visibly pale phenotype of 4‐week‐old laf6 plants to a WT appearance (Figure 4a), and this was confirmed by an analysis of chlorophyll levels. As shown in Figure 4(c), chlorophyll was significantly reduced in the laf6 mutant in agreement with Møller et al. (2001), but levels of both chlorophyll a and chlorophyll b were similar to WT levels in the 35S::SUFB/laf6‐4 and ‐7 lines. Identical results were observed in 7‐day‐old seedlings (Figure S4).
Figure 4.
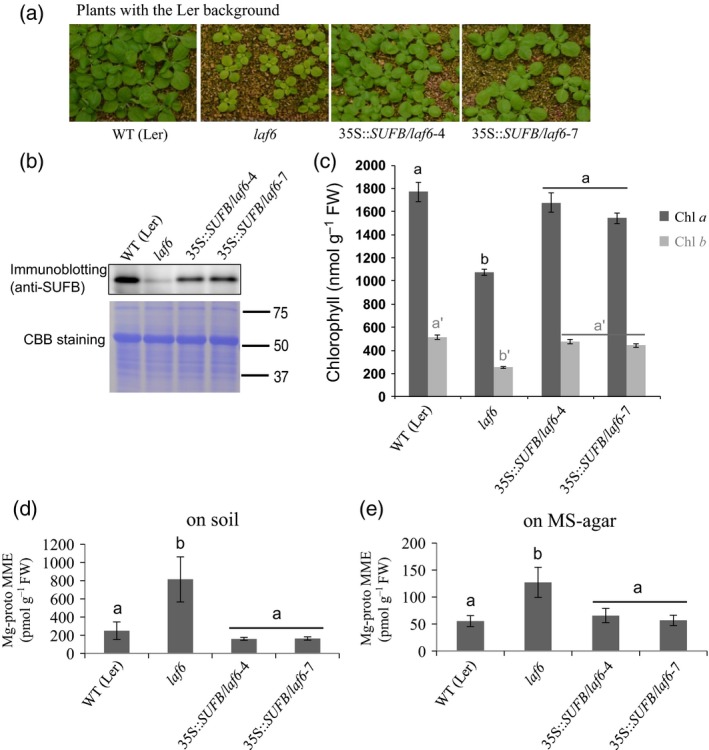
Complementation of laf6 with SUFB.
(a) Comparison of laf6 and SUFB overexpressing lines in a laf6 background grown on soil for 3 weeks under long‐day conditions.
(b) SUFB protein levels in the mutant and transgenic lines. Total protein extracts (8 μg protein) of developing leaves were analyzed by immunoblotting using anti‐SUFB antiserum (upper row) and the membrane was subsequently stained by Coomassie Brilliant Blue (CBB) as a loading control (bottom row).
(c) Chlorophyll a and b content of developing leaves of mutant and transgenic plants.
(d) Mg‐protoporphyrin IX monomethylester (Mg‐proto MME) content in developing leaves of 4‐week‐old plants grown on soil under long‐day conditions.
(e) The Mg‐proto MME content in 7‐day‐old seedlings grown on 1/2 MS medium under long‐day conditions.
Data points represent the mean ± SD of four biological replicates. Letters in black or grey above each bar indicate significant differences (P < 0.05) by Tukey's multiple‐comparison test.
Next we examined the chlorophyll biosynthesis intermediates Proto IX, Mg‐proto and Mg‐proto MME. In agreement with what we observed in the hmc1 mutant and SUFB RNAi lines (Figure 2), the laf6 mutant also showed elevated levels of Mg‐proto MME in 4‐week‐old plants (Figure 4d) and in seedlings (Figure 4e). This aspect of the laf6 phenotype was fully complemented by overexpression of SUFB (Figure 4d,e). Similarly to other SUFB‐deficient lines, laf6 also appeared to have slightly lower levels of Proto IX and Mg‐proto; however, this was not significant in this case (Figure S5).
Proto IX accumulation and long hypocotyl in far‐red‐light phenotypes of laf6 are not complemented by overexpression of SUFB
The laf6 phenotype was originally characterized by an accumulation of Proto IX in dark‐grown and FR light‐grown seedlings (Møller et al., 2001). We therefore measured the levels of Proto IX and other chlorophyll precursors in 7‐day‐old WT and SUFB‐deficient etiolated seedlings (Figure 5). First, we confirmed SUFB protein levels in dark‐grown seedlings of all the lines used. As shown in Figure 5(a), the laf6 mutant contained reduced amounts of SUFB compared with WT (Ler) and the two complemented lines over‐accumulated SUFB. Only a small decrease in SUFB was observed in hmc1, while in the SUFB RNAi lines, SUFB protein levels were severely reduced by Dex treatment. These results are consistent with those seen in 4‐week‐old plants (Figures 1b and 4b).
Figure 5.
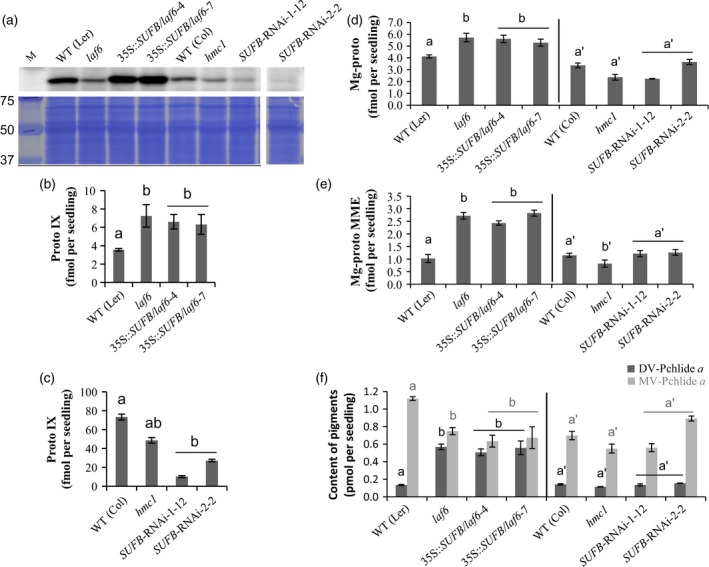
The phenotype of dark‐grown SUFB‐deficient seedlings.
Comparison of 7‐day‐old etiolated seedlings of conditional SUFB‐silenced lines, hmc1, laf6 and laf6 complemented lines expressing SUFB grown on 1/2 MS medium with 0.8% agar and 1% sucrose.
(a) Levels of SUFB protein in mutant and transgenic seedlings. Total protein extracts (16 μg protein) were analyzed by immunoblotting using anti‐SUFB antiserum (upper row) and the membrane was subsequently stained by Coomassie Brilliant Blue (CBB) as a loading control (lower row). (b, c) Protoporphyrin IX (Proto IX) content in etiolated seedlings in the Ler (b) and Col (c) backgrounds. Data points represent the mean ± SD of three biological replicates. Letters above each bar indicate significant differences (P < 0.05) by Tukey's multiple‐comparison test. (d) Mg‐proto IX (Mg‐proto) and (e) Mg‐proto IX monomethyl ester (Mg‐proto MME) contents of mutant and transgenic seedlings. Data points represent the mean ± SD of three biological replicates. Letters in black (Mg‐proto) or in grey (Mg‐proto MME) above each bar indicate significant differences (P < 0.05) by Tukey's multiple‐comparison test. (f) 3,8‐Divinyl protochlorophyllide a (DV‐Pchlide a) or monovinyl protochlorophyllide a (MV‐Pchlide a) contents in mutant and transgenic seedlings. Data points represent the mean ± SD of three biological replicates. Letters in black (DV‐Pchlide a) or in grey (MV‐Pchlide a) above each bar indicate significant differences (P < 0.05) by Tukey's multiple‐comparison test.
As reported by Møller et al. (2001), dark‐grown laf6 seedlings showed elevated levels of Proto IX compared with WT (Ler) seedlings (Figure 5b). In addition, Mg‐proto, Mg‐proto MME (Figure 5d,e) and DV‐Pchlide (Figures 5f and S6) all showed increased levels in laf6 compared with WT (Ler). In contrast, neither the hmc1 mutant nor the SUFB RNAi lines showed elevated chlorophyll precursors in dark‐grown seedlings (Figures 5c–f and S6). In fact, hmc1 and the SUFB‐silenced etiolated seedlings accumulated less Proto IX than did WT (Col) seedlings (Figure 5c). In addition, overexpression of SUFB did not rescue the increase in chlorophyll precursors seen in laf6 (Figures 5b,d–f and S6). Taken together, these results show that the excessive accumulation of Proto IX, Mg‐proto, Mg‐proto MME and DV‐Pchlide in etiolated laf6 seedlings is not the consequence of altered SUFB levels in this mutant.
The laf6 mutant was originally isolated from a screen for mutants with long hypocotyl after growth under FR light (Møller et al., 2001). To investigate this response and further explore the link between Fe–S cluster biosynthesis and phytochrome signaling, the response of the SUFB mutant and transgenic lines to FR light was examined. As expected, laf6 had a longer hypocotyl than the WT when grown under FR light irradiation (Figure 6a). However, this response was not rescued in the SUFB‐complemented lines, and the SUFB RNAi lines did not show enhanced hypocotyl elongation compared with WT (Col) seedlings on Dex (Figure 6a). These results show that the reduced responsiveness toward FR light in laf6 is not correlated with SUFB levels in seedlings.
Figure 6.
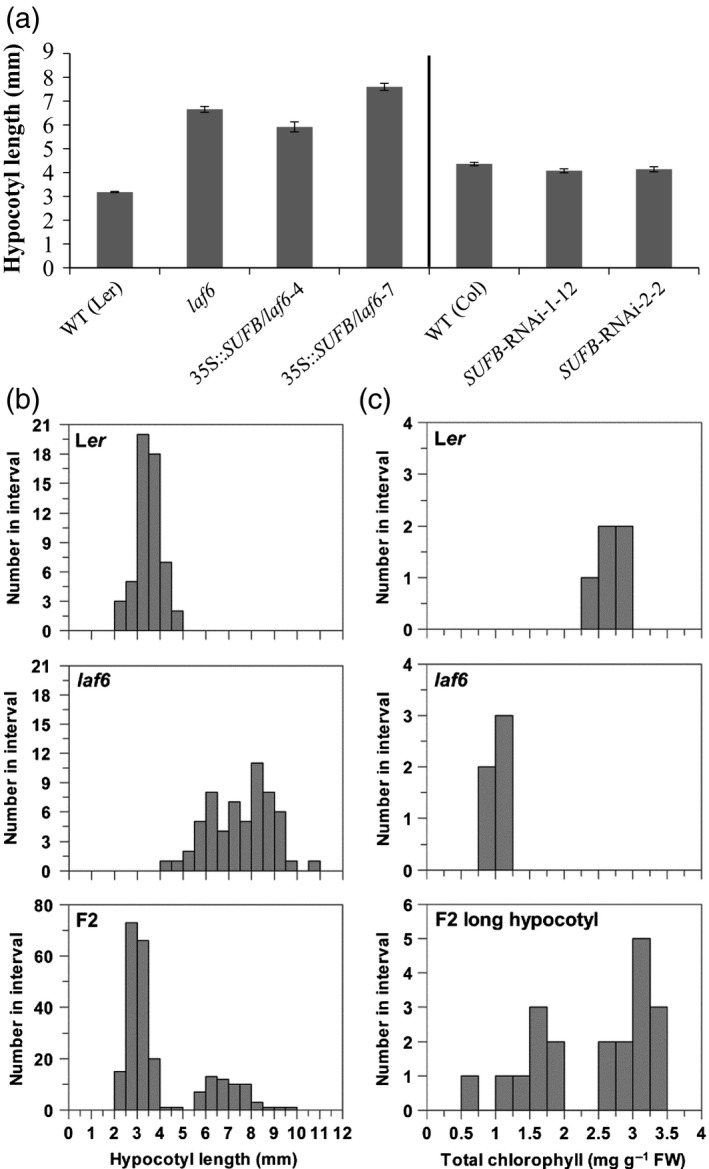
Segregation of the laf6 long‐hypocotyl phenotype.
(a) Hypocotyl length of laf6, laf6 expressing SUFB and conditional SUFB‐deficient seedling grown for 6 days under far‐red (FR) light. Data shown are mean ± SD of four biological replicates.
(b, c) Segregation of the long‐hypocotyl and pale leaf phenotypes of back‐crossed laf6 seedlings. (b) Histogram of hypocotyl lengths of Ler, laf6 and the F2 generation (Ler × laf6) seedlings after 6 days’ growth in FR light.
(c) Histogram of the foliar chlorophyll content of the parent lines and 20 randomly selected F2 seedlings with long hypocotyls after 6 days under FR. Following the FR treatment, all seedlings were supplemented with 3% sucrose and recovered in low‐intensity white light (WL) (4 days at 5 μmol m−2 sec−1 followed by 2 days at 25 μmol m−2 sec−1) before being transferred to WL (100 μmol m−2 sec−1) for 11 days.
The long hypocotyl phenotype of laf6 is due to a second, segregating mutation
The results in our hypocotyl elongation assays and on the accumulation of Proto IX and DV‐Pchlide suggest that the laf6 mutant has an additional mutation causing that phenotype independently of its effect on SUFB protein levels. To examine this possibility, we analyzed the segregation of laf6 phenotypes after backcrossing laf6 with WT (Ler). Seedlings in the F2 generation were analyzed initially for hypocotyl length in FR light and subsequently for chlorophyll biosynthesis in white light (Figure 6b,c). F2 seedlings segregated into two populations with longer (>5.5 mm) and shorter (<5.0 mm) hypocotyls. Of 234 seedlings measured, 176 (75.2%) had short hypocotyls while 58 (24.8%) had long hypocotyls, a ratio indicating that hypocotyl length was controlled by a single recessive locus (Figure 6b). Twenty seedlings from the population with long hypocotyls were then grown under continuous white light and chlorophyll levels determined. Again two populations were observed with 12 (60%) showing higher chlorophyll levels [>2.5 mg g−1 fresh weight (FW)] similar to the level seen in WT seedlings and 8 (40%) with lower chlorophyll levels (<2.0 mg g−1 FW) consistent with laf6 seedlings (Figure 6c). These results indicate that the long‐hypocotyl phenotype under FR light and the low‐chlorophyll phenotype under white light are controlled by two independent genes. This is consistent with our data showing that the long‐hypocotyl phenotype is not due to SUFB deficiency.
The laf6 mutant shows an accumulation of Proto IX and other chlorophyll precursors in dark‐grown seedlings, and we speculated that the long‐hypocotyl phenotype might be linked to a deficiency in synthesis of the phytochrome chromophore phytochromobilin, which is synthesized from heme (Frankenberg‐Dinkel and Terry, 2009; see Figure S1). This was initially examined using the chromophore analogue phycocyanobilin (PCB; Elich et al., 1989) and it was concluded that laf6 was not compromised in phytochrome chromophore synthesis (Møller et al., 2001). However, it was subsequently shown that the altered chromophore structure of PCB cannot support a FR high‐irradiance response leading to inhibition of hypocotyl elongation (Hanzawa et al., 2002), so we re‐addressed this question by feeding the chromophore precursor biliverdin IXα. However, as shown in Figure S7, biliverdin IXα did not rescue the long‐hypocotyl phenotype of laf6, but could rescue a hy1 mutant control.
Discussion
Iron–sulfur clusters are required for chlorophyll synthesis
Mutants lacking SUFB are reduced in PSI subunit accumulation and have reduced chlorophyll levels (Hu et al., 2017). However, it was unclear whether the reduction in chlorophyll was a consequence of a reduced accumulation of photosynthetic proteins or whether chlorophyll synthesis was directly affected. Here we show that a reduction of SUFB protein levels results in excessive accumulation of the chlorophyll precursor Mg‐proto MME and an overall reduction in chlorophyll levels. Both the laf6 and hmc1, the SUFB‐deficient mutant alleles showed this phenotype as did the SUFB RNAi knock‐down lines. Moreover, the SUFC and SUFD RNAi lines both accumulated Mg‐proto MME, suggesting that all subunits of the SUFBC2D complex are necessary for optimal chlorophyll synthesis and that inhibition of Fe–S cluster biosynthesis is the critical factor. To date no chlorophyll a biosynthesis enzymes have been shown to be Fe–S proteins, although two enzymes involved in chlorophyll metabolism, 7‐hydroxymethyl‐chlorophyll a reductase and pheophorbide a oxygenase, contain Fe–S clusters (Pruzinská et al., 2003; Meguro et al., 2011; Wang and Liu, 2016) and the substrates of both of these enzymes also accumulate in the hmc1 mutant (Nagane et al., 2010) (see Figure S1). It is also reported that siroheme biosynthesis requires an Fe–S protein, sirohydrochlorin ferrochelatase (Saha et al., 2012), although the effect of Fe–S deficiency on siroheme biosynthesis is not clear at present. Fe–S proteins, such as ferredoxin, could also be involved, but this has only been established for chlorophyll a oxygenase (Oster et al., 2000) and heme oxygenase (Muramoto et al., 2002). Mutants in both of these enzymes are pale in color (Chory et al., 1989; Terry, 1997; Espineda et al., 1999) and thus could be candidate targets for an impact of Fe–S cluster deficiency.
In the current study we have demonstrated that Mg‐proto MME accumulates in SUFB‐deficient mutants. Accumulation of Mg‐proto MME is seen in mutants that lack Mg‐proto MME cyclase activity (Tottey et al., 2003; Peter et al., 2009; Hollingshead et al., 2012), and we therefore propose that Mg‐proto MME cyclase activity is impaired in SUFB‐deficient plants, as the protein level of cyclase is unaffected. Based on genetic and biochemical analysis in barley (Rzeznicka et al., 2005), Mg‐proto MME cyclase is predicted to contain three subunits, although only two, YCF54 and CLH27, have been identified (Tottey et al., 2003; Albus et al., 2012; Hollingshead et al., 2012; Bollivar et al., 2014). However, neither of these subunits have Fe–S cluster‐binding motifs (Tottey et al., 2003; Hollingshead et al., 2012). It is therefore possible that the third as yet unidentified cyclase subunit contains an Fe–S cluster. Alternatively, other Fe–S proteins may function to accept electrons from the cyclase, as the cyclase reaction requires an electron acceptor. We have previously observed that treatment of methylviologen, an inhibitor of PSI, causes accumulation of Mg‐proto MME in cucumber seedlings (Aarti et al., 2006). Therefore, it is possible that PSI, which contains three Fe–S clusters, is itself required for the cyclase reaction. Finally, it is possible that the regulation of Mg‐proto MME cyclase is indirectly impaired in SUFB‐deficient plants. Little is known about the regulation of Mg‐proto MME cyclase, except for the involvement of NADPH‐dependent thioredoxin reductase C (NTRC). A mutant lacking this enzyme accumulates Mg‐proto MME (Stenbaek et al., 2008), thus it is possible that the SUFB‐deficient plants analyzed in this study are also impaired in the redox network in which NTRC is involved.
The phenotype of laf6 is partially due to second site mutations
When chlorophyll precursors were measured in dark‐grown seedlings we observed differences between the laf6 allele and other SUFB‐deficient lines. The hmc1 mutant and SUFB RNAi lines showed moderate decreases in Proto IX, Mg‐proto and Mg‐proto MME, although this was not significant in all cases, and they did not have reduced Pchlide levels. In contrast, the laf6 mutant accumulated Proto IX, Mg‐proto, Mg‐proto MME and DV‐Pchlide and showed a decrease in MV‐Pchlide in dark‐grown seedlings. The accumulation of Proto IX was consistent with a previous report (Møller et al., 2001). It is difficult to explain this set of results, but one possibility is that the conversion of DV‐Pchlide into MV‐Pchlide is impaired in the laf6 background and the general increase in porphyrins could be due to a regulatory effect such as product inhibition for enzymes prior to DV‐Pchlide in the pathway. Importantly, with regard to the function of SUFB, the effect of the laf6 mutation on porphyrin accumulation in dark‐grown seedlings could not be rescued by overexpression of SUFB, in contrast to the light‐dependent phenotypes of laf6. These results therefore indicate that the increase in chlorophyll intermediates in dark‐grown laf6 seedlings is caused by a second site mutation in the laf6 background.
In the original description of laf6, the accumulation of Proto IX was linked to a long‐hypocotyl phenotype under FR light (Møller et al., 2001). The authors had speculated that perturbations to tetrapyrrole synthesis might lead to an inhibition of phytochrome chromophore synthesis. We also tested this hypothesis, partly because more recent evidence indicated that the original control used could not have been successful (Hanzawa et al., 2002), but saw no evidence that the long‐hypocotyl phenotype was caused by chromophore deficiency. None of the other SUFB‐deficient lines tested had a long hypocotyl in FR light, and the phenotype was not complemented by SUFB expression. Moreover, segregation analysis following a backcross to WT seedlings demonstrated that the long‐hypocotyl phenotype of laf6 was separable from the chlorophyll‐deficient phenotype of SUFB‐deficient plants, and therefore also due to a second site mutation. It is not known whether the accumulation of porphyrins in dark‐grown seedlings and the long‐hypocotyl phenotype in FR light are linked. Both processes are regulated by phytochrome (Whitelam et al., 1993; McCormac and Terry, 2002), but while hypocotyl elongation is inhibited by phytochrome, porphyrin synthesis is promoted.
In conclusion, we have shown that deficiency in Fe–S cluster biosynthesis leads to reduced chlorophyll accumulation and we have identified Mg‐proto MME cyclase activity as the most likely target of inhibition. This is the first step in understanding the role of Fe–S cluster biosynthesis in chlorophyll metabolism and provides important clues about the mechanism of Mg‐proto MME cyclase. In addition, we have resolved the discrepancy between the phenotypes of different SUFB mutant alleles and demonstrated that the long‐hypocotyl phenotype of laf6 is unrelated to deficiency in SUFB. Together, these results help clarify the role of Fe–S clusters in plant development.
Experimental Procedures
Plant materials
The Arabidopsis thaliana laf6 and hmc1 mutants were described by Møller et al. (2001) and Nagane et al. (2010), respectively. Sterilized Arabidopsis seeds were sown on half‐strength Murashige and Skoog (1/2 MS) medium containing 1% (w/v) sucrose and 0.8% (w/v) agar or soil. Seeds were kept in the dark at 4°C for 3 days to induce uniform germination. The plants were subsequently grown under long‐day (16‐h light/8‐h dark) growth conditions under fluorescent light (70–90 μmol m−2 sec−1 at 23°C). For pigment and immunoblotting analyses, either developing leaves of 4‐week‐old plants or the whole seedlings of 7‐day‐old plants were harvested. For experiments under FR light (6 μmol m−2 sec−1) and in the dark, seedlings were grown for 7 days on 1/2 MS agar plates at 23°C.
For segregation experiments, surface sterilized seeds (Ler, laf6, F2‐1 and F2‐2) were sown on 1/2MS in 1% (w/v) agar, pH 5.8. After a 3‐day cold treatment (in the dark at 4°C), seeds were transferred to white light (WL; 100 μmol m−2 sec−1, 23°C) for 2 h, then incubated in the dark (23°C) for 1 day. Seedlings were then grown for 6 days in FR light (23°C). After the FR treatment, seedlings were gently flattened onto the surface of the medium and photographed for analysis of hypocotyl length. During this process, plates were kept in very low‐intensity WL as much as possible. Twenty randomly selected seedlings displaying a long‐hypocotyl phenotype in FR light were then transferred to a separate plate. All seedlings were then supplemented with filter‐sterilized 3% (w/v) sucrose and recovered in low‐intensity WL (4 days at 5 μmol m−2 sec−1 followed by 2 days at 25 μmol m−2 sec−1) before being transferred to WL (100 μmol m−2 sec−1) for 11 days. Chlorophyll assays were performed on five seedlings of each parent line, as well as the 20 randomly selected seedlings with long hypocotyls after the FR treatment.
Cloning and Arabidopsis transformation
For laf6 complementation, a complementary DNA (cDNA) covering the coding region of Arabidopsis SUFB was cloned from Arabidopsis (Col ecotype) into pEarleyGate100 (Earley et al., 2006) as described in the companion paper (Hu et al., 2017). In brief, to generate Dex‐inducible RNAi constructs, two 300‐bp regions from the coding sequence of either SUFB, SUFD or SUFC were amplified by PCR using the primer sets described in Hu et al. (2017). The PCR products were cloned into the pENTR4 dual selection vector (ThermoFisher Scientific, http://www.thermofisher.com), the resulting entry clones were recombined with the pOpOff2 (kan) vector (Wielopolska et al., 2005) by LR Clonase II (ThermoFisher Scientific) recombination reactions. the whole construct was introduced into Agrobacterium tumefaciens strain GV3101 and used to infect Arabidopsis with the floral dip method (Clough and Bent, 1998). Two‐week‐old seedlings of transformants with the pEarleyGate100 vector were selected by spraying with BASTA and transformants containing the pOpOff2 (kan) vector were selected on 50‐μm kanamycin on 1/2 MS plates. Third‐generation homozygous plants were employed for further analysis.
SUFB, SUFC and SUFD RNAi induction
Dex was first dissolved in ethanol to a concentration of 20 mm, and then diluted using 0.02% Tween‐20 aqueous solution to give a final concentration of 10 μm. To induce RNAi‐mediated gene silencing, 3‐week‐old plants grown in soil were sprayed with 10 μm Dex solution. In control experiments, plants were sprayed with the aqueous solution without Dex, but containing the same concentration of ethanol and Tween‐20. To induce SUFB‐silencing in plants grown on 1/2 MS medium, ethanol, with or without Dex, was added to autoclaved 1/2 MS medium (at about 55°C) to a final concentration of 10 μm. It is noted that the effects of RNAi suppression were only visible in developing leaves in mature 3‐ or 4‐week‐old plants. Therefore, only developing leaves were harvested for protein and pigment analyses.
Pigment analysis
For the segregation analysis, leaf tissue was weighed and then extracted in ice‐cold 80% acetone. Samples were centrifuged at 16 100 g for 5 min, and the absorbance of the supernatant was determined at 647 nm and 663 nm using a U‐2001 spectrophotometer (Hitachi, http://www.hitachi.com/). Total chlorophyll content (μg mL−1) was calculated by combining the chlorophyll a (12.25A663–2.79A647) and chlorophyll b (21.5A647–5.1A663) values before conversion to mg g−1 FW. For other experiments, chlorophyll and its intermediates were extracted from leaf tissue by homogenization with acetone, which was pre‐cooled to −30°C to suppress endogenous chlorophyllase activity (Hu et al., 2013). Extracts were subsequently centrifuged for 5 min at 20 000 g at 4°C, and the supernatant was analyzed by HPLC using a symmetry C8 column (150 mm long, 4.6 mm ID; Waters, http://www.waters.com/) according to the method of Zapata et al. (2000). Chlorophyll concentrations were estimated from the absorption monitored at 410 nm. Chlorophyll a and b standards were purchased from Juntec Co. Ltd ( http://juntec.co.jp/) and pheophorbide a was purchased from Wako Pure Chemical Industries, Ltd ( http://www.wako-chem.co.jp/english/). A fluorescence detector (Hitachi) was used for the detection of Proto IX, Mg‐proto, and Mg‐proto MME a after separation by HPLC. Proto IX was detected at 634 nm following excitation at 400 nm, Mg‐proto and Mg‐proto MME were detected at 600 nm after excitation at 417 nm, while DV‐Pchlide a and MV‐Pchlide a were quantified by measuring absorbance at 439 nm.
Immunoblot analysis
Total protein was extracted from leaves using 10 volumes (v/w) of protein extraction buffer containing 50 mm 2‐amino‐2‐(hydroxymethyl)‐1,3‐propanediol (TRIS)‐HCl (pH 8.0), 12% (w/v) sucrose (Suc), 2% (w/v) lithium lauryl sulfate and 1.5% (w/v) dithiothreitol. The protein concentration of samples was determined using the Rc‐Dc protein assay kit (Bio‐Rad, http://www.bio-rad.com/). Before SDS‐PAGE separation, all samples were mixed with an equal volume of 2× urea buffer containing 10 mm TRIS‐HCl (pH 8.0), 10% (w/v) Suc, 2% (w/v) SDS, 1 mm EDTA, 4 mm dithiothreitol, 0.04% (w/v) bromophenol blue and 10 m urea. Samples were then separated on a 14% polyacrylamide gel and electro‐blotted to polyvinylidene difluoride (PVDF) membranes. SUFB protein was detected using an anti‐SUFB antiserum raised against recombinant Arabidopsis SUFB protein expressed in E. coli (Rossetta DE3, Merck, http://www.merck.com/). The antibody raised against Arabidopsis CHL27 was a kind gift from Professor Sabeeha Merchant (University of California, Los Angeles, CA, USA).
Statistical analysis
Samples from two ecotypes (Ler and Col) were analyzed separately. The WT, mutants and the transgenic lines in each ecotype were first analyzed as explanatory valuables by generalized linear mixed‐effects models (GLMMs). Two SUFB‐complementation lines (35S::SUFB/laf6‐4 and ‐7) and two conditional SUFB‐RNAi lines (SUFB‐RNAi‐1‐12 and 2‐2) in Figures 2(b), 3, 4, S2 and S4 were nested and treated as random effects variables. After confirming the significance of the GLMMs (P < 0.05), multiple pairwise comparison was made with the Tukey contrasts fit for each result by using the ‘glht()’ function of the ‘multicomp’ package (Hothorn et al., 2008) of R (v.3.2.2; R Core Team, 2015), which allows simultaneous tests for outputs of mixed‐effects models.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Figure S1. Tetrapyrrole biosynthetic pathway.
Figure S2. Phenotype of 7‐day‐old SUFB‐deficient seedlings grown on 1/2 MS medium under long‐day conditions.
Figure S3. Analysis of chlorophyll biosynthetic intermediates in SUFB‐deficient plants.
Figure S4. Complementation of laf6 with SUFB.
Figure S5. The protoporphyrin IX (a) and Mg‐protoporphyrin IX (b) content of the developing leaves of 4‐week‐old laf6‐ and SUFB‐overexpressing lines in a laf6 background grown on soil under long‐day conditions.
Figure S6. Detection of protochlorophyllide a in 7‐day‐old etiolated mutant and transgenic seedlings with altered SUFB levels.
Figure S7. The effects of 0.1 mm biliverdin IXα on hypocotyl length under far‐red light of 6‐day‐old WT, laf6, hy1 and phyA seedlings.
Acknowledgements
We thank Ms Junko Kishimoto for her help in drawing figures and to Mr Daniel Trim for performing the phytochrome chromophore feeding experiment. We are grateful to Professor Sabeeha Merchant (University of California, Los Angeles, CA, USA) for her kind gift of anti‐Mg‐proto MME cyclase antiserum and to Professor Simon Møller (University of Stavanger, Norway; currently at St John's University, New York, USA) for laf6 mutant seeds. This work was supported by a Biotechnology and Biological Sciences Research Council grant BB/J018139/1 to MJT, by the CREST program, Japan Science and Technology Agency to AT and by JSPS KAKENHI grant no. 16H0655416 to RT.
References
- Aarti, P.D. , Tanaka, R. and Tanaka, A. (2006) Effects of oxidative stress on chlorophyll biosynthesis in cucumber (Cucumis sativus) cotyledons. Physiol. Plant. 128, 186–197. [Google Scholar]
- Albus, C.A. , Salinas, A. , Czarnecki, O. et al. (2012) LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. 160, 1923–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk, J. and Pilon, M. (2011) Ancient and essential: the assembly of iron‐sulfur clusters in plants. Trends Plant Sci. 16, 218–226. [DOI] [PubMed] [Google Scholar]
- Balk, J. and Schaedler, T.A. (2014) Iron cofactor assembly in plants. Annu. Rev. Plant Biol. 65, 125–153. [DOI] [PubMed] [Google Scholar]
- Bollivar, D. , Braumann, I. , Berendt, K. , Gough, S.P. and Hansson, M. (2014) The Ycf54 protein is part of the membrane component of Mg‐protoporphyrin IX monomethyl ester cyclase from barley (Hordeum vulgare L.). FEBS J. 281, 2377–2386. [DOI] [PubMed] [Google Scholar]
- Chahal, H.K. , Dai, Y. , Saini, A. , Ayala‐Castro, C. and Outten, F.W. (2009) The SufBCD Fe‐S scaffold complex interacts with SufA for Fe‐S cluster transfer. Biochemistry, 48, 10644–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J. , Peto, C.A. , Ashbaugh, M. , Saganich, R. , Pratt, L. and Ausubel, F. (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell, 1, 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cornah, J.E. , Terry, M.J. and Smith, A.G. (2003) Green or red: what stops the traffic in the tetrapyrrole pathway? Trends Plant Sci. 8, 224–230. [DOI] [PubMed] [Google Scholar]
- Couturier, J. , Touraine, B. , Briat, J.‐F. , Gaymard, F. and Rouhier, N. (2013) The iron‐sulfur cluster assembly machineries in plants: current knowledge and open questions. Front. Plant Sci. 4, e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. and Pikaard, C.S. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Elich, T.D. , McDonagh, A.F. , Palma, L.A. and Lagarias, J.C. (1989) Phytochrome chromophore biosynthesis. Treatment of tetrapyrrole‐deficient Avena explants with natural and non‐natural bilatrienes leads to formation of spectrally active holoproteins. J. Biol. Chem. 264, 183–189. [PubMed] [Google Scholar]
- Espineda, C.E. , Linford, A.S. , Devine, D. and Brusslan, J.A. (1999) The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA, 96, 10507–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg‐Dinkel, N. and Terry, M.J. (2009) Synthesis and role of bilins in photosynthetic organisms In Tetrapyrroles: Birth, Life and Death (Martin J., Warren M.J. and Smith A.G., eds). New York, NY: Springer, pp. 208–220. [Google Scholar]
- Hanzawa, H. , Shinomura, T. , Inomata, K. , Kakiuchi, T. , Kinoshita, H. , Wada, K. and Furuya, M. (2002) Structural requirement of bilin chromophore for the photosensory specificity of phytochromes A and B. Proc. Natl. Acad. Sci. USA, 99, 4725–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorth, E. , Hadfi, K. , Zauner, S. and Maier, U.G. (2005) Unique genetic compartmentalization of the SUF system in cryptophytes and characterization of a SufD mutant in Arabidopsis thaliana . FEBS Lett. 579, 1129–1135. [DOI] [PubMed] [Google Scholar]
- Hollingshead, S. , Kopečná, J. , Jackson, P.J. , Canniffe, D.P. , Davison, P.A. , Dickman, M.J. , Sobotka, R. and Hunter, C.N. (2012) Conserved chloroplast open‐reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J. Biol. Chem. 287, 27823–27833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn, T. , Bretz, F. and Westfall, P. (2008) Simultaneous inference in general parametric models. Biom. J. 50, 346–363. [DOI] [PubMed] [Google Scholar]
- Hu, X. , Tanaka, A. and Tanaka, R. (2013) Simple extraction methods that prevent the artifactual conversion of chlorophyll to chlorophyllide during pigment isolation from leaf samples. Plant Methods, 9, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X. , Kato, Y. , Sumida, A. , Tanaka, A. and Tanaka, R. (2017) The SUFBC2D complex is required for the biogenesis of all major classes of plastid Fe‐S proteins. Plant J. (in press). [DOI] [PubMed] [Google Scholar]
- Lezhneva, L. , Amann, K. and Meurer, J. (2004) The universally conserved HCF101 protein is involved in assembly of [4Fe‐4S]‐cluster‐containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37, 174–185. [DOI] [PubMed] [Google Scholar]
- McCormac, A.C. and Terry, M.J. (2002) Light‐signalling pathways leading to the co‐ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana . Plant J. 32, 549–559. [DOI] [PubMed] [Google Scholar]
- Meguro, M. , Ito, H. , Takabayashi, A. , Tanaka, R. and Tanaka, A. (2011) Identification of the 7‐hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis . Plant Cell, 23, 3442–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, N. , Tanaka, R. , Tanaka, A. , Masuda, T. and Nagatani, A. (2008) The steady‐state level of Mg‐protoporphyrin IX is not a determinant of plastid‐to‐nucleus signaling in Arabidopsis . Proc. Natl. Acad. Sci. USA, 105, 15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, N. , Tanaka, R. , Grimm, B. , Masuda, T. , Moulin, M. , Smith, A.G. , Tanaka, A. and Terry, M.J. (2010) The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 15, 488–498. [DOI] [PubMed] [Google Scholar]
- Møller, S.G. , Kunkel, T. and Chua, N.H. (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev. 15, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto, T. , Tsurui, N. , Terry, M.J. , Yokota, A. and Kohchi, T. (2002) Expression and biochemical properties of a ferredoxin‐dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiol. 130, 1958–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagane, T. , Tanaka, A. and Tanaka, R. (2010) Involvement of AtNAP1 in the regulation of chlorophyll degradation in Arabidopsis thaliana . Planta, 231, 939–949. [DOI] [PubMed] [Google Scholar]
- Oster, U. , Tanaka, R. , Tanaka, A. and Rüdiger, W. (2000) Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana . Plant J. 21, 305–310. [DOI] [PubMed] [Google Scholar]
- Outten, F.W. , Wood, M.J. , Munoz, F.M. and Storz, G. (2003) The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe‐S cluster assembly in Escherichia coli . J. Biol. Chem. 278, 45713–45719. [DOI] [PubMed] [Google Scholar]
- Peter, E. , Salinas, A. , Wallner, T. , Jeske, D. , Dienst, D. , Wilde, A. and Grimm, B. (2009) Differential requirement of two homologous proteins encoded by sll1214 and sll1874 for the reaction of Mg protoporphyrin monomethylester oxidative cyclase under aerobic and micro‐oxic growth conditions. Biochim. Biophys. Acta, 1787, 1458–1467. [DOI] [PubMed] [Google Scholar]
- Pruzinská, A. , Tanner, G. , Anders, I. , Roca, M. and Hörtensteiner, S. (2003) Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske‐type iron‐sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA, 100, 15259–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2015). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Rzeznicka, K. , Walker, C.J. , Westergren, T. , Kannangara, C.G. , von Wettstein, D. , Merchant, S. , Gough, S.P. and Hansson, M. (2005) Xantha‐l encodes a membrane subunit of the aerobic Mg‐protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc. Natl. Acad. Sci. USA, 102, 5886–5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, K. , Webb, M.E. , Rigby, S.E.J. , Leech, H.K. , Warren, M.J. and Smith, A.G. (2012) Characterization of the evolutionarily conserved iron–sulfur cluster of sirohydrochlorin ferrochelatase from Arabidopsis thaliana . Biochem. J. 444, 227–237. [DOI] [PubMed] [Google Scholar]
- Schlicke, H. , Hartwig, A.S. , Firtzlaff, V. , Richter, A.S. , Glässer, C. , Maier, K. , Finkemeier, I. and Grimm, B. (2014) Induced deactivation of genes encoding chlorophyll biosynthesis enzymes disentangles tetrapyrrole‐mediated retrograde signaling. Mol. Plant, 7, 1211–1227. [DOI] [PubMed] [Google Scholar]
- Sheftel, A. , Stehling, O. and Lill, R. (2010) Iron‐sulfur proteins in health and disease. Trends Endocrinol. Metab. 21, 302–314. [DOI] [PubMed] [Google Scholar]
- Stenbaek, A. , Hansson, A. , Wulff, R.P. , Hansson, M. , Dietz, K.‐J. and Jensen, P.E. (2008) NADPH‐dependent thioredoxin reductase and 2‐Cys peroxiredoxins are needed for the protection of Mg‐protoporphyrin monomethyl ester cyclase. FEBS Lett. 582, 2773–2778. [DOI] [PubMed] [Google Scholar]
- Tanaka, R. and Tanaka, A. (2007) Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346. [DOI] [PubMed] [Google Scholar]
- Terry, M.J. (1997) Phytochrome chromophore‐deficient mutants. Plant, Cell Environ. 20, 740–745. [Google Scholar]
- Tottey, S. , Block, M.A. , Allen, M. , Westergren, T. , Albrieux, C. , Scheller, H.V. , Merchant, S. and Jensen, P.E. (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA, 100, 16119–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touraine, B. , Boutin, J.P. , Marion‐Poll, A. , Briat, J.F. , Peltier, G. and Lobréaux, S. (2004) Nfu2: a scaffold protein required for [4Fe‐4S] and ferredoxin iron‐sulphur cluster assembly in Arabidopsis chloroplasts. Plant J. 40, 101–111. [DOI] [PubMed] [Google Scholar]
- Van Hoewyk, D. , Abdel‐Ghany, S.E. , Cohu, C.M. , Herbert, S.K. , Kugrens, P. , Pilon, M. and Pilon‐Smits, E.A.H. (2007) Chloroplast iron‐sulfur cluster protein maturation requires the essential cysteine desulfurase CpNifS. Proc. Natl. Acad. Sci. USA, 104, 5686‐5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. and Liu, L. (2016) Crystal structure and catalytic mechanism of 7‐hydroxymethyl chlorophyll a reductase. J. Biol. Chem. 291, 13349–13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G.C. , Johnson, E. , Peng, J. , Carol, P. , Anderson, M.L. , Cowl, J.S. and Harberd, N.P. (1993) Phytochrome A null mutants of Arabidopsis display a wild‐type phenotype in white light. Plant Cell, 5, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielopolska, A. , Townley, H. , Moore, I. , Waterhouse, P. and Helliwell, C. (2005) A high‐throughput inducible RNAi vector for plants. Plant Biotechnol. J. 3, 583–590. [DOI] [PubMed] [Google Scholar]
- Wollers, S. , Layer, G. , Garcia‐Serres, R. , Signor, L. , Clemancey, M. , Latour, J.‐M. , Fontecave, M. and Ollagnier de Choudens, S. (2010) Iron‐sulfur (Fe‐S) cluster assembly: the SufBCD complex is a new type of Fe‐S scaffold with a flavin redox cofactor. J. Biol. Chem. 285, 23331–23341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X.M. and Møller, S.G. (2004) AtNAP7 is a plastidic SufC‐like ATP‐binding cassette / ATPase essential for Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA, 101, 9143–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X.M. , Adams, S. , Chua, N. and Møller, S.G. (2005) AtNAP1 represents an atypical SufB protein in Arabidopsis plastids. J. Biol. Chem. 280, 6648–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe, T. , Morimoto, K. , Kikuchi, S. , Nishio, K. , Terashima, I. and Nakai, M. (2004) The Arabidopsis chloroplastic NifU‐like protein CnfU, which can act as an iron‐sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell, 16, 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata, M. , Rodríguez, F. and Garrido, J.L. (2000) Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and phridine‐containing mobile phases. Mar. Ecol. Prog. Ser. 195, 29–45. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Tetrapyrrole biosynthetic pathway.
Figure S2. Phenotype of 7‐day‐old SUFB‐deficient seedlings grown on 1/2 MS medium under long‐day conditions.
Figure S3. Analysis of chlorophyll biosynthetic intermediates in SUFB‐deficient plants.
Figure S4. Complementation of laf6 with SUFB.
Figure S5. The protoporphyrin IX (a) and Mg‐protoporphyrin IX (b) content of the developing leaves of 4‐week‐old laf6‐ and SUFB‐overexpressing lines in a laf6 background grown on soil under long‐day conditions.
Figure S6. Detection of protochlorophyllide a in 7‐day‐old etiolated mutant and transgenic seedlings with altered SUFB levels.
Figure S7. The effects of 0.1 mm biliverdin IXα on hypocotyl length under far‐red light of 6‐day‐old WT, laf6, hy1 and phyA seedlings.


