Abstract
Small molecule inhibitors, such as lapatinib, are effective against breast cancer in clinical trials, but tumor cells ultimately acquire resistance to the drug. Maintaining sensitization to drug action is essential for durable growth inhibition. Recently, adaptive reprogramming of signaling circuitry has been identified as a major cause of acquired resistance. We developed a computational framework using a Bayesian statistical approach to model signal rewiring in acquired resistance. We used the p1-model to infer potential aberrant gene-pairs with differential posterior probabilities of appearing in resistant-vs-parental networks. Results were obtained using matched gene expression profiles under resistant and parental conditions. Using two lapatinib-treated ErbB2-positive breast cancer cell-lines: SKBR3 and BT474, our method identified similar dysregulated signaling pathways including EGFR-related pathways as well as other receptor-related pathways, many of which were reported previously as compensatory pathways of EGFR-inhibition via signaling cross-talk. A manual literature survey provided strong evidence that aberrant signaling activities in dysregulated pathways are closely related to acquired resistance in EGFR tyrosine kinase inhibitors. Our approach predicted literature-supported dysregulated pathways complementary to both node-centric (SPIA, DAVID, and GATHER) and edge-centric (ESEA and PAGI) methods. Moreover, by proposing a novel pattern of aberrant signaling called V-structures, we observed that genes were dysregulated in resistant-vs-sensitive conditions when they were involved in the switch of dependencies from targeted to bypass signaling events. A literature survey of some important V-structures suggested they play a role in breast cancer metastasis and/or acquired resistance to EGFR-TKIs, where the mRNA changes of TGFBR2, LEF1 and TP53 in resistant-vs-sensitive conditions were related to the dependency switch from targeted to bypass signaling links. Our results suggest many signaling pathway structures are compromised in acquired resistance, and V-structures of aberrant signaling within/among those pathways may provide further insights into the bypass mechanism of targeted inhibition.
Introduction
Cell signaling pathways transduce input signals from extracellular to intracellular environments and determine various cell activities, including cell growth, proliferation, differentiation, migration, and apoptosis [1, 2]. Perturbation of a signaling network may occur when there are genetic alterations, such as DNA mutations and/or amplifications/deletions of a genomic region, or changes in gene expression (GE) [3, 4]. For example, the amplification or over-expression of the ErbB2 (HER2/neu) oncogene, that enhances various growth-related signaling activities [5] from receptor-level to effector-level [4], is commonly found in about 25% of breast cancer patients. In the majority of cancers, aberrant activities in signaling pathways are involved in various stages of tumor progression and metastasis [6–9].
Drugs targeting a signaling network, such as EGFR signaling pathway, often become ineffective as acquired resistance develops in cancer cells [10]. Primary reasons for acquired resistance to EGFR family receptor targeted therapies include: secondary mutations of targeted genes (e.g., the EGFR T790M mutation [11]), transcriptional and post-translational up-regulation of RTKs (Receptor Tyrosine Kinases) both within the receptor-family (e.g. ERBB3/HER3 [12, 13]) and other kinases (i.e. IGF1R, MET, FGFR2, FAK, SRC family kinases [14–16]), the over-expression of ABC transporters [3], and the re-activation of targeted pathways [5]. Moreover, tumor cells induce adaptive responses to targeted therapies [5] by rewiring in such a way that the adaptive signaling bypasses the inhibiting effects of initial treatments [4, 10, 17–19]. Therefore, rewiring of signaling networks plays a vital role as a non-genetic mechanism of acquired resistance [3, 14, 17, 18, 20]; targeting of which has the potential to improve the response durability of single kinase inhibitors [4, 5, 21]. However, reprogramming of signaling activities in acquired resistance inherently imposes increased uncertainties in the network structure when compared with their sensitive counterparts.
The functionality of biological networks is determined by their underlying architecture. Thus understanding, characterising, and analysing network structures are very important tasks in the field of systems biology [22]. Statistical modeling approaches offer a great deal of flexibility in terms of scalability and the number of local features that can be incorporated [22]. Moreover, as in other biological networks, signaling activities predicted using signaling data may be unreliable, whereas some crucial signaling links may not be predicted [23]. Measurements of the signaling activities often yield noisy data. Therefore, for such data-driven signaling networks a statistical modeling approach such as exponential random graph models (ERGMs) or p* can be a suitable choice [22, 23]. The p1-model, a special class of ERGMs which was originally proposed by Holland and Leinhardt [24], models the probability of an edge formation in the observed network based on network statistics (e.g. node degree) and associating model parameters with those statistics [22, 23].
Measuring the probabilistic nature of pair-wise relationships is an important aspect of modeling a gene-gene relationship network. Particularly in cancer drug resistance, some relationships between gene-pairs may evolve in the resistant conditions to compensate for the inhibiting effects of the drugs used [10, 19]. Some gene-pairs may have higher probabilities of evolving correlations in resistant conditions than in sensitive conditions. Simultaneously, some gene-pairs having high correlations in sensitive conditions may become loosely correlated (or even independent) in resistant conditions. For example, Komurov et al. reported that genes of the glucose-deprivation response network are up-regulated in lapatinib- (an EGFR/HER2 dual inhibitor) resistant conditions, thus providing an EGFR-independent mechanism of glucose uptake in cancer cells [19]. ErbB2-positive cancer cells largely depend on EGFR/ErbB2 signaling for their glucose uptake [19] which was recently reported as a major factor in oncogenic KRAS pathway mutations [25, 26]. Lapatinib mediates down-regulation of cell cycle machinery and up-regulation of cell cycle inhibitory complexes that are downstream of EGFR/ErbB2 signaling [19]. Moreover, the inhibitory effect of lapatinib on EGFR/ErbB2 signaling in the sensitive condition was found to be associated with glucose starvation of cancer cells, and thus induced cancer cell death [19]. However, in resistant conditions, up-regulation of activities involved in the glucose deprivation response network (and other hypoglycemic response networks) played an important role as a compensatory mechanism of glucose uptake in cancer cells for which tumors ultimately relapsed. Therefore, it can be hypothesized that genes involved in the process of cell proliferation and survival may evolve, in resistant conditions, to be highly correlated with the genes in the glucose deprivation response network in order to establish an alternate mechanism of glucose uptake in cancer cells, even though the inhibiting effects of lapatinib abrogated their dependencies on EGFR/ErbB2 signaling in sensitive conditions (See Fig 1 of [20].) Therefore, studying systematic characterizations of such differential dependencies among gene-pairs in resistant-vs-sensitive conditions, and their combined roles on particular genes’ dysregulations (in resistant-vs-sensitive) may reveal novel insights into mechanisms of acquired resistance.
Moreover, Komurov et al. [19] suggested that the drug resistance mechanism more likely occurs downstream of growth factor-mediated signaling pathways, such as Ras signaling, PI3K/AKT signaling, mTOR signaling, and others. However, an enormous number of diverse effector pathways may be involved in this process, making the prediction of biologically plausible hypotheses a challenging task. New computational approaches are needed to resolve such challenges in identifying the mechanistic underpinnings of acquired resistance.
Gene dysregulation is associated with aberrant signaling activities that are crucial for both cell growth and apoptosis in breast cancer [27]. For example, dysregulation [28] and/or mutation [28, 29] of apoptosis-related genes may overcome the initial response to apoptotic stimuli, thereby conferring resistance to apoptosis. Sharifnia et al. recently reported that several kinases and kinase-related genes from the Src family (e.g. FGFR1, FGFR2 and MOS) can compensate the loss of EGFR activity across multiple EGFR-dependent models [30]. Using unbiased gene-expression profiles of cells, their study revealed that over-expression of these EGFR-bypass genes plays a critical role in EGFR-independent activation of the MEK-ERK and PI3K-AKT signaling pathways in EGFR-mutant NSCLC cells. Recently, differential dependencies/associations were used to model rewiring in biological networks [31, 32]. Therefore, we hypothesize that differential associations between genes identified by modeling network reprogramming in resistant-vs-sensitive conditions could potentially explain gene dysregulation in acquired resistance.
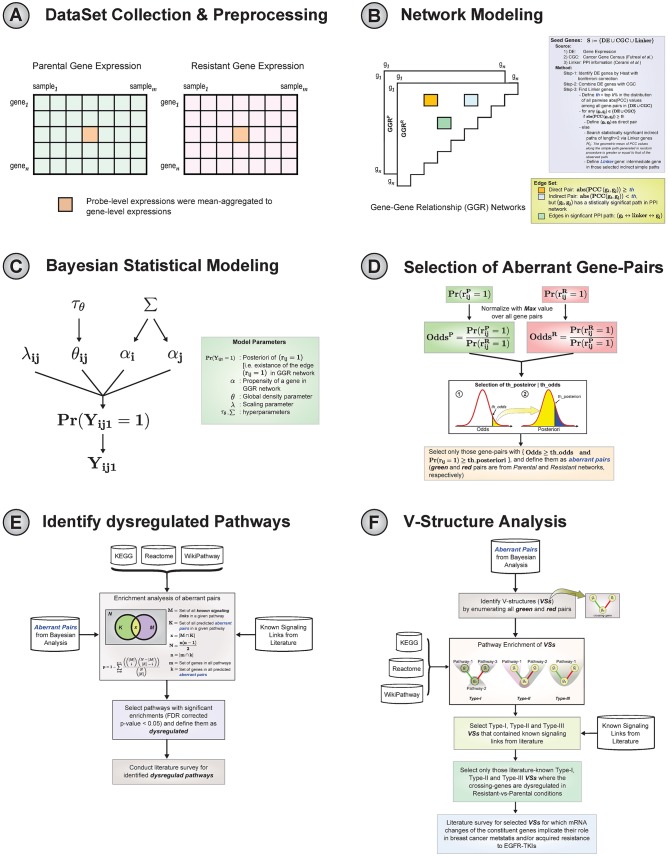
In this study, we propose a computational framework to identify dysregulated signaling pathways in resistant-vs-sensitive conditions, and a possible mechanism of gene dysregulation in acquired resistance. The schematic diagram of our proposed framework is shown in Fig 1. We used two breast cancer cell-lines, SKBR3 and BT474, each having gene expression values measured under matched lapatinib-sensitive (parental) and lapatinib-resistant conditions. A gene-gene relationship network was constructed for each gene expression dataset by combining data-driven and protein-protein interaction (PPI) information indicative of both direct and indirect relationships between gene-pairs. Then we applied a fully Bayesian approach involving the p1-model to infer gene-pairs with differential posterior probabilities between these two conditions. Next, statistically significant dysregulated signaling pathways from KEGG, Reactome, and WikiPathway were identified by enriching putative aberrant pairs using literature curated signaling links. Finally, by proposing a novel pattern of aberrant pairs, called a V-structure, we identified possible mechanisms of dysregulation in resistant-vs-sensitive conditions that may be crucial for breast cancer metastasis and/or EGFR-TKI resistance. We hope such patterns revealed using our framework will lead to further insights into aberrant signaling activities in acquired resistance.
Fig 1. Schematic diagram of our proposed framework to identify and analyse aberrant signaling pathways in acquired resistance.
(A) Gene expression datasets of breast cancer cell-lines for both parental and resistant conditions. (B) Two gene-gene relationship networks (GGR) were built from gene expression datasets of breast cancer cell-lines in Parental and resistant conditions. (C) & (D) A fully Bayesian approach was applied for detecting putative aberrant gene-pairs involved in acquired resistance. (E) Using the putative aberrant gene-pairs and a literature-curated signaling network, a statistical test was conducted to identify dysregulated pathways in acquired resistance. (F) Applying the known aberrant signaling links (from literature), we identify and explain the role of a proposed novel structure of aberrant pairs: V-structure (VS) in breast cancer metastasis and/or in developing acquired resistance to EGFR-TKIs.
Results
A framework for identifying putative aberrant gene-pairs in acquired resistance
We developed a computational framework exploiting Bayesian statistical modeling to identify putative aberrant signaling links involved in acquired resistance. In this study, we hypothesized that aberrant signaling can be detected as differential probabilities of occurrence of gene-pairs in resistant-vs-parental conditions. Thus, after building gene-gene relationship networks individually from both parental and resistant conditions, a comparative study of edge probabilities in those two networks may reveal aberrant relationships due to acquired resistance.
Our framework constructs a gene-gene relationship network, GGR: = (S, R) by combining GE and PPI datasets, where S is a set of seed genes and R is a set of pair-wise gene relationships (Fig 1). Table 1 shows primary statistics for the GGR networks of both SKBR3 (GSE38376) and BT474 (GSE16179) cell-lines. For SKBR3 cell-lines (Parental and Resistant), we selected 897 seed genes comprised of 345 differentially expressed (DE) genes (Bonferroni corrected p-value ≤ 0.01), 370 genes from the Cancer Gene Census (CGC), and 502 and 479 linker genes from Resistant and Parental cell-lines, respectively. For BT474 cell-lines, we found 875 distinct seed genes comprised of 354 DE genes (Bonferroni corrected p-value ≤ 0.05), 357 CGC genes, and 477 and 489 linker genes from Resistant and Parental cell-lines, respectively. Note that to find DE genes in SKBR3 and BT474 cell-lines, two different p-value thresholds: 0.01 and 0.05 were used, respectively. This was done for two reasons: firstly, because the computational cost of using a conventional threshold of 0.05 with SKBR3 was prohibitive, and secondly, to ensure the numbers of DE genes in the two different cell-lines were comparable, and similarly for the sizes of the seed gene sets [for details see S1 Text].
Table 1. Primary statistics of Gene-Gene Relationship (GGR) network construction for both SKBR3 and BT474 cell-lines.
| Cell Line | Cell Condition | # of DE Genes | # of CGC Genes | # of DE ∪ CGC Genes | # of All Pairs | # of Linker Genes | # of Total Seed Genes | # of combined Seed Genes | # of Direct Pairs | # of Indirect Pairs | # of PPI Pairs | # of Total Links |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SKBR3 | Resistant | 345 | 370 | 704 | 247456 | 502 | 1262 | 897 | 49492 | 1440 | 1757 | 52560 |
| Parental | 479 | 1245 | 1393 | 1758 | 52510 | |||||||
| BT474 | Resistant | 354 | 357 | 698 | 243253 | 477 | 1100 | 875 | 48651 | 1572 | 1895 | 51998 |
| Parental | 489 | 1101 | 1517 | 1951 | 51972 |
Our approach constructs a GGR in a series of stages: an initial set of genes is obtained by combining DE and CGC genes. Edges are added corresponding to direct relationships between pairs of these genes. We then search for indirect relationships among gene-pairs for which direct relationships couldn’t be found, and where indirect relationships are found the linker genes and the edges connecting them are added to the network. For the SKBR3 cell-line, the initial gene set contained 704 genes obtained by combining 345 DE and 370 CGC genes, whereas for the BT474 cell-line, the initial gene set contained 698 genes obtained by combining 354 DE and 357 CGC genes. To define direct relationships among the genes in the initial sets, we chose the top 20% from the ranked list of all pair-wise absolute Pearson Correlation Coefficients (PCC). Thus, we identified 49,492 (in both parental and resistant condition) and 48,651 (in both parental and resistant condition) direct relationships in SKBR3 and BT474 cell-lines, respectively. We justified this choice of threshold by applying an approach proposed by Elo et al. which analyses the topological properties of a co-expression network in order to find an optimal cutoff value [33] [for details see S1 Text]. In searching for indirect relationships, we found that 502 and 479 linker genes connect 1,440 and 1,393 distinct gene-pairs (for which direct relationships were not found) with the help of 1,757 and 1,758 distinct PPI links, for SKBR3 resistant and parental cell-lines, respectively. Similarly, for BT474 Resistant and Parental cell-lines, 477 and 489 linker genes connect 1,572 and 1,517 distinct indirect gene-pairs along with 1,895 and 1,951 distinct PPI links, respectively. In both datasets (SKBR3 and BT474), to build two GGR matrices for resistant and parental conditions with similar sets of genes, we constructed the final set of seed genes as an intersection of the two individual seed gene sets for Resistant and Parental conditions. Hence, 502 and 479 linker genes from SKBR3 resistant and parental conditions were combined with 704 (DE ∪ CGC) genes to form 1,262 and 1,245 seed genes, respectively, and then finding an intersection of these two sets yielded a set of 897 genes. Similarly, combining 698 (DE ∪ CGC) with 477 and 489 linker genes from BT474 resistant and parental genes produced 1100 and 1101 seed genes, respectively, and intersecting these resulted in a final set of 875 genes. At the end of this process, the SKBR3 resistant and parental GGR networks contained 897 distinct seed genes (DE ∪ CGC ∪ Linker) with 52,560 and 52,510 gene-gene relationships (direct ∪ indirect ∪ PPI), respectively, and the BT474 Resistant and Parental GGR network contained 875 distinct seed genes with 51,998 and 51,972 gene-gene relationships, respectively. Note that for both SKBR3 and BT474 cell-lines, although the total number of final seed genes is the same for both resistant and parental conditions, their respective GGR networks may contain different numbers of gene-gene relationships.
After building the GGR networks for both resistant and parental conditions and separately, we conducted Bayesian inference of parameters using the p1-model to estimate posterior probabilities of gene-gene relationships in each network. We used a WinBUGS script used in our previous work [10] for this inference. We ran the MCMC (Markov Chain Monte Carlo) method for 15,000 iterations, where the first 10,000 iterations were considered as ‘burn-in’, and the next 5,000 iterations were used for sampling. Time-series plots indicated that all parameters converged within the first few thousand iterations (data not shown). In both networks, the posterior probability of each edge was estimated to be the proportion of the 5,000 sampled networks in which that edge was present.
We identified a gene pair (genei, genej) as putatively aberrant if its posterior probabilities and of appearing in each network (resistant and parental networks, respectively) are significantly different. To determine which gene-pairs had this characteristic, we calculated two odds ratios—OddsR and OddsP—as shown in Eqs (3) and (4) for each gene-pair (genei, genej). Note that since the two posterior probabilities used in these odds ratios may lie in different ranges, we normalized their values by dividing by their respective maximum values over all the gene-pairs in the respective sets. We then used two thresholds to define significance: first, we constructed the empirical distribution of odds ratios and chose only those gene-pairs which had odds ratios among the top 20%. For SKBR3 cell-lines, these threshold values were 2.53 and 1.66 for resistant and parental conditions, respectively, and for BT474 these values were 12.028 and 2.115, respectively. Next, we constructed empirical distributions of the posterior probabilities of the previously selected gene-pairs, and chose only those gene-pairs whose posterior probabilities were in the top 50% in their respective distributions. For SKBR3 resistant and parental cell-lines, these thresholds of posterior probabilities were 0.212 and 0.252, respectively, and for BT474 cell lines, 0.177 and 0.304, for resistant and parental conditions, respectively. More detailed explanations regarding these two types of thresholds are provided in the Supplementary Methods section in S1 Text. Thus, our framework finally selected 80,372 and 76,476 aberrant gene-pairs for SKBR3 and BT474 cell-lines, respectively, and we hypothesized that these aberrant gene-pairs have the potential to explain the mechanism of acquired resistance in breast cancer. Lists of all identified putative aberrant gene-pairs for both SKBR3 and BT474 cell-lines are shown in S1 Table.
Comparing posterior probabilities to correlation coefficients
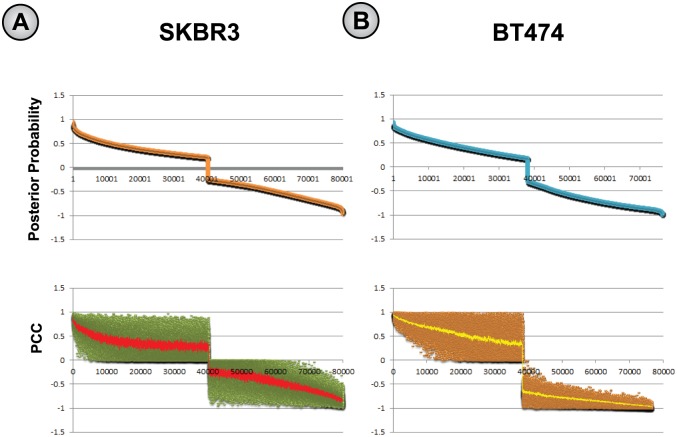
To investigate the robustness of our approach, we compared the posterior probabilities with the initial PCC (Pearson Correlation Coefficient) values for each of the putative aberrant gene pairs as shown in Fig 2. We treated the posterior probabilities of the red gene-pairs [see Methods] as positive values and the posterior probabilities of the green gene-pairs [see Methods] as negative, and plotted their sorted values in descending order (Fig 2). Next, we constructed a scatter plot with corresponding absolute PCC values for each of these gene-pairs, sorted based on posterior probabilities. We added a trendline using a moving average with window size 25, to investigate whether this trendline was in any way similar to the trend observed in the posterior probabilities. Interestingly, for both SKBR3 and BT474 cell-lines, the trendlines of PCC values revealed a visually similar pattern to that of the corresponding posterior probability values. This confirms our expectation that our Bayesian analysis is sensitive to a signal in the PCC values that would be otherwise difficult to detect.
Fig 2. Comparing the posterior probabilities of putative aberrant gene-pairs with corresponding PCC (Pearson Correlation Coefficient) values that were defined among genes prior to the Bayesian analyses, (A) for SKBR3 and (B) BT474 cell-lines.
The first figures in (A) and (B) show the sorted posterior probability values of the putative aberrant gene-pairs in descending order, and the second figures of (A) and (B) show the scatter plot of their corresponding PCC values. Note that for both the graphs in (A) and (B) the rank of ordered aberrant pairs is shown in X-axis, and the posterior probabilities and PCC values of red gene-pairs are shown in Positive Y-axis and those of green pairs are shown in Negative Y-axis, correspondingly. A trendline (red or yellow trendlines for the SKBR3 and BT474 cell-lines, respectively) is drawn for each of the scatter plots (in (A) and (B)) by using a moving average with a window size set to 25. For both SKBR3 and BT474, these trendlines clearly show the similarity of the signal contained in the PCC values (defined prior to Bayesian analyses) and the pattern of changes in a posteriori values (resulting from Bayesian analyses), and demonstrates the robustness of Bayesian statistical modeling for selecting putative aberrant gene-pairs involved in acquired resistance.
Many crucial signaling pathways are significantly enriched with aberrant gene-pairs in acquired resistance
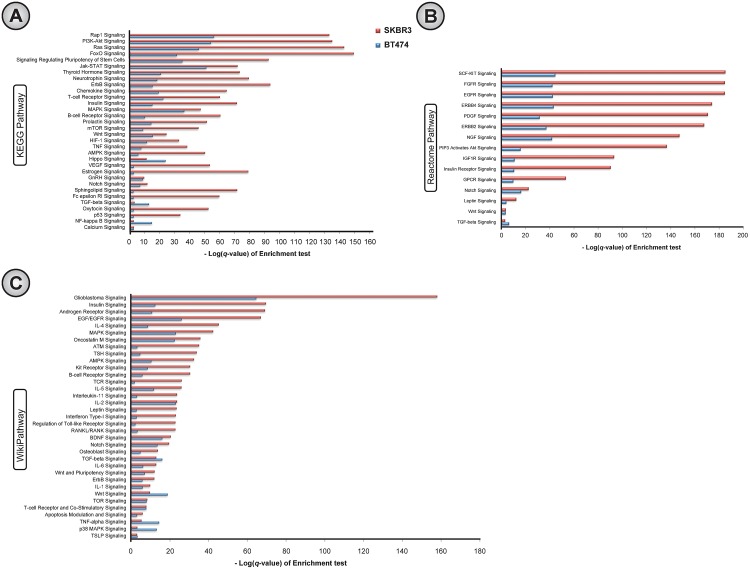
To measure the significance of signaling pathways in terms of aberrant signaling activities in acquired resistance, we conducted a hypergeometric test. In this test, we measured how significant was the overlap between the set of literature-supported signaling links [34] found in a particular signaling pathway with the set of putative aberrant gene-pairs in the same pathway. We identified all the signaling pathways from KEGG, Reactome, and WikiPathway databases for which the corresponding q-value (FDR corrected p-value) from the above hypergeometric test was < 0.05 in both SKBR3 and BT474 cell-lines as is shown in Fig 3. For both SKBR3 and BT474 cell-lines, 71.11% (32 out of 45), 62.5% (15 out of 24), and 57.38% (35 out of 61) signaling pathways from KEGG, Reactome, and WikiPathways, respectively, were found to be significantly enriched with aberrant signaling gene-pairs in acquired resistance (Fig 3). Again, for all corresponding KEGG, Reactome, and WikiPathway databases, such high percentages of enriched signaling pathways found in both SKBR3 and BT474 cell-lines indicates that our framework is consistent in terms of finding aberrant gene-pairs in both cell-lines. Complete enrichment results of this hypergeometric test are reported in S2 Table.
Fig 3. Analysis of dysregulated pathways by conducting pathway enrichment test of aberrant gene-pairs with known signaling links [34] involved in acquired resistance in SKBR3 and BT474 breast cancer cell-lines.
Enrichments of all signaling pathways in (A) KEGG, (B) Reactome, and (C) WikiPathway.
We conducted a literature survey for the putative dysregulated signaling pathways, and found that the aberrant activities in most of these pathways are strongly associated with acquired resistance to EGFR tyrosine kinase inhibitors (EGFR-TKIs) [18]. EGFR (also known as HER1, or ErbB1) and EGFR 2 (also known as HER2/neu, or ErbB2) are cell surface transmembrane proteins, and members of the HER family of receptors. EGFR (in KEGG, Reactome, and WikiPathway) and ErbB2 (in Reactome) are reported to be frequently mutated and/or over-expressed in various types of cancer resulting in aberrant activities contributing to abnormal cell growth, survival, migration, and differentiation [35, 36]. However, over-expression and secondary mutations of both EGFR [11, 37–39] and ErbB2 [40] are associated with acquired resistance to EGFR-TKI. Moreover, being key components of cell signaling systems, these RTKs control major downstream signaling pathways, i.e. Ras/Raf/MAPK (in KEGG and WikiPathway), PI3K-Akt (in KEGG and Reactome), FoxO (in KEGG), and Jak-STAT (in KEGG) that are crucial for cancer cell growth and survival [3]. Moreover, as these downstream signaling pathways further regulate multiple downstream effector pathways (related to cell growth and survival), aberrant re-activation of those pathways provide a common mechanism to compensate for inhibition of targeted pathways, thereby conferring acquired resistance to EGFR-TKIs [4, 41, 42]. Interestingly, these signaling pathways (i.e. Ras, PI3K-Akt, FoxO, Jak-Stat signaling) were found as the top-most in the list of aberrant signaling pathways in both datasets (SKBR3 and BT474) based on the above hypergeometric test using KEGG database as shown in Fig 3. For ErbB4 signaling (in Reactome), recently it has been reported that in ErbB2-positive breast cancer cell-lines, ErbB4 was up-regulated at the protein level in vitro and re-activated PI3K-Akt signaling in resistant conditions compared to the sensitive condition, and the knock-down of ErbB4 induced apoptosis in both the lapatinib-resistant and trastuzumab-resistant cell-lines [43]. Rap1 (in KEGG) and ras (in KEGG) signaling are activated by lung cancer oncogene CRKL whose focal amplification (secondary mutation) was reported to be associated with acquired resistance to EGFR inhibitor [44]. Again, signals for cell proliferation and survival from activated AKT may transduce through several phosphorylated transcription factors, such as FoxO (in KEGG) [45], which indicates that the dysregulation of FoxO signaling pathway (in KEGG) may potentially be associated with resistance to EGFR-TKIs.
Our previous study found cross-talks between EGFR signaling and pathways triggered by other types of receptors, e.g. Notch, Wnt, IGF1R, GPCR, etc. which contributed to acquired resistance to lapatinib (an EGFR/Her2 dual inhibitor) [10]. Here, we also found these pathways showing significant aberrant activities in acquired resistance to lapatinib [Fig 3]. The activation of IGF1R signaling (in KEGG, Reactome, and WikiPathway) is commonly reported to induce acquired resistance to EGFR-TKIs by many studies [46, 47], and its inhibition could down-regulate PI3K-Akt signaling, eventually inhibiting cell growth, providing co-inhibition of EGFR and IGF1R signaling a clinical success [3]. Similarly, the Notch signaling pathway (in KEGG, Reactome, and WikiPathway) cross-talks with EGFR signaling in breast cancer, thus maintaining the cancer cell growth signal through MAPK and PI3K-Akt signaling [48]. It is suggested that an improved drug-sensitivity could be achieved by down-regulating the Notch signaling pathway with specific inhibitors [49, 50]. Again, genes involved in Wnt signaling (in KEGG, Reactome, and WikiPathway) were up-regulated in the resistant condition in both breast and colon cancer when compared to the sensitive condition [51, 52], thus contributing to acquired resistance to EGFR-TKIs [51].
Targeting angiogenesis is another important aspect of anticancer therapies [53], as aberrant vascularity and hypoxia are directly associated with tumor growth and survival [3]. In our analysis, we found aberrant angiogenic pathways including signaling by Vascular Endothelial Growth Factors (VEGFs) (in KEGG), Fibroblast Growth Factors (FGFs), and Platelet-Derived Growth Factors (PDGFs). It has been reported that the VEGF/VEGFR-2 feed-forward loop increases VEGF secretion in lung cancer via mTOR-dependent regulation that is required for the activation of downstream signaling [54], and the over-expression of VEGFR-1 reduces EGFR-TKIs sensitivity in different human cancer cells [3, 55]. Alternate activation of the FGFR signaling pathway (in Reactome) through the over-expressions of FGFR1 and FGF2 acts as a compensating mechanism for EGFR-TKIs [56] by maintaining signals for cell survival and proliferation in the downstream signaling pathways [4]. Again, it has been recently reported that, in PDGFR signaling (in Reactome), transcriptional de-repression of PDGFR-β contributed to compensating for the effects of EGFR-TKIs in EGFR-mutant glioblastoma via an mTORC1- and extracellular signal regulated kinase-dependent mechanism [21].
The hippo signaling pathway (in KEGG) is associated with cell proliferation, apoptosis, organ size control, and stem cell self renewal [57]. YAP is a transcription co-activator and oncoprotein [58], and plays a central role in cancer-related activities of the hippo signaling pathway [57]. Huang et al. have recently reported that down-regulating YAP expression in various cell-lines can improve the sensitivity of erlotinib (an EGFR-TKI) and cetuximab (anti-EGFR drug) [59]. We found the gene-pair AKT2:MYC as a signaling cross-talk between EGFR/ErbB and the TGF-β signaling pathway (in KEGG, Reactome, and WikiPathway) in our previous study [10]. Recently, it has been reported that combined inhibition of EGFR-TKIs (erlotinib) and TGF-β type I receptor inhibitor may improve sensitivity of EGFR-TKIs in lung cancer without EGFR T790M mutation [60].
For both SKBR3 and BT474 cell-lines, the primary findings in this study with supporting references are summarized in Tables 2 and 3. In this table, for each aberrant pathway, we also show what percentages of predicted gene-pairs from Bayesian analysis were previously defined as direct relationships, indirect relationships, and PPI during the network modeling. It is apparent that substantial proportions of predicted pairs came from direct and indirect relationships in both SKBR3 and BT474 cell-lines. This also indicates the robustness of our Bayesian modeling in inferring gene-pair relationships. Note that in the above calculation, if a predicted pair was defined both as direct and PPI, or both as indirect and PPI, then we counted that as direct or indirect, respectively, since that prediction for that particular pair was made by our framework. Again, some of the predicted pairs (by Bayesian modeling) may not be defined as direct or indirect previously, because the definitions of the terms predicted (based on posterior probability from Bayesian modeling), direct, and indirect were based on thresholds calculated from the distributions of corresponding values [see Methods]. Thus, the enrichment test with literature supported gene-dependencies [34] along with the evidences from the above literature survey confirm that our framework is able to identify significantly dysregulated signaling pathways that have key associations with acquired resistance in cancer.
Table 2. Summary of predicted dysregulated EGFR and its downstream signaling pathways from KEGG, Reactome and WikiPathway databases in acquired resistance in both SKBR3 and BT474 cell-lines.
| Aberrant Pathways in EGFR-TKIs Resistancek,r,w | % of Direct Pair(s,b)k,r,w | % of Indirect Pair(s,b)k,r,w | % of PPI Pair(s,b)k,r,w | # of Enriched Pair(s,b)k,r,w | Enrichment q-value(s,b)k,r,w | Literature References |
|---|---|---|---|---|---|---|
| EGFR and downstream pathways | ||||||
| EGFR signaling | (53.12%, 71.05%)k | (18.75%, 13.16%)k | — | (6, 32)k | (5.1e-16, 1.5e-94)k | [11, 37–39] |
| (30.43%, 71.57%)r | (8.7%, 6.86%)r | (__, 0.49%)r | (18, 73)r | (3.2e-43, 1.7e-185)r | ||
| (56%, 71.79%)w | (12%, 7.69%)w | (__, 0.85%)w | (2, 4)w | (1.0e-26, 1.3e-67)w | ||
| ErbB2 signaling | (33.96%, 74.12%)r | (7.55%, 4.12%)r | (__, 0.59%)r | (15, 64)r | (7.8e-38, 2.5e-168)r | [40] |
| ErbB4 signaling | (29.09%, 72.19%)r | (9.09%, 4.73%)r | (__, 0.59%)r | (17, 65)r | (6.6e-44, 7.5e-175)r | [43] |
| Ras signaling | (34.62%, 66.05%)k | (11.54%, 6.17%)k | (__, 0.62%)k | (22, 60)k | (6.5e-47, 6.9e-144)k | [4, 41–44] |
| MAPK signaling | (35.82%, 60.32%)k | (8.96%, 7.94%)k | — | (19, 23)k | (4.4e-37, 4.2e-48)k | [3, 4] |
| (31.82%, 48.05%)w | (9.09%, 12.99%)w | — | (12, 19)w | (8.7e-24, 5.1e-43)w | ||
| PI3K-Akt signaling | (35.27%, 61.85%)k | (10.62%, 5.69%)k | (__, 0.95%)k | (34, 75)k | (5.4e-55, 7.2e-136)k | [3, 4] |
| (26.67%, 73.45%)r | (6.67%, 4.42%)r | (__, 0.88%)r | (6, 46)r | (1.5e-16, 1.4e-137)r | ||
| Jak-Stat signaling | (25.49%, 71.19%)k | (19.61%, 13.56%)k | (__, 1.69%)k | (20, 7)k | (6.8e-52, 8.6e-73)k | [3, 4] |
| Rap1 signaling | (25%, 61.03%)k | (11%, 8.09%)k | (1%, 0.74%)k | (25, 53)k | (4.4e-57, 5.5e-134)k | [44] |
| FoxO signaling | (48.15%, 78.45%)k | (7.41%, 2.76%)k | (1.85%, 1.1%)k | (12, 54)k | (3.1e-32, 2.7e-150)k | [45] |
k KEGG
r Reactome
w WikiPathway
s SKBR3
b BT474;
Table 3. Summary of predicted dysregulated signaling pathways from KEGG, Reactome and WikiPathway databases that plays a role as compensatory pathway of EGFR/HER2 inhibition in acquired resistance in both SKBR3 and BT474 cell-lines.
| Aberrant Pathways in EGFR-TKIs Resistancek,r,w | % of Direct Pair(s,b)k,r,w | % of Indirect Pair(s,b)k,r,w | % of PPI Pair(s,b)k,r,w | # of Enriched Pair(s,b)k,r,w | Enrichment q-value(s,b)k,r,w | Literature References |
|---|---|---|---|---|---|---|
| Compensating Pathways of EGFR/HER2 inhibition | ||||||
| Notch signaling | (40%, 75%)k | (__, 25%)k | — | (2, 3)k | (8.6e-08, 1.7e-12)k | [48–50] |
| (46.15%, 71.43%)r | (7.69%, 4.76%)r | — | (5, 7)r | (5.2e-17, 3.6e-23)r | ||
| (35%, 70.37%)w | (__, 7.41%)w | — | (5, 7)w | (3.2e-14, 3.2e-20)w | ||
| Wnt signaling | (25%, 50%)k | (12.5%, 28.57%)k | — | (6, 8)k | (3.4e-16, 3.2e-25)k | [51, 52] |
| (21.88%, 66.67%)r | (3.12%, 3.33%)r | — | (2, 2)r | (2.9e-04, 2.7e-04)r | ||
| (25%, 55.56%)w | (12.5%, 11.11%)w | — | (3, 6)w | (1.7e-19, 2.1e-10)w | ||
| Insulin Receptor/IGF1R signaling | (40%, 70.49%)k | (13.33%, 9.84%)k | — | (6, 25)k | (7.9e-16, 3.1e-72)k | [3, 10, 46, 47] |
| (29.41%, 87.93%)r | (5.88%, 3.45%)r | — | (4, 30)r | (1.3e-11, 6.6e-94)r | ||
| (35.9%, 80%)w | (12.82%, 9.33%)w | (__, 1.33%)w | (6, 28)w | (4.1e-13, 2.9e-70)w | ||
| VEGFR signaling | (40%, 81.82%)k | (__, 4.55%)k | — | (1, 15)k | (1.9e-03, 3.6e-54)k | [3, 55] |
| FGFR signaling | (32.05%, 71.14%)r | (8.97%, 6.97%)r | (__, 0.5%)r | (18, 72)r | (7.6e-43, 1.7e-185)r | [4, 56] |
| PDGFR signaling | (40.78%, 71.35%)r | (3.88%, 3.78%)r | (__, 0.54%)r | (15, 67)r | (2.2e-32, 1.9e-171)r | [21] |
| Others | ||||||
| Hippo signaling | (41.46%, 72.22%)k | (12.2%, 11.11%)k | — | (9, 4)k | (1.0e-24, 6.7e-12)k | [59] |
| TGF-β signaling | (22.22%, 100%)k | (11.11%, __)k | — | (4, 1)k | (1.1e-13, 5.0e-04)k | [10, 60] |
| (50%, 50%)r | (25%, __)r | — | (2, 1)r | (4.4e-07, 7.7e-04)r | ||
| (54.55%, 30%)w | (18.18%, 40%)w | — | (5, 4)w | (1.1e-16, 1.2e-13)w |
k KEGG
r Reactome
w WikiPathway
s SKBR3
b BT474;
Comparing with our previous study
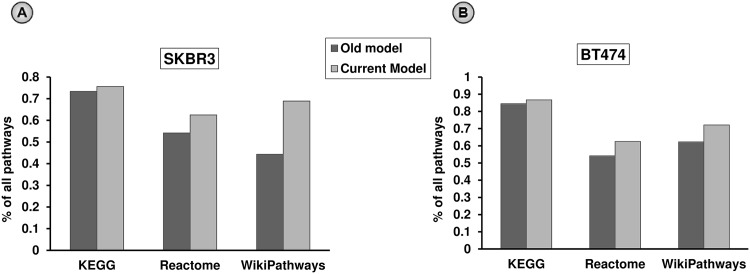
To compare the performances of our current framework with our previous one [10], we investigated which of the two frameworks identify a greater number of dysregulated signaling pathways from KEGG, Reactome, and WikiPathway databases, since we used similar gene expression datasets (SKBR3 and BT474) in both approaches. We conducted a hypergeometric test to measure the statistical significance of the overlap between the aberrant pairs and known signaling links [34]. For that purpose, we defined the aberrant pairs in our previous approach [10] with oddsP and oddsR > 10.0, and posterior probabilities, and > 0.5. We found that greater percentages of pathways from KEGG, Reactome, and WikiPathway databases were found as perturbed (dysregulated) in acquired resistance when the current approach was used compared to the old one [Fig 4].
Fig 4. Performance comparison between the current model and our previous model [10] in terms of detecting perturbed signaling in acquired resistance.
Percentages of signaling pathways detected as perturbed in acquired resistance by our current and old models in all KEGG, Reactome and WikiPathway databases: (A) in SKBR3, and (B) in BT474 cell-lines. For both the cell-lines, the performances using KEGG and Reactome pathways are comparable in both approaches, whereas our current model outperforms the old model for pathways from WikiPathway database.
One of the main differences between these two approaches was in the definitions of the set of edges in GGR network models: the current approach used direct pairs and non-direct pairs (indirect pairs and PPI pairs), whereas the old approach only used direct pairs [10]. Therefore, we conducted two experiments to investigate the importance of non-direct pairs in the new model. First, in aberrant signaling pathways that were detected by our current but not the previous model, we observed what percentages of enriched links (i.e. aberrant pairs found as known signaling links) were previously defined as non-direct (indirect and PPI) pairs in our current model. In both SKBR3 and BT474 cell-lines, we found that all such dysregulated pathways from KEGG, Reactome, and WikiPathway databases contained high percentages of non-direct (indirect and PPI) enriched links [S3 Table]. Second, in aberrant signaling pathways that were detected by both of our current and previous models and were ranked (based on enrichment q-values) high in the current model but low in the previous model, we observed what percentages of enriched links were previously defined as non-direct in our current model. Considering the rank difference ≥ 10 (an empirical cutoff threshold), we found that aberrant pathways from KEGG, Reactome, and WikiPathway databases that showed such behavior in both SKBR3 and BT474 cell-lines, also contained high percentages of non-direct (indirect and PPI) enriched links [S4 Table]. Therefore, we claim that our current model demonstrate enhanced performances in detecting dysregulated signaling pathways in acquired resistance compared with our previous model.
Comparing with other methods
Next, we compared our framework with other published methods in terms of identifying the aberrant signaling pathways, specifically SPIA [61], DAVID [62], GATHER [63], ESEA [64] and PAGI [65]. The first three methods (i.e. SPIA, DAVID, and GATHER) are node-centric methods, where the role of differentially expressed (DE) genes was the key to identifying dysregulated pathways. However, ESEA and PAGI are edge-centric methods, where topological information regarding pathway structures was significantly exploited. All of these methods use GE datasets, except DAVID and GATHER which take a list of DE genes as input and identify aberrant pathways, or pathways enriched with given DE genes, respectively. For this comparative analysis, we used KEGG signaling pathways only, and for all the methods default configurations were applied unless specified otherwise.
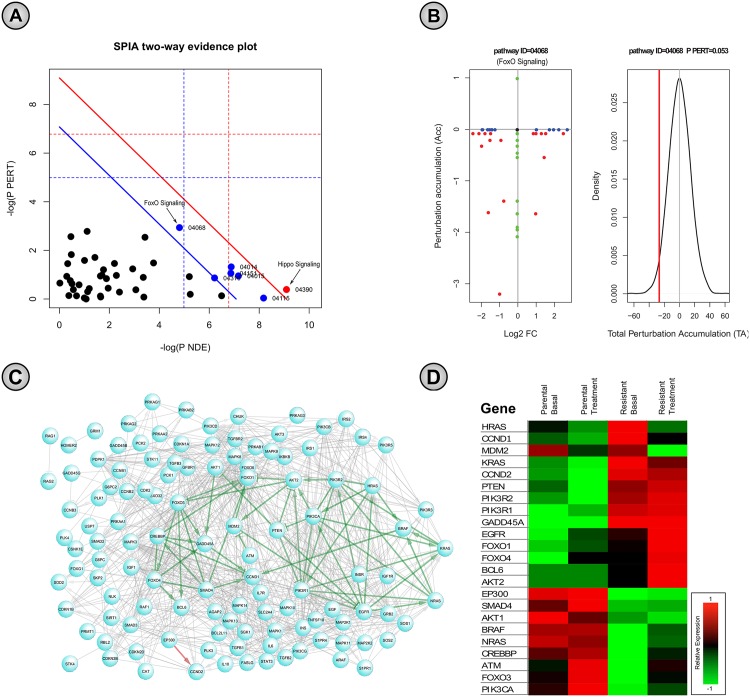
The SPIA method combines classical enrichment analysis and actual aberrant activities by analysing Cancer-Vs-Normal GE samples [61], and ranks corresponding signaling pathways by calculating a global pathway significance p-value, called pG. The global p-value (pG) is obtained by combining the perturbation probability (p-value: pPERT) and the probability of over-representation of DE genes (using log fold-change) in a particular pathway (p-value: pNDE) by using either Fisher’s method or the normal inversion method [61]. Here, we conducted the same analysis but with Resistant-vs-Parental GE samples aiming to capture the aberrant activities responsible for acquired resistance. In the case of multi-probe sets for the same gene, we used the most significant probe to get a single log2 fold-change value per gene. For SKBR3 cell-lines, we found 5 signaling pathways as significant (raw pG-value ≤ 0.05) including Ras signaling, PI3K-Akt signaling, Rap1 signaling, hippo signaling, thyroid hormone signaling, and TGF-β signaling pathways. Interestingly, we found that the significance (−log(q-values)) of aberrant pathways found by our approach is strongly correlated with the global p-values (pG) found by SPIA analysis, both for all pathways (-0.4) and for above 5 signaling pathways only (-0.928). This indicates, in SKBR3 cell-lines, the signaling pathways from our framework with high enrichment of aberrant gene-pairs in acquired resistance are also consistent with the results from SPIA in terms of identifying aberrant activities. Again, for BT474, we found 12 signaling pathways with significant aberration (raw pG-value ≤ 0.05), i.e. hippo, p53, Ras, Rap1, PI3K-Akt, FoxO, Wnt, neurotrophin, insulin, estrogen, ErbB, and MAPK signaling pathways. Moreover, among these 12 signaling pathways, the first 6 had FDR-corrected pG ≤ 0.05, among which hippo signaling pathway had Bonferroni-corrected pG ≤ 0.05, as shown in Fig 5A. Among these 12 significantly dysregulated pathways in BT474 cell-line, we chose FoxO signaling to investigate further, since it was found highly perturbed by both SPIA (pPERT = 0.053) and our methods (enrichment q-value = 2.7 × 10−150). We observed perturbation plots for this signaling pathway (KEGG pathway ID = 04068), in which perturbations of all genes were plotted as a function of their initial log2 fold-change Fig 5B. Here, non-DE genes were assigned 0 for their log2 fold-change. However, many genes were identified as DE, since their absolute log2 fold-change values were mostly ∼2. Again, compared to the null distribution of net accumulated perturbation values, the observed value was also found significant as shown with the red vertical line in Fig 5B. Next, we also drew the network view of the FoxO signaling pathway, where the nodes were the constituent genes (from KEGG), and the edges were the known signaling links from the literature [34]. Here, we found 54 known signaling links that were also identified as aberrant gene-pairs by our method. Next, we plotted the heatmap of the expression values of the genes in these 54 known aberrant signaling links, where each expression value was the mean of all three replicates [66], z-transformed, and normalized with absolute max value (of the z-scores across the particular gene). Here, this heatmap not only shows the differential expression of the genes in aberrant gene-pairs but also indicates the similarities of their expression changes within this signaling pathway, which is a marker of aberrant activities in a modular way. Such differential gene expression in resistant-vs-parental conditions may indicate that pathway dysregulation within the signaling circuitry can be mediated by the corresponding aberrant gene-pairs.
Fig 5. Detection of perturbed pathways with SPIA method.
(A) Two-way evidence plot for all 45 KEGG pathways for BT474 cell-line is drawn. Here, pathways are represented with dots and the pathways with red dots and blue dots correspond to perturbed pathways with FDR-corrected and Bonferroni-corrected global p-value, pG < 0.05, respectively. (B) Next, the perturbation plot for FoxO signaling pathway (KEGG pathway ID = 04068) was also observed, since it contains the lowest perturbation p-value among all, pPERT = 0.053. In this plot, perturbation of all genes in the FoxO signaling pathway are shown as a function of their initial log2 fold-change (lower-left panel), where each dot indicates a gene in the pathway, and non-differentially expressed genes are assigned 0 as their log2 fold-change value. The null distribution and the observed net accumulated perturbation (red line) are shown in the lower-right panel. (C) Network view of FoxO signaling pathway for BT474 cell-line, where nodes are the constituent genes and the edges are known links collected from literature [34]. Here, green and red edges are the aberrant gene-pairs found by our method. (D) The heatmap of the genes’ expression in aberrant gene-pairs found by our method in FoxO signaling network for BT474 cell-line.
As DAVID and GATHER both take as input a list of presumably differentially expressed genes for their pathway enrichments, we used the list of 703 and 683 distinct genes in the list of aberrant gene-pairs which were found by our framework from SKBR3 and BT474 cell lines, respectively. To detect statistically significant pathways using DAVID and GATHER we select those for which the raw p-values of their enrichment were < 0.05. For SKBR3 cell-line, DAVID and GATHER identified 15 and 5 signaling pathways as statistically significant, respectively. Again, for BT474 cell-lines they found 13 and 4 pathways as significant, respectively.
For both ESEA and PAGI analyses, we used our Resistant and Parental GE datasets for both SKBR3 and BT474 cell-lines. For both analyses, we used the default running parameters, except for the parameter nperm (the number of permutations) which was set to 1000. Both of these methods used a built-in set of topological structures of pathways from known pathway databases including KEGG. After running these methods with our GE datasets, if the identified signaling pathways had nominal p-value < 0.05, then we considered them as significantly dysregulated in resistant-vs-parental conditions. Thus, in the SKBR3 cell-line, we found 4 and 15 significantly dysregulated signaling pathways by ESEA and PAGI methods, respectively. For the BT474 cell-line, we found 2 and 12 signaling pathways significantly dysregulated by ESEA and PAGI, respectively.
All the dysregulated KEGG signaling pathways identified by any of these six methods are listed in Table 4. Some pathways were found consistently as dysregulated in both SKBR3 and BT474 cell-lines, but none were common to all six methods. However, our method identifies 33 KEGG pathways in both SKBR3 and BT474 cell-lines among which 17 were also identified by at least one of the other five methods (including both node-centric and edge-centric methods), for example MAPK, insulin (in DAVID, GATHER, and PAGI), ErbB, Wnt, B-cell receptor, Neurotrophin (in DAVID and PAGI), p53 (in DAVID and ESEA), and Jak-Stat signaling (in DAVID and GATHER). Moreover, our method identifies some novel dysregulated pathways in both SKBR3 and BT474 cell-lines which were not detected by any other methods. These pathways include Hif-1, AMPK, TNF and calcium signaling, which were reported to be involved in lapatinib-resistance in ErbB2-positive breast cancer cell-lines [4, 19, 67, 68]. Thus, the comparative identification of dysregulated pathways in resistant-vs-parental conditions in both SKBR3 and BT474 cell-lines indicates that our method is not only comparable to others but also able to detect novel findings which were validated by literature evidence.
Table 4. Comparative identification of pathway dysregulation in all 45 KEGG signaling pathways in resistant-vs-parental conditions in both SKBR3 and BT474 cell-lines.
‘S’ for SKBR3 cell-line, and ‘B’ for BT474 cell-line.
| Pathway | SPIA | DAVID | GATHER | ESEA | PAGI | Our Method |
|---|---|---|---|---|---|---|
| MAPK signaling | B | SB | SB | SB | SB | |
| Insulin signaling | B | SB | SB | SB | SB | |
| ErbB signaling | B | SB | SB | SB | ||
| p53 signaling | B | SB | SB | B | SB | |
| Wnt signaling | B | SB | SB | SB | ||
| Jak-Stat signaling | SB | SB | SB | |||
| B-cell receptor signaling | SB | SB | SB | |||
| Neorotrophin signaling | B | SB | SB | SB | ||
| Ras signaling | SB | SB | ||||
| Rap1 signaling | SB | SB | ||||
| Chemokine signaling | SB | S | SB | |||
| mTOR signaling | SB | B | SB | |||
| PI3K-Akt signaling | SB | SB | ||||
| TGF-beta signaling | S | S | SB | SB | ||
| VEGF signaling | SB | S | S | SB | ||
| Hippo signaling | SB | SB | ||||
| Fc epsilon RI signaling | SB | SB | ||||
| Calcium signaling | SB | |||||
| NF-kappa B signaling | S | SB | ||||
| HIF-1 signaling | SB | |||||
| FoxO signaling | B | S | SB | |||
| Phosphatidylinositol | SB | B | ||||
| signaling system | ||||||
| Sphingolipid signaling | SB | |||||
| AMPK signaling | SB | |||||
| Notch signaling | S | B | SB | |||
| Toll-like receptor signaling | SB | S | B | |||
| T-cell receptor signaling | B | S | SB | |||
| TNF signaling | SB | |||||
| GnRH signaling | B | SB | ||||
| Estrogen signaling | B | SB | ||||
| Prolactin signaling | SB | |||||
| Thyroid hormone signaling | S | SB | ||||
| Oxytocin signaling | SB | |||||
| Epithelial cell signaling in | B | |||||
| Helicobacter pylori infection | ||||||
| PPAR signaling | S | |||||
| cGMP-PKG signaling | B | |||||
| cAMP signaling | B | |||||
| Adrenergic signaling | B | |||||
| in cardiomyocytes | ||||||
| Hedgehog signaling | S | |||||
| signaling pathways regulating | SB | |||||
| pluripotency of stem cells | ||||||
| NOD-like receptor signaling | S | S | ||||
| RIG-I-like receptor signaling | S | |||||
| Adipocytokine signaling | B | S | ||||
| Glucagon signaling | B |
V-structures can explain the role of aberrant signaling in acquired resistance
The importance of V-structures
To investigate the potential of the putative aberrant gene-pairs to characterise acquired resistance, we hypothesized that genes become dysregulated in acquired resistance because of the compensating effect of aberrant signaling that evolves in resistant-vs-parental conditions. In the simplest cases, this will involve both red and green aberrant edges incident upon a particular dysregulated gene. To investigate this hypothesis, we identified all genes with at least two aberrant links to observe which of two possible architecture types are associated with a larger number of dysregulated genes: 1) both red and green aberrant edges incident upon a gene (forming V-structures—see Methods for the definition), or 2) only red or only green aberrant edges incident upon a gene. Next, we identified the dysregulated genes among these for which the following was true: a gene is over-/under-expressed (in any patient sample) in PT-vs-PB conditions, but respectively under-/over-expressed in both RB-vs-PB and RT-vs-PB conditions, where PB, PT, RB and RT stand for ‘Parental Basal’, ‘Parental Treatment’, ‘Resistant Basal’ and ‘Resistant Treatment’, respectively. The rationale for using only such combinations is as follows. Both expression datasets of SKBR3 (GSE38376) [19] and BT474 (GSE16179) [66] cell-lines contain steady-state measurements of signaling activities, for both parental and resistant conditions. Therefore, we hypothesized that the expression changes of dysregulated genes in PT-vs-PB conditions may indicate the sensitivity of Lapatinib drug (EGFR/HER2 dual inhibitor) in the parental (sensitive) conditions whereas the opposite changes in expressions in both RB-vs-PB and RT-vs-PB conditions may indicate two things: 1) the cell-line had already became resistant to the drug for which the tumorigenic phenotype of cancer cells relapsed in the resistant condition (RB-vs-PB), and 2) the resistance characteristics of the cell-line persisted even with further treatment with lapatinib (RT-vs-PB). For each comparison, we examined the log2 of fold-change values, and the treatment and basal doses were 1.0 μM and 0 μM, respectively. We found that, for both SKBR3 and BT474 cell-lines, higher percentages of dysregulated genes were identified with both green and red aberrant signaling links compared to those with only a single type of incident edge (either red or green). For SKBR3 and BT474 cell-lines we identified 111 and 108 genes with degree ≥ 2, respectively. For the SKBR3 cell-line, 90 of the 111 genes had only one type of aberrant signaling link incident upon them, out of which 48 showed dysregulation (53.3%), whereas the remaining 21 of the 111 genes had both red and green aberrant signaling links, out of which 13 genes were dysregulated (62%). Similarly, for BT474 cell-lines, among the 108 genes with degree ≥ 2, 78 out of 102 (76%) of genes with only one type of aberrant link and 6 out of 6 (100%) of genes with both types of aberrant signaling links, exhibited dysregulation. These results suggest that for a dysregulated gene in resistant-vs-parental conditions, the expression changes that occur upon treatment in parental conditions are likely to be compensated by aberrant signaling link(s) that evolved in resistant conditions. Therefore, the initial effect of inhibitors on oncogene(s)/tumor suppressor gene(s) becomes abrogated by restoring their tumorigenic phenotype once the cell acquires resistance to that inhibitor. This experiment demonstrates that V-structures can explain an interesting mechanism of acquired resistance in cell-lines by associating the dysregulated gene(s) with both red and green aberrant signaling links.
Type-II and Type-III V-structures provide a possible mechanism of gene dysregulation in acquired resistance
From the list of all putative aberrant gene-pairs (after Bayesian analysis), we enumerated all possible V-structures. We first listed all of the genes in red aberrant pairs, and separately listed all of the genes in green aberrant pairs. We then identified the genes common to both lists, which we termed crossing-genes. Next, we aggregated aberrant gene-pairs incident upon crossing-genes and enumerated all possible pairs of a red and green edge incident upon that gene. Thus, we found 23,156 distinct Type-I V-structures [see Methods for Type-I, Type-II and Type-III V-structure definitions] in SKBR3 cell-lines using signaling pathways from KEGG, Reactome, and WikiPathway, out of which 53 V-structures were found in the literature-curated signaling network [34]. Similarly for BT474, there were 5,271 distinct Type-I V-structures in all KEGG, Reactome, and WikiPathway signaling pathways, and 11 of them overlapped with the literature-curated network [34]. For Type-II V-structures in SKBR3 and BT474 cell-lines, 1,525 and 263 distinct V-structures were found in all KEGG, Reactome and WikiPathway databases, respectively, out of which 29 and 4 V-structures were found in the literature-curated network [34], respectively. For Type-III V-structures in SKBR3 and BT474 cell-lines, 940 and 376 distinct V-structures were found in all KEGG, Reactome, and WikiPathway databases, respectively, where 18 and 10 V-structures overlapped with the literature-curated signaling network [34]. A summary of these results for SKBR3 and BT474 cell-lines is provided in S5 and S6 Tables, respectively. Note that Type-I and Type-II V-structures have the potential to explain the role of signaling cross-talks in acquired resistance, but here we focus on Type-II V-structures only, since we have already investigated the role of signaling cross-talks in acquired resistance in our previous study [10] which are the similar kind of Type-I V-structures.
We investigated whether Type-II and Type-III V-structures can provide insights of a possible mechanism of acquired resistance in cancer cell-lines, focusing on the dysregulations of the crossing-genes in resistant-vs-parental conditions and its association with the GE changes of the other two genes in a particular V-structure. Our rationale was that the dysregulation of a crossing-gene may provide an indication that significant changes evolved in resistant-vs-parental conditions are associated with acquired resistance of cell-lines to a particular inhibitor. Moreover, significant GE changes in either of the two other genes (in the V-structure) would indicate that their differential associations with crossing-gene(s) may disrupt their functional coherence in signaling activities [30]. Therefore, we considered the above-mentioned 13 and 6 dysregulated genes in SKBR3 and BT474, respectively, for further analyses in which gene-pairs in corresponding V-structures overlapped with known signaling links [34]. Among the 13 dysregulated genes in SKBR3 cell-lines, 8 genes (CTNNB1, TP53, MYC, RAC2, LCK, PIK3R1, PIK3CA, and TGFBR2) were found in 22 (out of 29) literature-supported Type-II V-structures and 4 genes (CTNNB1, TP53, MYC, and PIK3CA) were found in 9 (out of 18) literature-supported Type-III V-structures (S5 Table). Similarly, among 6 dysregulated genes in BT474 cell-lines, 3 genes (CTNNB1, LEF1, and TP53) were found in 4 (out of 4) literature-supported Type-II V-structures and 4 genes (MET, TP53, CTNNB1, and LEF1) were found in 10 (out of 10) literature-supported Type-III V-structures (S6 Table). In Fig 6A, we show the network-view of the literature-supported Type-II V-structures incident upon the 8 and 4 dysregulated genes in SKBR3 and BT474 cell-lines, respectively, along with their annotated signaling pathways. Similarly, Fig 6B shows the Type-III literature-supported V-structures in both SKBR3 and BT474 cell-lines. Next, for each of the genes in the selected V-structures in Fig 6 we observed gene expression differences among all four conditions: PB (Parental Basal: 0 μM), PT (Parental Treatment: 1.0 μM), RB (Resistant Basal: 0 μM), and RT (Resistant Treatment: 1 μM) using both two-tailed paired t-tests and one-way ANOVA tests. For these statistical tests we used the mean expression value of all three replicates. In the t-tests, we compared the mean expression of all PT, RB and RT conditions with the mean of PB. Additionally, we also compared the mean of the RT condition with the means of the PT and RB conditions to observe 1) how a gene is behaving differently upon treatment in resistant-vs-parental conditions (RT-vs-PT), and 2) its expression changes upon treatment from its Resistant basal condition (RT-vs-RB). Moreover, one-way ANOVA tests (with the mean of PB as the control condition for the multiple comparison test) may indicate the significance of overall changes in all four groups. All of these statistical tests were done using GraphPad Prism 6.0 software. Concurrently, we also surveyed the literature to determine whether the observed significance of expression changes in resistant-vs-parental conditions were also supported by the literature. We found literature evidence (Fig 6C) supporting a role in breast cancer metastasis and/or in developing acquired resistance to EGFR-TKIs for the SMAD4 − TGFBR2 − RPS6KA2 (Type-II) V-structure in SKBR3, and SMAD4 − LEF1 − CCND2 (Type-II) and PTEN − TP53 − DDB2 (Type-III) V-structures in BT474 cell lines, respectively. Below we discuss these three V-structures in more detail.
-
SMAD4 − TGFBR2 − RPS6KA2 (in SKBR3): TGFBR2 encodes a transmembrane protein which has been reported as a potent inhibitor of tumor growth and proliferation in breast epithelial cells, and loss of its function has also been associated with tumor malignancies [69]. Moreover, mRNA expression of TGFBR2 was reported to be significantly down-regulated in many tumorigenic cell-lines including SKBR3 and BT474 compared to the non-tumorigenic MCF-10F cell-lines [69]. This indicates the tumor-suppressing role of the TGFBR2 gene, and the reduction of its mRNA level may confer a resistance to targeted inhibitors by relapsing tumor growth and proliferation. In the GE dataset for the SKBR3 cell-line, the TGFBR2 gene was down-regulated in PT-vs-PB conditions without significance, but in resistant conditions it showed significant down-regulation compared to parental conditions (RB-vs-PB: p-value = 0.0003; RT-vs-PB: p-value = 0.002; RT-vs-PT: p-value = 0.001). A one-way ANOVA test also found the overall GE changes to be significant: Sidak corrected p-value = 0.0021. Thus, both literature evidence and GE data suggest an association of mRNA down-regulation of TGFBR2 gene with lapatinib resistance in SKBR3 cell-lines.
RPS6KA2 (RSK3) encodes one of the members of the ribosomal S6 kinase which mediates resistance to PI3K pathway inhibitors in breast cancer [70]. RTK (Receptor Tyrosine Kinase) signaling induces the Ras and PI3K pathways, but upon lapatinib treatment such RTK signaling pathways are disrupted, downstream effectors (e.g. mTOR) are abrogated, and eventually Ras and PI3K signaling become inhibited [20]. Over-expression of RSK3 attenuates the apoptotic response and up-regulates protein translation, and thus promotes cell survival and proliferation under conditions of PI3K/mTOR blockade [70]. Moreover, lapatinib down-regulates the Akt pathway in both SKBR3 and BT474 cell-lines [71]. We observed significant and consistent over-expression of RSK3 mRNA in resistant condition compared to parental conditions in our SKBR3 cell-line dataset (RB-vs-PB: p-value = 0.011; RT-vs-PB: p-value = 0.0046; RT-vs-PT: p-value = 0.011; RT-vs-RB: p-value = 0.003). Overall expression changes were also found significant: Sidak corrected p-value = 0.0011. Therefore, both literature evidence and our experimental data strongly suggest that RSK3 over-expression is associated with lapatinib resistance via a PI3K/mTOR signaling blockade.
SMAD4 is a downstream mediator of TGF-β [72] which plays an important role both in tumor suppression and progression in breast cancer [72, 73]. Liu et al. reported that SMAD4 expression was decreased in breast cancer cells compared to adjacent normal breast epithelial tissue [72]. Moreover, SMAD4 is sensitive to lapatinib according to the COSMIC database [74] with no mutational signature in breast cancer cell-lines. In our GE dataset of SKBR3 cell-lines, SMAD4 expression was up-regulated in PT-vs-PB, but was down-regulated in the RB-vs-PB condition, and again up-regulated in the RT-vs-PB condition. Note that however, that none of these comparisons were statistically significant in t-tests at the 0.05 level, and the one-way ANOVA also did not detect significant differences (Sidak corrected p-value = 0.101). Interestingly, both SMAD4 and TGFBR2 mRNA expression changes in PT-vs-PB conditions were non-significant; however, in resistant conditions (RB and RT) both TGFBR2 and RPS6KA2 showed significant changes in mRNA level compared to parental conditions (PB and PT). This may indicate the dependency switch of TGFBR2 from SMAD4 to RPS6KA2 in resistant-vs-parental conditions.
TGFBR2 phosphorylates SMAD4 in the TGF-β signaling [34, 75], and both of their mRNA changes in parental conditions (PT-vs-PB) were non-significant. However, TGFBR2 is an upstream kinase that phosphorylates RPS6KA2 [34, 75], and both of their mRNA changes in resistant conditions were very significant compared to parental conditions. Thus, we hypothesize that the gene dysregulation of TGFBR2 in acquired resistance can be explained by its significant association with RPS6KA2 which evolved in resistant conditions compared to parental conditions.
-
SMAD4 − LEF1 − CCND2 (in BT474): LEF1 plays an oncogenic role in breast cancer, since both mRNA and protein expression of this gene were found to be higher in breast cancer cell-lines compared to normal cells [76]. A high level of LEF1 was also found in HER2 expressing BT474 cell-lines [77], where HER2-activated β-catenin plays a crucial role in producing an increase in the downstream target LEF1 [76]. Increased expression of LEF1 drives cells towards resistance to TGF-β-induced growth inhibitory activities [78]. In our GE datasets of BT474 cell-lines, LEF1 mRNA expressions were significantly increased in resistant conditions compared to the parental basal condition (RB-vs-PB: p-value = 0.0178; RT-vs-PB: p-value = 0.003). Interestingly, over-expression of LEF1 was even more significant in resistant-vs-parental conditions in the presence of lapatinib (RT-vs-PT: p-value < 0.0001). Moreover, overall expression changes were also proved to be significant by one-way ANOVA test (Sidak corrected p-value = 0.004). Thus, the experimental data and the literature evidences support a role of LEF1 gene in lapatinib resistance in the BT474 cell-lines.
CCND2 is involved in the cell cycle process, and is a regulatory subunit of a complex formed with CDK4 or CDK6 that is required for cell cycle G1/S transition [79]. Although CCND2 over-expression is found in ovarian, testicular [79] and gastric cancer [80], little is known about its role in breast cancer especially in the presence of lapatinib. In the GE data for the BT474 cell-line, CCND2 mRNA expression was significantly down-regulated in the PT-vs-PB condition (p-value = 0.024), and this possibly indicates the association of its mRNA down-regulation with lapatinib sensitivity in lapatinib-sensitive BT474 cell-lines. We investigated whether this behaviour is coherent with the literature. Schmidt et al. reported that both mRNA and protein expression of CCND1 and CCND2 were down-regulated when FOXO3A induced the process of cell cycle arrest [81]. Such inhibition of CCND1 and CCND2 perturbs CDK4 functionality to inactivate the S-phase repressor Rb [81]. Moreover, Hegde et al. reports that mRNA expression of FOXO3 and CCND1 were significantly up- and down-regulated, respectively, in both SKBR3 and BT474 cell-lines (lapatinib-sensitive) in response to lapatinib treatment [71]. To explain the above-mentioned down-regulation of CCND2, we observed FOXO3, CCND1 and RB1 mRNA changes in PT-vs-PB conditions (in BT474 datasets), to determine whether these are coherent with the above literature findings. In SKBR3 cell-lines, FOXO3 was significantly up-regulated (p-value = 0.0028) and CCND1 was significantly down-regulated (p-value = 0.0029). In BT474 cell-lines, 2 out of 3 replicates showed a similar pattern of mRNA changes for these two genes (FOXO3 and CCND1) (p-values = 0.042 and 0.017, respectively) as in SKBR3 cell-lines. In BT474 cell-lines RB1 mRNA expression was found slightly up-regulated in PT-vs-PB conditions. Moreover, CCND2 mRNA expressions are up-regulated in both resistant conditions (RB-vs-PB and RT-vs-PB) compared to the parental basal condition. The above experimental data may indicate the possible reason for CCND2 down-regulation in lapatinib-sensitive BT474 cell-lines with lapatinib treatment, and its mRNA up-regulation in both resistant conditions (RB-vs-PB and RT-vs-PB) could possibly be due to acquired resistance of BT474 cell-lines to lapatinib.
SMAD4 expression was reported to be decreased in breast cancer cells [72], and the COSMIC database [74] reports SMAD4 as sensitive to lapatinib in the BT474 cell-line along with other EGFR-TKI, BIBW2992 and erlotinib [74] with IC50 effect = 0.225 (p-value = 0.000014) and with significant mutational signature in skin cancer, but none in breast cancer cell-lines. However, in the GE data for the BT474 cell-line, mRNA expression of SMAD4 was up-regulated in PT-vs-PB conditions, but was down-regulated in resistant-vs-parental conditions, with or without lapatinib treatment (RB-vs-PB and RT-vs-PT), indicating its sensitivity to lapatinib in parental conditions. Note that we observed no significant changes using a one-way ANOVA test (Sidak corrected p-value = 0.1212).
SMAD4 binds to LEF1 [82], and the changes in expression of both of their mRNAs indicate sensitivity to lapatinib treatment in parental conditions (PT-vs-PB). Again, LEF1 regulates the transcription of CCND2 gene in the Wnt signaling pathway [83], and both genes exhibited up-regulation in resistant conditions compared to parental conditions. Thus, we can hypothesize that the dysregulation of the LEF1 gene can be explained by its differential associations with SMAD4 and CCND2 mRNA changes in resistant-vs-parental conditions.
-
PTEN − TP53 − DDB2 (in BT474): PTEN is one of the most commonly mutated tumor suppressor genes, and the loss of its mRNA and protein expression are found in many metastatic malignancies including breast cancer [84]. PTEN modulates lapatinib sensitivity [85], and its loss acts as a marker of poor lapatinib response [58, 86, 87]. In the GE dataset for the BT474 cell-line, no mutation has been detected for PTEN and TP53 in their corresponding DNA sequences between parental and resistant conditions as reported in the original article associated with this dataset [66], and PTEN expression was up-regulated even in resistant-vs-parental conditions with or without lapatinib (RB-vs-PB, and RT-vs-PB), but the overall mRNA changes were not significant as tested with the one-way ANOVA test (p-value = 0.264). TP53 is another well known tumor suppressor gene, and its inhibition greatly inhibits apoptosis as p53 up-regulates several pro-apoptotic gene products including Puma, Noxa, Apaf-1, and Bax [88]. The loss of p53 is consistently associated with the acquired resistance of EGFR inhibitors cetuximab and erlotinib [89]. However, more experimental evidence is required to claim that p53 loss can be a predictive feature of acquired resistance to EGFR inhibitors [90]. In the GE dataset for the BT7474 cell-line, TP53 expression was significantly decreased in both RB-vs-PB (p-value = 0.013) and RT-vs-PB (p-value = 0.025) conditions, and the overall changes were statistically significant (Sidak corrected p-value = 0.01). For the DDB2 gene, its under-expression is correlated with poor outcome in ovarian cancer [91]. In breast cancer, although DDB2 showed putative oncogenic behaviour by promoting cell-cycle progression [92], it was not over-expressed in ER-negative breast cancer cells [92, 93], e.g. SKBR3 [93]. Moreover, DDB2 is down-regulated in lapatinib-resistant cell-lines [94]. This suppression was induced by the over-expression of the hepatitis B viral-encoded X protein (HBX) in the p53/lincRNA-p21 axis and IKK-dependent manner [94]. In our GE dataset for the BT474 cell-line, DDB2 was significantly down-regulated in resistant-vs-parental conditions (RT-vs-PB: p-value = 0.002) and the over-all changes were significant as well (Sidak corrected p-value = 0.046).
p53 up-regulates or enhances PTEN transcription [95–97], and we found both genes’ mRNA changes in parental conditions (PT-vs-PB) to be non-significant. Moreover, p53 transcriptionally regulates DDB2 expression in a cell cycle-dependant manner [98, 99], and both of their mRNA changes were found to be significant, showing similar phenotypes in resistant-vs-parental conditions. Thus we can claim that the switch in dependency of TP53 from PTEN to DDB2 (in PTEN − TP53 − DDB2) can be a possible mechanism of TP53 dysregulation in acquired resistance.
Fig 6. The role of literature-supported Type-II and Type-III V-structures (VSs) in explaining gene dysregulation in acquired resistance.
(A) Network views of Type-II VSs along with their pathway annotations in SKBR3 and BT474 cell-lines. (B) Network views of Type-III VSs in SKBR3 and BT474 cell-lines. Note that VSs shown here are only those for which the crossing-genes were found as up- or down-regulated in PT-vs-PB conditions, but oppositely regulated in both RB-vs-PB and RT-vs-PB conditions. Nodes are genes, and the edges are known signaling links [34] that were also found as aberrant gene-pairs identified by our framework. Note that the width of edges is proportional to the posterior probability of corresponding pairs. Furthermore, for three VSs shown in (A) and (B) (right panels), mRNA changes for their constituent genes were found in the literature, implicating their role in breast cancer metastasis and/or in developing acquired resistance in EGFR-TKIs. (C) Above three VSs with their corresponding posterior probabilities, odds, and literature references of gene-pair associations for each of the red and green pairs. Statistical significance tests were done using t-tests and one-way ANOVA with multiple corrections (Sidak method). All the mRNA values were normalized by corresponding PB expression values in all three replicates. Significance was indicated by * (p-value < 0.05), ** (p-value < 0.005), and so on.
Gene dysregulation plays an important role in developing acquired resistance to EGFR-TKIs in breast cancer [28–30, 100]. Here, along with literature-supported gene-gene associations in V-structures (Fig 6C), we demonstrated that the switch in dependency from the targeted signaling link involving green gene-pair (with the inhibitor) to the bypass signaling link involving red gene-pair (evolved in resistant conditions) is a possible mechanism mediating the dysregulation of crossing-genes in acquired resistance.
Discussion
In this study, we proposed a computational framework that models signal rewiring by systematically characterizing potential aberrant signaling in acquired resistance. We hypothesized that an aberrant signaling link involved in acquired resistance may have differential probabilities of appearing (either higher, or lower) in resistant-vs-parental networks, where in each network, nodes were genes and the edges were the relationships among genes. In this gene-gene relationship network, called GGR, we considered both direct and indirect correlations (via linker genes) among genes for defining the edges that combine both data-driven (from gene expression) and topological (from PPI) information about gene-pairs. Note that the PPI edges in the statistically significant paths [see Methods], defining indirect relationships among gene-pairs for which direct relationships were not found, were also added to the final edge set [Table 1]. The rationale for including those PPI edges was: 1) to retain precise information regarding how indirect relationships were constructed, and 2) to better model the data-driven signaling networks (resistant and parental GGR networks) for the Bayesian statistical analysis (using p1-model) of their respective global structure formation. We used a fully Bayesian statistical model: a special class of Exponential Random Graph Model, called p1-model for inferring aberrant gene-pairs with differential posterior probabilities in resistant-vs-parental GGR networks, where these networks were constructed from matched gene expression values of resistant and parental conditions, respectively. When selecting aberrant gene-pairs, we chose the thresholds for Odds and posterior probabilities from their frequency distributions, sequentially. Firstly, we chose the gene-pairs with top 20% of odd-ratio values from two distribution individually (oddR and oddP) by ensuring their mutual exclusivity after selection, and termed them as red and green, respectively. Then, we further filter red and green pairs with top 50% of their respective posterior probability values. Note that before calculating the Odds values, we normalized both posterior probabilities (from resistant and parental conditions) with their corresponding max values over all gene-pairs, individually, in order to achieve same scaling. All other model parameters in p1-model were estimated with Gibbs sampling [see Methods].
After detecting putative aberrant pairs in resistant-vs-parental conditions, we analyzed them in two-ways, 1) Identifying potentially dysregulated pathways in acquired resistance, 2) Identifying their roles in explaining a possible mechanism of acquired resistance via dysregulation of crucial genes. Using two lapatinib-treated breast cancer cell-lines: SKBR3 and BT474, our method was able to predict similar pathways as dysregulated. The rationale for using these datasets for our experiments was that—to the best of our knowledge—these are only datasets available for responsive and resistant lapatinib-treated ERBB2-positive breast cancer cell-lines. Our results suggested that signal rewiring is a major event in acquired resistance since we found a range of dysregulated pathways in both SKBR3 and BT474 cell-lines including EGFR-related pathways (e.g. EGFR, ErbB2, PI3K-Akt, Mapk, Jak-Stat, FoxO signaling, etc.) as well as other receptor-related pathways (e.g. Notch, Wnt, insulin, PDGFR, FGFR, VEGFR signaling, etc). Although there may be some false-positives in those results, we found literature evidence from Huang et al. [3] that aberrant signaling in most of our predicted dysregulated pathways were actually related with acquired resistance in EGFR-TKIs. Furthermore, our predictions of network re-adjustment in multiple signaling pathways were also consistent with the results recently published by Stuhlmiller et al. [5]. Their study suggested that multiple heterogenous kinases (e.g. DDR1, FGFRs, IGFI1, MET, etc.) compensate for the ErbB2 inhibition by kinome re-programming induced by lapatinib [5], which provides an indication that aberrant signaling activities in those kinase-related pathways are crucial for such bypass mechanism. Note that since the pathway annotations are still incomplete, we used three pathway databases here: KEGG, Reactome, and WikiPathways to define constituent genes of signaling pathways individually. However, to maintain the same true-relationship among those constituent genes we used literature-supported signaling links (collected from online resources of Wang Lab [34]) since it is the largest manually curated human signaling network as reported.
Gene dysregulation plays crucial roles in acquired resistance by mediating both uncontrolled cell-growth and disrupted apoptosis [27–29]. Here, to evaluate the potentialities of identified aberrant signaling, we conducted an analysis which demonstrated that the greater number of dysregulated genes were found in resistant-vs-parental conditions when they were incident with both red and green-types of aberrant pairs (V-structures) compared to those with single type only (either red, or green). Manual literature survey also validated some of the V-structures, such as SMAD4 − TGFBR2 − RPS6KA2, SMAD4 − LEF1 − CCND2, and PTEN − TP53 − DDB2, as consistent with our hypothesis. Thus, we claim that a mechanism of dependency shift from targeted signaling (by inhibitor) towards bypass signaling can potentially cause dysregulation of shared genes (crossing-genes). Similar idea of dependency switch was recently reported by Sharifnia et al. [30] that EGFR-dependent status of downstream signaling nodes can be modified by other over-expressed kinase-related genes that shared them (downstream signaling nodes) with EGFR-dependant signaling. However, to the best of our knowledge, our study is the first to emphasise the compensating effects of aberrant signaling upon mRNA expression changes of crucial genes by examining the dependency switch from targeted signaling to bypass signaling.
We included all the available genes from the Cancer Gene Census (CGC) into the list of seed genes in our framework for which gene expression data was available (see Methods): 370 and 357 genes in SKBR3 and BT474 cell-lines, respectively. Cancer genes are crucial for mediating various cancer related activities and many are hub genes in mammalian signaling networks [101]. Therefore, they are very important in terms of signaling network formation, an aspect which we examine in this study by statistical models (i.e. p1-model). Note that we combined cancer genes with a set of differentially expressed (DE) genes even though some may not be differentially expressed. However, cancer genes can still be important in network-based analyses of studies comparing two conditions (i.e. resistant-vs-parental). For example, in a network-based classification of breast cancer patients, Chuang et al. [102] reported that the subnetworks which can classify metastatic and non-metastatic patients contain genes playing a central role connecting DE genes even though those cancer genes were non-DE themselves [103]. Moreover, we intend to include all CGC genes, not just those ones that are breast cancer related, since no classifications are perfect, and the census is continuously being updated [104]. CGC genes are selected based on the mutational profiles of cancer patients [105], hence their transcriptional profiles may also reveal additional insights into the mechanisms of aberrant signaling activities in acquired resistance. To investigate the influence of CGC genes in our framework, we observed all the genes involved in all the V-Structures (VSs) of aberrant pairs (Type-I, Type-II and Type-III VSs) found in pathways from KEGG, Reactome and WikiPathway databases [See S5 Table]. We found that many of the genes involved in VSs overlapped with genes from CGC, where most of those cancer genes were not identified as DE genes during the formation of the seed gene list [see Methods] [S7 Table]. Thus, we claim that CGC genes were very important in the network-based analyses of our framework.
In this paper, we considered only gene expression values for modeling gene-gene relationship networks (GGR). However, we look forward to adapting other appropriate high-throughput datasets, such as miRNA expression, methylation, copy number aberration, and phosphorylation datasets into our framework in order to better model gene-gene dependencies in resistant-vs-parental conditions to reflect greater mechanistic insights. Moreover, the V-structures we have examined in our current study can be called first-order V-Structures since they involve only a single aberrant edge of each type (red and green). In future we intend to examine the role of higher order V-structures in acquired resistance.
Materials and methods
Literature and database search
Our research hypothesis was primarily focused on studying the acquired resistance mechanisms of HER2-positive breast cancer cells to lapatinib (an EGFR/HER2 dual inhibitor). Therefore, we conducted a literature survey in Pubmed database using keywords: ‘lapatinib’, ‘acquired resistance’, and ‘breast cancer’, which lead us to find two articles: Komurov et al. [19] and Liu et al. [66]. Both of these articles studied the resistance mechanisms of HER2-positive breast cancer cell-lines by analysing gene expression datasets of lapatinib-treated sensitive (parental) and resistant conditions. To find these gene expression datasets, we also searched GEO (Gene Expression Omnibus) database with the same keywords as above and found two data collections with accession IDs: GSE38376 and GSE16179, respectively. Detailed technical descriptions of cell-line preparation and dataset generation were reported in their respective original articles. The first dataset (GSE38376) included SKBR3 parental and resistant (SKBR3-R) cell-lines, and the second dataset (GSE16179) included BT474 parental and resistant (BT474-J4) cell-lines. In both of these datasets, expression values of both parental and resistant samples were measured first in basal condition (0 μM), and then in treatment conditions (0.1 μM and 1.0 μM for GSE38376; 1.0 μM only for GSE16179). For both GSE38376 (SKBR3) and GSE16179 (BT474), we converted probe-level expression values into gene-level values using the corresponding annotation files: GPL6947 (Illumina HumanHT-12 V3.0 expression beadchip) and GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array), respectively, which were also collected from GEO database. For some genes, multiple probes were mapped to a single gene, and we averaged the GE values of such probes to obtain the final GE values of the corresponding gene. Next, for each collection (GSE38376 and GSE16179) we built two data matrices, one from the parental and another from the resistant GE dataset, where rows were labeled with gene symbols and columns were labeled with samples under different treatment conditions. A protein-protein interaction dataset was obtained from Cerami et al. [106]. For the enrichment analysis, we collected gene sets of all 1) the 24 signaling pathways from Reactome [107] (downloaded at 19/05/2014), 2) 45 signaling pathways from KEGG [83, 108] (downloaded at 12/05/2015), and 3) 61 signaling pathways from WikiPathway [109] (downloaded at 16/10/2014) databases. Each signaling pathway downloaded from these databases was encoded as tab-delimited lists of gene symbols. For KEGG signaling pathways, we built a parser program that extracted gene names from the web-responses after making HTTP web-requests to KEGG server using a list of IDs corresponding to signaling pathways.
Constructing gene-gene relationship network
We denote the gene-gene relationship network as GGR:= (S, R) for each GE data matrix. Here, S is the set of seed genes, which is the union of a set of differentially expressed (DE) genes, a set of cancer genes collected from the Cancer Gene Census (CGC) [105], and a set of linker genes (see below) selected from the PPI network. R is the set of edges defined among the genes in the set S. The sets S and R were constructed as follows.
Defining S: The seed genes
We built the set S cumulatively; first a set of DE genes was identified by differential expression analysis of parental and resistant GE data using a two-tailed pooled Student’s t-test. For this test, significant p-values were identified using the Bonferroni correction method, and genes with such corrected p-values ≤ threshold (see Results) were selected as differentially expressed. Next, we added CGC genes for which corresponding GE data was available. The rationale for such inclusion is that CGC genes are well known to be hub genes in mammalian cellular signaling networks [101] where they play key regulatory roles in various cancer related activities. In the process of finding indirect relationships among (DE ∪ CGC) genes, a set of intermediate genes from the PPI network was identified, which we defined as linker genes (see next section). The final set of seed genes consisted of (DE ∪ CGC ∪ Linker) genes.
Defining R: The edges
To identify interacting gene pairs, all pair-wise absolute Pearson Correlation Coefficients (PCCs) were calculated for expression levels of the genes in the (DE ∪ CGC) gene set. The value demarcating the top 20% of absolute PCCs was selected as the threshold for defining direct relationships among the genes in the above set. That is, for each gene pair (genei, genej), if the corresponding PCC value was above the threshold then the pair was considered to have a direct relationship, and hence added into the set of edges, R.
Otherwise, a gene pair was said to have an indirect relationship if there was at least one statistically significant simple path in the PPI network between genei and genej via an intermediate gene (called a linker gene). Here, we imposed a path-length threshold of 2 and restricted to paths in the PPI network, otherwise considering all the remaining genes as possible intermediates would convert this searching procedure into an NP-hard problem. Simple paths of length 2 [for details see S1 Text] connecting a given pair (genei, genej) in the PPI network were considered statistically significant if one can reject the following null hypothesis: the geometric mean of pairwise PCC values of constituent edges in the path is distributed as for paths of length 2 between these genes generated by a randomized procedure. Random paths of the form genei → linker → genej were generated by replacing linker with any other gene in the network except genei, genej and any gene on a path of length 2 connecting these genes in the PPI network. To evaluate the PCC for a random path, we used the same expression values for the genes as in the observed case. Paths were considered significant if the probability of generating a path using above randomized procedure with a geometric mean of pairwise PCC values greater than or equal to that observed for the PPI network was ≤0.05 (an empirical p-value). PPI edges comprising statistically significant simple paths were added to the set R. The set of edges R was finally composed of direct relationships, indirect relationships, and PPI edges of statistically significant simple paths, which are used for identifying those indirect relationships [see Discussion].
Bayesian statistical modeling of GGR network
Exponential Random Graph Models (ERGMs) are parametric probability distributions over spaces of networks [24] that have been successfully used to evaluate probabilities of the presence of each edge in a network [23, 24]. Here, in order to infer edge probabilities in a gene-gene relationship network, we used the p1-model, a special class of ERGM introduced by Holland and Leinhardt [24]. The p1-model has previously been used by Bulashevska et al. [23] to model human protein-protein interaction networks. In this approach, edge probabilities are evaluated by summarizing topological properties of networks in a parametric form and associating them with sufficient statistics [23, 24]. The definition of the p1-model for a directed graph is contained in the original article [24]. An equivalent log-linear formulation was proposed by Fienberg and Wasserman [110], in which each directed edge was assigned four Bernoulli variables Yij00, Yij01, Yij10 and Yij11. Since our GGR network is an undirected graph, the model can be simplified by using only two Bernoulli variables Yij0 and Yij1 defined as follows:
where, the binary outcome uij = 1 if genei interacts with genej in GGR, and uij = 0 otherwise. Under this simplified model, the posterior probability of an edge connecting genei and genej is given by:
| (1) |
| (2) |
for i < j. Here, θ is the global density parameter, αi is the expansiveness/attractiveness of genei, and λij is the scaling parameter ensuring . We hypothesized that some aberrant gene-pairs involved in acquired resistance may have unusually high probability of appearing in Resistant-vs-Parental conditions, whereas other pairs may have unusually low probabilities. Hence, we used two Yk data matrices, and , from GGR matrices of Resistant and Parental samples, respectively. Note that it is possible to replace the expansiveness and attractiveness parameters by a single parameter α that represents the propensity of a gene to be connected in an undirected network.
We used a fully Bayesian approach, both for modeling the network parameters and their estimation. To estimate the model parameters, we used Gibbs sampling, a Markov Chain Monte Carlo (MCMC) method implemented in WinBUGS [111] which allows users to construct complex Bayesian models in a simple manner. We constructed a hierarchical Bayesian model in which the model parameters were further defined as dependent upon hyperparameters as follows:
We assigned the density parameter θ a normal prior distribution with mean zero and standard deviation σθ. (In fact, this was implemented in WinBUGS using the precision parameter ). Next, the parameter τθ was assigned a gamma prior distribution with hyperparameters a0 = 0.001 and b0 = 0.001. We set a0 = 0.001 and b0 = 0.001 to express large uncertainty regarding the value , following [23]. For the propensity parameters and , we selected the above prior following Adam et al. [112].
Robust selection of aberrant gene-pairs
One of our primary hypotheses in this study is that aberrant gene-pairs involved in network re-wiring in drug-resistance are likely to have high probabilities of occurring in one network (resistant or parental) but low probabilities in the other network. To determine which gene-pairs exhibit this pattern, we calculated two odds ratios defined in the following equations:
| (3) |
| (4) |
where, and are defined for resistant and parental networks, respectively, and their corresponding posterior probabilities are estimated using MCMC sampling. Before calculating these ratios, we normalized the posterior probabilities by their respective maximum values over all gene-pairs, since two values ( and ) may not be in the same scale. For the sake of brevity we refer to these ratios as odds ratios, but they are more appropriately called normalized odds ratios.
Our intention is to identify gene-pairs for which only one of the two odds ratios (Eqs (3) and (4)) is very high. Additionally, we require that both posterior probabilities exceed a minimum threshold, since very small denominators can yield high odds ratio scores even if the edge has low probability in both networks. We therefore defined two thresholds, one for odds ratio values and another for posterior probabilities. We examined the distributions of all oddsR and oddsP values and set a threshold demarcating the top 20%. Next, we examined the distribution of posterior probabilities for gene-pairs exceeding the odds ratio threshold and set a second threshold to demarcate the top 50%. Finally, we chose only those gene-pairs that had posterior probabilities above that threshold, and identified as putative aberrant gene-pairs that were potentially involved in network rewiring in acquired resistance.
Edges were then grouped into two types: gene-pairs for which the oddsR scores and the were greater than their respective thresholds in Eq (3) were defined as red pairs, whereas gene-pairs for which the oddsP scores and the were greater than their respective thresholds in Eq (4) were defined as green pairs.
Enrichment of aberrant gene-pairs using known signaling links
Putative aberrant gene-pairs from the above Bayesian analyses were then further filtered by comparing them to another set of known (true) signaling links from the literature. For that purpose, we obtained a signaling network from the online resources of Wang Lab [34], which is claimed as the largest manually curated signaling network available to date. This network has more than 6,000 proteins and ∼63,000 binary relations defined, including activations, inhibitions and physical interactions. Note that signaling pathways from KEGG, Reactome, and WikiPathway databases were merely genesets, and to define true signaling links among the genes within those genesets we considered the signaling links from Wang Lab [34]. Next, to find dysregulated signaling pathways from KEGG, Reactome, and WikiPathway databases, we searched for significant overlaps between the set of true signaling links and the set of putative aberrant gene-pairs, for the genesets in a specific pathway. To this end, we designed a hypergeometric test as follows:
| (5) |
where N is the number of distinct gene-pairs contained in all of the signaling pathways (from a particular pathway database) and all the predicted aberrant gene-pairs, M is the set of all known signaling links in a given pathway, K is the set of aberrant gene-pairs predicted by our framework, and x = |M ∩ K|. After measuring p-values using Eq (5), a False Discovery Rate (FDR) multiple comparison correction technique was conducted to obtain q-values. Signaling pathways with q-value <0.05 were considered to be significantly dysregulated in acquired drug resistance. A similar gene-pair enrichment test, called Edge Set Enrichment Analysis (ESEA) using the weighted Kolmogorov-Smirnov statistic was recently proposed by Han et el. for detecting dysregulated pathways in the context of gene expression datasets [113].
Identifying V-structures
To investigate the role of signaling rewiring in acquired drug resistance, we searched for a configuration of edges that we call a V-structure (Fig 1F). A V-structure consists of three genes connected by one red edge and one green edge. One gene, called a crossing-gene, is connected to both of the other genes, to one by a red edge and to the other by a green edge. Thus V-structures involve both types of aberrant pairs: one gene-pair present only in Resistant conditions, and another gene-pair present only in Parental conditions, with the crossing-gene common to both pairs. Our rationale is that the compensatory kinases may switch the oncogenic-addiction of cancer-related (growth/survival) genes to overcome their dependencies upon their primary driver kinases (e.g. EGFR/HER2) that were initially targeted in parental conditions with inhibitors [19, 30], thereby relapsing into their tumorigenic roles in acquired resistance. We hypothesise that crossing-genes that are dysregulated restore their metastatic phenotype (i.e. up- or down-regulation of oncogenes or tumor suppressor genes, respectively) in resistant conditions by forming a V-structure in the rewired signaling network.
Therefore, we define a V-structure to be a pair of aberrant gene-pairs (gi, gk) and (gj, gk) such that (gi, gk) are connected by a green edge and (gj, gk) are connected by a red edge. To identify V-structures, first we identified the set of common genes in the two mutually exclusive sets of aberrant gene-pairs (red and green gene-pairs). This set of common genes are the crossing-genes (see Fig 1). Next, we observed and enumerated all the gene-pairs (red and green) incident on each of the crossing-genes, and enumerated all of the possible pairings of one red and one green edge to form a V-structure.
Pathway configurations of V-structures: Type-I, Type-II, and Type-III configurations
Next, for each V-structure, we identified signaling pathways from KEGG, Reactome, and WikiPathway databases that contained at least one gene in that V-structure. We classified V-structures into three sub-types based on their configurations relative to these pathways. Firstly, Type-I V-structures are those in which all three member genes belong to different signaling pathways. Type-II V-structures are those in which the two aberrant gene-pairs in a particular V-structure are from two different signaling pathways, with the crossing-gene common to both pathways. Type-III V-structures are those in which all three genes are from the same signaling pathway. Note that Type-I and Type-II V-structures may represent signaling pathway cross-talks that play crucial roles in acquired drug-resistance. In our previous study, we investigated and explained the concept of Type-I V-structures, their involvement in the cross-talk between EGFR/ErbB and other signaling pathways, and their contribution to lapatinib resistance [10]. Type-III V-structures can explain the aberrant co-regulation of genes within a single pathway. We observed and analysed all the V-structures that overlap with the literature curated signaling network [34].
Supporting information
Supplementary Methods.
(PDF)
List of identified putative aberrant gene-pairs (for both SKBR3 and BT474) cell-lines in acquired resistance.
(XLSX)
Full results of pathway enrichment tests of identified aberrant gene-pairs in acquired resistance from KEGG, Reactome, and WikiPathway databases for both SKBR3 and BT474 cell-lines.
(XLSX)
Comparing our current model with the previous model by observing the percentages of non-direct (indirect and PPI pair) enriched links (aberrant pairs as known signaling links) in the aberrant signaling pathways from KEGG, Reactome, and WikiPathway databases that were detected by our current but not the previous model, for both SKBR3 and BT474 cell-lines.
(XLSX)
Comparing our current model with the previous model by observing the percentages of non-direct (indirect and PPI pair) enriched links (aberrant pairs as known signaling links) in the aberrant signaling pathways from KEGG, Reactome, and WikiPathway databases that were detected by both of our current and previous models, and were ranked (based on enrichment q-value) high in the current model but low in the previous model, for both SKBR3 and BT474 cell-lines.
(XLSX)
Summary of Type-I, Type-II and Type-III enrichment of V-structures in KEGG, Reactome, and WikiPathway databases in SKBR3 cell-line.
(XLSX)
Summary of Type-I, Type-II and Type-III enrichment of V-structures in KEGG, Reactome, and WikiPathway databases in BT474 cell-line.
(XLSX)
CGC genes in all the Type-I, Type-II and Type-III V-structures in SKBR3 and BT474 cell-lines.
(XLSX)
Acknowledgments
We thank Dr. Tianhai Tian for his initial support in this project.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by Monash International Postgraduate Research Scholarship and Monash Graduate Scholarship at the Monash University, Australia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brivanlou AH, Darnell JE. Signal transduction and the control of gene expression. Science. 2002;295(5556):813–818. 10.1126/science.1066355 [DOI] [PubMed] [Google Scholar]
- 2. Sarkar S, Mandal M. Growth factor receptors and apoptosis regulators: signaling pathways, prognosis, chemosensitivity and treatment outcomes of breast cancer. Breast Cancer (Auckl). 2009;3:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang L, Fu L. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharmaceutica Sinica B. 2015;5(5):390–401. 10.1016/j.apsb.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamaguchi H, Chang SS, Hsu JL, Hung MC. Signaling cross-talk in the resistance to HER family receptor targeted therapy. Oncogene. 2014;33(9):1073–1081. 10.1038/onc.2013.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stuhlmiller TJ, Miller SM, Zawistowski JS, Nakamura K, Beltran AS, Duncan JS, et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell Rep. 2015;11(3):390–404. 10.1016/j.celrep.2015.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clark GJ, Der CJ. Aberrant function of the Ras signal transduction pathway in human breast cancer. Breast Cancer Res Treat. 1995;35(1):133–144. 10.1007/BF00694753 [DOI] [PubMed] [Google Scholar]
- 7. Shi I, Sadraei N Hashemi, Duan ZH, Shi T. Aberrant signaling pathways in squamous cell lung carcinoma. Cancer Inform. 2011;10:273–285. 10.4137/CIN.S8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mao H, Lebrun DG, Yang J, Zhu VF, Li M. Deregulated signaling pathways in glioblastoma multiforme: molecular mechanisms and therapeutic targets. Cancer Invest. 2012;30(1):48–56. 10.3109/07357907.2011.630050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCleary-Wheeler AL, McWilliams R, Fernandez-Zapico ME. Aberrant signaling pathways in pancreatic cancer: a two compartment view. Mol Carcinog. 2012;51(1):25–39. 10.1002/mc.20827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Azad A, Lawen A, Keith J. Prediction of signaling cross-talks contributing to acquired drug resistance in breast cancer cells by Bayesian statistical modeling. BMC Syst Biol. 2015;9(1):2 10.1186/s12918-014-0135-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR Mutation and Resistance of Non-Small-Cell Lung Cancer to Gefitinib. New England Journal of Medicine. 2005;352(8):786–792. 10.1056/NEJMoa044238 [DOI] [PubMed] [Google Scholar]
- 12. Amin DN, Sergina N, Ahuja D, McMahon M, Blair JA, Wang D, et al. Resiliency and vulnerability in the HER2-HER3 tumorigenic driver. Sci Transl Med. 2010;2(16):16ra7 10.1126/scitranslmed.3000389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci USA. 2011;108(12):5021–5026. 10.1073/pnas.1016140108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Azuma K, Tsurutani J, Sakai K, Kaneda H, Fujisaka Y, Takeda M, et al. Switching addictions between HER2 and FGFR2 in HER2-positive breast tumor cells: FGFR2 as a potential target for salvage after lapatinib failure. Biochem Biophys Res Commun. 2011;407(1):219–224. 10.1016/j.bbrc.2011.03.002 [DOI] [PubMed] [Google Scholar]
- 15. Huang C, Park CC, Hilsenbeck SG, Ward R, Rimawi MF, Wang YC, et al. ß1 integrin mediates an alternative survival pathway in breast cancer cells resistant to lapatinib. Breast Cancer Res. 2011;13(4):R84 10.1186/bcr2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rexer BN, Arteaga CL. Intrinsic and acquired resistance to HER2-targeted therapies in HER2 gene-amplified breast cancer: mechanisms and clinical implications. Crit Rev Oncog. 2012;17(1):1–16. 10.1615/CritRevOncog.v17.i1.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kolch W, Halasz M, Granovskaya M, Kholodenko BN. The dynamic control of signal transduction networks in cancer cells. Nat Rev Cancer. 2015;15(9):515–527. 10.1038/nrc3983 [DOI] [PubMed] [Google Scholar]
- 18. Logue JS, Morrison DK. Complexity in the signaling network: insights from the use of targeted inhibitors in cancer therapy. Genes Dev. 2012;26(7):641–650. 10.1101/gad.186965.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Komurov K, Tseng JT, Muller M, Seviour EG, Moss TJ, Yang L, et al. The glucose-deprivation network counteracts lapatinib-induced toxicity in resistant ErbB2-positive breast cancer cells. Molecular Systems Biology. 2012;8(1). 10.1038/msb.2012.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Locasale JW. Metabolic rewiring drives resistance to targeted cancer therapy. Mol Syst Biol. 2012;8:597 10.1038/msb.2012.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akhavan D, Pourzia AL, Nourian AA, Williams KJ, Nathanson D, Babic I, et al. De-Repression of PDGFR-beta Transcription Promotes Acquired Resistance to EGFR Tyrosine Kinase Inhibitors in Glioblastoma Patients. Cancer Discovery. 2013;3(5):534–547. 10.1158/2159-8290.CD-12-0502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saul ZM, Filkov V. Exploring biological network structure using exponential random graph models. Bioinformatics. 2007;23(19):2604–2611. 10.1093/bioinformatics/btm370 [DOI] [PubMed] [Google Scholar]
- 23. Bulashevska S, Bulashevska A, Eils R. Bayesian statistical modelling of human protein interaction network incorporating protein disorder information. BMC Bioinformatics. 2010;11:46 10.1186/1471-2105-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holland PW, Leinhardt S. An Exponential Family of Probability Distributions for Directed Graphs. Journal of the American Statistical Association. 1981;76(373):33–50. 10.2307/2287042 [DOI] [Google Scholar]
- 25. Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325(5947):1555–1559. 10.1126/science.1174229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roberts CG, Millar EK, O’Toole SA, McNeil CM, Lehrbach GM, Pinese M, et al. Identification of PUMA as an estrogen target gene that mediates the apoptotic response to tamoxifen in human breast cancer cells and predicts patient outcome and tamoxifen responsiveness in breast cancer. Oncogene. 2011;30(28):3186–3197. 10.1038/onc.2011.36 [DOI] [PubMed] [Google Scholar]
- 28. Ng CP, Bonavida B. A new challenge for successful immunotherapy by tumors that are resistant to apoptosis: two complementary signals to overcome cross-resistance. Adv Cancer Res. 2002;85:145–174. 10.1016/S0065-230X(02)85005-9 [DOI] [PubMed] [Google Scholar]
- 29. Debatin KM. Apoptosis pathways in cancer and cancer therapy. Cancer Immunol Immunother. 2004;53(3):153–159. 10.1007/s00262-003-0474-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharifnia T, Rusu V, Piccioni F, Bagul M, Imielinski M, Cherniack AD, et al. Genetic modifiers of EGFR dependence in non-small cell lung cancer. Proc Natl Acad Sci USA. 2014;111(52):18661–18666. 10.1073/pnas.1412228112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tian Y, Zhang B, Hoffman EP, Clarke R, Zhang Z, Shih IeM, et al. Knowledge-fused differential dependency network models for detecting significant rewiring in biological networks. BMC Syst Biol. 2014;8:87 10.1186/s12918-014-0087-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jung S, Kim S. EDDY: a novel statistical gene set test method to detect differential genetic dependencies. Nucleic Acids Res. 2014;42(7):e60 10.1093/nar/gku099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Elo LL, Jarvenpaa H, Oresic M, Lahesmaa R, Aittokallio T. Systematic construction of gene coexpression networks with applications to human T helper cell differentiation process. Bioinformatics. 2007;23(16):2096–2103. 10.1093/bioinformatics/btm309 [DOI] [PubMed] [Google Scholar]
- 34.Wang E. Human signaling network; 2014. Database: Cancer Systems Biology and Bioinformatics. Available from: http://www.cancer-systemsbiology.org/dataandsoftware.htm
- 35. Dhomen NS, Mariadason J, Tebbutt N, Scott AM. Therapeutic targeting of the epidermal growth factor receptor in human cancer. Crit Rev Oncog. 2012;17(1):31–50. 10.1615/CritRevOncog.v17.i1.40 [DOI] [PubMed] [Google Scholar]
- 36. Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23(11):2445–2459. 10.1200/JCO.2005.11.890 [DOI] [PubMed] [Google Scholar]
- 37. Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4(10):e315 10.1371/journal.pmed.0040315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Toyooka S, Date H, Uchida A, Kiura K, Takata M. The Epidermal Growth Factor Receptor D761Y Mutation and Effect of Tyrosine Kinase Inhibitor. American Association for Cancer Research. 2007;13(11):3431–3431. 10.1158/1078-0432.CCR-07-0070 [DOI] [PubMed] [Google Scholar]
- 39. Bean J, Riely GJ, Balak M, Marks JL, Ladanyi M, Miller VA, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res. 2008;14(22):7519–7525. 10.1158/1078-0432.CCR-08-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Landi L, Cappuzzo F. HER2 and lung cancer. Expert Review of Anticancer Therapy. 2013;13(10):1219–28. 10.1586/14737140.2013.846830 [DOI] [PubMed] [Google Scholar]
- 41. Luo M, Fu LW. Redundant kinase activation and resistance of EGFR-tyrosine kinase inhibitors. Am J Cancer Res. 2014;4(6):608–628. [PMC free article] [PubMed] [Google Scholar]
- 42. Bertotti A, Burbridge MF, Gastaldi S, Galimi F, Torti D, Medico E, et al. Only a subset of Met-activated pathways are required to sustain oncogene addiction. Sci Signal. 2009;2(102):er11 [DOI] [PubMed] [Google Scholar]
- 43. Canfield K, Li J, Wilkins OM, Morrison MM, Ung M, Wells W, et al. Receptor tyrosine kinase ERBB4 mediates acquired resistance to ERBB2 inhibitors in breast cancer cells. Cell Cycle. 2015;14(4):648–655. 10.4161/15384101.2014.994966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung HW, Du J, Boehm JS, He F, Weir BA, Wang X, et al. Amplification of CRKL induces transformation and EGFR inhibitor resistance in human non small cell lung cancers. Cancer Discovery. 2011;. [DOI] [PMC free article] [PubMed]
- 45. Azuaje F, Tiemann K, Niclou SP. Therapeutic control and resistance of the EGFR-driven signaling network in glioblastoma. Cell Commun Signal. 2015;13:23 10.1186/s12964-015-0098-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118(7):2609–2619. 10.1172/JCI34588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peled N, Wynes MW, Ikeda N, Ohira T, Yoshida K, Qian J, et al. Insulin-like growth factor-1 receptor (IGF-1R) as a biomarker for resistance to the tyrosine kinase inhibitor gefitinib in non-small cell lung cancer. Cell Oncol (Dordr). 2013;36(4):277–288. 10.1007/s13402-013-0133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baker AT, Zlobin A, Osipo C. Notch-EGFR/HER2 Bidirectional Crosstalk in Breast Cancer. Front Oncol. 2014;4:360 10.3389/fonc.2014.00360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Z, Li Y, Ahmad A, Azmi AS, Banerjee S, Kong D, et al. Targeting Notch signaling pathway to overcome drug resistance for cancer therapy. Biochim Biophys Acta. 2010;1806(2):258–267. 10.1016/j.bbcan.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Al-Hussaini H, Subramanyam D, Reedijk M, Sridhar SS. Notch signaling pathway as a therapeutic target in breast cancer. Mol Cancer Ther. 2011;10(1):9–15. 10.1158/1535-7163.MCT-10-0677 [DOI] [PubMed] [Google Scholar]
- 51. Loh YN, Hedditch EL, Baker LA, Jary E, Ward RL, Ford CE. The Wnt signalling pathway is upregulated in an in vitro model of acquired tamoxifen resistant breast cancer. BMC Cancer. 2013;13:174 10.1186/1471-2407-13-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chikazawa N, Tanaka H, Tasaka T, Nakamura M, Tanaka M, Onishi H, et al. Inhibition of Wnt signaling pathway decreases chemotherapy-resistant side-population colon cancer cells. Anticancer Res. 2010;30(6):2041–2048. [PubMed] [Google Scholar]
- 53. Ballas MS, Chachoua A. Rationale for targeting VEGF, FGF, and PDGF for the treatment of NSCLC. Onco Targets Ther. 2011;4:43–58. 10.2147/OTT.S18155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chatterjee S, Heukamp LC, Siobal M, Schottle J, Wieczorek C, Peifer M, et al. Tumor VEGF:VEGFR2 autocrine feed-forward loop triggers angiogenesis in lung cancer. The Journal of Clinical Investigation. 2013;123(4):1732–1740. 10.1172/JCI65385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bianco R, Rosa R, Damiano V, Daniele G, Gelardi T, Garofalo S, et al. Vascular Endothelial Growth Factor Receptor-1 Contributes to Resistance to Anti-Epidermal Growth Factor Receptor Drugs in Human Cancer Cells. Clinical Cancer Research. 2008;14(16):5069–5080. 10.1158/1078-0432.CCR-07-4905 [DOI] [PubMed] [Google Scholar]
- 56. Azuma K, Kawahara A, Sonoda K, Nakashima K, Tashiro K, Watari K, et al. FGFR1 activation is an escape mechanism in human lung cancer cells resistant to afatinib, a pan-EGFR family kinase inhibitor. Oncotarget. 2014;5(15). 10.18632/oncotarget.1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11(12):1444–1450. 10.1038/ncb1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhao Y, Yang X. The Hippo pathway in chemotherapeutic drug resistance. Int J Cancer. 2015;137(12):2767–2773. 10.1002/ijc.29293 [DOI] [PubMed] [Google Scholar]
- 59. Huang JM, Nagatomo I, Suzuki E, Mizuno T, Kumagai T, Berezov A, et al. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene. 2013;32(17):2220–2229. 10.1038/onc.2012.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Serizawa M, Takahashi T, Yamamoto N, Koh Y. Combined treatment with erlotinib and a transforming growth factor-beta type I receptor inhibitor effectively suppresses the enhanced motility of erlotinib-resistant non-small-cell lung cancer cells. J Thorac Oncol. 2013;8(3):259–269. 10.1097/JTO.0b013e318279e942 [DOI] [PubMed] [Google Scholar]
- 61. Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim Js, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. 10.1093/bioinformatics/btn577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang daW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 63. Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22(23):2926–2933. 10.1093/bioinformatics/btl483 [DOI] [PubMed] [Google Scholar]
- 64. Han J, Shi X, Zhang Y, Xu Y, Jiang Y, Zhang C, et al. ESEA: Discovering the Dysregulated Pathways based on Edge Set Enrichment Analysis. Sci Rep. 2015;5:13044 10.1038/srep13044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Han J, Li C, Yang H, Xu Y, Zhang C, Ma J, et al. A novel dysregulated pathway-identification analysis based on global influence of within-pathway effects and crosstalk between pathways. J R Soc Interface. 2015;12(102):20140937 10.1098/rsif.2014.0937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu L, Greger J, Shi H, Liu Y, Greshock J, Annan R, et al. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Res. 2009;69(17):6871–6878. 10.1158/0008-5472.CAN-08-4490 [DOI] [PubMed] [Google Scholar]
- 67. Karakashev SV, Reginato MJ. Hypoxia/HIF-alpha induces lapatinib resistance in ERBB2-positive breast cancer cells via regulation of DUSP2. Oncotarget. 2015;6(4):1967–1980. 10.18632/oncotarget.2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xia W, Bacus S, Husain I, Liu L, Zhao S, Liu Z, et al. Resistance to ErbB2 tyrosine kinase inhibitors in breast cancer is mediated by calcium-dependent activation of RelA. Mol Cancer Ther. 2010;9(2):292–299. 10.1158/1535-7163.MCT-09-1041 [DOI] [PubMed] [Google Scholar]
- 69. Lynch MA, Petrel TA, Song H, Knobloch TJ, Casto BC, Ramljak D, et al. Responsiveness to transforming growth factor-beta (TGF-beta)-mediated growth inhibition is a function of membrane-bound TGF-beta type II receptor in human breast cancer cells. Gene Expr. 2001;9(4-5):157–171. 10.3727/000000001783992560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Serra V, Eichhorn PJ, Garcia-Garcia C, Ibrahim YH, Prudkin L, Sanchez G, et al. RSK3/4 mediate resistance to PI3K pathway inhibitors in breast cancer. J Clin Invest. 2013;123(6):2551–2563. 10.1172/JCI66343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hegde PS, Rusnak D, Bertiaux M, Alligood K, Strum J, Gagnon R, et al. Delineation of molecular mechanisms of sensitivity to lapatinib in breast cancer cell lines using global gene expression profiles. Mol Cancer Ther. 2007;6(5):1629–1640. 10.1158/1535-7163.MCT-05-0399 [DOI] [PubMed] [Google Scholar]
- 72. Liu NN, Xi Y, Callaghan MU, Fribley A, Moore-Smith L, Zimmerman JW, et al. SMAD4 is a potential prognostic marker in human breast carcinomas. Tumour Biol. 2014;35(1):641–650. 10.1007/s13277-013-1088-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Deckers M, van Dinther M, Buijs J, Que I, Lowik C, van der Pluijm G, et al. The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006;66(4):2202–2209. 10.1158/0008-5472.CAN-05-3560 [DOI] [PubMed] [Google Scholar]
- 74. Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–961. 10.1093/nar/gks1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hu J, Rho HS, Newman RH, Zhang J, Zhu H, Qian J. PhosphoNetworks: a database for human phosphorylation networks. Bioinformatics. 2014;30(1):141–142. 10.1093/bioinformatics/btt627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Khalil S, Tan GA, Giri DD, Zhou XK, Howe LR. Activation status of Wnt/beta-catenin signaling in normal and neoplastic breast tissues: relationship to HER2/neu expression in human and mouse. PLoS ONE. 2012;7(3):e33421 10.1371/journal.pone.0033421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lamb R, Ablett MP, Spence K, Landberg G, Sims AH, Clarke RB. Wnt pathway activity in breast cancer sub-types and stem-like cells. PLoS ONE. 2013;8(7):e67811 10.1371/journal.pone.0067811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sasaki T, Suzuki H, Yagi K, Furuhashi M, Yao R, Susa S, et al. Lymphoid enhancer factor 1 makes cells resistant to transforming growth factor beta-induced repression of c-myc. Cancer Res. 2003;63(4):801–806. [PubMed] [Google Scholar]
- 79. Maglott DR, Katz KS, Sicotte H, Pruitt KD. NCBI’s LocusLink and RefSeq. Nucleic Acids Res. 2000;28(1):126–128. 10.1093/nar/28.1.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Takano Y, Kato Y, Masuda M, Ohshima Y, Okayasu I. Cyclin D2, but not cyclin D1, overexpression closely correlates with gastric cancer progression and prognosis. J Pathol. 1999;189(2):194–200. [DOI] [PubMed] [Google Scholar]
- 81. Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, et al. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol. 2002;22(22):7842–7852. 10.1128/MCB.22.22.7842-7852.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Qin H, Chan MW, Liyanarachchi S, Balch C, Potter D, Souriraj IJ, et al. An integrative ChIP-chip and gene expression profiling to model SMAD regulatory modules. BMC Syst Biol. 2009;3:73 10.1186/1752-0509-3-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. 10.1002/0470857897.ch8 [DOI] [PubMed] [Google Scholar]
- 84. Kechagioglou P, Papi RM, Provatopoulou X, Kalogera E, Papadimitriou E, Grigoropoulos P, et al. Tumor suppressor PTEN in breast cancer: heterozygosity, mutations and protein expression. Anticancer Res. 2014;34(3):1387–1400. [PubMed] [Google Scholar]
- 85. Eichhorn PJ, Gili M, Scaltriti M, Serra V, Guzman M, Nijkamp W, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68(22):9221–9230. 10.1158/0008-5472.CAN-08-1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bouchalova K, Cizkova M, Cwiertka K, Trojanec R, Friedecky D, Hajduch M. Lapatinib in breast cancer—the predictive significance of HER1 (EGFR), HER2, PTEN and PIK3CA genes and lapatinib plasma level assessment. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2010;154(4):281–288. 10.5507/bp.2010.043 [DOI] [PubMed] [Google Scholar]
- 87. Du G, Bian L, Wang T, Xu X, Zhang S, Guo Y, et al. [PTEN loss correlates withthe clinical efficacy of lapatinib in HER2 positive metastatic breast cancer with trastuzumab-resistance]. Zhonghua Yi Xue Za Zhi. 2015;95(28):2264–2267. [PubMed] [Google Scholar]
- 88. Carpenter RL, Lo HW. Regulation of Apoptosis by HER2 in Breast Cancer. Journal of Carcinogenesis & Mutagenesis. 2013;0(0):1–7. 10.4172/2157-2518.S7-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Huang S, Benavente S, Armstrong EA, Li C, Wheeler DL, Harari PM. p53 modulates acquired resistance to EGFR inhibitors and radiation. Cancer Res. 2011;71(22):7071–7079. 10.1158/0008-5472.CAN-11-0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Boeckx C, Baay M, Wouters A, Specenier P, Vermorken JB, Peeters M, et al. Anti-epidermal growth factor receptor therapy in head and neck squamous cell carcinoma: focus on potential molecular mechanisms of drug resistance. Oncologist. 2013;18(7):850–864. 10.1634/theoncologist.2013-0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Han C, Zhao R, Liu X, Srivastava A, Gong L, Mao H, et al. DDB2 suppresses tumorigenicity by limiting the cancer stem cell population in ovarian cancer. Mol Cancer Res. 2014;12(5):784–794. 10.1158/1541-7786.MCR-13-0638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ennen M, Klotz R, Touche N, Pinel S, Barbieux C, Besancenot V, et al. DDB2: a novel regulator of NF-kB and breast tumor invasion. Cancer Res. 2013;73(16):5040–5052. 10.1158/0008-5472.CAN-12-3655 [DOI] [PubMed] [Google Scholar]
- 93. Kattan Z, Marchal S, Brunner E, Ramacci C, Leroux A, Merlin JL, et al. Damaged DNA binding protein 2 plays a role in breast cancer cell growth. PLoS ONE. 2008;3(4):e2002 10.1371/journal.pone.0002002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.He YH. Involvement of DDB2 Down-regulation in Lapatinib-induced Cross-resistance to Chemotherapy. Ph.D. Thesis, National Digital Library of Theses and Dissertations in Taiwan; 2012. Available from: http://handle.ncl.edu.tw/11296/ndltd/06833793361974888604
- 95. Nieto-Sampedro M, Valle-Argos B, Gomez-Nicola D, Fernandez-Mayoralas A, Nieto-Diaz M. Inhibitors of Glioma Growth that Reveal the Tumour to the Immune System. Clin Med Insights Oncol. 2011;5:265–314. 10.4137/CMO.S7685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wee KB, Surana U, Aguda BD. Oscillations of the p53-Akt network: implications on cell survival and death. PLoS ONE. 2009;4(2):e4407 10.1371/journal.pone.0004407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–730. 10.1038/nature03918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sun NK, Sun CL, Lin CH, Pai LM, Chao CC. Damaged DNA-binding protein 2 (DDB2) protects against UV irradiation in human cells and Drosophila. J Biomed Sci. 2010;17:27 10.1186/1423-0127-17-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Carson C, Omolo B, Chu H, Zhou Y, Sambade MJ, Peters EC, et al. A prognostic signature of defective p53-dependent G1 checkpoint function in melanoma cell lines. Pigment Cell Melanoma Res. 2012;25(4):514–526. 10.1111/j.1755-148X.2012.01010.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A, Michalowski A, et al. A gene expression signature of acquired chemoresistance to cisplatin and fluorouracil combination chemotherapy in gastric cancer patients. PLoS ONE. 2011;6(2):e16694 10.1371/journal.pone.0016694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Awan A, Bari H, Yan F, Moksong S, Yang S, Chowdhury S, et al. Regulatory network motifs and hotspots of cancer genes in a mammalian cellular signalling network. IET Syst Biol. 2007;1(5):292–297. 10.1049/iet-syb:20060068 [DOI] [PubMed] [Google Scholar]
- 102. Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140 10.1038/msb4100180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Guney E, Sanz-Pamplona R, Sierra A, Oliva B. In: Azmi AS, editor. Understanding Cancer Progression Using Protein Interaction Networks. Dordrecht: Springer Netherlands; 2012. p. 167–195. [Google Scholar]
- 104.Wang E. COSMIC: Cancer Gene census; 2017. Available from: http://cancer.sanger.ac.uk/census/
- 105. Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. 10.1038/nrc1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS ONE. 2010;5(2):e8918 10.1371/journal.pone.0008918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472–477. 10.1093/nar/gkt1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102(43):15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, et al. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40(Database issue):D1301–1307. 10.1093/nar/gkr1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wasserman S, Pattison P. Logit models and logistic regressions for social networks: I. An introduction to Markov graphs andp. Psychometrika. 1996;61(3):401–425. 10.1007/BF02294547 [DOI] [Google Scholar]
- 111. Lunn DJ, Thomas A, Best N, Spiegelhalter D. WinBUGS—A Bayesian Modelling Framework: Concepts, Structure, and Extensibility. Statistics and Computing. 2000;10(4):325–337. 10.1023/A:1008929526011 [DOI] [Google Scholar]
- 112. Adams S, Carter N, Hadlock C, Haughton D, Sirbu G. Change in connectivity in a social network over time: A bayesian perspective. Connections. 2008;28:17–27. [Google Scholar]
- 113. Han J, Shi X, Zhang Y, Xu Y, Jiang Y, Zhang C, et al. ESEA: Discovering the Dysregulated Pathways based on Edge Set Enrichment Analysis. Sci Rep. 2015;5:13044 10.1038/srep13044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods.
(PDF)
List of identified putative aberrant gene-pairs (for both SKBR3 and BT474) cell-lines in acquired resistance.
(XLSX)
Full results of pathway enrichment tests of identified aberrant gene-pairs in acquired resistance from KEGG, Reactome, and WikiPathway databases for both SKBR3 and BT474 cell-lines.
(XLSX)
Comparing our current model with the previous model by observing the percentages of non-direct (indirect and PPI pair) enriched links (aberrant pairs as known signaling links) in the aberrant signaling pathways from KEGG, Reactome, and WikiPathway databases that were detected by our current but not the previous model, for both SKBR3 and BT474 cell-lines.
(XLSX)
Comparing our current model with the previous model by observing the percentages of non-direct (indirect and PPI pair) enriched links (aberrant pairs as known signaling links) in the aberrant signaling pathways from KEGG, Reactome, and WikiPathway databases that were detected by both of our current and previous models, and were ranked (based on enrichment q-value) high in the current model but low in the previous model, for both SKBR3 and BT474 cell-lines.
(XLSX)
Summary of Type-I, Type-II and Type-III enrichment of V-structures in KEGG, Reactome, and WikiPathway databases in SKBR3 cell-line.
(XLSX)
Summary of Type-I, Type-II and Type-III enrichment of V-structures in KEGG, Reactome, and WikiPathway databases in BT474 cell-line.
(XLSX)
CGC genes in all the Type-I, Type-II and Type-III V-structures in SKBR3 and BT474 cell-lines.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.