Abstract
The seven members of the tumor necrosis factor receptor (TNF‐R)‐associated factor (TRAF) family of intracellular proteins were originally discovered and characterized as signaling adaptor molecules coupled to the cytoplasmic regions of receptors of the TNF‐R superfamily. Functionally, TRAFs act both as a scaffold and/or enzymatic proteins to regulate activation of mitogen‐activated protein kinases (MAPKs) and transcription factors of nuclear factor‐κB family (NF‐κB). Given the wide variety of stimuli intracellularly conveyed by TRAF proteins, they are physiologically involved in multiple biological processes, including embryonic development, tissue homeostasis, and regulation of innate and adaptive immune responses. In the last few years, it has become increasingly evident the involvement of TRAF7, the last member of the TRAF family to be discovered, in the genesis and progression of several human cancers, placing TRAF7 in the spotlight as a novel tumor suppressor protein. In this paper, we review and discuss the literature recently produced on this subject. J. Cell. Physiol. 232: 1233–1238, 2017. © 2016 The Authors. Journal of Cellular Physiology Published by Wiley Periodicals, Inc.
The seven TRAF proteins were originally discovered as signal transducing components of the TNF‐R superfamily members (Ha et al., 2009; Xie, 2013). Upon binding of their respective ligands, these receptors transmit in the cell a wide range of different extracellular signals that regulate important and fundamental biological processes, including embryonic development and morphogenesis, the innate and acquired immune responses, cell survival and proliferation, tissues homeostasis, and stress responses (Ha et al., 2009; Xie, 2013).
Structurally, TRAF proteins share a typical modular organization of conserved domains. All family members, with the exception of TRAF1, contain a RING finger domain localized at their amino‐terminus, followed by one or more zinc finger domains (Fig. 1). The RING finger domain is common to many E3 ubiquitin ligases, constituting the core of the ubiquitin ligase catalytic domain. The carboxy‐terminus portion of TRAF proteins is made of the extensive TRAF domain, which mediates oligomerization reactions and interactions with partner proteins (Ha et al., 2009; Xie, 2013). The last member of the TRAF family identified, TRAF7, lacks the TRAF domain, which is replaced by seven repeats of the WD domain (Zotti et al., 2012).
Figure 1.
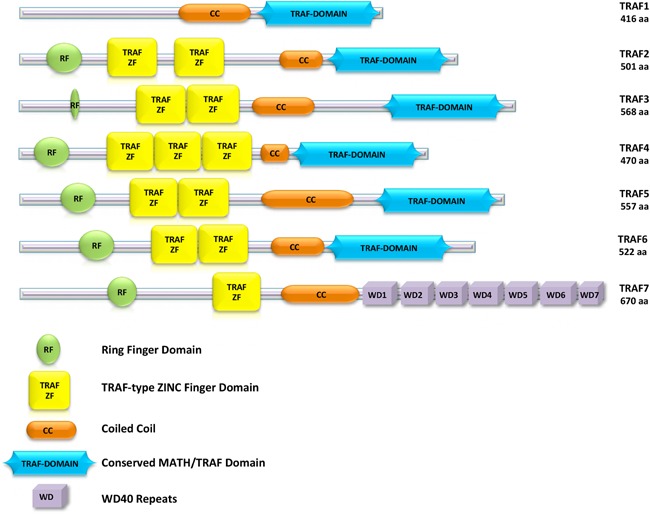
Domain organization of the seven TRAF proteins. Amino acids numbering refers to the human proteins.
In the context of TNF‐R signaling, TRAF proteins are primarily involved in regulating the activation state of mitogen‐activated protein kinases (MAPKs) and NF‐κB. For this, TRAF proteins function as molecular scaffolds that assist the ligand‐induced recruitment of other signaling proteins to the TNF‐R signaling complexes. Moreover, as the signaling pathways controlled by TRAF proteins are often regulated through ubiquitination events, several family members, including TRAF2, 3, 5, and 6, also act as E3 ubiquitin ligase enzymes (Ha et al., 2009; Xie, 2013).
Some excellent works have been published on the role of TRAF proteins in the signal transduction pathways activated by members of the TNF‐R superfamily, to which we refer for further reading (Ha et al., 2009; Xie, 2013). In these pages, we will focus on the experimental evidences now existing pointing to a role for TRAF proteins, particularly TRAF7, in the genesis and progression of several types of cancer.
TRAF7
TRAF7 was originally discovered in a screening aiming at the definition of a protein–protein interaction network around known and candidate components of the TNFα/NF‐κB pathway (Bouwmeester et al., 2004). Using an integrated approach comprising tandem affinity purification and liquid‐chromatography mass spectrometry, TRAF7 was identified with TAP‐tagged MEKK3, a MAPKKK required for the TNF‐α‐induced activation of NF‐κB (Yang et al., 2001). Independently, TRAF7 was also identified using bioinformatics tools in a search for sequences containing a TRAF‐like RING finger domain (Xu et al., 2004). As other TRAF family members, TRAF7 contains an amino‐terminal RING finger domain (aa 125–160), followed by an adjacent zinc finger domain (aa 221–287). However, instead of the classical TRAF domain, the carboxy terminus of TRAF7 contains seven WD40 repeats (Bouwmeester et al., 2004; Xu et al., 2004). Due to the lack of a canonical carboxy‐terminal TRAF domain, some authors question whether TRAF7 is a “proper” TRAF protein. As mentioned before, TRAF7 has been identified for its association with MEKK3. However, while other TRAF family members interact with various signaling molecules, including protein kinases, through the TRAF domain, the interaction of TRAF7 with MEKK3 is mediated by the WD40 repeats‐containing region of the protein (Bouwmeester et al., 2004). Functionally, TRAF7 and MEKK3 cooperate in the signal transduction pathways that lead to activation of JNK and p38 MAP kinases following TNFα stimulation (Bouwmeester et al., 2004; Xu et al., 2004; Scudiero et al., 2012). In addition to AP1, the main transcription factors activated by JNK, TRAF7 also positively regulates the transcriptional activity of CHOP/gadd153, a transcription factor activated following exposure to different cellular stresses, such as UV light, genotoxic agents, and protein misfolding in the endoplasmic reticulum (Xu et al., 2004). Similarly to TRAF2, TRAF3, and TRAF6 (Karin and Gallagher, 2009; Häcker et al., 2011; Xie, 2013), the RING domain of TRAF7 possesses an E3 ubiquitin ligase activity (Bouwmeester et al., 2004). In addition to auto‐ubiquitination (Bouwmeester et al., 2004), TRAF7 also promotes ubiquitination of several cellular targets, including the adapter molecule NF‐κB essential modulator (NEMO) (Zotti et al., 2011), the p65 subunit of NF‐κB transcription factor (Tsikitis et al., 2010; Zotti et al., 2011), the anti‐apoptotic protein c‐FLIP (Scudiero et al., 2012) and the tumor suppressor protein p53 (Wang et al., 2013). Ubiquitin possesses seven lysines (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) and the consequence of an ubiquitination reaction depends on the Lys‐type of ubiquitin linkage (Kulathu and Komander, 2012). Indeed, TRAF7 is able to catalyze different Lys‐types of ubiquitinations. In fact, TRAF7 promotes predominantly Lys29‐linked polyubiquitination of NEMO, p65, and c‐FLIP (Zotti et al., 2011; Scudiero et al., 2012), and this type of ubiquitination is associated with lysosomal degradation of the target proteins (Chastagner et al., 2006; Zotti et al., 2011; Scudiero et al., 2012). On the other hand, TRAF7 promotes Lys48‐linked ubiquitination of p53 (Wang et al., 2013), which is usually followed by the proteosomal degradation of the ubiquitinated protein. Along with ubiquitination reactions, TRAF7 also binds to and stimulates sumoylation of the proto‐oncogene product c‐Myb (Morita et al., 2005), a transcription factor that regulates proliferation and differentiation of hematopoietic cells (Mucenski et al., 1991).
TRAF proteins in tumors
Having a primary role in the regulation of the acquired and innate immune responses, it is not surprising that the first evidence for an involvement of TRAF proteins in tumors came from the molecular analysis of specific B cells malignancies. For instance, deletions of TRAF2 (Keats et al., 2007; Demchenko et al., 2010) and bi‐allelic or mono‐allelic deletion of TRAF3, often associated with epigenetic alteration, have been detected in multiple myeloma samples (Annunziata et al., 2007; Keats et al., 2007; Demchenko et al., 2010). Homozygous deletions of the TRAF3 locus have also been observed in cases of chronic lymphocytic leukemias (Nagel et al., 2009), Waldenström's macroglobulinemia—a distinct B‐cell neoplasm characterized by a lymphoplasmacytic infiltrate in bone marrow and IgM paraprotein production (Braggio et al., 2009), and classical Hodgkin lymphoma (Otto et al., 2012). In addition to the above mentioned inactivating mutations of TRAF2 found in B cell malignancies, a significant fraction of diffuse large B‐cell lymphoma (DLBL) carry somatic activating mutations in TRAF2 (Compagno et al., 2009). These mutations produce TRAF2 molecules with considerably enhanced ability to activate NF‐κB, which catalyzes transcription of cytoprotective and anti‐apoptotic genes. Thus, in the same cell type, the B lymphocytes, TRAF2 can function as both oncogene and tumor suppressor gene. In the same study by Compagno et al. (2009), mutations of TRAF5 were identified even with higher frequency compared to TRAF2 (5% TRAF5 vs. 3% TRAF2). However, the effect of these TRAF5 mutations on NF‐κB activation, or other signaling pathways, has not been investigated yet.
While TRAF2 and TRAF3 have been involved in several lymphoid tumors, TRAF4 and TRAF6 are involved in a broader spectrum of cancers. Indeed, TRAF4 cDNA was initially isolated from metastatic breast cancer (Régnier et al., 1995; Tomasetto et al., 1995), where the TRAF4 gene was amplified. In a subsequent larger study including epithelial tumors, TRAF4 was found overexpressed in approximately 60% of lung, breast and ovarian, 44% of bladder, and 28% of colon carcinomas (Camilleri‐Broët et al., 2007). Moreover, TRAF4 overexpression was not only most commonly observed in epithelial tumors (48%), but also in 31% of melanomas, 21% of neurogenic tumors, and 17% of lymphomas (Camilleri‐Broët et al., 2007). Thus, TRAF4 protein overexpression appears to be a common feature of several human cancers.
Frequent amplification of the TRAF6 locus, with concomitant mRNA overexpression, has been reported in lung cancers (Starczynowski et al., 2011), colon cancer (Sun et al., 2014), and osteosarcoma samples (Meng et al., 2012). Expression of TRAF6 was found up‐regulated also in pancreatic cancer tissues, resulting in deregulated expression of multiple genes involved in cell growth, apoptosis, and migration (Rong et al., 2014).
TRAF7 in meningioma
Meningiomas are tumors originating from the meningeal coverings of the brain and the spinal cord (Mawrin and Perry, 2010). They represent one of the most frequent primary brain tumor, accounting for about one third of all cerebral nervous system tumors. The WHO currently recognizes 15 different variants of meningioma (Louis et al., 2007), classified in three grades of malignancy. The secretory subtype of meningioma, which represents approximately 3% of all meningiomas, follows a more aggressive clinical course due to increased brain swelling (Probst‐Cousin et al., 1997; Mawrin and Perry, 2010; Mawrin et al., 2015). A large fraction of all sporadic meningiomas (40–60%) display recurrent genetic alterations resulting in inactivating mutations in the NF2 gene locus located on chromosome 22 (Rouleau et al., 1993; Trofatter et al., 1993; Ruttledge et al., 1994; Peyre and Kalamarides, 2014; Domingues et al., 2015). The rate of NF2 mutations is similar in menigiomas of different WHO grades, suggesting that NF2 inactivation is critical for tumor initiation, rather than progression (Mawrin and Perry, 2010; Choy et al., 2011). The NF2 locus encodes for merlin, a pleiotropic protein involved in cytoskeleton organization and cell contact inhibition (Pecina‐Slaus, 2013), which also regulates transmembrane receptor accumulation and receptor signaling, including Erb receptors, platelet‐derived growth factor receptor, insulin‐like growth factor 1 receptor and vascular endothelial growth factor receptors (Lallemand et al., 2009). Besides these roles in cytoskeleton organization and receptor signaling, merlin also regulates ubiquitination events, by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus (Li et al., 2010).
Several recent genome‐wide studies of meningiomas (Brastianos et al., 2013; Clark et al., 2013; Reuss et al., 2013; Abedalthagafi et al., 2016; Clark et al., 2016) have led to the discovery of driver TRAF7 mutations implicated in meningioma tumorigenesis. Overall, these studies show that mutations in TRAF7 are observed in nearly one‐fourth of the meningiomas studied, and in all meningiomas of the secretory subtype. Interestingly, TRAF7 mutations are exclusive of NF2 mutations—suggesting that the two proteins act along the same pathway—and often co‐occur with mutations in KLF4 or in AKT1. KLF4 is a transcription factor that regulates differentiation in a variety of different cell types and its expression is essential to reprogram adult cells in pluripotent adult stem cells (Takahashi et al., 2007), whereas AKT1 is serine‐threonine kinase involved in proliferation, and is a well known oncogene (Vivanco and Sawyers, 2013). Also, whereas the co‐occurring mutations in KLF4 and AKT1 are always the same (K409Q for KLF4 and E17K for AKT1), the TRAF7 mutations found in the various samples are distributed along the whole protein, with a marked predilection for the WD40 repeats (Fig. 2). For instance, the AKT1E17K mutation found in meningiomas often occurs in various other human cancers, including breast, colon, and ovarian cancers (Carpten et al., 2007).
Figure 2.
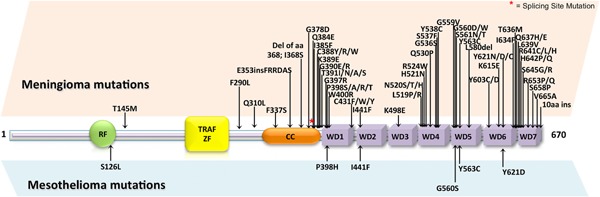
TRAF7 mutations in human cancers. In the domain structure of TRAF7, on the top are indicated the mutations found in meningiomas, on the bottom the mutations found in mesothelioma.
Specifically, combination of KLF4 and TRAF7 mutations was only found in virtually 100% of secretory meningiomas (Clark et al., 2013). Moreover, meningiomas arising in specific anatomical positions are characterized by mutations in the same genes. In fact, tumors with TRAF7/AKT1/KLF4 mutations predominantly localize medially in the skull base, whereas meningiomas with NF2 mutations predominantly localize in the hemispheres or laterally in the skull base (Clark et al., 2013).
TRAF7 in mesothelioma
Malignant mesothelioma (MM) is an aggressive cancer that results from unregulated proliferation of the mesothelial cells lining the pleural, peritoneal, and pericardial cavities (Yang et al., 2008). The vast majority of MMs is associated to exposure to mineral fibers, particularly asbestos and erionite. MMs are histologically classified in the three subtypes epithelial, biphasic (or mixed), and sarcomatoid. Malignant pleural mesothelioma (MPM) is the most common type of MM, accounting for approximately 70% of all cases. Unfortunately, MPM is commonly diagnosed at advanced stages and its survival rate is lower than 12 months (Yang et al., 2008). The genes whose role has been well established in MPM are CDKN2A, NF2 (i.e., the same gene mutated in meningiomas), and BAP1 (Cheng et al., 1994; Bianchi et al., 1995; Bott et al., 2011; Guo et al., 2015). The CDKN2A locus encodes for several proteins, the most well‐studied are the p16(INK4a) and the p14(ARF) proteins, which both function as tumor suppressors (Zhao et al., 2016). BAP1 (BRCA1‐associated protein 1) also is a tumor suppressor gene, and it encodes for a deubiquitinating enzyme that regulates key cellular pathways, including cell cycle, cellular differentiation, transcription, and DNA damage response (Wang et al., 2016). In a recent sequencing work of transcriptomes and exomes from 216 MPMs, TRAF7 was found among the significantly mutated genes (5/216; 2,3%) (Bueno et al., 2016). As in the case of meningiomas, most of the MMs mutations observed in TRAF7 (4/5) were localized at the carboxy‐terminal WD40 repeats (Fig. 2). Mutations in TRAF7 often co‐occurred with mutations of p53, but, similarly to what observed in meningiomas, rarely with mutations of NF2, which was found mutated in 19% of the samples. From the histological point of view, TRAF7 was found mutated in the epithelial and biphasic subtypes of MPM, but not in the sarcomatoid subtype. In addition to MPM, a somatic mutation of TRAF7 has been reported in a case of papillary peritoneal mesothelioma, where NF2 was not mutated as well (Yu et al., 2011).
Conclusion
Recent data generated through deep sequencing of primary tumors have clearly indicated the involvement of TRAF7 in the genesis of various human cancers. Specifically, TRAF7 is mutated in about 25% of meningiomas and significantly, although at a lower rate, in human mesotheliomas. An inactivating mutation of TRAF7 has also been reported in a case of Merkel cell carcinoma, a rare and aggressive cutaneous neuroendocrine carcinoma (Goh et al., 2015). It is certainly interesting the fact that mutations in TRAF7 in human meningioma and mesothelioma cancers are alternative to mutations in NF2, a gene which, strikingly, also shows a high rate of mutation in these two types of tumors. This evidence suggests that TRAF7 and NF2 may act on a common pathway. However, merlin, the product of the NF2 gene, is a multifaceted protein, functioning as a scaffold protein that connects cytoskeleton dynamics to signaling transduction pathways that control cell death and proliferation, including those triggered by tyrosine kinase receptors, small GTPases, mammalian target of rapamycin (mTOR), PI3K/Akt, and hippo pathways (Petrilli and Fernández‐Valle, 2016). Thus, in this variety of different functions it is not obvious to distinguish which of them is/are shared by TRAF7 and NF2 in promoting cancer development. Most of the TRAF7 mutations identified in human tumors localize at the WD40 repeats region and at the neighboring coiled coil domain, that is, the regions through which TRAF7 establishes interactions with several molecular partners (Zotti et al., 2012). For example, the coiled coil domain of TRAF7 is involved in the interaction with NEMO (Zotti et al., 2011), an adapter molecule essential for activation of the canonical pathway of NF‐κB, a transcription factor that by promoting expression of anti‐apoptotic and proliferative genes is known to be generally hyperactive in the vast majority of human cancers (Karin, 2006; Wu et al., 2015). Generally, TRAF7 exerts a negative control on NF‐κB activity, by promoting ubiquitination and lysosomal degradation of NEMO and that of the p65 subunit of NF‐κB (Zotti et al., 2011). TRAF7 also promotes ubiquitination and lysosomal degradation of c‐FLIP (Scudiero et al., 2012), a very well known inhibitor of apoptosis by interfering with caspases activation in tumor cells (Safa and Pollok, 2011; Shirley and Micheau, 2013) whose stability is known to be controlled by TRAF proteins (Guiet et al., 2002). Consistent with its role as a crucial negative regulator of the apoptotic pathway, c‐FLIP is found overexpressed in many different types of cancer, including MPM (Rippo et al., 2004).
Interestingly, the tumor suppressor protein p53 is another target of the ubiquitinating activity of TRAF7, and TRAF7‐mediated ubiquitination of p53 plays a critical role in breast cancer development and progression (Wang et al., 2013). The WD40 repeats seem to be the main hotspot for TRAF7 mutations found in human cancers. WD40 repeats‐containing proteins are generally abundantly present in eukaryote genomes—the human genome encodes for 349 predicted WD40 repeat‐containing proteins—and are characterized by the presence of repeating units with 40–60 variable residues that ended with tryptophan (W) and aspartate (D) dipeptides (Zhang and Zhang, 2015). For instance, the co‐presence of a RING finger domain and WD40 repeats allows to classify TRAF7 as a member of a smaller family of E3 ubiquitin ligases named RFWD proteins, which includes COP1 and RFWD3 (Marine, 2012). RFWD proteins target transcriptional regulators such as p53, ETV, and ETS, and are known to be involved in tumorigenesis as COP1 is a tumor suppressor and RFWD3 is a susceptibility locus in myeloma and testicular cancer (Marine, 2012; Chung et al., 2013; Mitchell et al., 2016).
The basic function of WD40 proteins is to serve as scaffold platforms for protein–protein and protein–DNA interactions. In fact, some functionally relevant molecular interactions are established by TRAF7 through its carboxy‐terminal WD40 repeats, including association with MEKK3. Indeed, co‐expression of MEKK3 with TRAF7 results in activation of the JNK and p38 MAP kinases pathways (Bouwmeester et al., 2004; Xu et al., 2004), which constitute a pivotal signaling pathway in cytokine‐ and stress‐induced apoptosis. Coherently with its positive role in activation of p38 and JNK pathways, and its negative role in regulating NF‐κB activation and c‐FLIP expression level, TRAF7 promotes apoptotic pathways. However, which of the multiple functions performed by TRAF7 is compromised as a result of mutations found in different human tumors remains still to be defined. In this regard, the next coming years will definitely witness profound advances in our understanding of the biology of TRAF7, and will clarify whether TRAF7 can be considered as a potential molecular target for the therapeutic approach of the diverse human cancers in which this protein is involved.
Conflicts of Interest: None of the authors has a conflict of interest to disclose.
Literature Cited
- Abedalthagafi M, Bi WL, Aizer AA, Merrill PH, Brewster R, Agarwalla PK, Listewnik ML, Dias‐Santagata D, Thorner AR, Van Hummelen P, Brastianos PK, Reardon DA, Wen PY, Al‐Mefty O, Ramkissoon SH, Folkerth RD, Ligon KL, Ligon AH, Alexander BM, Dunn IF, Beroukhim R, Santagata S. 2016. Oncogenic PI3. Neuro Oncol 18:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, Dave S, Hurt EM, Tan B, Zhao H, Stephens O, Santra M, Williams DR, Dang L, Barlogie B, Shaughnessy JD, Jr. , Kuehl WM, Staudt LM. 2007. Frequent engagement of the classical and alternative NF‐kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12:115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi AB, Mitsunaga SI, Cheng JQ, Klein WM, Jhanwar SC, Seizinger B, Kley N, Klein‐Szanto AJ, Testa JR. 1995. High frequency of inactivating mutations in the neurofibromatosis type 2 gene (NF2) in primary malignant mesotheliomas. Proc Natl Acad Sci USA 92:10854–10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, Creaney J, Lake RA, Zakowski MF, Reva B, Sander C, Delsite R, Powell S, Zhou Q, Shen R, Olshen A, Rusch V, Ladanyi M. 2011. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 43:668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti‐Furga G. 2004. A physical and functional map of the human TNF‐alpha/NF‐kappa B signal transduction pathway. Nat Cell Biol 6:97–105. [DOI] [PubMed] [Google Scholar]
- Braggio E, Keats JJ, Leleu X, Van Wier S, Jimenez‐Zepeda VH, Valdez R, Schop RF, Price‐Troska T, Henderson K, Sacco A, Azab F, Greipp P, Gertz M, Hayman S, Rajkumar SV, Carpten J, Chesi M, Barrett M, Stewart AK, Dogan A, Bergsagel PL, Ghobrial IM, Fonseca R. 2009. Identification of copy number abnormalities and inactivating mutations in two negative regulators of nuclear factor‐kappaB signaling pathways in Waldenström's macroglobulinemia. Cancer Res 69:3579–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brastianos PK, Horowitz PM, Santagata S, Jones RT, McKenna A, Getz G, Ligon KL, Palescandolo E, Van Hummelen P, Ducar MD, Raza A, Sunkavalli A, Macconaill LE, Stemmer‐Rachamimov AO, Louis DN, Hahn WC, Dunn IF, Beroukhim R. 2013. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet 45:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, Gnad F, Nguyen TT, Jaiswal BS, Chirieac LR, Sciaranghella D, Dao N, Gustafson CE, Munir KJ, Hackney JA, Chaudhuri A, Gupta R, Guillory J, Toy K, Ha C, Chen YJ, Stinson J, Chaudhuri S, Zhang N, Wu TD, Sugarbaker DJ, de Sauvage FJ, Richards WG, Seshagiri S. 2016. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions, and splicing alterations. Nat Genet 48:407–416. [DOI] [PubMed] [Google Scholar]
- Camilleri‐Broët S, Cremer I, Marmey B, Comperat E, Viguié F, Audouin J, Rio MC, Fridman WH, Sautès‐Fridman C, Régnier CH. 2007. TRAF4 overexpression is a common characteristic of human carcinomas. Oncogene 26:142–147. [DOI] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker‐Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. 2007. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448:439–444. [DOI] [PubMed] [Google Scholar]
- Chastagner P, Israel A, Brou C. 2006. Itch/AIP4 mediates Deltex degradation through the formation of K29‐linked polyubiquitin chains. EMBO Rep 7:1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JQ, Jhanwar SC, Klein WM, Bell DW, Lee WC, Altomare DA, Nobori T, Olopade OI, Buckler AJ, Testa JR. 1994. P16 alterations and deletion mapping of 9p21‐p22 in malignant mesothelioma. Cancer Res 54:5547–5551. [PubMed] [Google Scholar]
- Choy W, Kim W, Nagasawa D, Stramotas S, Yew A, Gopen Q, Parsa AT, Yang I. 2011. The molecular genetics and tumor pathogenesis of meningiomas and the future directions of meningioma treatments. Neurosurg Focus 30:E6. [DOI] [PubMed] [Google Scholar]
- Chung CC, Kanetsky PA, Wang Z, Hildebrandt MA, Koster R, Skotheim RI, Kratz CP, Turnbull C, Cortessis VK, Bakken AC, Bishop DT, Cook MB, Erickson RL, Fosså SD, Jacobs KB, Korde LA, Kraggerud SM, Lothe RA, Loud JT, Rahman N, Skinner EC, Thomas DC, Wu X, Yeager M, Schumacher FR, Greene MH, Schwartz SM, McGlynn KA, Chanock SJ, Nathanson KL. 2013. Meta‐analysis identifies four new loci associated with testicular germ cell tumor. Nat Genet 45:680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark VE, Erson‐Omay EZ, Serin A, Yin J, Cotney J, Ozduman K, Avşar T, Li J, Murray PB, Henegariu O, Yilmaz S, Günel JM, Carrión‐Grant G, Yilmaz B, Grady C, Tanrikulu B, Bakircioğlu M, Kaymakçalan H, Caglayan AO, Sencar L, Ceyhun E, Atik AF, Bayri Y, Bai H, Kolb LE, Hebert RM, Omay SB, Mishra‐Gorur K, Choi M, Overton JD, Holland EC, Mane S, State MW, Bilgüvar K, Baehring JM, Gutin PH, Piepmeier JM, Vortmeyer A, Brennan CW, Pamir MN, Kiliç T, Lifton RP, Noonan JP, Yasuno K, Günel M. 2013. Genomic analysis of non‐NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 339:1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark VE, Harmancı AS, Bai H, Youngblood MW, Lee TI, Baranoski JF, Ercan‐Sencicek AG, Abraham BJ, Weintraub AS, Hnisz D, Simon M, Krischek B, Erson‐Omay EZ, Henegariu O, Carrión‐Grant G, Mishra‐Gorur K, Durán D, Goldmann JE, Schramm J, Goldbrunner R, Piepmeier JM, Vortmeyer AO, Günel JM, Bilgüvar K, Yasuno K, Young RA, Günel M. 2016. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet 48:1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Dalla‐Favera R, Pasqualucci L. 2009. Mutations of multiple genes cause deregulation of NF‐kappaB in diffuse large B‐cell lymphoma. Nature 459:717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM. 2010. Classical and/or alternative NF‐kappaB pathway activation in multiple myeloma. Blood 115:3541–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues P, González‐Tablas M, Otero Á, Pascual D, Ruiz L, Miranda D, Sousa P, Gonçalves JM, Lopes MC, Orfao A, Tabernero MD. 2015. Genetic/molecular alterations of meningiomas and the signaling pathways targeted. Oncotarget 6:10671–10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G, Walradt T, Markarov V, Blom A, Riaz N, Doumani R, Stafstrom K, Moshiri A, Yelistratova L, Levinsohn J, Chan TA, Nghiem P, Lifton RP, Choi J. 2015. Mutational landscape of MCPyV‐positive and MCPyV‐negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget 7:3403–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiet C, Silvestri E, De Smaele E, Franzoso G, Vito P. 2002. C‐FLIP efficiently rescues TRAF‐2‐/‐ cells from TNF‐induced apoptosis. Cell Death Differ 9:138–144. [DOI] [PubMed] [Google Scholar]
- Guo G, Chmielecki J, Goparaju C, Heguy A, Dolgalev I, Carbone M, Seepo S, Meyerson M, Pass HI. 2015. Whole‐exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res 75:264–269. [DOI] [PubMed] [Google Scholar]
- Ha H, Han D, Choi Y. 2009. TRAF‐mediated TNFR‐family signaling. Curr Protoc Immunol 87:11.9D.1 11.9D.19. [DOI] [PubMed] [Google Scholar]
- Häcker H, Tseng PH, Karin M. 2011. Expanding TRAF function: TRAF3 as a tri‐faced immune regulator. Nat Rev Immunol 11:457–468. [DOI] [PubMed] [Google Scholar]
- Karin M. 2006. Nuclear factor‐kappaB in cancer development and progression. Nature 441:431–436. [DOI] [PubMed] [Google Scholar]
- Karin M, Gallagher E. 2009. TNFR signaling: Ubiquitin‐conjugated TRAFfic signals control stop‐and‐go for MAPK signaling complexes. Immunol Rev 228:225–240. [DOI] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, Braggio E, Henry T, Zhu YX, Fogle H, Price‐Troska T, Ahmann G, Mancini C, Brents LA, Kumar S, Greipp P, Dispenzieri A, Bryant B, Mulligan G, Bruhn L, Barrett M, Valdez R, Trent J, Stewart AK, Carpten J, Bergsagel PL. 2007. Promiscuous mutations activate the noncanonical NF‐kappaB pathway in multiple myeloma. Cancer Cell 12:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y, Komander D. 2012. Atypical ubiquitylation—The unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 13:508–523. [DOI] [PubMed] [Google Scholar]
- Lallemand D, Manent J, Couvelard A, Watilliaux A, Siena M, Chareyre F, Lampin A, Niwa‐Kawakita M, Kalamarides M, Giovannini M. 2009. Merlin regulates transmembrane receptor accumulation and signaling at the plasma membrane in primary mouse Schwann cells and in human schwannomas. Oncogene 28:854–865. [DOI] [PubMed] [Google Scholar]
- Li W, You L, Cooper J, Schiavon G, Pepe‐Caprio A, Zhou L, Ishii R, Giovannini M, Hanemann CO, Long SB, Erdjument‐Bromage H, Zhou P, Tempst P, Giancotti FG. 2010. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell 140:477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis D, Ohgaki H, Wiestler O, Cavenee W. 2007. World Health Organization classification of tumours of the central nervous system In: Bosman F, Jaffe E, Lakhani S, Ohgaki H, editors. World Health Organization classification of tumours, 4th edn Lyon: IARC. [Google Scholar]
- Marine JC. 2012. Spotlight on the role of COP1 in tumorigenesis. Nat Rev Cancer 12:455–464. [DOI] [PubMed] [Google Scholar]
- Mawrin C, Perry A. 2010. Pathological classification and molecular genetics of meningiomas. J Neurooncol 99:379–391. [DOI] [PubMed] [Google Scholar]
- Mawrin C, Chung C, Preusser M. 2015. Biology and clinical management challenges in meningioma. Am Soc Clin Oncol Educ Book e106–e115. [DOI] [PubMed] [Google Scholar]
- Meng Q, Zheng M, Liu H, Song C, Zhang W, Yan J, Qin L, Liu X. 2012. TRAF6 regulates proliferation, apoptosis, and invasion of osteosarcoma cell. Mol Cell Biochem 371:177–186. [DOI] [PubMed] [Google Scholar]
- Mitchell JS, Li N, Weinhold N, Försti A, Ali M, van Duin M, Thorleifsson G, Johnson DC, Chen B, Halvarsson BM, Gudbjartsson DF, Kuiper R, Stephens OW, Bertsch U, Broderick P, Campo C, Einsele H, Gregory WA, Gullberg U, Henrion M, Hillengass J, Hoffmann P, Jackson GH, Johnsson E, Jöud M, Kristinsson SY, Lenhoff S, Lenive O, Mellqvist UH, Migliorini G, Nahi H, Nelander S, Nickel J, Nöthen MM, Rafnar T, Ross FM, da Silva Filho MI, Swaminathan B, Thomsen H, Turesson I, Vangsted A, Vogel U, Waage A, Walker BA, Wihlborg AK, Broyl A, Davies FE, Thorsteinsdottir U, Langer C, Hansson M, Kaiser M, Sonneveld P, Stefansson K, Morgan GJ, Goldschmidt H, Hemminki K, Nilsson B, Houlston RS. 2016. Genome‐wide association study identifies multiple susceptibility loci for multiple myeloma. Nat Commun 7:12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Kanei‐Ishii C, Nomura T, Ishii S. 2005. TRAF7 sequesters c‐Myb to the cytoplasm by stimulating its sumoylation. Mol Biol Cell 16:5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Jr. , Potter SS. 1991. A functional c‐myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65:677–689. [DOI] [PubMed] [Google Scholar]
- Nagel I, Bug S, Tönnies H, Ammerpohl O, Richter J, Vater I, Callet‐Bauchu E, Calasanz MJ, Martinez‐Climent JA, Bastard C, Salido M, Schroers E, Martin‐Subero JI, Gesk S, Harder L, Majid A, Dyer MJ, Siebert R. 2009. Biallelic inactivation of TRAF3 in a subset of B‐cell lymphomas with interstitial del(14)(q24.1q32.33). Leukemia 23:2153–2155. [DOI] [PubMed] [Google Scholar]
- Otto C, Giefing M, Massow A, Vater I, Gesk S, Schlesner M, Richter J, Klapper W, Hansmann ML, Siebert R, Küppers R. 2012. Genetic lesions of the TRAF3 and MAP3K14 genes in classical Hodgkin lymphoma. Br J Haematol 157:702–708. [DOI] [PubMed] [Google Scholar]
- Pecina‐Slaus N. 2013. Merlin, the NF2 gene product. Pathol Oncol Res 19:365–373. [DOI] [PubMed] [Google Scholar]
- Petrilli AM, Fernández‐Valle C. 2016. Role of c/NF2 inactivation in tumor biology. Oncogene 35:537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyre M, Kalamarides M. 2014. Molecular genetics of meningiomas: Building the roadmap towards personalized therapy. Neurochirurgie pii: S0028‐3770(14)00113‐1. [DOI] [PubMed] [Google Scholar]
- Probst‐Cousin S, Villagran‐Lillo R, Lahl R, Bergmann M, Schmid KW, Gullotta F. 1997. Secretory meningioma: Clinical, histologic, and immunohistochemical findings in 31 cases. Cancer 79:2003–2015. [DOI] [PubMed] [Google Scholar]
- Régnier CH, Tomasetto C, Moog‐Lutz C, Chenard MP, Wendling C, Basset P, Rio MC. 1995. Presence of a new conserved domain in CART1, a novel member of the tumor necrosis factor receptor‐associated protein family, which is expressed in breast carcinoma. J Biol Chem 270:25715–25721. [DOI] [PubMed] [Google Scholar]
- Reuss DE, Piro RM, Jones DT, Simon M, Ketter R, Kool M, Becker A, Sahm F, Pusch S, Meyer J, Hagenlocher C, Schweizer L, Capper D, Kickingereder P, Mucha J, Koelsche C, Jäger N, Santarius T, Tarpey PS, Stephens PJ, Andrew Futreal P, Wellenreuther R, Kraus J, Lenartz D, Herold‐Mende C, Hartmann C, Mawrin C, Giese N, Eils R, Collins VP, König R, Wiestler OD, Pfister SM, von Deimling A. 2013. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol 125:351–358. [DOI] [PubMed] [Google Scholar]
- Rippo MR, Moretti S, Vescovi S, Tomasetti M, Orecchia S, Amici G, Catalano A, Procopio A. 2004. FLIP overexpression inhibits death receptor‐induced apoptosis in malignant mesothelial cells. Oncogene 23:7753–7760. [DOI] [PubMed] [Google Scholar]
- Rong Y, Wang D, Wu W, Jin D, Kuang T, Ni X, Zhang L, Lou W. 2014. TRAF6 is over‐expressed in pancreatic cancer and promotes the tumorigenicity of pancreatic cancer cells. Med Oncol 31:260. [DOI] [PubMed] [Google Scholar]
- Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang‐Xuan K, Demczuk S, Desmaze C, Plougastel B, Pulst SM, Lenoir G, Bijlsma, Fashold R, Dumanski J, De Jong P, Parry D, Elridge R, Aurias A, Delattre O, Thomas G. 1993. Alteration in a new gene encoding a putative membrane‐organizing protein causes neurofibromatosis type 2. Nature 363:515–521. [DOI] [PubMed] [Google Scholar]
- Ruttledge MH, Sarrazin J, Rangaratnam S, Phelan CM, Twist E, Merel P, Delattre O, Thomas G, Nordenskjöld M, Collins VP, Dumanski JP, Rouleau GA. 1994. Evidence for the complete inactivation on the NF2 gene in the majority of sporadic meningiomas. Nat Genet 6:180–184. [DOI] [PubMed] [Google Scholar]
- Safa AR, Pollok KE. 2011. Targeting the anti‐apoptotic protein c‐FLIP for cancer therapy. Cancers (Basel) 3:1639–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudiero I, Zotti T, Ferravante A, Vessichelli M, Reale C, Masone MC, Leonardi A, Vito P, Stilo R. 2012. Tumor necrosis factor (TNF) receptor‐associated factor 7 is required for TNFα‐induced Jun NH2‐terminal kinase activation and promotes cell death by regulating polyubiquitination and lysosomal degradation of c‐FLIP protein. J Biol Chem 287:6053–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley S, Micheau O. 2013. Targeting c‐FLIP in cancer. Cancer Lett 332:141–150. [DOI] [PubMed] [Google Scholar]
- Starczynowski DT, Lockwood WW, Deléhouzée S, Chari R, Wegrzyn J, Fuller M, Tsao MS, Lam S, Gazdar AF, Lam WL, Karsan A. 2011. TRAF6 is an amplified oncogene bridging the RAS and NF‐κB pathways in human lung cancer. J Clin Invest 121:4095–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Li X, Fan L, Wu G, Li M, Fang J. 2014. TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int J Biochem Cell Biol 53:195–201. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. [DOI] [PubMed] [Google Scholar]
- Tomasetto C, Régnier C, Moog‐Lutz C, Mattei MG, Chenard MP, Lidereau R, Basset P, Rio MC. 1995. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11‐q21.3 region of chromosome 17. Genomics 28:367–376. [DOI] [PubMed] [Google Scholar]
- Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, Haase V, Ambrose CM, Munroe D, Bove C, Haines JL, Martuza RL, MacDonald ME, Seizinger BR, Short MP, Buckler AJ, Gusella JF. 1993. A novel moesin‐, ezrin‐, radixin‐like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 72:791–800. [DOI] [PubMed] [Google Scholar]
- Tsikitis M, Acosta‐Alvear D, Blais A, Campos EI, Lane WS, Sánchez I, Dynalacht BD. 2010. Traf7, a MyoD1 transcriptional target, regulates nuclear factor‐κB activity during myogenesis. EMBO Rep 11:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. 2013. The phosphatidylinositol 3‐kinase AKT pathway in human cancer. Nat Rev Cancer 2:489–501. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang L, Zhang S, Qu G, Zhang D, Li S, Liu S. 2013. Downregulation of ubiquitin E3 ligase TNF receptor‐associated factor 7 leads to stabilization of p53 in breast cancer. Oncol Rep 29:283–287. [DOI] [PubMed] [Google Scholar]
- Wang A, Papneja A, Hyrcza M, Al‐Habeeb A, Ghazarian D. 2016. Gene of the month: BAP1. J Clin Pathol 69:750–753. [DOI] [PubMed] [Google Scholar]
- Wu D, Wu P, Zhao L, Huang L, Zhang Z, Zhao S, Huang J. 2015. NF‐κB expression and outcomes in solid tumors: A systematic review and meta‐analysis. Medicine (Baltimore) 94:e1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P. 2013. TRAF molecules in cell signaling and in human diseases. J Mol Signal 8:7–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Li LY, Shu HB. 2004. TRAF7 potentiates MEKK3‐induced AP1 and CHOP activation and induces apoptosis. J Biol Chem 279:17278–17282. [DOI] [PubMed] [Google Scholar]
- Yang J, Lin Y, Guo Z, Cheng J, Huang J, Deng L, Liao W, Chen Z, Liu Z, Su B. 2001. The essential role of MEKK3 in TNF‐induced NF‐kappaB activation. Nat Immunol 2:620–624. [DOI] [PubMed] [Google Scholar]
- Yang H, Testa JR, Carbone M. 2008. Mesothelioma epidemiology, carcinogenesis, and pathogenesis. Curr Treat Options Oncol 9:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Chan‐On W, Teo M, Ong CK, Cutcutache I, Allen GE, Wong B, Myint SS, Lim KH, Voorhoeve PM, Rozen S, Soo KC, Tan P, Teh BT. 2011. First somatic mutation of E2F1 in a critical DNA binding residue discovered in well‐differentiated papillary mesothelioma of the peritoneum. Genome Biol 12:R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang F. 2015. The multifunctions of WD40 proteins in genome integrity and cell cycle progression. J Genomics 3:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Choi BY, Lee MH, Bode AM, Dong Z. 2016. Implications of genetic and epigenetic alterations of CDKN2A (p16(INK4a)) in cancer. EBioMedicine 8:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotti T, Vito P, Stilo R. 2012. The seventh ring: Exploring TRAF7 functions. J Cell Physiol 227:1280–1284. [DOI] [PubMed] [Google Scholar]
- Zotti T, Uva A, Ferravante A, Vessichelli M, Scudiero I, Ceccarelli M, Vito P, Stilo R. 2011. TRAF7 protein promotes Lys‐29‐linked polyubiquitination of IkappaB kinase (IKKgamma)/NF‐kappaB essential modulator (NEMO) and p65/RelA protein and represses NF‐kappaB activation. J Biol Chem 286:22924–22933. [DOI] [PMC free article] [PubMed] [Google Scholar]


