Abstract
Background
Lentiviral vectors have been successfully used for human skin cell gene transfer studies. Defining the selection of integration sites for retroviral vectors in the host genome is crucial in risk assessment analysis of gene therapy. However, genome-wide analyses of lentiviral integration sites in human keratinocytes, especially after prolonged growth, are poorly understood.
Material/Methods
In this study, 874 unique lentiviral vector integration sites in human HaCaT keratinocytes after long-term culture were identified and analyzed with the online tool GTSG-QuickMap and SPSS software.
Results
The data indicated that lentiviral vectors showed integration site preferences for genes and gene-rich regions.
Conclusions
This study will likely assist in determining the relative risks of the lentiviral vector system and in the design of a safe lentiviral vector system in the gene therapy of skin diseases.
MeSH Keywords: Keratinocytes, Lentivirus, Virus Integration
Background
Gene transfer that is mediated by retroviral vectors has been successfully and extensively undertaken. However, gene transfer technology carries the risk of insertional mutagenesis that results from proviral integration [1]. Several studies have found that premalignant clonal proliferation of T cells or T cell acute lymphoblastic leukemia occurs in some patients with severe combined immunodeficiency (SCID) and Wiskott-Aldrich syndrome (WAS) treated with a γ-retroviral vector (MLV). This procedure is often associated with vector-mediated insertional activation of the LMO2 oncogene [2]. Thus, studies on the selection of integration sites for retroviral or gene therapeutic vectors in the host genome are particularly important [3].
Previous studies have shown that integration site selection of some retroviruses or retroviral vectors is not random. Different retroviruses or retroviral vectors have different integration preferences in human and animal genomes [4,5]: (I) those found within genes (i.e., transcription unit-like lentiviruses such as HIV, SIV, EIAV, and FIV); (II) those found near transcription start sites and CpG islands (γ-retroviruses such as MLV, XMRV, PERV, MSCV, FV and HERV); and (III) those that display only weak preferences for transcription units, transcription start sites and CpG islands or that are randomly dispersed (α-retroviruses such as ASLV; β-retroviruses such as MMTV; and δ-retroviruses such as HTLV-1 and BLV). Of course, host cells may also affect integration site selection of the retrovirus or retroviral vectors. For example, Mitchell et al. [6] found that tissue-specific transcription resulted in tissue-specific integration that was targeted by the HIV-1-based vector in the human lymphoid SupT1 cell-line, human peripheral blood mononuclear cells (PBMCs), and IMR-90 lung fibroblasts.
Lentiviral vectors are a very potent and versatile class of retroviral vectors that are derived from HIV, SIV, EIAV, and FIV, among others, or a number of ex vivo or in vivo gene transfer applications into dividing and non-dividing cells, and are promising candidates for use in the gene therapy of human skin inherited diseases [7]. However, the characteristics of lentiviral integration site selection in the genomes of human skin cells, particularly in the keratinocyte genome, have until now been poorly defined.
It might be more advantageous to study vector integration site selection in host cells after prolonged growth. Thus, it may be necessary to explore the prolonged genomic toxicity of retroviral vectors. It has been demonstrated that mouse bone marrow cells containing lentiviral vector genetic integration sites become progressively less common after prolonged growth [8]. However, an unresolved question concerns integration sites and their relevance in human keratinocytes. To address this issue, we identified 874 HIV-based lentiviral vector integration sites in HaCaT human keratinocytes after prolonged growth, and evaluated the distribution of integrants in relation to normal healthy genes, cancer genes, transcription start sites, CpG islands, and repetitive elements. These data will strengthen our capacity to study the relative risk of the lentiviral vector system in the setting of skin cell gene therapy.
Material and Methods
Cell culture and vector preparation
In our laboratory, HaCaT human keratinocytes were cultured as a monolayer at 37°C in a 5% CO2/95% air atmosphere in 25-cm2 culture flasks with defined keratinocyte-SFM culture medium (Gibco-BRL, USA) supplemented with penicillin (100 IU ml−1) and streptomycin sulfate (100 μg ml−1). The 293FT cell line (Invitrogen) was maintained as a monolayer at 37°C in a 5% CO2/95% air atmosphere in 25-cm2 culture flasks in a standard culture medium of DMEM (Gibco-BRL, USA) supplemented in 7% FBS, 2 mML-glutamine, and antibiotics (50 U ml−1 penicillin and 50 mg ml−1 streptomycin sulfate).
To produce lentiviral vectors, 293FT cells were co-transfected with the following 3 plasmids by using the calcium phosphate method: I) a plasmid that was encoded by the HIV-1-based lentiviral SIN vector segment (pHSER-EF1α-GFP, which was described previously [9], a kind gift of Dr. Guangqian Zhou, Queen’s University Belfast, Belfast, UK); II) the packaging construct (pSPAXI, preserved by our laboratory); and III) the envelope protein-producing construct (pMGID, preserved by our laboratory). Forty-eight hours after transfection, the viral supernatant was harvested, centrifuged to pellet cellular debris, and filtered through a 0.45-μm filter unit. The vector titer was determined by transduction of 3.0×104 HaCaT cells with dose-dependent quantities of vector supernatant and polybrene (8 μg ml−1). Cells were collected 96 h post-transduction and analyzed by fluorescence-activated cell sorting for expression of green fluorescent protein (GFP).
Lentiviral vector gene transfer and keratinocyte clone screening
In these studies, 3.0×104 HaCaT cells at 60–80% confluence were incubated with 1.88×105 infection units per ml of the lentiviral SIN vector supernatant. Cells were incubated with the supernatant for 48 h in the presence of polybrene (8 μg ml−1). Transduction efficiencies of ≥80% were achieved. Transduced HaCaT cells were trypsinized and then seeded into a 96-well plate with defined keratinocyte-SFM culture medium (Gibco, USA) containing G418 (500 μg ml−1) by limiting dilution. After 2 weeks of continuous culture, the concentration of G418 in the medium was changed to 200 μg ml−1. Then, a typical single-cell clone appeared in 1 well after 5–8 weeks of continuous culture. This GFP-positive keratinocyte clone was selected and expanded in a 24-well plate with defined keratinocyte-SFM (Gibco, USA), and then sub-cultured to a passage of 48 for further analysis when the cells had grown in static culture for about 6 months.
Lentiviral insertion analysis and statistical measurements
Proviral integration sites were cloned by ligation-mediated PCR (LM-PCR) as described previously [10]. Briefly, genomic DNA was purified from 1 to 5×106 cells that were digested with MseI and PstI to prevent amplification of internal 3′LTR fragments and ligated to an MseI double-strand linker. LM-PCR was performed with nested primers (Supplementary Table 1) specific for the LTR and the linker. PCR products without purification were directly shotgun cloned by using the TOPO TA Cloning Kit for Sequencing (Invitrogen, USA) and transformed into TOP10-competent cells to form libraries of integration junctions, which were then sequenced to saturation by the GS-FLX Genome Sequencer (Roche/454 Life Sciences) pyrosequencing platform following the manufacturer’s instructions.
Sequences were trimmed to remove the linker and viral DNA sequences using the software program Primer Premier 6.0 and mapped onto the human genome (Ens62, Apr 2011, GRCh37.p3/HG19) using the online tool GTSG-QuickMap [11], which is a system that can automatically identify genuine integration sites (target set) and calculate the frequencies of integration within or near various genomic features of interest (genes, transcription start sites, and CpG islands). In addition, GTSG-QuickMap spontaneously generated a reference set of 1 million random integration sites as the matched control. SPSS software (Version 18.0, SPSS, USA), and chi-square analysis was then used to compare the integration frequencies of the target versus control set. Differences of P<0.05 were considered statistically significant.
Results
We sequenced 7017 amplified junction sequences, and only 874 of these were mapped to unique locations in the human genome. Other raw (unmapped) sequences were redundant and excluded from further analysis.
Chromosomal distribution of lentiviral vector integration sites
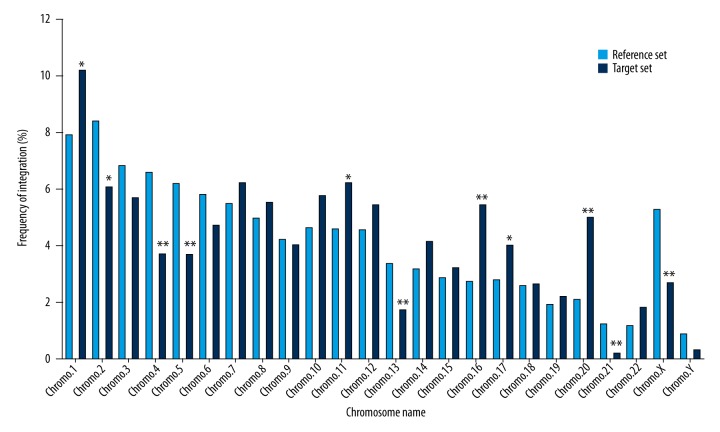
The results revealed that all integration sites were broadly distributed among autosomes and sex chromosomes (Figure 1). On chromosomes 1, 11, 16, 17, and 20, a significantly higher frequency of integration was observed, whereas lower frequencies of integration than that seen in the matched control were found on chromosomes 2, 4, 5, 13, 21, and X (P<0.05; Figure 1). The distribution of integration events was not significantly different among the remaining chromosomes (Figure 1). Observations indicated that lentiviral vector integration in human keratinocyte genome favors chromosome 1, 11, 16, 17, and 20, but not that of chromosome 2, 4, 5, 13, 21, and X.
Figure 1.
Integration sites of the lentiviral vector in the chromosomes of human keratinocyte clones. The lentiviral vector integration sites (target set, n=874) were plotted as the percentage of all integration sites in different chromosomes, and compared with the matched random control (reference set, n=1,000,000) using the χ2 analysis test. * Represents a significance difference (P<0.05) for the χ2 test as compared to the reference set. ** Represents a significance difference (P<0.01) for the χ2 test as compared to the reference set.
Distribution of lentiviral vector integration sites within known genes and near transcription start sites
As shown in Table 1, the percentage of lentiviral vector integration sites located within markedly exceeded that of simulated integration sites. Moreover, significantly higher frequencies of integration were found within introns. Lentiviral vector integration displayed a strong preference within 7 different windows (+5 kb, ±5 kb, +50 kb, − 50 kb, ± 50 kb, + 5~+50 kb, and −5~−50 kb) of transcription start sites, with a significantly higher frequency (Table 1; P<0.05) than that nominally expected from random distribution. However, no differences from the simulated integration sites were found within the other window (−5 kb) of the transcription start sites (Table 1). Thus, more accurately, lentiviral vectors preferentially integrate within introns of genes and at the 5–50 kb upstream region of transcriptional start sites in the human keratinocyte genome.
Table 1.
Lentiviral vector integration profiles within genes and near transcription start sitesa.
| Distance to genes or TSS | Integration frequency (%) of the reference set (n=1,000,000) | Integration frequency (%) of the target set (n=874) | P value |
|---|---|---|---|
| Within genes | 44.61 (446080) | 49.77 (435) | 0.002** |
| Introns | 42.12 (421233) | 46.80 (409) | 0.005** |
| Exons | 2.48 (24847) | 2.97 (26) | 0.352 |
| Within +5 kb of TSS | 6.18 (61849) | 8.01 (70) | 0.025* |
| Within −5 kb of TSS | 4.24 (42405) | 5.03 (44) | 0.244 |
| Within ±5 kb of TSS | 10.43 (104254) | 13.04 (114) | 0.011* |
| Within +50 kb of TSS | 44.61 (446080) | 49.77 (435) | 0.002** |
| Within −50 kb of TSS | 25.22 (252156) | 33.98 (297) | 0.000** |
| Within ±50 kb of TSS | 69.82 (698236) | 83.75 (732) | 0.000** |
| Within +5 kb ~ +50 kb of TSS | 38.42 (384231) | 41.76 (365) | 0.042* |
| Within −5 kb ~ −50 kb of TSS | 20.98 (209751) | 28.95 (253) | 0.000** |
Different distances to the transcriptional start sites (TSS);
‘+’ – downstream of TSS, ‘−’ – upstream of TSS.
Represented a significant difference (P<0.05) for the χ2 test as compared with the reference set.
Represented a significant difference (P<0.01) for the χ2 test as compared to the reference set.
Distribution of lentiviral vector integration sites with respect to CpG islands
We found that the proportions of lentiviral vector integrations located within the 0–5 kb and 5–50 kb upstream/downstream region of the CpG islands were significantly greater than those expected for random datasets (Table 2; P<0.01); whereas the relative frequencies of lentiviral vector integrations were indistinguishable from those for random integrations within CpG islands and the 50–250 kb upstream/downstream region of CpG islands (Table 2). On the basis of this analysis, lentiviral vectors exhibit a marked ability to integrate into the 0–50 kb upstream/downstream region from CpG islands.
Table 2.
Lentiviral vector integration into/near CpG islands.
| Distance to CpG islands | Integration frequency (%) of reference set (n=1,000,000) | Integration frequency (%) of target set (n=874) | P value |
|---|---|---|---|
| Within CpG islands | 0.84 (8384) | 0.46 (4) | 0.217 |
| Within ±(0~5) kb of CpG islands | 7.18 (71763) | 11.78 (103) | 0.000** |
| Within ±(5~50) kb of CpG islands | 33.53 (335321) | 41.30 (361) | 0.000** |
| Within ±(50~250) kb of CpG islands | 37.24 (372426) | 33.98 (297) | 0.050 |
Significantly different at P<0.01 for the χ2 analysis as compared to the reference set.
‘+’ – downstream of CpG islands, ‘−’ – upstream of CpG islands.
Distribution of lentiviral vector integration sites within repetitive elements
Among the analyzed elements, only the percentage of lentiviral vector integration sites within the SINEs was significantly higher than that observed for the reference set (Figure 2; P<0.05). Conversely, lentiviral integrants were under-represented in the LINEs and LTRs, with frequencies that fell below those found by random integration (Figure 2; P<0.01). Thus, it is evident that lentiviral vector integration favors the SINEs and do not favor LINEs and LTRs in the genome of human keratinocytes.
Figure 2.
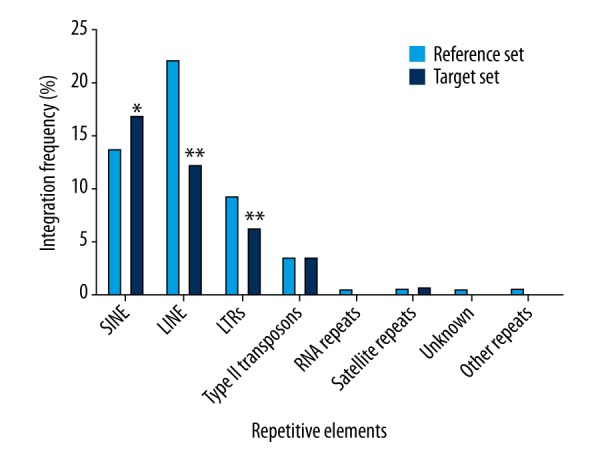
The characteristics of lentiviral vector integration sites within repetitive elements. The lentiviral vector integration sites (target set, n=874) were plotted as the integration frequency in different repetitive elements, and compared with the matched random control (reference set, n=1,000,000) using the χ2 test. * Represents a significant difference (P<0.05) for the χ2 test as compared to the reference set. ** Represents the significant difference (P<0.01) for the χ2 test as compared to the reference set.
Distribution of lentiviral vector integration sites within cancer genes and those positioned near their transcription start sites
As shown in Table 3, 13 direct hits to the cancer genes were found. The integration frequency within the 50–250 kb upstream/downstream region of transcription start sites of the selected cancer genes was significantly higher than that found by random distribution. Nevertheless, lentiviral vector integrants were found at approximately the same frequency as those identified by randomly generated sites within the other windows [±(0~5) kb, ±(5~50) kb] of transcription start sites of the cancer genes (Table 4). In summary, we found remarkable integration preference by lentiviral vectors in the 50–250 kb upstream/downstream region of transcription start sites of certain cancer-associated genes in our keratinocyte assay.
Table 3.
Lentiviral vector integrations to known cancer genesa.
| Locus | Name | Integration type | Exon/Intron number | Distance to TSSb (bp) |
|---|---|---|---|---|
| RUNXBP2 | MYST3, MYST histone acetyltransferase 3 | Intron | 12 | −105240 |
| MYH9 | Myosin, heavy chain 9, non-muscle | Intron | 2; 24 | −15425; −91289 |
| HCMOGT-1 | SPECC1, sperm antigen with calponin homology and coiled-coil domains 1 | Intron | 3 | −162419 |
| GRAF | ARHGAP26, Rho GTPase activating protein 26 | Exon | 2 | −103063; −103053 |
| FANCC | Fanconi anemia, complementation group C | Intron | 8 | −176695; +176695 |
| EGFR | Epidermal growth factor receptor | Intron | 1 | −83030 |
| BCL11B | B cell CLL/lymphoma 11B (zinc finger protein) | Intron | 3 | −93863 |
| LPP | LIM domain containing preferred translocation partner in lipoma | Intron | 6; 7 | −535722; −555412 |
| LASP1 | LIM and SH3 protein 1 | Intron | 4 | −31265 |
Cancer genes from the Ensembl database (http://www.ensembl.org/index.html);
Distance of the provirus relative to the transcription start site (TSS) of the cancer gene; and orientation of the provirus relative to the orientation of the cancer gene is also shown (‘+’ – forward; and ‘−’ – reverse).
Table 4.
The lentiviral vector integration profile within cancer genes and positioned near their transcription start sites (TSS).
| Distance to cancer genes or TSS | Integration frequency (%) of reference set (n=1,000,000) | Integration frequency (%) of target set (n=874) | P value |
|---|---|---|---|
| Within cancer genes | 1.49 (14877) | 1.49 (13) | 0.999 |
| Within ±(0~5) kb of TSS of cancer genes | 0.16 (1577) | 0.00 (0) | 0.240 |
| Within ±(5~50) kb of TSS of cancer genes | 1.37 (13666) | 1.37 (12) | 0.987 |
| Within ±(50~250) kb of TSS of cancer genes | 5.98 (59798) | 8.35 (73) | 0.003** |
The P value was determined by a χ2 test compared to the reference set.
Represents a significant difference (P<0.01) for the χ2 test as compared to the reference set.
‘+’ – downstream of TSS, ‘−’ – upstream of TSS.
Discussion
We performed a genome-wide analysis of lentiviral integration site selection in HaCaT human keratinocytes after prolonged growth. We found that lentiviral vectors showed integration site preferences for genes and gene-rich regions.
Our study demonstrates that 49.77% of integration sites resided in genes, which represents a significant departure from random placement (49.77% vs. 44.61%, P<0.01). However, this integration frequency was relatively lower than that found (not less than 50%) in most of the published reports of lentiviral vectors [12]. This indicates that the lentiviral vector may have a reduced genotoxic profile in human keratinocytes after prolonged growth.
Within known genes, integration was favored in introns over exons (46.80% vs. 42.12%, P<0.01; 2.97% vs. 2.48%; P>0.05), which was similar to the results of a previous study [13]. Moreover, the sequences of introns do not represent junk DNA, but actually play a key role in gene expression and regulation [14]. Thus, whether the expression of these genes is disturbed will form the basis of subsequent studies from our laboratory.
In addition, we chose a conservative window size of 50 kb upstream and downstream of the transcription start sites for analysis. The data showed that there was enrichment for lentiviral vector integration sites within 5–50 kb upstream of the transcription start sites (28.95% vs. 20.98%; P<0.01). This was an unexpected finding. In general, integration was annotated as TSS-proximal when it occurred within a distance of ±2.5 kb from the TSS of any known gene, and was considered as intragenic when it occurred within a distance inside a known gene >2.5 kb from the TSS, and was considered intergenic in all other cases [15]. Therefore, this distance (5–50 kb from the upstream region of TSS) may still reside in genes or extend to intergenic regions. In any case, genes, but not transcription start sites, are favored targets for lentiviral vector integration in the human keratinocyte genome.
Our study also shows that the ±50 kb region from the CpG islands was favored by the lentiviral vector. For CpG islands, these regions commonly correspond to gene regulatory regions containing clustered transcription factor binding sites, and many of them are within 10 kb of the gene [6]. In other words, CpG islands are more frequent in gene-rich regions. HIV integration extends from being disfavored at short distances (less than 1 kb) to being favored at longer distances (more than 10 kb) [6]. It is possible of course, that this distance from the CpG islands and the 5–50 kb upstream region of transcription start sites may have also overlapped. Genes are favored targets for lentiviral vector integration. Thus, it is clear that this area is a focus of integration. Our integration data also suggested that lentiviral vector integration was strongly favored in SINEs and disfavored in LINEs and LTRs (gene-dense regions are rich in SINEs, sparse in LINEs and LTRs [16]), and showed no preference for cancer-associated genes and their transcriptional start sites. A similar preference for these genomic features was previously reported [13,17,18].
Taken together, these findings show some unique integration features of lentiviral vectors in the human keratinocyte genome. This means that the details of genetic integration, transcription start sites, and CpG islands are not fully parallel with those of other cell types previously reported. The exact mechanism remains unclear, and additional research is clearly warranted. Many host factors may influence integration site profiles, such as growth time, cell cycle, and cellular proteins [6,8,12,19,20]. Cells containing integration sites in genes become less common after prolonged growth, which suggests negative selection [8]. Fortunately, in our study, no malignant clone emerged over the extended culture period, indicating both negative selection and lack of oncogene activation, raising the question of further safety. Cell cycle status can determine lentiviral integration in actively transcribed and developmentally-related genes [19]. To date, LEDGF/p75 is one of the most intensively researched cellular proteins, which can recruit the lentiviral pre-integration complex (PIC) to transcriptional units, thereby promoting integration efficiency and dictating lentiviral integration site selection [21]. LEDGF/p75 can be truncated by deleting the N-terminal chromatin-reading PWWP-domain, and replacing this domain with alternative pan-chromatin binding peptides. Expression of these LEDGF-hybrids in LEDGF-depleted cells can result in more randomly distributed lentiviral integration throughout the host-cell genome [22].
Conclusions
Our findings offer new data showing the pattern of lentiviral vector integration in the genome of human keratinocytes. This work will lay the foundation for further research aimed at determining and then establishing the biosafety of the lentiviral vector system and in designing a safer lentiviral vector system in the context of specific gene therapy for skin diseases.
Supplementary File
Supplementary Table 1.
Primers used for ligation-mediated PCR (LM-PCR) in this study.
| Name | Sequence (5′-3′) |
|---|---|
| MseI linker primer | GTAATACGACTCACTATAGGGC |
| MseI linker nested primer | AGGGCTCCGCTTAAGGGAC |
| 5′LTR primer | GAGGGATCTCTAGTTACCAGAGTCACA |
| 5′LTR nested primer | AGCCAGAGAGCTCCCAGGCTCAGATC |
Footnotes
Conflict of interest
None.
Source of support: This work was supported by grants from the National Natural Science Foundation of China (No. 81071575) and the Science and Technology Innovation Plan of Southwest Hospital (No. SWH2016JCZD-06)
References
- 1.Vargas JE. Retroviral vectors and transposons for stable gene therapy: Advances, current challenges and perspectives. J Transl Med. 2016;14(1):288. doi: 10.1186/s12967-016-1047-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cicalese MP, Aiuti A. Clinical applications of gene therapy for primary immunodeficiencies. Hum Gene Ther. 2015;26(4):210–19. doi: 10.1089/hum.2015.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doi K, Takeuchi Y. Gene therapy using retrovirus vectors: Vector development and biosafety at clinical trials. Uirusu. 2015;65(1):27–36. doi: 10.2222/jsv.65.27. [DOI] [PubMed] [Google Scholar]
- 4.Murakami H, Yamada T, Suzuki M, et al. Bovine leukemia virus integration site selection in cattle that develop leukemia. Virus Res. 2011;156(1–2):107–12. doi: 10.1016/j.virusres.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Nowrouzi A, Glimm H, Kalle CV, et al. Retroviral vectors: Post entry events and genomic alterations. Viruses. 2011;3(5):429–55. doi: 10.3390/v3050429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell RS, Beitzel BF, Schroder AR, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2(8):e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgiadis C, Syed F, Petrova A, et al. Lentiviral engineered fibroblasts expressing codon-optimized COL7A1 restore anchoring fibrils in RDEB. J Invest Dermatol. 2015;136(1):284–92. doi: 10.1038/JID.2015.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ronen K, Negre O, Roth S, et al. Distribution of lentiviral vector integration sites in mice following therapeutic gene transfer to treat β-thalassemia. MolTher. 2011;19(7):1273–86. doi: 10.1038/mt.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, Li SS, Segersvärd R, et al. Hypoxia inducible factor-1 mediates effects of insulin on pancreatic cancer cells and disturbs host energy homeostasis. Am J Pathol. 2007;170(2):469–77. doi: 10.2353/ajpath.2007.060489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciuffi A, Barr SD. Identification of HIV integration sites in infected host genomic DNA. Methods. 2011;53(1):39–46. doi: 10.1016/j.ymeth.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Appelt JU, Giordano FA, Ecker M, et al. QuickMap: A public tool for large-scale gene therapy vector insertion site mapping and analysis. Gene Ther. 2009;16(7):885–93. doi: 10.1038/gt.2009.37. [DOI] [PubMed] [Google Scholar]
- 12.Desfarges S, Ciuffi A. Retroviral integration site selection. Viruses. 2010;2(1):111–30. doi: 10.3390/v2010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schröder ARW, Shinn P, Chen H, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(110):521–29. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 14.Patrushev LI, Kovalenko TF. Functions of noncoding sequences in Mammalian genomes. Biochemistry. 2015;79(13):1442–69. doi: 10.1134/S0006297914130021. [DOI] [PubMed] [Google Scholar]
- 15.Moiani A, Miccio A, Rizzi E, et al. Deletion of the LTR enhancer/promoter has no impact on the integration profile of MLV vectors in human hematopoietic progenitors. PLoS One. 2013;8(1):e55721. doi: 10.1371/journal.pone.0055721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith MY, Evans CA, Holt RA. The sequence of the human genome. Science. 2001;291(5507):428–36. [Google Scholar]
- 17.Ciuffi A, Mitchell RS, Hoffmann C, et al. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol Ther. 2006;13(2):366–73. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Wang GP, Levine BL, Binder GK, et al. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. MolTher. 2009;17(5):844–50. doi: 10.1038/mt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papanikolaou E, Paruzynski A, Kasampalidis I, et al. Cell cycle status of CD34(+) hemopoietic stem cells determines lentiviral integration in actively transcribed and development-related genes. Mol Ther. 2015;23(4):683–96. doi: 10.1038/mt.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debyser Z, Christ F, De RJ, et al. Host factors for retroviral integration site selection. Trends BiochemSci. 2015;40(2):108–16. doi: 10.1016/j.tibs.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Lesbats P, Engelman AN, Cherepanov P. Retroviral DNA Integration. Chem Rev. 2016;116(20):12730–57. doi: 10.1021/acs.chemrev.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vranckx LS, Demeulemeester J, Debyser Z, et al. Towards a safer, more randomized lentiviral vector integration profile exploring artificial LEDGF chimeras. PLoS One. 2016;11(10):e0164167. doi: 10.1371/journal.pone.0164167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Primers used for ligation-mediated PCR (LM-PCR) in this study.
| Name | Sequence (5′-3′) |
|---|---|
| MseI linker primer | GTAATACGACTCACTATAGGGC |
| MseI linker nested primer | AGGGCTCCGCTTAAGGGAC |
| 5′LTR primer | GAGGGATCTCTAGTTACCAGAGTCACA |
| 5′LTR nested primer | AGCCAGAGAGCTCCCAGGCTCAGATC |