Abstract
Flight represents a key trait in most insects, being energetically extremely demanding, yet often necessary for foraging and reproduction. Additionally, dispersal via flight is especially important for species living in fragmented landscapes. Even though, based on life‐history theory, a negative relationship may be expected between flight and immunity, a number of previous studies have indicated flight to induce an increased immune response. In this study, we assessed whether induced immunity (i.e. immune gene expression) in response to 15‐min forced flight treatment impacts individual survival of bacterial infection in the Glanville fritillary butterfly (Melitaea cinxia). We were able to confirm previous findings of flight‐induced immune gene expression, but still observed substantially stronger effects on both gene expression levels and life span due to bacterial infection compared to flight treatment. Even though gene expression levels of some immunity‐related genes were elevated due to flight, these individuals did not show increased survival of bacterial infection, indicating that flight‐induced immune activation does not completely protect them from the negative effects of bacterial infection. Finally, an interaction between flight and immune treatment indicated a potential trade‐off: flight treatment increased immune gene expression in naïve individuals only, whereas in infected individuals no increase in immune gene expression was induced by flight. Our results suggest that the up‐regulation of immune genes upon flight is based on a general stress response rather than reflecting an adaptive response to cope with potential infections during flight or in new habitats.
Keywords: gene expression, immune response, insect flight, Melitaea cinxia
Introduction
Parasites and pathogens represent a strong selection pressure to the host, as they are ubiquitous and can cause substantial fitness costs (Decaestecker et al., 2007; Mone et al., 2010). Therefore, the evolution of the immune system is a crucial factor in the life of any species. The investment of an organism in its immune defence depends on several factors such as the risk of an attack and the efficiency of the defence but also on the costs associated with the activation of the immune system (Zuk & Stoehr, 2002). Further, immunity investment might be affected by individual's body condition or nutritional status (Klemola et al., 2007; Valtonen et al., 2009; Srygley & Lorch, 2011).
Another key life‐history trait in many organisms is dispersal, playing a major role in population dynamics, as it is a prerequisite for spreading of individuals and hence of gene flow among populations (Clobert et al., 2012). Dispersal includes several functions, such as escape from unfavourable conditions or habitats, avoidance of kin competition or inbreeding, but it also distributes offspring into different locations and different environmental conditions (Matthysen, 2012). In many insects, flight is a key prerequisite for dispersal.
As both flight and activation of immunity are energetically demanding, potential trade‐offs between them may be expected (Bonte et al., 2012). Studies with crickets (Gryllus texensis) and bumblebees (Bombus terrestris), for example, have shown reduced immune defence after foraging or tethered flight (Köning & Schmid‐Hempel, 1995; Adamo et al., 2008), potentially due to energetic costs of flight. However, positive correlations between flight and immunity have also been observed (Snoeijs et al., 2004; Suhonen et al., 2010). For example, in the great tit (Parus major) immigrants have higher humoral immune response (Snoeijs et al., 2004). The positive relationship between immunity and flight may be an adaptive response allowing individuals to cope with a potentially increasing infection risk due to dispersal, for example, because entering a new habitat may entail different quality or quantity of pathogens. In such case, flight‐induced immune activation should increase individual's survival to pathogens. Alternatively, up‐regulation of immunity genes may solely reflect a general stress response due to the wearisome and stressful act of flight.
In insects, the immune system is triggered by surface particles of pathogens that are able to bind to secreted but also membrane‐bound receptors that can be found in the haemolymph (Yoshida et al., 1996). Two main pathways are part of the insect immune system, the Toll and the IMD pathway, of which the latter responds to gram‐negative and the former to gram‐positive bacteria and fungi (reviewed in Lemaitre & Hoffmann, 2007). Upon receptor binding, the Spätzle protein gets activated via a proteolytic cascade which then binds to the Toll receptor on the cell surface. Contrarily, antigens are able to bind directly to cell surface receptors in the IMD pathway to then transmit the signal inside the cell. In both cases, the intracellular signalling cascade leads to the activation of transcription factors (Dorsal and Relish for Toll and IMD, respectively) that alter gene expression of different immune genes (Hoffmann, 2003). Different proteins and molecules will be expressed in the fat body and secreted into the haemolymph, for example antimicrobial peptides (AMPs) and serpins.
A general stress response has previously been shown to interact with the immune response in many insect species (Adamo, 2008, 2012). This connection has either evolved independently in different phyla or represents a conserved connection (Adamo, 2008) and seems to be crucial for survival in many species. The NF‐kB system, one of the key regulators of the innate immune system, for example, is closely connected to oxidative stress and inflammation (Salminen et al., 2008). During acute stress (fight‐or‐flight), different stress hormones are released, of which in insects the most important are octopamine and adipokinetic hormone (Orchard et al., 1993). Both hormones trigger the release of lipids from the fat body to optimize the body for a fight‐or‐flight reaction. Immune challenge likewise leads to the increase in octopamine to increase the availability of energy‐rich compounds (Adamo, 2010). The liberation of lipids might result in a shift of molecular resources away from immunity into the fight‐or‐flight response, as both pathways rely on apolipophorin III, a lipoprotein that is responsible for lipid transport (Adamo et al., 2008). This protein has both storage and immune function, as it is able to bind bacterial lipoteichoic acid (Kim et al., 2004; Ma et al., 2006). Stress hormones tend to also increase immune responses such as increased phagocytosis and phenoloxidase response (Baines et al., 1992; Goldsworthy et al., 2002), most likely due to stress‐hormone receptors on haemocytes (Adamo, 2008; Kim et al., 2009; Huang et al., 2012). In larvae of the greater wax moth (Galleria mellonella), acute stress had an immune‐enhancing effect even 24 h after a 2‐min stress event (Mowlds et al., 2008).
In this study, we aimed to disentangle why individuals would invest in an up‐regulation of the immune system upon flight, and more specifically whether the activation is based on a general stress response or on an adaptive response that may have evolved along with a higher infection risk when dispersing to new environments. We are using the Glanville fritillary (Melitaea cinxia) as a study system, which has a classic metapopulation structure in the Åland Islands in the south‐west of Finland. The metapopulation is characterized by annual extinctions and recolonizations of local populations, making dispersal essential for population viability in a highly fragmented landscape (Hanski, 1999a). Flight in this species is energetically demanding and might impact the individual's condition, therefore placing dispersing individuals at a higher risk of infections. Dispersing individuals may also be facing different or more infections by parasites and pathogens in the habitat matrix or in the new habitat patches they disperse to. The energetic demands of flight might further increase pathogen exposure by increased food uptake after flight events. Previous studies in this species have shown that forced flight provokes an activation of the immune system, measured as higher encapsulation rate (Saastamoinen & Rantala, 2013). In addition, immune genes are up‐regulated upon forced flight treatment (Kvist et al., 2015).
We infected adult butterflies with a bacterial strain right after a forced flight treatment to try to tease apart whether induced immunity upon flight mitigates individuals to overcome infection, and hence show similar or longer life span than those without flight treatment. As an immune response, we assessed gene expression of seven immune genes that have been previously shown to be expressed upon forced flight treatment similar to that used in the present experiment (Kvist et al., 2015), and which are known to be expressed upon infection with bacteria. A similar pattern to that in life span should be visible in the gene expression if the adaptive response hypothesis is true, hence showing equally high or even higher expression levels for the flight treatment in comparison with control individuals when facing an infection. The alternative hypothesis is that flight‐induced immune activation is due to a general stress response, in which case we expect no positive effect on survival or immune gene expression due to flight, and might in fact expect a negative effect if the stress is severe enough. We were aiming to cover a wide range of immune genes, including recognition proteins, antimicrobial peptides, and receptors and proteins involved in both the Toll and IMD pathway to see whether the observed responses were general or pathway‐ or gene‐specific. Whereas the receptor PGRP‐LC and the AMP attacin are part of the IMD pathway, pelle is involved in the Toll pathway. We furthermore included lysozyme, prophenoloxidase (proPO), serpin and β‐1,3‐glucan recognition protein in our study, based on these criteria.
Materials and methods
Study system
The Glanville fritillary butterfly, Melitaea cinxia (Melitaeini: Nymphalidae), is present in Finland only on the Åland Islands south‐west of mainland Finland where it has a classic metapopulation (Hanski, 1999b). The species is characterized by a univoltine life cycle in Finland. Larvae feed for five instars on one of two host plants (Plantago lanceolata and Veronica spicata) before they spend the winter in diapause in a silken web. In the spring, larvae continue feeding until pupation occurs in May following by a flight season from June to mid‐July, with males emerging about 2–3 days earlier than females (Boggs & Nieminen, 2004). Adults feed on nectar.
In general, this species is relatively sedentary (based on mark–release–recapture studies), although many individuals move between the small meadows at some point during the adult stage (Kuussaari et al., 1996). Flight typically consists of short flight bouts and rapid take‐offs in case of males that locate females by the ‘perching’ tactic, in which they establish a mating territory waiting for a suitable female that they defend from intruding males by chasing them away. In addition, males might fly more continuously around the habitat in search of females (Boggs & Nieminen, 2004). The average flight distance experimentally measured was about 32 m and mean lifetime dispersal distance several hundreds of metres with longest dispersal events of 1–2 km (Kuussaari et al., 1996; Niitepõld et al., 2011) and the longest recorded colonization distance of 4–5 km (Van Nouhuys & Hanski, 2002). Females show higher rates of dispersal compared to males that might remain in the natal population (Kuussaari et al., 1996).
Experimental design
Larvae were collected from 58 different populations (one to five individuals per population, on average 2.07) in Åland in the spring 2015 and fed ad libitum with P. lanceolata until pupation. Upon eclosion, 386 adult butterflies from different populations (families) were randomly divided into a flight treatment and control group, with an equal sex distribution. Individuals were kept in cages (40 × 50 cm) with no more than 40 individuals per cage and fed on 20% honey:water solution. Butterflies were kept at room temperature to discourage flight activity and food uptake on the second day of eclosion to standardize nutritional state as well as activity. On the third day after eclosion, butterflies in the flight treatment were placed into a cylindrical plastic chamber and allowed to acclimatize to the chamber before the treatment. Individuals were forced to fly actively for 15 min by gently tapping or shaking the chamber whenever the butterfly landed. The temperature during flight was maintained at 30 °C. This treatment reflects the general flight metabolic rate measurement assay often used in this species (Niitepõld et al., 2009). The individuals not included in the flight treatment were not flown but otherwise treated equally (i.e. placed in the chamber with the same temperature). The 3‐day‐old adults from both control and flight treatment groups were then randomly divided across three different immunity treatments: naïve, injection of 2 μL PBS into the thorax (wounding control) or injection of 2 μL of a 5‐mg mL−1 lyophilized Micrococcus luteus (ATCC No. 4698; Sigma‐Aldrich) solution into the thorax. The butterfly was spanned with a net on a soft sponge with ventral side up to ensure that it is not able to move. Through a small hole in the net, the thorax is accessible for the injection with a Hamilton syringe. Here as well, naïve individuals were placed on the sponge under the net, even though not injected. After the different immune treatments, butterflies were provided with 20% honey:water solution and kept in standardized conditions, avoiding dehydration or other stress that might influence gene expression. Individuals from the three different immunity treatment groups were randomly divided into (1) measurement of life span or (2) assessing immune gene expression and therefore RNA sampling. Individuals whose life span was assessed were provided with 20% honey:water solution, and survival was checked daily.
RNA sampling
Twenty hours after the flight treatment, individuals that had been randomly chosen for RNA sampling were killed by flash‐freezing them in liquid nitrogen. Individuals across all treatments (flight and control and the three immune treatments) have been used for RNA sampling. Based on the 20‐h incubation, samples were taken between 8 and 12 am. Control individuals that did not experience flight treatment were similarly sampled 20 h after placing them once into the flight chamber. All samples were stored at −80 °C until RNA extraction from the thorax.
RNA extraction and reverse transcription
Total RNA was extracted from the frozen thorax using TRIzol reagent (Life Technologies Corporation, Carlsbad, CA, USA) followed by extraction with acid–phenol:chloroform:isoamyl alcohol (24 : 24 : 1, pH 5) and chloroform. Precipitation of the RNA was performed using isopropanol, washed with 75% ethanol, air‐dried and resuspended in 35–50 μL MQ water. RNA quantity and quality were checked using NanoDrop (Thermo Fischer Scientific Inc., Waltham, MA, USA). Samples were stored at −80 °C until further usage. Potential contaminations of genomic DNA in the RNA samples were removed using DNase I (Thermo Fischer Scientific Inc.). The samples were then reverse‐transcribed to cDNA using iScript™ cDNA Synthesis Kit (Bio‐Rad Laboratories, Hercules, CA, USA) according to the manufacturer's instructions.
Quantitative real‐time PCR (qPCR)
Primers were designed with Primer3 (Rozen & Skaletsky, 2000) for seven immune response genes: lysozyme C (MCINX003391), prophenoloxidase (proPO; MCINX013403), attacin (MCINX009397), peptidoglycan recognition protein LC (PGRP‐LC; MCINX014869), β‐1,3‐glucan recognition protein (βGRP; MCINX012854), serpin 3a (MCINX005220) and pelle (MCINX001775); and three endogenous control genes: mitochondrial ribosomal protein L37 (MCINX003184) and S24 (MCINX003139) and histone variant H2A.Z (MCINX016093). All primers were ordered from Oligomer (Oligomer Oy, Helsinki, Finland). The sequences can be found in the supporting information (Appendix S1). Amplification efficiencies (E) of the primer pairs were determined with five dilutions (1 : 1, 1 : 5, 1 : 25, 1 : 125, 1 : 625) of template cDNA, where E = 10‐1/slope. The qPCR was performed with three technical replicates, one water control and a plate control sample in a 384‐well plate with 10 μL volume, using C1000™ Thermal Cycler (Bio‐Rad Laboratories). All samples were tested for genomic DNA contamination with ‐RT controls prior to qPCR. Each reaction used 1 μL of the 1 : 5 diluted cDNA, 5 μL of SYBR® Green containing master mix (iQ™ SYBR® Green Supermix for qPCR; Bio‐Rad Laboratories), 3 μL of nuclease‐free water and 0.5 μL of each primer (10 μm).
Statistical analysis
Immune gene expression for each sample was calculated relative to the geometric mean of the three reference genes. For each sample, the mean from the three technical replicates was used, with the exception of removing a possible outlier. Raw C t values for all genes and technical replicates can be found in the supporting information (Appendix S2). A linear mixed‐model approach (R 3.1.2 for Windows; The R Project for Statistical Computing; lmer from package lme4; Bates et al., 2015) was used to analyse the effects of flight treatment and infection on immune gene expression, using bacterial treatment, flight treatment, sex and gene as fixed factors and family (population) as a random term. In a subsequent analysis (due to three‐way interaction between flight treatment, bacterial treatment and gene) to explore the effect for every gene separately, an independent model for each of the immune genes was conducted with bacterial treatment, flight treatment and sex as fixed factors and family (population) as a random term. Post hoc analysis was performed to explore paired comparisons of the different treatment groups. The model with the lowest Akaike information criterion (AIC) value was chosen as the best fitting model, and the model fit was further assessed using the conditional R 2 (sem.model.fits from package piecewiseSEM; Lefcheck & Freckleton, 2016). AIC values and R 2 of the final models are shown in Table 1 and in Appendix S3 for the initial full models.
Table 1.
Relative expression levels and fold increase for the used immune genes divided by the different treatment groups
| Gene | Flight treatment | Immune treatment | Relative expression difference (log2) | Fold increase | SE | P |
|---|---|---|---|---|---|---|
| Lysozyme | Control | PBS | 0.46 | 1.38 | 0.24 | 0.94 |
| AIC = 414.58; R 2 = 0.03 | Bacteria | −0.02 | 0.99 | 0.49 | 1.0 | |
| Flight | Naïve | 0.49 | 1.40 | 0.3 | 0.91 | |
| PBS | 0.62 | 1.54 | 0.29 | 0.79 | ||
| Bacteria | 0.14 | 1.10 | 0.31 | 0.99 | ||
| β‐1,3‐glucan recognition protein | Control | PBS | 1.29 | 2.45 | 0.35 | 0.12 |
| AIC = 418.84; R 2 = 0.7 | Bacteria | 4.48 | 22.32 | 0.27 | < 0.00001 | |
| Flight | Naïve | 1.46 | 2.75 | 0.3 | 0.049 | |
| PBS | 1.67 | 3.18 | 0.25 | 0.02 | ||
| Bacteria | 4.71 | 26.17 | 0.44 | < 0.00001 | ||
| proPO | Control | PBS | 0.32 | 1.25 | 0.26 | 0.99 |
| AIC = 419.61; R 2 = 0.06 | Bacteria | 1.28 | 2.43 | 0.45 | 0.11 | |
| Flight | Naïve | 0.47 | 1.39 | 0.24 | 0.94 | |
| PBS | 0.1 | 1.07 | 0.29 | 0.99 | ||
| Bacteria | 0.45 | 1.37 | 0.26 | 0.99 | ||
| Serpin | Control | PBS | 0.5 | 1.41 | 0.22 | 0.92 |
| AIC = 421.55; R 2 = 0.13 | Bacteria | 1.29 | 2.45 | 0.49 | 0.11 | |
| Flight | Naïve | 0.29 | 1.22 | 0.49 | 0.99 | |
| PBS | 0.9 | 1.87 | 0.12 | 0.47 | ||
| Bacteria | 1.66 | 3.16 | 0.2 | 0.01 | ||
| Attacin | Control | PBS | 6.41 | 85.04 | 0.52 | < 0.00001 |
| AIC = 572.59; R 2 = 0.53 | Bacteria | 9.08 | 541.19 | 0.73 | < 0.00001 | |
| Flight | Naïve | 2.99 | 7.94 | 0.77 | 0.04 | |
| PBS | 4.26 | 19.16 | 0.66 | 0.001 | ||
| Bacteria | 8.18 | 290.02 | 0.85 | < 0.00001 | ||
| Pelle | Control | PBS | 1.22 | 2.33 | 0.24 | 0.16 |
| AIC = 423.07; R 2 = 0.53 | Bacteria | 4.65 | 25.11 | 0.37 | < 0.00001 | |
| Flight | Naïve | 1.9 | 3.73 | 0.32 | 0.003 | |
| PBS | 2.61 | 6.11 | 0.46 | < 0.0001 | ||
| Bacteria | 3.82 | 14.12 | 0.32 | < 0.00001 | ||
| PGRP‐LC | Control | PBS | 0.73 | 1.66 | 0.12 | 0.14 |
| AIC = 312.23.07; R 2 = 0.14 | Bacteria | 0.66 | 1.58 | 0.25 | 0.22 | |
| Flight | Naïve | 0.81 | 1.75 | 0.18 | 0.07 | |
| PBS | 0.93 | 1.91 | 0.15 | 0.02 | ||
| Bacteria | 0.72 | 1.65 | 0.18 | 0.14 |
Expression levels are calibrated to naïve individuals without flight treatment, and sexes are pooled. Significant effects are highlighted in bold and calculated with Post hoc analysis (Tukey honest significant differences).
The effect of flight or infection on the life span was analysed using Poisson distribution with glmmPQL to handle overdispersion (package MASS; Venables & Ripley, 2002), using bacterial and flight treatment and sex as fixed factors and family (population) as a random term. Backward model selection was used by starting with a full model including all meaningful second‐order interactions and sequentially eliminating nonsignificant interaction terms (P > 0.05) that did not improve the model.
Results
Immune gene expression
We found a significant three‐way interaction between flight treatment, bacterial treatment and gene (χ 2 1,776 = 40.56, P < 0.0001; AIC = 3119.78, R 2 = 0.65; Appendix S4) that was further explored with a gene‐by‐gene analysis. A significant increase in four of the seven immune genes in the bacteria‐exposed groups relative to the naïve groups was observed (P < 0.003 for all; Fig. 1; Table 1). A strong up‐regulation was detected for attacin, showing on average 540‐fold increase (log2FC = 9.08) in expression in the bacteria‐exposed group compared to naïve individuals. Pelle and βGRP likewise showed a strong up‐regulation with on average 22‐ to 26‐fold increase (log2FC = 4.65 & 4.48) in expression in the bacteria‐exposed group compared to naïve individuals. A moderate up‐regulation was detected for serpin with on average 2.5‐fold increase (log2FC = 1.29) in expression due to bacterial injection. Wounding itself led to an increase in expression levels for attacin and pelle only (P < 0.02 for both; Fig. 1; Table 1), with on average 85‐ and two‐fold increase (log2FC = 6.41 and 1.22) compared to the naïve group, respectively. In addition, we found a significant interaction between bacterial treatment and sex for βGRP (bacterial treatment*sex: χ 2 1,110 = 6.63, P = 0.036; Appendix S5), showing higher expression levels for the bacterial treatment for females. Finally, flight treatment provoked an increase in expression levels for βGRP and PGRP‐LC (βGRP: χ 2 1,110 = 11.58, P = 0.0007; PGRP‐LC: χ 2 1,110 = 4.29, P = 0.038). The flight treatment elevated the expression of attacin and pelle in the naïve samples, whereas in the infected samples no such elevation was observed (bacterial treatment*flight treatment: attacin: χ 2 1,110 = 14.79, P = 0.0006; pelle: χ 2 1,110 = 16.24, P = 0.0003; Appendix S5). If anything, in the infected individuals the expression was slightly reduced by the flight treatment.
Figure 1.
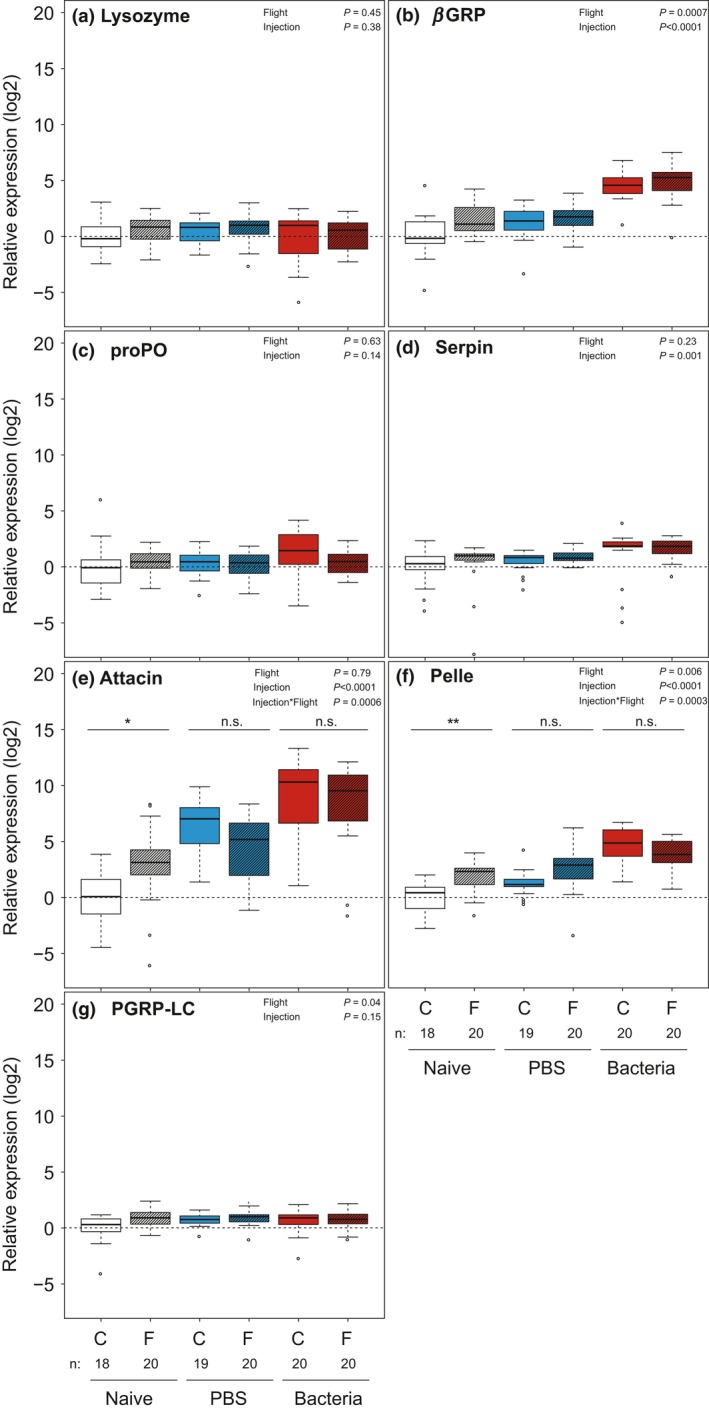
Immune gene expression in the Glanville fritillary butterfly. Shown are the relative expression levels (log2‐transformed) of the seven tested immune genes of naïve (white), PBS‐injected (blue; wounding with 2 μL PBS) and bacteria‐injected (red; 2 μL of 5‐mg mL−1 Micrococcus luteus in PBS) individuals divided into control (C) and flight (F) treatment groups. Expression levels are calibrated to the naïve individuals that did not experience flight treatment. Sexes are combined, as no sex difference was observed. The P‐values for the effect of flight and immune treatment and their interaction, whenever significant, are presented. The interactions between flight and immune treatment found for attacin and pelle are indicated with asterisks and lines based on the Post hoc test performed.
There were no significant changes for lysozyme and proPO. All results of the initial models used for the gene‐by‐gene analysis can be found in Appendix S3.
Life span
Males lived longer than females (t 1,171 = 2.11, P = 0.037; Fig. 2). Life span was significantly reduced in both sexes by the bacterial infection treatment (by almost 66%, P < 0.00001; Fig. 2) compared to naïve individuals (naïve: 24.9 (± 1.6) and 23.7 (± 1.6); bacteria: 11.9 (± 1.5) and 8.3 (± 1.4); life span in days for males and females, respectively). Injection of PBS had no significant effect on life span in either sex (P > 0.1; males: 23.1 (± 1.7) and females: 19.4 (± 1.5) days), showing that even though infection was not performed under sterile conditions, the wounding itself did not significantly affect individual life span. Flight treatment did not influence life span (t 1,171 = −1.12, P = 0.26; Fig. 2).
Figure 2.
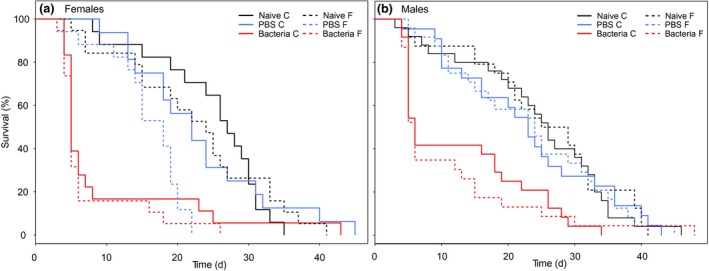
Survival of the adult (a) females and (b) males in days. Solid lines indicate individuals without flight treatment (C), whereas dashed lines indicate those with flight treatment (F) prior to injection. Naïve group is presented in black (n = 17 (C) and n = 19 (F) for females and n = 25 (C) and n = 24 (F) for males) in comparison with the group injected with 2 μL PBS in blue (n = 16 (C) and n = 17 (F) for females and n = 22 (C) and n = 24 (F) for males) and the group injected with 2 μL of a 5‐mg mL−1 Micrococcus luteus solution in PBS in red (n = 18 (C) and n = 19 (F) for females and n = 24 (C) and n = 23 (F) for males). Only bacterial injection resulted in a significant reduction of the life span for both sexes.
Discussion
In this study, we aimed to disentangle whether the commonly observed positive relationship between flight and immune response also previously found in our study species (Saastamoinen & Rantala, 2013; Kvist et al., 2015) is a by‐product of a general stress response or potentially reflects an adaptive response that evolved along with higher infection risk due to dispersal events. We found a significant increase in expression levels in four of the seven tested immune genes as well as reduced life span based on bacterial infection. Flight itself led to an increase in expression levels of some of the immune genes; however, it had no influence on individual survival upon infection. Furthermore, the up‐regulation of immune genes after flight treatment was far lower than that induced by infection with bacteria or wounding. Our findings therefore suggest that the increased immune gene expression upon flight is most likely due to a general stress response rather than an adaptive response to possible upcoming infections.
As we are unaware of any specific natural pathogen affecting the Glanville fritillary butterfly, we used a simple and rather general bacterial strain in our study. The used strain successfully infects our species and leads to reduced survival and increased immune gene expression in both sexes. Most strains of Micrococcus luteus are gram‐positive (Madigan et al., 2015) and hence should trigger the Toll pathway. The increased expression of pelle upon bacterial injection supports the activation of this pathway. As β‐1,3‐glucan is an antigen on the surface of gram‐positive bacteria, the increased expression levels for β‐1,3‐glucan recognition protein are likewise expected for this bacterial strain (Jiang et al. 2004). Also, peptidoglycan recognition proteins (PGRP) are known to be strongly up‐regulated upon bacterial infection in the fruit fly (Drosophila melanogaster; Zaidman‐Remy et al., 2011) and in the silkworm (Bombyx mori; Tanaka et al., 2008). In contrast, we did not observe a significant increase in expression of PGRP‐LC upon bacterial infection. PGRP‐LC is a receptor in the IMD pathway and therefore theoretically not affected by gram‐positive bacteria. However, this receptor detects peptidoglycan of bacteria, and generally, both pathways do interact in case of an infection (Lemaitre & Hoffmann, 2007). The investigated time frame of the expression levels may have been too short to detect a significant up‐regulation of PGRP‐LC. Lysozyme is an enzyme damaging the cell wall of both gram‐positive and gram‐negative bacteria and potentially activates the Toll pathway by the release of components of the bacterial cell wall (Dunn, 1986; Hultmark, 2003). We did not detect changes in expression of lysozyme activity induced by our treatments. There are several lysozyme genes in M. cinxia as in most organisms of which some are more specific to the bacterial cell wall. Potentially, the chosen gene in this study was not strongly affected by our immune treatment. In general, up‐regulation of lysozyme should last long as it has been even detected 120 h after bacterial infection in the mosquito Anopheles gambiae (Kajla et al. 2010), suggesting that the timing is not the issue. Prophenoloxidase represents the main regulator of the melanization pathway in insects (Cerenius & Söderhall, 2004). As for lysozyme activity, no changes in expression of proPO upon bacterial infection after 20 h were observed. As intermediate products of the melanin synthesis are toxic (Cerenius & Söderhall, 2004), melanization is a tightly regulated process in insects. The pathway needs to be suppressed quickly to avoid possible damage to own tissue such as cuticles and wings or in severe cases even the death of the individual (De Gregorio et al., 2002). Certain inhibitors exist to prevent extended expression of the melanization pathway. Serine protease inhibitors, also known as serpins, represent such molecules and are able to bind to the prophenoloxidase‐activating enzyme (PPAE) that normally converts inactive prophenoloxidase to the active form phenyloxidase (De Gregorio et al., 2002). The here tested serpin gene was significantly up‐regulated upon bacterial infection. Together with the observation that proPO did not show any increase in expression, we hypothesize that increased levels of serpin have led to the inactivation of PPAE to avoid potential tissue damage. Thus, an earlier time point likely would show an increase in proPO due to bacterial infection and potentially a slightly lower level of serpin.
The flight treatment itself resulted in higher expression levels for two of the tested immune genes, βGRP and PGRP‐LC. Somewhat surprisingly, expression of these genes was not elevated by flight in the previous study of the same species (Kvist et al., 2015). Consistent with the previous study, no increase in expression due to flight was found 20 h after the flight treatment for proPO and lysozyme. Most of the used genes in this study are known to be strongly up‐regulated due to infection. However, flight might not trigger such expression levels, as it reflects cellular stress and is therefore less specific. Interestingly, we found a significant interaction between flight and bacterial treatment influencing the expression levels of attacin and pelle. For both of these genes, flight did induce higher expression levels but only in naïve individuals. In individuals that were infected with bacteria, flight either caused no effect or even reduced the expression levels. This slight decrease in expression of attacin and pelle due to the flight treatment in infected individuals might indicate a resource trade‐off, and the infected individuals simply cannot further induce their immune gene expression when flying. Similarly, a trade‐off between flight and immunity has been observed, for example, in the migratory monarch butterfly (Danaus plexippus), where such a decrease in number of haemocytes was found in individuals that experienced tethered flight assay compared to a control group (Fritzsche McKay et al., 2016). The increase in the expression levels for attacin and pelle due to the flight treatment in the naïve individuals is in accordance with the previous study (Kvist et al., 2015). It was suggested that forced flight potentially shifted molecular resources (such as apolipophorin III) away from immune response towards flight‐related functions in case of an infection (Adamo et al., 2008). Notably, however, in the present study the expression level induced by flight in combination with infection is still higher than that of flight treatment on the naïve individual. We therefore hypothesize that the elevated immune gene expression upon flight in the Glanville fritillary butterfly is acting in maintaining the maximal immune defence while at the same time optimizing the individuals for a fight‐or‐flight response.
We only used one bacterial strain in the present study, and potentially, the results may be different if a different strain, for example more specific to Lepidoptera, would have been used. Similarly, injection of bacteria into the haemocoel is not a natural way of infection in the wild. However, butterflies do experience injuries, for example due to predators, which might allow pathogens to enter directly into the haemocoel and provoke a faster and more drastic immune response. Our injection treatment therefore mimics such an event in nature. To ensure that our effect was not just provoked by wounding alone, we included a wounding control in the experimental set‐up. Wounding led to an increased expression of two immune genes that were likewise triggered by the bacterial treatment (see Fig. 1), as indicated by the PBS treatment. However, the up‐regulation was substantially higher with bacterial infection compared to wounding only, suggesting that introduction of bacteria definitely provoked a stronger immune response and led to an infection. The additional life span data confirm that wounding itself might activate the immune response but does not significantly reduce life span, which occurred upon bacterial infection. Finally, the flight treatment used in this study does not reflect a natural dispersal event; however, experimentally induced flight can produce important knowledge on the costs and benefits of flight (Chapman et al., 2015). As gene expression levels change readily, we treated individuals equally in terms of handling, acclimation and feeding across all treatments.
Although we do not provide a complete picture of the effects of infection and flight on immunity, our study provides interesting new insights in trying to understand why different organisms in some cases up‐regulate their immune system upon or during flight or dispersal. Even though previous studies have shown that immunity is often up‐regulated during flight, these studies have not looked at whether that up‐regulation actually benefits individuals in case of infection. We found no indication that the up‐regulation of immunity genes due to flight would help individuals to recover or survive from infection. On the contrary, we detect a potential trade‐off between flight and immunity for two genes, attacin and pelle. Applying our findings to other systems, especially those with known natural pathogens, would be highly interesting. Here, the monarch butterfly (D. plexippus) and the protozoan Ophryocystis elektroscirrha are interesting candidates. Further studies in the Glanville fritillary butterfly, especially within the metapopulation framework, would be useful. For example, comparing the relationship between dispersal and immunity among individuals from old and newly established populations or from continuous vs. fragmented landscapes that are known to differ in dispersal ability (Saastamoinen, 2007; Somervuo et al., 2014) would be highly relevant. Similarly, further experiments investigating the immune response in combination with disease models assessing epidemiology in this metapopulation could bring new insights into disease dynamics in the wild.
Ethics statement
The Glanville fritillary butterfly is not classified as an endangered or protected species. No permits are required for the collection of individuals in the Åland Islands.
Funding
This study was funded by grants from the European Research Council (Independent Starting grant META‐STRESS; 637412) and the Academy of Finland (Decision numbers 273098 and 265641) to MS.
Competing interest
The authors declare that they have no competing interests.
Authors’ contribution
LW, JK and MS designed the study; LW performed the experiment; LW analysed the data; and LW, JK and MS wrote the manuscript.
Supporting information
Appendix S1 Primer sequences.
Appendix S2 Raw C t values.
Appendix S3 Results of all initial models used in the gene expression analysis.
Appendix S4 Supplementary results for the general model analysis for the gene expression and combined sex analysis for life span.
Appendix S5 Supplementary figures to visualize interactions for the gene expression.
Acknowledgment
We acknowledge Suvi Ikonen and Kati Schenk for their help with the experiment, Mikko Frilander for helpful discussions, Kristjan Niitepõld for comments on the manuscript and Toshka Nyman for RNA extractions.
Data deposited at Dryad: doi: 10.5061/dryad.38c5g
References
- Adamo, S.A. 2008. Norepinephrine and octopamine: linking stress and immune function across phyla. Invertebrate Surviv. J. 5: 12–19. [Google Scholar]
- Adamo, S.A. 2010. Why should an immune response activate the stress response? Insights from the insects (the cricket Gryllus texensis). Brain Behav. Immun. 24: 194–200. [DOI] [PubMed] [Google Scholar]
- Adamo, S.A. 2012. The effects of the stress response on immune function in invertebrates: an evolutionary perspective on an ancient connection. Horm. Behav. 62: 324–330. [DOI] [PubMed] [Google Scholar]
- Adamo, S.A. , Roberts, J.L. , Easy, R.H. & Ross, N.W. 2008. Competition between immune function and lipid transport for the protein apolipophorin III leads to stress‐induced immunosuppression in crickets. J. Exp. Biol. 211: 531–538. [DOI] [PubMed] [Google Scholar]
- Baines, D. , DeSantis, T. & Downer, R.G.H. 1992. Octopamine and 5‐hydroxytryptamine enhance the phagocytic and nodule formation activities of cockroach (Periplaneta americana) haemocytes. J. Insect Physiol. 38: 905–914. [Google Scholar]
- Bates, D. , Maechler, M. , Bolker, B.M. & Walker, S. 2015. Fitting linear mixed‐effects models using lme4. J. Stat. Softw. arXiv:1406.5823 [Google Scholar]
- Boggs, C.L. & Nieminen, M. 2004. Checkerspot Reproductive Biology. Oxford University Press, Oxford. [Google Scholar]
- Bonte, D. , Van Dyck, H. , Bullock, J.M. , Coulon, A. , Delgado, M. , Gibbs, M. , et al. 2012. Costs of dispersal. Biol. Rev. Camb. Philos. Soc. 87: 290–312. [DOI] [PubMed] [Google Scholar]
- Cerenius, L. & Söderhall, K. 2004. The prophenoloxidase‐activating system in invertebrates. Immunol. Rev. 198: 116–126. [DOI] [PubMed] [Google Scholar]
- Chapman, J.W. , Reynolds, D.R. & Wilson, K. 2015. Long‐range seasonal migration in insects: mechanisms, evolutionary drivers and ecological consequences. Ecol. Lett. 18: 287–302. [DOI] [PubMed] [Google Scholar]
- Clobert, J. , Baguette, M. , Benton, T.G. & Bullock, J.M. 2012. Dispersal Ecology and Evolution. Oxford University Press, Oxford, UK. [Google Scholar]
- De Gregorio, E. , Han, S.J. , Lee, W.‐J. , Baek, M.‐J. , Osaki, T. , Kawabata, S.‐I. , et al. 2002. An immune‐responsive serpin regulates the melanization cascade in drosophila. Dev. Cell 3: 581–592. [DOI] [PubMed] [Google Scholar]
- Decaestecker, E. , Gaba, S. , Raeymaekers, J.A. , Stoks, R. , Van Kerckhoven, L. , Ebert, D. & De Meester, L. 2007. Host‐parasite ‘Red Queen’ dynamics archived in pond sediment. Nature 450: 870–873. [DOI] [PubMed] [Google Scholar]
- Dunn, P.E. 1986. Biochemical aspects of insect immunity. Annu. Rev. Entomol. 31: 321–339. [Google Scholar]
- Fritzsche McKay, A. , Ezenwa, V.O. & Altizer, S. 2016. Unravelling the costs of flight for immune defenses in the migratory monarch butterfly. Integr. Comp. Biol. 56: 278–289. [DOI] [PubMed] [Google Scholar]
- Goldsworthy, G. , Opoku‐Ware, K. & Mullen, L. 2002. Adipokinetic hormone enhances laminarin and bacterial lipopolysaccharide‐induced activation of the prophenoloxidase cascade in the African migratory locust, Locusta migratoria. J. Insect Physiol. 48: 601–608. [DOI] [PubMed] [Google Scholar]
- Hanski, I. 1999a. Habitat connectivity, habitat continuity, and metapopulations in dynamic landscapes. Oikos 87: 209–219. [Google Scholar]
- Hanski, I. 1999b. Metapopulation Ecology. Oxford University Press, New York. [Google Scholar]
- Hoffmann, J.A. 2003. The immune response of Drosophila. Nature 426: 33–38. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Wu, S.F. , Li, X.H. , Adamo, S.A. & Ye, G.Y. 2012. The characterization of a concentration‐sensitive alpha‐adrenergic‐like octopamine receptor found on insect immune cells and its possible role in mediating stress hormone effects on immune function. Brain Behav. Immun. 26: 942–950. [DOI] [PubMed] [Google Scholar]
- Hultmark, D. 2003. Drosophila immunity: paths and patterns. Curr. Opin. Immunol. 15: 12–19. [DOI] [PubMed] [Google Scholar]
- Jiang, H. , Ma, C. , Lu, Z.‐Q. & Kanost, M.R. 2004. β‐1,3‐Glucan recognition protein‐2 (βGRP‐2) from Manduca sexta: an acute‐phase protein that binds β‐1,3‐glucan and lipoteichoic acid to aggregate fungi and bacteria and stimulate prophenoloxidase activation. Insect Biochem. Mol. Biol. 34: 89–100. [DOI] [PubMed] [Google Scholar]
- Kajla, M.K. , Andreeva, O. , Gilbreath, T.M. & Paskewitz, S.M. 2010. Characterization of expression, activity and role in antibacterial immunity of Anopheles gambiae lysozyme c‐1. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G.S. , Nalini, M. , Kim, Y. & Lee, D.W. 2009. Octopamine and 5‐hydroxytryptamine mediate hemocytic phagocytosis and nodule formation via eicosanoids in the beet armyworm, Spodoptera exigua . Arch. Insect Biochem. Physiol. 70: 162–176. [DOI] [PubMed] [Google Scholar]
- Kim, H.J. , Je, H.J. , Park, S.Y. , Lee, I.H. , Jin, B.R. , Yun, H.K. , et al. 2004. Immune activation of apolipophorin‐III and its distribution in hemocyte from Hyphantria cunea . Insect Biochem. Mol. Biol. 34: 1011–1023. [DOI] [PubMed] [Google Scholar]
- Klemola, N. , Klemola, T. , Rantala, M.J. & Ruuhola, T. 2007. Natural host‐plant quality affects immune defence of an insect herbivore. Entomol. Exp. Appl. 123: 167–176. [Google Scholar]
- Kuussaari, M. , Nieminen, M. & Hanski, I. 1996. An experimental study of migration in the glanville fritillary butterfly Melitaea cinxia . J. Anim. Ecol. 65: 791–801. [Google Scholar]
- Kvist, J. , Mattila, A.L. , Somervuo, P. , Ahola, V. , Koskinen, P. , Paulin, L. , et al. 2015. Flight‐induced changes in gene expression in the Glanville fritillary butterfly. Mol. Ecol. 24: 4886–4900. [DOI] [PubMed] [Google Scholar]
- Köning, C. & Schmid‐Hempel, P. 1995. Foraging activity and immunocompetence in workers of the bumble bee, Bombus terrestris L. Proc. Biol. Sci. 260: 225–227. [Google Scholar]
- Lefcheck, J.S. & Freckleton, R. 2016. piecewiseSEM: piecewise structural equation modelling in r for ecology, evolution, and systematics. Methods Ecol. Evol. 7: 573–579. [Google Scholar]
- Lemaitre, B. & Hoffmann, J. 2007. The host defense of Drosophila melanogaster . Annu. Rev. Immunol. 25: 697–743. [DOI] [PubMed] [Google Scholar]
- Ma, G. , Hay, D. , Li, D. , Asgari, S. & Schmidt, O. 2006. Recognition and inactivation of LPS by lipophorin particles. Dev. Comp. Immunol. 30: 619–626. [DOI] [PubMed] [Google Scholar]
- Madigan, M.T. , Martinko, J.M. , Bender, K.S. , Buckley, D.H. & Stahl, D.A. 2015. Brock Biology of Microorganisms, 14th edn Pearson, Boston. [Google Scholar]
- Matthysen, E. 2012. Multicausality of Dispersal: A Review. Oxford University Press, Oxford, UK. [Google Scholar]
- Mone, Y. , Gourbal, B. , Duval, D. , Du Pasquier, L. , Kieffer‐Jaquinod, S. & Mitta, G. 2010. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PLoS Negl. Trop Dis. 4: e813. doi: 10.1371/journal.pntd.0000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowlds, P. , Barron, A. & Kavanagh, K. 2008. Physical stress primes the immune response of Galleria mellonella larvae to infection by Candida albicans . Microb. Infect. 10: 628–634. [DOI] [PubMed] [Google Scholar]
- Niitepõld, K. , Mattila, A.L.K. , Harrison, P.J. & Hanski, I. 2011. Flight metabolic rate has contrasting effects on dispersal in the two sexes of the Glanville fritillary butterfly. Oecologia 165: 847–854. [DOI] [PubMed] [Google Scholar]
- Niitepõld, K. , Smith, A.D. , Osborne, J.L. , Reynolds, D.R. , Carreck, N.L. , Martin, A.P. , et al. 2009. Flight metabolic rate and Pgi genotype influence butterfly dispersal rate in the field. Ecology 90: 2223–2232. [DOI] [PubMed] [Google Scholar]
- Orchard, I. , Ramirez, J.‐M. & Lange, A.‐B. 1993. A multifunctional role for octopamine in locust flight. Annu. Rev. Entomol. 38: 227–249. [Google Scholar]
- Rozen, S. & Skaletsky, H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Saastamoinen, M. 2007. Mobility and lifetime fecundity in new versus old populations of the Glanville fritillary butterfly. Oecologia 153: 569–578. [DOI] [PubMed] [Google Scholar]
- Saastamoinen, M. & Rantala, M.J. 2013. Influence of developmental conditions on immune function and dispersal‐related traits in the Glanville fritillary (Melitaea cinxia) butterfly. PLoS One 8: e81289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen, A. , Huuskonen, J. , Ojala, J. , Kauppinen, A. , Kaarniranta, K. & Suuronen, T. 2008. Activation of innate immunity system during aging: NF‐kB signaling is the molecular culprit of inflamm‐aging. Ageing Res. Rev. 7: 83–105. [DOI] [PubMed] [Google Scholar]
- Snoeijs, T. , Van de Casteele, T. , Adriaensen, F. , Matthysen, E. & Eens, M. 2004. A strong association between immune responsiveness and natal dispersal in a songbird. Proc. Biol. Sci. 271(Suppl 4): S199–S201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervuo, P. , Kvist, J. , Ikonen, S. , Auvinen, P. , Paulin, L. , Koskinen, P. , et al. 2014. Transcriptome analysis reveals signature of adaptation to landscape fragmentation. PLoS One 9: e101467. doi: 10.1371/journal.pone.0101467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srygley, R.B. & Lorch, P.D. 2011. Weakness in the band: nutrient‐mediated trade‐offs between migration and immunity of Mormon crickets, Anabrus simplex . Anim. Behav. 81: 395–400. [Google Scholar]
- Suhonen, J. , Honkavaara, J. & Rantala, M.J. 2010. Activation of the immune system promotes insect dispersal in the wild. Oecologia 162: 541–547. [DOI] [PubMed] [Google Scholar]
- Tanaka, H. , Ishibashi, J. , Fujita, K. , Nakajima, Y. , Sagisaka, A. , Tomimoto, K. , et al. 2008. A genome‐wide analysis of genes and gene families involved in innate immunity of Bombyx mori . Insect Biochem. Mol. Biol. 38: 1087–1110. [DOI] [PubMed] [Google Scholar]
- Valtonen, T.M. , Kleino, A. , Rämet, M. & Rantala, M.J. 2009. Starvation reveals maintenance cost of humoral immunity. Evol. Biol. 37: 49–57. [Google Scholar]
- Van Nouhuys, S. & Hanski, I. 2002. Colonization rates and distances of a host butterfly and two specific parasitoids in a fragmented landscape. J. Anim. Ecol. 71: 639–650. [Google Scholar]
- Venables, W.N. & Ripley, B.D. 2002. Modern Applied Statistics with S, 4th edn Springer, New York. [Google Scholar]
- Yoshida, H. , Kinoshita, K. & Ashida, M. 1996. Purification of a peptidoglycan recognition protein from haemolymph from the silkworm Bombyx mori . J. Biol. Chem. 271: 13854–13860. [DOI] [PubMed] [Google Scholar]
- Zaidman‐Remy, A. , Poidevin, M. , Herve, M. , Welchman, D.P. , Paredes, J.C. , Fahlander, C. , et al. 2011. Drosophila immunity: analysis of PGRP‐SB1 expression, enzymatic activity and function. PLoS One 6: e17231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk, M. & Stoehr, A.M. 2002. Immune defense and host life history. Am. Nat. 160: S9–S22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Primer sequences.
Appendix S2 Raw C t values.
Appendix S3 Results of all initial models used in the gene expression analysis.
Appendix S4 Supplementary results for the general model analysis for the gene expression and combined sex analysis for life span.
Appendix S5 Supplementary figures to visualize interactions for the gene expression.


