Abstract
Photosynthesis in some phototrophic bacteria requires the PufX component of the reaction centre–light‐harvesting 1–PufX (RC‐LH1‐PufX) complex, which creates a pore for quinone/quinol (Q/QH 2) exchange across the LH1 barrier surrounding the RC. However, photosynthetic bacteria such as Thermochromatium (T.) tepidum do not require PufX because there are fewer carotenoid binding sites, which creates multiple pores in the LH1 ring for Q/QH 2 exchange. We show that an αTrp‐24→Phe alteration of the Rhodobacter (Rba.) sphaeroides LH1 antenna impairs carotenoid binding and allows photosynthetic growth in the absence of PufX. We propose that acquisition of PufX and confining Q/QH 2 traffic to a pore adjacent to the RC QB site is an evolutionary upgrade that allows increased LH1 carotenoid content for enhanced light absorption and photoprotection.
Keywords: bacterial photosynthesis; carotenoid; light‐harvesting; membrane protein; quinones, reaction centre
Abbreviations
BChl(s), bacteriochlorophyll(s)
Blc, Blastochloris
Crt, carotenoid
cytbc 1, cytochrome bc 1 complex
LH2, light‐harvesting LH2 complex
OD, optical density
Q/QH2, quinols/quinones
Rba., Rhodobacter
RC, reaction centre
RC‐LH1, reaction centre–light‐harvesting 1 complex
Rps., Rhodopseudomonas
Rsp., Rhodospirillum
T., Thermochromatium
WT, wild‐type
β‐DDM, β‐dodecylmaltoglucoside
The reaction centre–light‐harvesting 1 (RC‐LH1) complexes of purple phototrophic bacteria collect energy from sunlight, and in many bacteria, they also receive energy from LH2 antenna complexes, thereby increasing the light‐absorbing capacity of the photosystem. The RC traps this energy by initiating a series of electron transfers that eventually produce a quinol molecule, destined to leave the RC‐LH1 complex and diffuse to the cytochrome bc 1 (cytbc 1) complex 1. Here, the original excitation energy is converted to a proton motive force. In some bacteria such as Rhodospirillum (Rsp.) rubrum and Thermochromatium (T.) tepidum, the LH1 antenna ring completely surrounds the reaction centre 2, 3, 4, but in others, such as Rhodobacter (Rba.) capsulatus, Rba. sphaeroides and Rhodopseudomonas (Rps.) palustris, the LH1 ring is interrupted by a PufX/PufW polypeptide 5, 6, 7. The structure of the dimeric Rba. sphaeroides RC‐LH1‐PufX complex 8 shows that PufX opens a small channel in the LH1 ring that allows quinols/quinones (Q/QH2) to cross the LH1 barrier and gain access to the RC QB site 8. This structure accounts for the abolished photosynthetic growth of PufX‐minus mutants 9, 10, 11, 12, 13, 14, as it appears that the space in the LH1 ring occupied by PufX is filled by two extra LH1 subunits that block access to the RC QB site 15. However, many bacteria do not have a PufX yet they still photosynthesise; the recent structure of the T. tepidum RC‐LH1 complex shows how this is possible, and inspection of the LH1 ring structure revealed a series of small pores that were suggested to allow Q/QH2 exchange in the absence of PufX 4; a similar situation could apply to Rsp. rubrum, as proposed earlier 16.
If, in principle, Q/QH2 can cross the LH1 barrier directly in T. tepidum and Rsp. rubrum, why do bacteria such as Rba. sphaeroides need to incorporate PufX into their LH1 rings to create a quinone channel? The explanation is proposed to originate in the carotenoid (Crt) contents of bacteria with and without PufX; for example, the T. tepidum and Rsp. rubrum LH1 complexes have a carotenoid: bacteriochlorophyll (BChl) ratio of 1 : 2 4, 17 and the LH1 complex is composed of α1β1BChl2Crt1 units, whereas this ratio is 1 : 1 for the Rba. sphaeroides LH1 18 and the LH1 complex is assembled from α1β1BChl2Crt2 units 8. We suggest that these ‘extra’ LH1 carotenoids confer significant advantages in terms of light harvesting and photoprotection and, although they would likely block the quinone pores identified in the ‘low‐carotenoid’ T. tepidum LH1 complex 4, PufX alleviates this potential problem by preventing LH1 from fully encircling the RC. Thus, the pore created by PufX allows Q/QH2 exchange 8, and the efficiency and robustness of the antenna are increased by increasing the carotenoid content.
In order to test this proposition, we constructed a genomic point mutation that lowers the carotenoid content of LH1, and combined it with genomic deletion of the pufX gene. The PufX− strain DM1R, which harbours the LH1 αW‐24F mutation that lowers LH1 carotenoid content, grows photosynthetically, whereas the PufX− mutant control with normal LH1 carotenoids cannot do so. We suggest that partial loss of carotenoid from LH1 α1β1BChl2Crt units encircling the RC in the DM1R mutant opens some pores in LH1 for quinone exchange, mimicking the ‘low‐carotenoid’ LH1 complexes of Rsp. rubrum and T. tepidum to a limited extent, and enabling this strain to tolerate the loss of PufX.
Materials and methods
Rba. sphaeroides strains
Genomic mutations were constructed in various combinations in the Rba. sphaeroides 2.4.1 WT using the suicide vector pk18mobsacB 19 as described 20, 21. These mutations were as follows: (a) deletion of the pucBA1 and pucBA2 genes encoding polypeptides of the LH2 complex, (b) deletion of the pufX gene and (c) site‐specific mutation of pufA to convert αTrp‐24→Phe (the numbering of LH1 residues relates to the conserved His0 residue that binds bacteriochlorophyll). Separately, we constructed a control strain DX13 harbouring a R49LR53L variant of PufX, which yields monomeric PufX‐containing RC‐LH1 complexes (Qian et al., in preparation; Table 1).
Table 1.
Rba. sphaeroides strains used in this study
| Strain | Description | Phenotype | Reference |
|---|---|---|---|
| DO10 | Genomic deletion of pucBA1, pucBA2, pufX | LH2−, PufX− | This work |
| DM1R | Genomic deletion of pucBA1, pucBA2, pufX. Mutation in pufA giving W‐24F | LH2−, PufX−, LH1αW‐24F | This work |
| DX13 | Genomic deletion of pucBA1, pucBA2. Mutation in pufX giving R49L, R53L. | LH2−, monomeric RC‐LH1‐PufX complexes | Qian et al., in preparation |
| RCO1 | Strain DD13 (pucBA1::Sm R ; pufBALM::Kan R ) complemented in trans with pRKEH10D (pufLMX) | RC‐only | Jones et al. 1992 24 |
Photosynthetic and semi‐aerobic growth of Rba. sphaeroides strains
The cells were grown in M22+ medium supplemented with vitamins and 0.1% casamino acids. The cells were grown either semi‐aerobically in the dark, as 1.5 L cultures in 2‐L conical flasks in a shaking incubator at 34 °C, or photosynthetically at room temperature in stirred 1‐L Roux bottles at ~ 50 μE light intensity.
The photosynthetic growth curves were recorded for cultures in 15‐mL flat‐bottomed screw cap tubes sealed with Parafilm and filled to capacity, and with a micro stir bar. The cultures were illuminated by halogen light bulbs to give a minimum light intensity of 50 μE. Growth was monitored with a WPA Colourwave CO7000 Medical Colorimeter equipped with a 680‐nm filter. Four separate cultures of each strain were grown simultaneously and their optical density values averaged for each time point. Each inoculum, using a semi‐aerobically grown culture of each strain, was adjusted so that each culture started with the same number of cells.
Pigment extraction
Cells from 1 mL of an 80 mL semi‐aerobic culture of the relevant strains were pelleted in a microcentrifuge and extracted twice into 0.5 mL of acetone : methanol (7 : 2 v/v). Absorption spectra of pooled 1 mL samples of extracts were recorded using a Cary 60 spectrometer. The peak heights at 478 nm (carotenoids) and 770 nm (BChls) were measured; three replicates of three separate semi‐aerobic cultures were measured to obtain the final averaged data. All procedures were conducted under dim light conditions.
Membrane preparation and low‐temperature spectroscopy
Membranes were prepared as described in Olsen et al. 22 and kept at −20 °C prior to use. Low‐temperature (80K) absorbance spectra were measured using a DN10 Cryostat (Oxford Instruments, Abingdon, UK) mounted in a Cary 60 spectrophotometer (Agilent, Stockport, UK).
Fractionation of photosynthetic complexes on sucrose density gradients
Membranes were fractionated as described in 21. Samples were solubilised in 3% β‐DDM, fractionated on a discontinuous sucrose gradient containing 20, 21.25, 22.5, 23.75 and 25% sucrose in 20 mm HEPES and 0.03% β‐DDM, then centrifuged in a Beckman SW41 Ti rotor at 27 000 rpm (90 000 g) for 40 h.
Results
Photosynthetic growth rates of DM1R, and PufX+ and PufX− control strains
In order to lower the carotenoid content of the Rba. sphaeroides LH1 complex, thereby producing at least a few of the pores observed in the low‐carotenoid T. tepidum RC‐LH1 complex 4, the LH2−, PufX− strain DM1R was constructed in which αTrp‐24 at the N terminus of the LH1α polypeptide was changed to Phe (LH1αW‐24F). This mutation is similar to the αTrp‐24 to Tyr alteration in Rba. capsulatus that lowered the carotenoid content of LH1 by 60% 23. In order to simplify spectral analyses, all strains harboured genomic deletions of pucBA1, pucBA2 that remove LH2 complexes.
DM1R was tested for photosynthetic growth against the two LH2− control strains: DO10, which is the same as DM1R, that is PufX− but with normal LH1 complexes, and DX13, which is PufX+ but with monomeric RC‐LH1‐PufX core complexes to match the monomeric RC‐LH1 core complexes in DM1R and DO10. Absorption of whole cells (not shown) showed that DM1R has 61% of the LH1 content of DO10, which likely arises from some impairment of LH1 assembly by the LH1αW‐24F alteration. DM1R grew more slowly than the positive control PufX+ strain DX13, likely because DX13 has an optimised quinone channel created by PufX, but significantly faster than the PufX− LH2− control strain DO10, which was photosynthetically inactive as expected (Fig. 1A). Equally inactive is the RC‐only (LH2‐minus, LH1‐minus) strain RCO1 24, which shows no photosynthetic growth within the same 24‐h period used in Fig. 1, and at the same 50 μE light intensity (results not shown).
Figure 1.
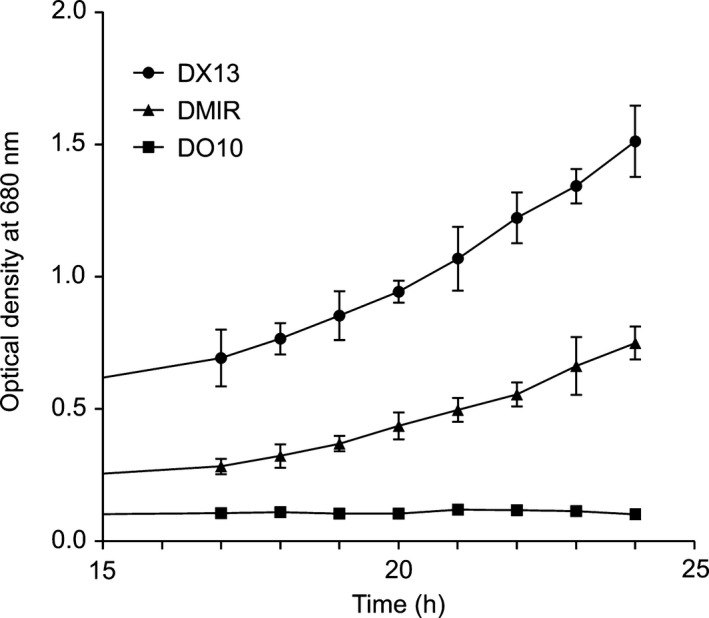
Photosynthetic growth curves of DM1R, DO10 and DX13 strains, with four biological replicates for each time point. DX13 is the positive control PufX+ strain; DM1R is PufX−, LH1αW‐24F; DO10 is a PufX− negative control.
Biochemical and spectroscopic analyses of the DM1R and DO10 strains
The ability of DM1R to grow photosynthetically could be explained if the LH1αW‐24F alteration destabilises the formation of a complete LH1 ring; then, missing LH1 α1β1 units could create gaps for Q/QH2 exchange. In order to investigate this point, we performed biochemical and spectroscopic analyses of the PufX− DM1R and DO10 strains. The low‐temperature (77K) absorption spectra in Fig. 2A were normalised for LH1 absorption, to emphasise the equivalence of the core complexes in terms of LH1 : RC stoichiometry (see below), and to reflect the presence of some RCs with no LH1, which appear in fractions B1 and B2 (Fig. 2B,C).
Figure 2.
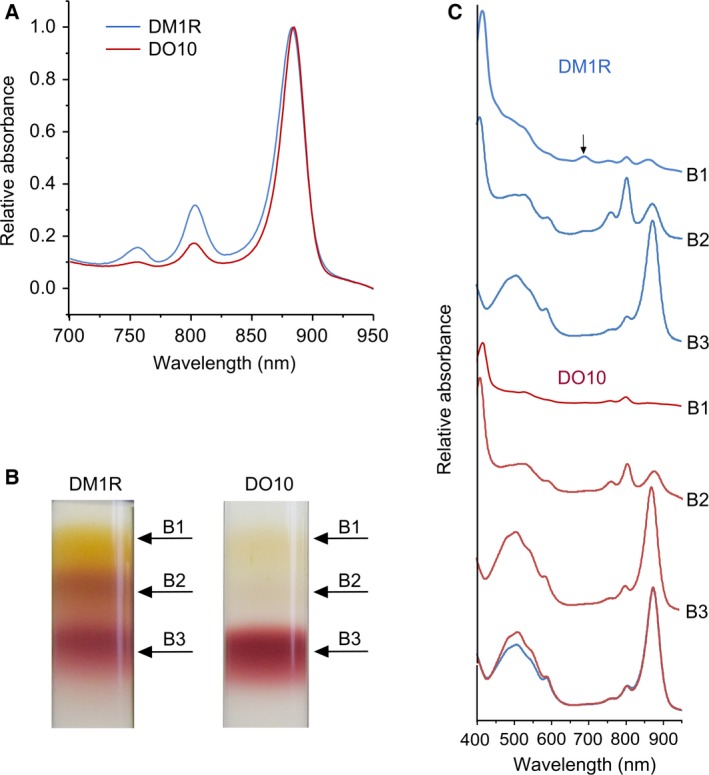
(A) Low‐temperature absorbance spectra of DM1R and DO10 membrane samples recorded at 77K, normalised at the LH1 absorption maximum near 875 nm. (B) Fractionation of detergent‐solubilised intracytoplasmic membranes from DM1R and DO10 strains on sucrose density gradients. (C) Room temperature absorbance spectra of the B1, B2 and B3 fractions from the gradients in panel B. The arrow in the DM1R B1 fraction indicates absorption at ~ 680 nm; overlaid spectra (bottom) compare the two B3 fractions.
To analyse differences between DM1R and DO10 strains in more detail, comparable amounts of DM1R and the DO10 control membranes, in terms of RC content, were solubilised with β‐dodecylmaltoglucoside (β‐DDM) and then fractionated by centrifugation through sucrose density step gradients. Figure 2B shows that β‐DDM releases some ‘free’ RCs from DM1R membranes, as shown by the distinctive absorption spectrum of these RC complexes (Fig. 2C,B1,B2, blue). The equivalent fractions from the DO10 control, barely visible in the images of the gradients (Fig. 2B), have a similar absorption spectrum (Fig. 2C,B1,B2, red). These spectra show that the limited impairment of LH1 assembly by the LH1αW‐24F alteration in DM1R has left some RCs with no encircling LH1 antenna. Another likely consequence of lessened LH1 assembly is seen in the B1 fractions; some RCs are present in each case, but relative to DO10 the B1 fraction in DM1R has more unattached carotenoid and also a small absorption peak at ~ 680 nm (Fig. 2C, arrowed), possibly from an intermediate of BChl biosynthesis.
If the LH1 ring was interrupted in DM1R, due to incomplete assembly of LH1 α1β1 subunits round the RC, there would be space for Q/QH2 exchange despite the loss of PufX and this would explain the photosynthetic growth of DM1R in Fig. 1A. However, the cores that do assemble in DM1R have a normal LH1 : RC stoichiometry, as shown by the close correspondence between the overlaid spectra of the B3 fractions for DM1R and the DO10 positive control (Fig. 2C, bottom). These two spectra have almost identical 874 : 805 nm (LH1 : RC) absorption ratios so these two PufX− mutants, DM1R and DO10, have a complete ring of LH1 subunits encircling the RC. The only difference between the RC‐LH1 complexes of DM1R and DO10 is seen in the 450‐ to 550‐nm carotenoid region where there is clear evidence for lowered carotenoid content, 14% lower when measured at 508 nm, as a consequence of the W‐24F alteration of LH1 in DM1R.
In summary, the lowered carotenoid content of the LH1 antenna (Fig. 2C, bottom spectra), a consequence of the LH1αW‐24F mutation, is the likely explanation for the acquisition of photosynthetic growth in the absence of PufX. The presence of LH1‐free RCs cannot account for the photosynthetic growth of the PufX− strain DM1R because the antenna‐free, RC‐only strain RCO1 is photosynthetically inactive under the same light conditions (results not shown).
The DM1R mutant has a lowered carotenoid content relative to LH1 BChl pigments
The appearance of more carotenoid in the B1 fraction in the DM1R gradient (Fig. 2B,C) shows that some of this pigment, destined for binding to LH1, is unattached to any complex. The presence of some ‘free’ carotenoid in the B1 fraction of the DO10 control shows that carotenoid biosynthesis is not perfectly coupled to complex assembly. In order to verify the loss of some carotenoid from the LH1 of the DM1R mutant (Fig. 2C), DM1R (LH1αW‐24F, PufX−) and the DO10 (WT LH1, PufX−) control were grown semi‐aerobically and the carotenoid and bacteriochlorophyll (BChl) pigments were extracted from whole cells with acetone/methanol. The 478 : 770 nm absorption ratios for three biological replicates of each strain were 1.38 ± 0.12 and 1.80 ± 0.08, respectively, so the carotenoid content of the DM1R strain is 23% lower with respect to DO10, on the basis of equal BChl content. The lower LH1 content per cell in DM1R, which is 61% of that in DO10, is taken into account by comparing 478 : 770 nm absorption ratios for pigments extracted from these two strains. Full occupancy of pigment binding sites in the RC‐LH1 complex of DO10 equates to 33 Crt (32 LH1 Crt and 1 RC Crt) and 36 BChl (32 LH1 BChl and 4 RC BChl). The appreciable population of LH1‐free RCs in DM1R, each with four BChl and one Crt, skews the overall BChl : Crt ratio towards a higher number. Conversely, the presence of more ‘free’ carotenoids in DM1R (Fig. 2B,C), which are included in the extractions of whole cells, leads to an overestimation of LH1‐bound carotenoids in DM1R. Overall, we conclude that the W‐24F alteration of LH1 in DM1R has impaired the binding of carotenoids, as also found in the earlier study 23 on a mutant of Rba. capsulatus harbouring an LH1αW‐24Y alteration.
Discussion
It has long been known that PufX− mutants of Rba. sphaeroides are photosynthetically inactive 9, 11, 12, 13, 14, 25 because they assemble an LH1 ring that completely encloses the RC 25, 26. This observation has been difficult to reconcile with bacteria with similarly closed LH1 antenna rings such as Rsp. rubrum, T. tepidum and B. viridis 2, 4, 27, 28, which do grow photosynthetically. Aird et al. 16 conducted a steered molecular dynamics simulation study of the LH1 complex of R. rubrum and proposed that the LH1 ring could allow quinones to move across the complex. Subsequently, the 3D structure of T. tepidum 4 provided a structural basis for this proposal by identifying pores in the LH1 ring for Q/QH2 exchange. In the present work, we set out to engineer a few such pores in the LH1 ring of Rba. sphaeroides, having first deleted the pufX gene in order to close the normal pore created by the PufX polypeptide 8. We were guided by the earlier mutagenesis work of Babst et al. 23, who showed that the LH1αW‐24Y mutation results in the loss of 40% of the carotenoids from the LH1 complex of Rba. capsulatus. Here, we show that alteration of LH1αW‐24 to F in the DM1R mutant, which lowers the cellular carotenoid content and also the level of carotenoids bound by the DM1R LH1 complex, restores some photosynthetic growth to a PufX− LH2− strain of Rba. sphaeroides.
The structural basis for lowered carotenoid levels in the LH1αW‐24F complex has not been established, as carotenoid binding sites are poorly defined in the Rba. sphaeroides RC‐LH1‐PufX complex. This mutation also affects the assembly of the LH1 complexes to a limited degree, which results in some ‘free’ RCs in the membrane with no surrounding LH1 ring; similar effects were observed in previous studies of LH1 assembly mutants of Rba. sphaeroides 21, 29. The PufX‐minus RC‐LH1 complexes in the DM1R mutant assemble closed LH1 α16β16 rings because co‐operative forces between the RC and LH1 α1β1BChl2 subunits drive the process of encircling RCs with LH1 α1β1BChl2Crt2 units to completion, at the expense of some ‘free’ RCs that have no LH1 subunits. This LH1 ring completion process is observed even under quite extreme circumstances; for example, only ~ 14% of the LH1α polypeptide survives deletion of the LH1 assembly factor LhaA yet complexes still assemble with a normal RC : LH1 stoichiometry, leaving a substantial population of ‘free’ RCs 21. In summary, we propose that the LH1 ring of the PufX− LH1αW‐24F mutant DM1R completely surrounds the RC, on the basis of the near‐identical RC : LH1 absorption ratios for DM1R and the control strain DO10 (Fig. 2C, bottom). The LH1‐free RCs in DM1R require no PufX to facilitate Q/QH2 turnover at the RC QB site, but they are unable to function at the low light intensities used because they have no antenna to collect and deliver excitation energy; this is why the control RC‐only strain RCO1 shows negligible photosynthetic growth. Thus, the observed growth rate of DM1R likely arises from the lowered carotenoid content of the RC‐LH1 complexes and the ensuing pores for Q/QH2 traffic, which are depicted in Fig. 3.
Figure 3.
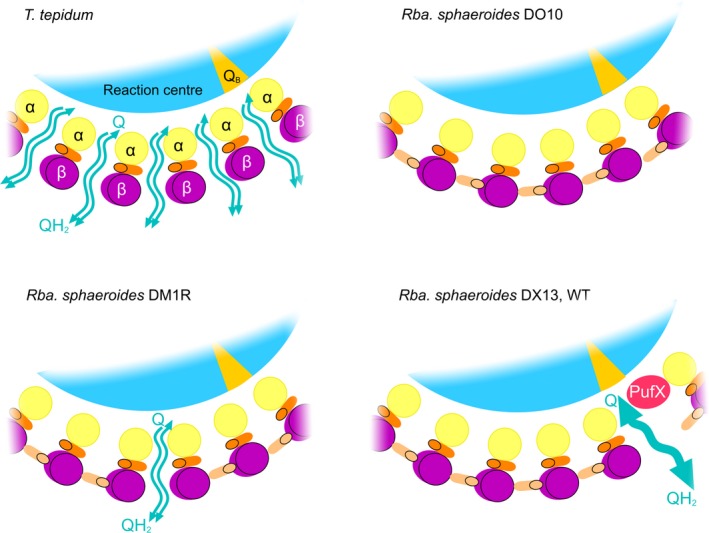
Schematic representation of part of the RC‐LH1 complex of T. tepidum, Rba. sphaeroides control strains DO10 and DM13, as well as the lowered LH1 carotenoid strain DM1R. The schematic depicts a section through the LH1 ring at the level of putative quinone pores between adjacent LH1 α1β1 BChl2 units. Carotenoids are depicted as foreshortened cylinders, viewed end‐on. For simplicity, BChls are not shown. In T. tepidum, only one of the two carotenoid binding sites is occupied for each α1β1 BChl2 LH1 unit; we propose that in Rba. sphaeroides the carotenoid (shown in dark orange) between the α and β LH1 apoproteins corresponds to the binding site seen in the T. tepidum structure. In Rba. sphaeroides strain DO10 (WT LH1, PufX−) full occupancy of carotenoid binding sites in LH1 prevents formation of T. tepidum‐like pores. Lowering the LH1 carotenoid content in strain DM1R impairs full occupancy of the carotenoid binding sites and passage of quinones through one or more pores is sufficient to permit some photosynthetic growth; 86% of the LH1 carotenoids are retained in DM1R with respect to DO10, so the assumption is made that most carotenoid binding sites are not affected by the alteration of LH1αW‐24 to F in the DM1R mutant. In the DX13 and WT strains, the PufX polypeptide creates a quinone pore, which allows the assembly of 14 α1β1 BChl2Crt2 units round the RC.
Phototrophic bacteria such as T. tepidum and Rsp. rubrum possess monomeric LH1 complexes that completely encircle the RC but small pores in each LH1 complex, created by having only a single carotenoid for each α1β1BChl2 subunit, likely allow Q/QH2 exchange and therefore turnover at the RC QB site 4, 16. Figure 3 shows schematic diagrams of a section of the LH1 ring of the control strain DO10, the reduced carotenoid mutant DM1R, T. tepidum and the core monomer mutant DX13, which in this context is equivalent to the WT. We illustrate the proposed gaps between LH1 α1β1BChl2Crt units that allow Q/QH2 exchange across the T. tepidum LH1 ring 4, and suggest that in Rba. sphaeroides the second carotenoid within each LH1 α1β1BChl2 unit effectively plugs these pores and prevents Q/QH2 exchange. This problem in the PufX− strain DO10 is partly alleviated by the ~ 14% reduction in the carotenoid content of LH1 in strain DM1R, which creates a few pores for passage of Q/QH2. The optimal configuration is found in DX13 and the WT, where Q/QH2 traffic is confined to a site adjacent to the RC QB site, and a doubling of carotenoid content provides enhanced light absorption by the LH1 ring.
In relation to monomeric cores lacking PufX, the dimeric RC‐LH1‐PufX complex of Rba. sphaeroides has several functional upgrades that improve its light‐harvesting and quinone exchange functions. First, PufX binds to the RC‐H subunit and creates a single, specialised channel for efficient quinone traffic across the LH1 ring at a strategic location, relatively close by the RC QB site 8. Second, the LH1 complex is freed from maintaining small pores for quinone exchange across LH1 α1β1BChl2Crt units, so allowing recruitment of more carotenoids that enhance the light‐harvesting and photoprotective functions of the LH1 complex. Photoprotective effects are initiated by the oxygen‐driven in situ modification of spheroidene to yield spheroidenone 30, 31, 32, activating an intermolecular charge‐transfer state that channels excitation energy to the LH1 BChls whilst optimising the triplet energy for singlet oxygen quenching 33. This highly effective photoprotective mechanism occurs even in LH1 complexes that have been removed from the native membrane 34. Third, PufX‐promoted dimerisation of the RC‐LH1‐PufX complex confers additional benefits by allowing excitation and quinone sharing, as originally proposed by Comayras et al. 35 and shown directly in the structure of the dimeric RC‐LH1‐PufX complex 8.
Acknowledgements
JDO, ECM, and CNH gratefully acknowledge financial support from the Biotechnology and Biological Sciences Research Council (BBSRC UK), Award Number BB/M000265/1. CNH was also supported by an Advanced Award 338895 from the European Research Council. This work was also supported as part of the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE‐SC 0001035. PARC's role was to provide partial support for CNH.
Author contributions
JDO and CNH designed the project. JDO and ECM carried out the experiments and analyzed the data. JDO and CNH wrote the manuscript.
Edited by Richard Cogdell
References
- 1. Cartron ML, Olsen JD, Sener M, Jackson PJ, Brindley AA, Qian P, Dickman MJ, Leggett GJ, Schulten K and Hunter CN (2014) Integration of energy and electron transfer processes in the photosynthetic membrane of Rhodobacter sphaeroides . Biochim Biophys Acta 1837, 1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jamieson SJ, Wang P, Qian P, Kirkland JY, Conroy MJ, Hunter CN and Bullough PA (2002) Projection Structure of the photosynthetic reaction centre‐antenna complex of Rhodospirillum rubrum at 8.5Å resolution. EMBO J 21, 3927–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fotiadis D, Qian P, Philippsen A, Bullough PA, Engel A and Hunter CN (2004) Structural analysis of the RC‐LH1 photosynthetic core complex of Rhodospirillum rubrum using atomic force microscopy. J Biol Chem 279, 2063–2068. [DOI] [PubMed] [Google Scholar]
- 4. Niwa S, Yu LJ, Takeda K, Hirano Y, Kawakami T, Wang‐Otomo ZY and Miki K (2014) Structure of the LH1‐RC complex from Thermochromatium tepidum at 3.0 Å. Nature 508, 228–232. [DOI] [PubMed] [Google Scholar]
- 5. Qian P, Hunter CN and Bullough PA (2005) The 8.5 Å projection structure of the core RC‐LH1‐PufX dimer of Rhodobacter sphaeroides . J Mol Biol 349, 948–960. [DOI] [PubMed] [Google Scholar]
- 6. Qian P, Bullough P and Hunter CN (2008) Three‐dimensional reconstruction of a membrane‐bending complex. J Biol Chem 283, 14002–14011. [DOI] [PubMed] [Google Scholar]
- 7. Roszak AW, Howard TD, Southall J, Gardiner AT, Law CJ, Isaacs NW and Cogdell RJ (2003) Crystal structure of the RC‐LH1 core complex from Rhodopseudomonas palustris . Science 302, 1969–1972. [DOI] [PubMed] [Google Scholar]
- 8. Qian P, Papiz MZ, Jackson PJ, Brindley AA, Ng I, Olsen JD, Dickman MJ, Bullough PA and Hunter CN (2013) The 3‐D structure of the Rhodobacter sphaeroides RC‐LH1‐PufX complex: dimerization and quinone channels promoted by PufX. Biochemistry 52, 7575–7585. [DOI] [PubMed] [Google Scholar]
- 9. Farchaus JW, Gruenberg H and Oesterhelt D (1990) Complementation of a reaction center‐deficient Rhodobacter sphaeroides pufLMX deletion strain in trans with pufBALM does not restore the photosynthesis‐positive phenotype. J Bacteriol 172, 977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lilburn TG, Haith CE, Prince RC and Beatty JT (1992) Pleiotropic effects of pufX gene deletion on the structure and function of the photosynthetic apparatus of Rhodobacter capsulatus . Biochim Biophys Acta 1100, 160–170. [DOI] [PubMed] [Google Scholar]
- 11. McGlynn P, Hunter CN and Jones MR (1994) The Rhodobacter sphaeroides PufX protein is not required for photosynthetic competence in the absence of a light harvesting system. FEBS Lett 349, 349–353. [DOI] [PubMed] [Google Scholar]
- 12. Barz WP, Francia F, Venturoli G, Melandri BA, Verméglio A and Oesterhelt D (1995) Role of the PufX protein in photosynthetic growth of Rhodobacter sphaeroides. 1. PufX is required for efficient light‐driven electron transfer and photophosphorylation under anaerobic conditions. Biochemistry 34, 15235–15247. [DOI] [PubMed] [Google Scholar]
- 13. Barz WP, Verméglio A, Francia F, Venturoli G, Melandri BA and Oesterhelt D (1995) Role of the PufX protein in photosynthetic growth of the Rhodobacter sphaeroides. 2. PufX is required for efficient ubiquinone/ubiquinol exchange between the reaction center QB site and the cytochrome bc 1 complex. Biochemistry 34, 15248–15258. [DOI] [PubMed] [Google Scholar]
- 14. Francia F, Wang J, Zischka H, Venturoli G and Oesterhelt D (2002) Role of the N‐ and C‐terminal regions of the PufX protein in the structural organization of the photosynthetic core complex of Rhodobacter sphaeroides . Eur J Biochem 269, 1877–1885. [DOI] [PubMed] [Google Scholar]
- 15. Siebert CA, Qian P, Fotiadis D, Engel A, Hunter CN and Bullough P (2004) The role of PufX in the molecular architecture of photosynthetic membranes in Rhodobacter sphaeroides . EMBO J 23, 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aird A, Wrachtrup J, Schulten K and Tietz C (2007) Possible pathway for ubiquinone shuttling in Rhodospirillum rubrum revealed by molecular dynamics simulation. Biophys J 92, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakagawa K, Suzuki S, Fujii R, Gardiner AT, Cogdell RJ, Nango M and Hashimoto H (2008) Probing the effect of the binding site on the electrostatic behavior of a series of carotenoids reconstituted into the light‐harvesting 1 complex from purple photosynthetic bacterium Rhodospirillum rubrum detected by Stark spectroscopy. J Phys Chem B 112, 9467–9475. [DOI] [PubMed] [Google Scholar]
- 18. Broglie RM, Hunter CN, Delepelaire P, Niederman RA, Chua N‐H and Clayton RK (1980) Isolation and characterization of the pigment‐protein complexes of Rhodopseudomonas sphaeroides by lithium dodecyl sulfate polyacrylamide gel electrophoresis. Proc Natl Acad Sci USA 77, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G and Pühler A (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene 145, 69–73. [DOI] [PubMed] [Google Scholar]
- 20. Chi SC, Mothersole DJ, Dilbeck P, Niedzwiedzki DM, Zhang H, Qian P, Vasilev C, Grayson KJ, Jackson PJ, Martin EC et al (2015) Assembly of functional photosystem complexes in Rhodobacter sphaeroides incorporating carotenoids from the spirilloxanthin pathway. Biochim Biophys Acta 1847, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mothersole DJ, Jackson PJ, Vasilev C, Tucker JD, Brindley AA, Dickman MJ and Hunter CN (2016) PucC and LhaA direct efficient assembly of the light‐harvesting complexes in Rhodobacter sphaeroides . Mol Microbiol 99, 307–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olsen JD, Sockalingum GD, Robert B and Hunter CN (1994) Modification of a hydrogen bond to a bacteriochlorophyll a molecule in the light‐harvesting 1 antenna of Rhodobacter sphaeroides . Proc Natl Acad Sci USA 91, 7124–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Babst M, Albrecht H, Wegmann I, Brunisholz R and Zuber H (1991) Single amino acid substitutions in the B870 α and β light‐harvesting polypeptides of Rhodobacter capsulatus. Structural and spectral effects. Eur J Biochem 202, 277–284. [DOI] [PubMed] [Google Scholar]
- 24. Jones MR, Visschers RW, van Grondelle R and Hunter CN (1992) Construction and characterisation of a mutant of Rhodobacter sphaeroides with the reaction centre as the sole pigment‐protein complex. Biochemistry 31, 4458–4465. [DOI] [PubMed] [Google Scholar]
- 25. McGlynn P, Westerhuis WHJ, Jones MR and Hunter CN (1996) Consequences for the organisation of reaction centre/LH1 complexes of Rhodobacter sphaeroides arising from deletion of amino acid residues from the C terminus of the LH1 polypeptide. J Biol Chem 271, 3285–3292. [DOI] [PubMed] [Google Scholar]
- 26. Walz T, Jamieson SJ, Bowers CM, Bullough PA and Hunter CN (1998) Projection structures of three photosynthetic complexes from Rhodobacter sphaeroides: LH2 at 6Å, LH1 and LH1‐RC at 25Å. J Mol Biol 282, 833–845. [DOI] [PubMed] [Google Scholar]
- 27. Stark W, Kühlbrandt W, Wildhaber H, Wehrli E and Mühlethaler K (1984) The structure of the photoreceptor unit of Rhodopseudomonas viridis . EMBO J 3, 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scheuring S, Seguin J, Marco S, Levy D, Robert B and Rigaud JL (2003) Nanodissection and high‐resolution imaging of the Rhodopseudomonas viridis photosynthetic core complex in native membranes by AFM. Proc Natl Acad Sci USA 100, 1690–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olsen JD, Adams PG and Hunter CN (2014) Aberrant assembly intermediates of the RC‐LH1‐PufX core complex of Rhodobacter sphaeroides imaged by atomic force microscopy. J Biol Chem 289, 29927–29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneour EA (1962) Carotenoid pigment conversion in Rhodopseudomonas spheroides . Biochim Biophys Acta 62, 534–540. [DOI] [PubMed] [Google Scholar]
- 31. Glaeser J and Klug G (2005) Photo‐oxidative stress in Rhodobacter sphaeroides: protective role of carotenoids and expression of selected genes. Microbiology 151, 1927–1938. [DOI] [PubMed] [Google Scholar]
- 32. Gerjets T, Steiger S and Sandmann G (2009) Catalytic properties of the expressed acyclic carotenoid 2‐ketolases from Rhodobacter capsulatus and Rubrivivax gelatinosus . Biochim Biophys Acta 1791, 125–131. [DOI] [PubMed] [Google Scholar]
- 33. Šlouf V, Chábera P, Olsen JD, Martin EC, Qian P, Hunter CN and Polívka T (2012) Photoprotection in a purple phototrophic bacterium mediated by oxygen‐dependent alteration of carotenoid excited‐state properties. Proc Natl Acad Sci USA 109, 8570–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Magis JG, Olsen JD, Reynolds NP, Leggett GJ, Hunter CN, Aartsma TJ and Frese RN (2011) Use of engineered unique cysteine residues to facilitate oriented coupling of proteins directly to a gold substrate. Photochem Photobiol 87, 1050–1057. [DOI] [PubMed] [Google Scholar]
- 35. Comayras R, Jungas C and Lavergne J (2005) Functional consequences of the organization of the photosynthetic apparatus in Rhodobacter sphaeroides – I. Quinone domains and excitation transfer in chromatophores and reaction center center‐antenna complexes. J Biol Chem 280, 11203–11213. [DOI] [PubMed] [Google Scholar]


