Abstract
Background
Retinoic Acid (RA), the active metabolite of Vitamin A, has been demonstrated to be important for growth and branching morphogenesis of mammalian embryonic salivary gland epithelium. However, it is not known whether RA functions directly within epithelial cells or in associated tissues that influence morphogenesis of salivary epithelium. Moreover, downstream targets of RA regulation have not been identified.
Results
Here we show that canonical RA signaling occurs in multiple tissues of embryonic mouse salivary glands, including epithelium, associated parasympathetic ganglion neurons, and non-neuronal mesenchyme. By culturing epithelium explants in isolation from other tissues we demonstrate that RA influences epithelium morphogenesis by direct action in that tissue. Moreover, we demonstrate that inhibition of RA signaling represses cell proliferation and expression of FGF10 signaling targets, and upregulates expression of basal epithelial keratins Krt5 and Krt14. Importantly, we show that the stem cell gene Kit is regulated inversely from Krt5/Krt14 by RA signaling.
Conclusions
RA regulates Krt5 and Krt14 expression independently of stem cell character in developing salivary epithelium. RA, or chemical inhibitors of RA signaling, could potentially be used for modulating growth and differentiation of epithelial stem cells for the purpose of re-populating damaged glands or generating bioengineered organs.
Keywords: Keratin, retinoid, Vitamin A, KRT5, progenitor, stem cell
INTRODUCTION
Salivary glands are important for human health. Loss of salivary gland function, which occurs frequently in patients suffering from the autoimmune condition Sjogren's syndrome, or in cancer patients treated with radiation to the head and neck, has devastating consequences for quality of life. Thus, knowledge of salivary gland biology is an important research goal.
Much insight about salivary gland organogenesis has been gained via study of submandibular salivary gland (SMG) morphogenesis in mouse embryos or by investigation of mouse salivary glands cultured ex vivo (reviewed in (Tucker, 2007; Knosp et al., 2012)) Previous research has highlighted the importance of interactions between different tissues of developing salivary glands during SMG morphogenesis. For example, at early stages of development, interactions between oral epithelium and underlying mesenchyme are critical for salivary gland formation (Kratochwil, 1969; Wells et al., 2013). At later stages of morphogenesis, neurons of the submandibular parasympathetic ganglion stimulate growth, branching and tubulogenesis of gland epithelium (Knox et al., 2010; Nedvetsky et al., 2014). Studies of mutant mice and experiments with tissue explants cultured ex vivo have demonstrated that signaling by growth factor FGF10 via its receptor FGFR2b is critical for growth and branching morphogenesis of embryonic salivary epithelium (De Moerlooze et al., 2000; Ohuchi et al., 2000; Entesarian et al., 2005; Jaskoll et al., 2005; Steinberg et al., 2005).
A major goal of salivary gland research is to identify the molecular regulation of epithelial progenitor cells that could contribute to regeneration of damaged glands or could be used to direct differentiation of stem cells to bioengineer replacement salivary epithelium. One pair of molecules proposed to mark salivary gland progenitor cells are the intermediate filament proteins cytokeratin 5 (KRT5) and KRT14 (Knox et al., 2010; Lombaert et al., 2011). Krt5 is expressed in the basal layer of developing SMG epithelium. Lineage tracing of cells expressing Krt5 early demonstrated that these cells give rise to most of the SMG epithelium, suggesting Krt5 marks multipotent cells with progenitor character (Knox et al., 2010). In addition to marking progenitor cells of salivary glands, KRT5 and KRT14 are present in basal progenitors cells in other epithelial organs, including trachea (Rock et al., 2009), prostate (Hudson et al., 2001), bladder (Colopy et al., 2014), and lung (Zuo et al., 2015). Although Krt5 expression is associated with progenitor character in salivary glands, the recent discovery that SMG acinar cells regenerate by self-duplication demonstrated that acinar epithelium does not renew from ductal Krt5+ cells (Aure et al., 2015).
An additional factor that is present in stem cells or progenitor cells of salivary epithelium is the receptor tyrosine kinase KIT. KIT is present in stem or progenitor cells of the hematopoetic system and many other tissues and organs (Ogawa et al., 1991; Broudy, 1997). In salivary glands, KIT+ epithelial progenitor cells are able to regenerate irradiated glands (Lombaert et al., 2008; Nanduri et al., 2013).
RA, the active metabolite of Vitamin A (all-trans-retinol), is a small lipid soluble molecule that regulates many aspects of embryogenesis and adult health (reviewed in (Clagett-Dame and Knutson, 2011)). Knowledge about how RA and related molecules may regulate embryonic morphogenesis of specific tissue types is needed for a basic understanding of developmental biology and because retinoids hold obvious potential to be used pharmacologically. Canonical RA signaling occurs through a family of ligand responsive nuclear receptors known as retinoic acid receptors (RAR), which bind to regulatory DNA elements known as RA response elements. Although canonical RA signaling through RAR has historically been considered primarily in terms of ligand-dependent activation, emerging evidence indicates that ligand-dependent repression by RAR is a common mechanism of RA-mediated gene regulation (Liu et al., 2014).
Analysis of Krt5 and Krt14 cis-regulatory elements indicates that RA signaling represses Krt5 expression in epidermal epithelial cells. RAR regulate Krt5 expression by binding to negative RA response elements upstream of the Krt5 promoter (Tomic et al., 1990; Ohtsuki et al., 1992; Radoja et al., 1997; Jho et al., 2001). In that context RAR bound to RA ligand suppress expression while unliganded RAR promote expression of Krt5 (Tomic-Canic et al., 1996).
We recently identified that RA is a critical regulator of mammalian salivary gland morphogenesis, and that blockage of RA signaling disrupts growth and branching morphogenesis of salivary epithelium (Wright et al., 2015). Our initial study was based on analyses of RA deficient mouse embryos and ex vivo culture of whole SMG. As such, it was not possible to discern whether RA influences epithelial growth and branching by direct action in epithelial cells, or if RA influences epithelial morphogenesis indirectly by regulation of a different tissue that is needed for epithelium development. Moreover, downstream target genes of RA regulation have not been investigated.
Here we report that RA signaling occurs in epithelial, neuronal, and mesenchymal tissues of the developing mouse SMG. By culturing isolated epithelial rudiments (ER) ex vivo in the presence or absence of a chemical RAR inhibitor, we show that RA signaling regulates growth and branching morphogenesis of epithelial tissue directly. We identify that the RA signaling pathway positively regulates cell proliferation and expression of the FGF10 signaling target Etv5 in cultured SMG epithelia. We further demonstrate that inhibition of RA signal in cultured ER is associated with dramatic upregulation of Krt5 and Krt14, but the upregulation of Krt5 and Krt14 does not correlate with altered expression of other salivary keratin genes or with stem cell markers. These findings demonstrate that RA signaling modulates differentiation of salivary epithelium by direct action within epithelial cells, negatively regulating expression of Krt5 and Krt14, and positively influencing cell proliferation, and expression of Etv5, and stem cell marker Kit.
RESULTS
RA signaling occurs in multiple tissues of developing SMG
In order to investigate the precise distribution of active RA signaling in developing SMG tissues we performed immunostaining on frontal sections of mouse embryos carrying the RARE-lacZ transgenic reporter (Rossant et al., 1991). This reporter expresses LacZ encoding β-galactosidase in response to canonical RA signaling through RAR. In embryos carrying this reporter, staining for β-galactosidase reveals RA signaling activity.
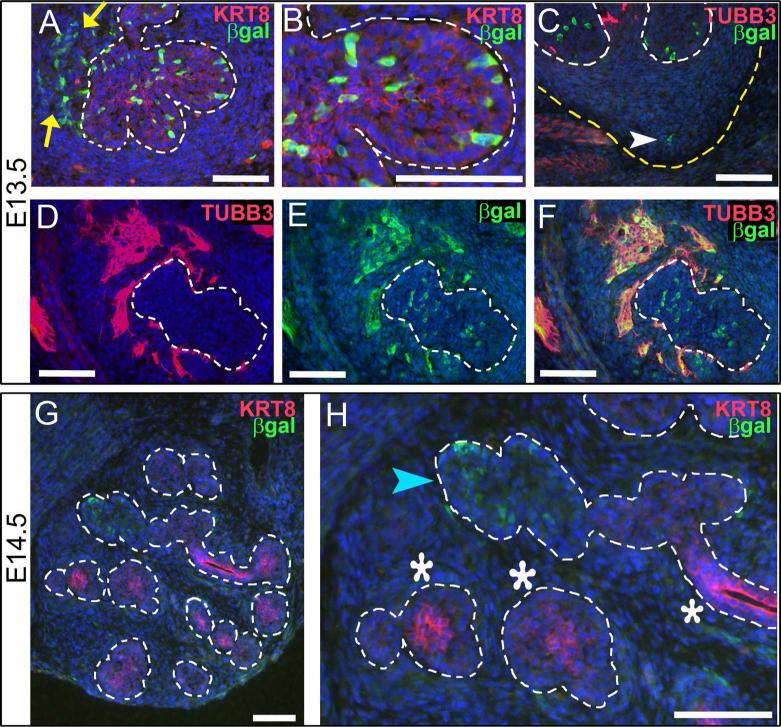
At embryonic day 13.5 (E13.5) we observed strong RA signaling in individual cells of the developing SMG (Fig. 1 A-F). Co-staining for KRT8, which is weakly expressed in the epithelium at this stage, reveals that cells of the SMG epithelium are positive for RA signaling (Fig. 1 A, B, within white dotted outline). RA signal-positive cells appear in a random mosaic pattern within the KRT8-positive epithelium. Counting the RA-positive cells in comparison to the number of DAPI-positive nuclei of the epithelium indicates that RA signal-positive cells comprise 10% of the epithelium (n= 4 sections). Cells positive for RA signaling are present within the basal layer of the epithelium, and also within the supra-basal compartment of the epithelium (Fig. 1 B).
Figure 1. RA signaling occurs in multiple tissues of developing SMG.
Immunostaining for β-galactosidase on cryosections from embryos carrying the RARE-LacZ reporter transgene reveals location of RA signaling in developing SMG at E13.5 (A-F) and E14.5 (G-H). (A, B) At E13.5 co-staining for KRT8 and β-galactosidase reveals numerous cells of the epithelium are positive for RA signaling (cells within dotted outline). Cells positive for RA signaling are also detected in non-epithelial cells, particularly in the area around the main duct (yellow arrows). Examination of an individual endbud (B, (detail of A)) reveals that RA positive cells are present in basal epithelium and also within interior epithelium that will later form lumens. Co-staining for neurons (TUBB3) and RA signaling (β-galactosidase) on a posterior section through the apex of the strawberry-shaped SMG (C) reveals RA signaling occurs in a small number of cells in non-neuronal mesenchyme in this region (white arrowhead). (D-F) Co-staining for neurons (TUBB3) and RA signaling (β-galactosidase) in sections containing the parasympathetic ganglion reveals that many neurons of the SMG parasympathetic ganglion are positive for RA signaling at E13.5. (G,H) By E14.5, the number of SMG cells positive for RA signaling is reduced relative to E13.5. (G) At E14.5 RA positive cells are present in only a few endbuds. (H) KRT8 is expressed strongly in some endbuds (white asterisks) and weakly in others (blue arrowhead). Endbuds with RA positive cells correspond to those with low level of KRT8 (blue arrowhead), while endbuds and ducts with higher level of KRT8 have little or no detectable RA signaling (white asterisks). White dotted lines, edge of epithelium; yellow arrows, non-epithelial RA positive cells in vicinity of main duct; yellow dotted line, edge of SMG mesenchyme; white arrowhead, RA signal positive cell in non-neuronal mesenchyme at tip of strawberry-shaped SMG; blue arrowhead, endbud positive for RA signaling with low KRT8; white asterisks, endbuds and ducts with no detectable RA signal with high KRT8. Scale bars = 100 μm.
Non-epithelial tissues of the developing SMG are also positive for RA signal. RA signal-positive cells are detected in non-neuronal mesenchyme at the tip of the developing SMG (Fig. 1 C, white arrowhead). Numerous RA signal-positive cells are also detected in the area surrounding the main duct epithelium, where the neurons of the parasympathetic ganglion are known to be located (Fig. 1 A, yellow arrows). Sections from RARE-lacZ reporter embryos were co-stained for TUBB3 and β-galactosidase to determine if RA signaling occurs within neurons of the SMG parasympathetic ganglion. Co-staining for the two markers reveals that strong RA signaling is present within many or most of the cell bodies of the developing SMG parasympathetic ganglion (Fig. 1 D-F). Taken together these data indicate that E13.5 SMG have active RA signaling in epithelial cells, in non-neuronal mesenchyme, and within the neurons the SMG parasympathetic ganglion.
We examined also the presence and distribution of RA signaling relative to KRT8 at later developmental stages. We observe KRT8 is preferentially localized to luminal epithelium of ducts in E14.5 SMG (Fig. 1 G, H), a distribution consistent with previous analyses of KRT8 localization in SMG and prostate (Hudson et al., 2001; Rebustini et al., 2007). Co-staining for β-galactosidase and KRT8 reveals that RA signaling is reduced at E14.5 relative to E13.5 (Fig. 1 G, H). Counting the number of cells positive for RA signaling relative to the number of DAPI-positive nuclei in the epithelium indicates that RA signal positive cells comprise less than 1% of epithelial cells at E14.5 (N=4 sections). At this stage the RA signaling-positive cells within the epithelium are limited to a few epithelial endbuds, particularly those with the lowest levels of KRT8 (Fig. 1 H, blue arrowhead). Endbuds and ducts with relatively high KRT8 have few or no cells positive for RA signaling (Fig. 1 H, white asterisks). RA signaling is also detected in a few scattered non-epithelial cells at this stage. At E15.5 RA signaling was not detected by β-galactosidase staining (data not shown). We have previously reported, based on β-galactosidase enzymatic staining, that RA signaling is downregulated in SMG after E13.5. The immunostain data presented here extend the initial observation, showing that epithelial regions retaining RA signaling the longest are KRT8-negative endbuds.
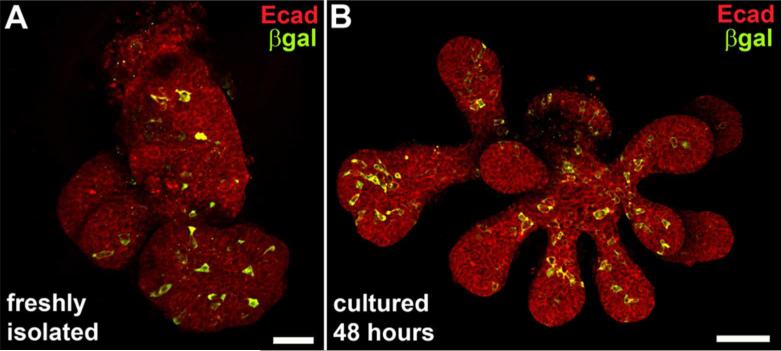
In order to visualize the overall distribution of RA signaling within the highly 3-dimensional structure of a developing SMG epithelium, we evaluated RA signaling in whole mount ER specimens. Isolated ER from E13.5 SMG of RARE-lacZ reporter embryos were immunostained as whole mount specimens for E-cadherin, which marks all epithelium, and for β-galactosidase, which marks the cells with active RA signaling. Confocal micrographs were collected through the entire specimen and Z-stacks of image planes were collapsed into a single image. The resulting whole mount images reveal RA signaling is active in a mosaic distribution of cells, and is present within the endbuds and main duct of the E13.5 SMG epithelium (Fig. 2 A). RA signal positive cells were not detected preferentially at any position or site.
Figure 2. Mosaic RA signaling in in ducts and endbuds persists during culture.
Confocal micrographs of whole mount epithelial tissue isolated from E13.5 SMG of RARE-lacZ reporter embryos immunostained for RA activity and epithelium reveals distribution of RA signaling in main duct and endbuds, and persistence of signaling in culture. (A) Staining freshly isolated ER for epithelium (E-cadherin) and RA signaling (β-galactosidase) reveals a mosaic pattern of RA signaling in main duct and endbuds. (B) RA signaling is detected in ER after 48 hours in culture in Matrigel. The mosaic distribution of RA positive cells in ducts and endbuds is similar to that observed in freshly isolated ER. Scale bars = 50μm with respect to a single image plane.
Because much analysis of salivary gland biology has been elucidated through the means of ex vivo cultured ER, we sought to determine if RA signaling occurred in that context. We therefore assessed RA signaling in isolated ER cultured from E13.5 RARE-lacZ reporter embryos. After two days in culture, ER display a mosaic pattern of active RA signaling (Fig. 2 B), which was similar to the distribution observed in freshly isolated ER (Fig. 2A). In both cases RA signal positive cells are present in ducts and endbuds.
Inhibition of RA signaling impairs branching morphogenesis, expression of FGF10 target Etv5, and cell proliferation in cultured ER
In order to determine if RA signaling influences SMG development by direct action within epithelial tissue we examined whether RA signaling is important for growth of isolated ER cultured ex vivo. We cultured isolated ER in the presence or absence of a chemical inhibitor of RA signaling, BMS 493, which is a pan-RAR inverse agonist. ER were isolated from E13.5 SMG by treatment with dispase I and microdissection. ER were cultured for 48 hours on filters supported over medium containing 5 μM BMS 493 or on control medium containing equivalent volume of the solvent DMSO.
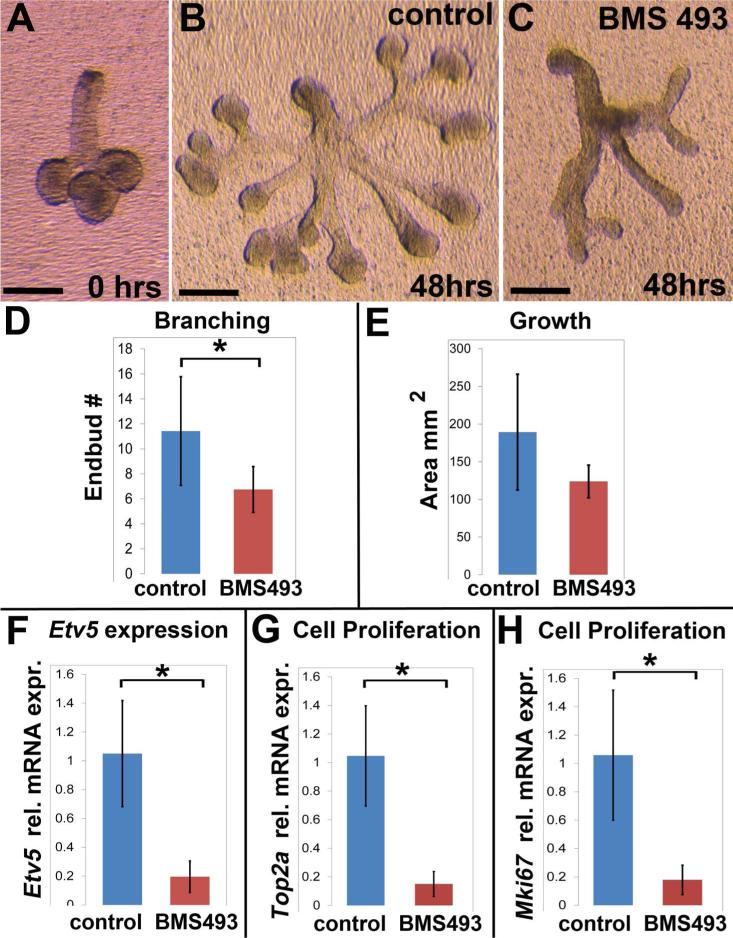
Prior to culture each isolated ER was a small compact structure with 3-6 small endbuds (Fig. 3 A). After 48 hours, ER that had been cultured on control medium grew robustly with extensive branches, large rounded endbuds, and elongated translucent ducts (Fig. 3 B). In contrast, ER that had been cultured on medium containing BMS 493 grew in an atypical manner (Fig, 3 C). BMS 493-treated ER had fewer branches and smaller endbuds, with ducts that were narrow and optically dense. Counting the number of endbuds revealed that ER grown on medium containing BMS 493 had significantly fewer endbuds, N=7 ER control, N=8 ER BMS 493, p ≤ 0.03 (Fig. 3 D). The amount of tissue growth for control and BMS 493-treated ER was assessed by tracing the outline of each specimen imaged at the end of the culture period and measuring the 2-dimensional area. As measured by area outline, tissue growth was not significantly different between control and BMS-493 treated ER specimens (Fig. 3 E). These data reveal that RA signaling is important for branching morphogenesis of ER cultured in isolation ex vivo, demonstrating that RA influences epithelial morphogenesis by direct action in epithelial tissue.
Figure 3. RA signaling regulates developmental growth of epithelium by direct action in epithelial tissue.
Inhibition of RA signaling by BMS 493 impedes ex vivo growth of isolated ER. (A) Freshly isolated E13.5 ER with 3-6 endbuds were placed in culture. (B) ER cultured for 48 hours on control medium grew well, branched, and formed numerous elongated translucent ducts. (C) ER cultured on medium containing BMS 493 had abnormal growth with fewer branches, smaller endbuds, and an optically dense appearance. (D) ER grown on BMS 493 had significantly fewer endbuds than those grown on control medium, Control ER N= 7, BMS 493 ER N=8, p≤0.03. (E) Amount of tissue growth, as assessed by outlined area, of ER cultured on BMS 493 medium was not significantly different from controls. (F) Expression of Etv5, a known target of FGF10 signaling, is significantly down-regulated in ER cultured on BMS 493 relative to control, N = 3 independent culture experiments with 6-8 ER/condition × 3 technical qPCR replicates, p = 0.01. (G) Expression of Top2a, which is expressed in the S-phase of the cell cycle, is significantly down-regulated in ER cultured on BMS 493 relative to control, N = 3 independent culture experiments with 6-8 ER/condition × 3 technical qPCR replicates, p = 0.03. (G) Expression of Mki67, which is preferentially expressed in the G2 phase of the cell cycle, is significantly down-regulated in ER cultured on BMS 493 relative to control, N = 3 independent culture experiments with 6-8 ER/condition × 3 technical qPCR replicates, p = 0.03. Error bars on histograms represent standard deviations. Scale bars = 200μm.
It is well established that branching morphogenesis of SMG epithelium requires signaling by the growth factor FGF10 through its receptor FGFR2b (Steinberg et al., 2005), and FGF10 is included as an additive to the ER culture medium. One downstream target of FGF10 signaling in epithelial tissues is Etv5 (Firnberg and Neubuser, 2002; Liu et al., 2003; Michos et al., 2010). In order to discern whether RA signaling impacts ER morphogenesis in a manner that is concordant with, or antagonistic to, the FGF10 pathway, we assessed expression of Etv5. Inhibition of RA signaling by treatment with BMS 493 caused a ≥ 5-fold downregulation of Etv5 relative to controls, N = 3 independent culture experiments with 6-8 ER/condition × 3 technical qPCR replicates, p = 0.01, (Fig. 3 F). These data demonstrate that inhibition of RA signaling in cultured ER reduces expression of a known downstream target of FGF10 signaling, indicating that RA signaling and FGF10 signaling function in the same direction to promote epithelium branching in this context.
Growth and branching of cultured SMG is associated with epithelial cell proliferation (Steinberg et al., 2005). Because treatment with BMS 493 impaired branching morphogenesis we sought to determine whether cell proliferation was also impacted by inhibition of RA signaling. To that end, we quantified expression of two genes whose expression is associated with cell proliferation. Top2a, which encodes DNA topoisomerase II, is enriched during the S-phase of the cell cycle (Goswami et al., 1996). The gene Mki67, which encodes the widely used proliferation marker antigen Ki67, is expressed preferentially in the G2 phase of the cell cycle (Ishida et al., 2001; Whitfield et al., 2002). We quantified expression of these two cell cycle genes by qPCR. Inhibition of RA signaling by treatment with BMS 493 caused a ≥ 5-fold downregulation of both Top2a and Mki67 relative to controls (Fig. 3 G, H). For both Top2a and Mki67 N = 3 independent culture experiments with 6-8 ER/condition × 3 technical qPCR replicates, p = 0.03. These data demonstrate that inhibition of RA signaling reduces cell proliferation in cultured SMG epithelium.
Block of RA signaling upregulates expression of KRT5
An important marker of basal epithelial progenitor cells in SMG ducts is KRT5. Krt5 gene expression has been shown to be negatively regulated by RA in epithelial cells of the epidermis (Ohtsuki et al., 1992; Tomic-Canic et al., 1996; Radoja et al., 1997). We therefore sought to determine if RA signaling regulates KRT5 in developing salivary epithelium. To that end we cultured ER in the presence or absence of RAR inhibitor BMS 493 and assessed the level and distribution of KRT5 protein by confocal microscopy. ER from E13.5 RARE-lacZ reporter embryos were isolated and cultured for 48 hours on medium containing BMS 493 or on control medium. Following culture, ER were fixed as whole mount specimen and immunostained for KRT8 to visualize epithelium, for β-galactosidase to visualize RA signaling activity, and for KRT5. Immunostained specimens were imaged by confocal microscopy.
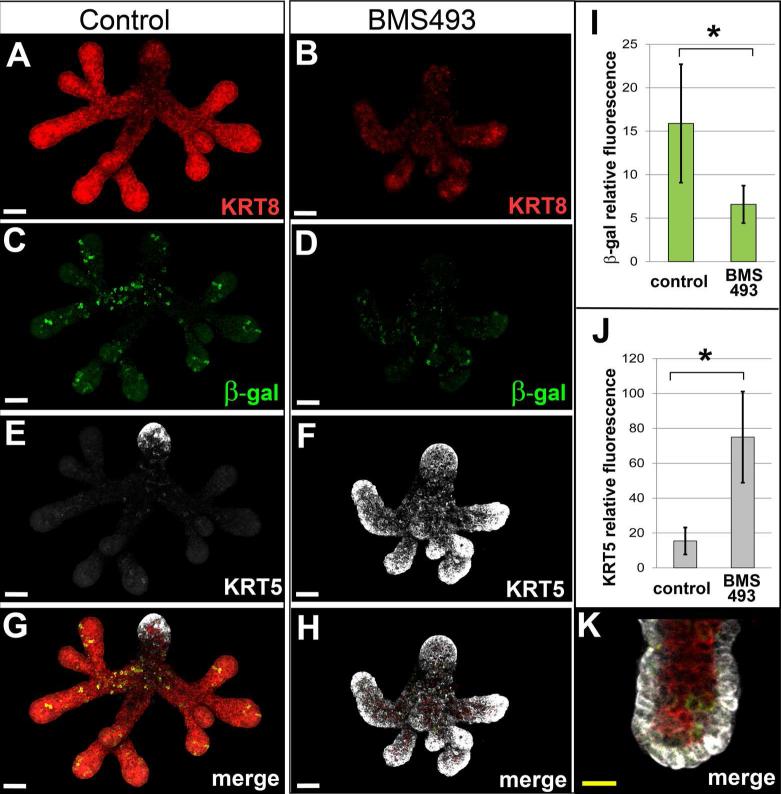
Consistent with our initial ER culture experiments (Fig. 3 A-E), ER from RARE-lacZ embryos cultured on BMS 493 exhibited abnormal growth and branching relative to specimens grown on control medium. BMS 493-treated ER had fewer endbuds, shorter branches and abnormal morphology (Fig. 4 A, B). Immunostaining for β-galactosidase to detect RA signaling validated that RA signaling was reduced in ER cultured on BMS 493 (Fig. 4 C, D). To quantify the reduction of RA signaling in cultured ER the sum of β-galactosidase immunofluorescent signal was measured using IMARIS image analysis software. BMS 493-treated ER had ≥ 2-fold reduction in the amount of β-galactosidase fluorescence relative to controls (N=3 ER/condition × 2 experiments, p ≤ 0.02), (Fig. 4 I).
Figure 4. Inhibition of RA signaling in cultured ER upregulates ductal cytokeratin KRT5.
Immunostain analysis reveals dramatically elevated level of KRT5 in RARE-LacZ reporter ER specimens cultured on medium containing BMS 493 relative to specimens grown on control medium. ER cultured for 48 hours on medium containing BMS 493 (B) were smaller with fewer branches and endbuds relative to their counterparts grown on control medium (A) as visualized by staining for KRT8. RA signaling, visualized by β-galactosidase fluorescence signal was reduced in ER cultured on BMS 493 (D) relative to control specimens (C). (I) The amount of RA signaling, as measured by the sum of relative fluorescence intensity signal for β-galactosidase, is reduced ≥2-fold, p = 0.02, N = 6 ER. KRT5 is dramatically upregulated in ER cultured on BMS 493 (F) relative to controls (E). For ER grown on control medium KRT5 signal is limited to a few cells at the tip of the main duct (E, G). In contrast, ER grown on medium containing BMS 493 had highly elevated KRT5 signal in all endbuds and ducts (F, H). (J) The amount of KRT5 protein, as measured by the sum of relative fluorescence intensity signal, was elevated ~ 5-fold in BMS 493-treated ER relative to control specimens, N = 6 ER, p=0.002. (K) Elevated KRT5 expression in BMS 493-treated ER was restricted to cells of the basal epithelium. White scale bars 50 μm, yellow scale bar = 20 μm.
ER cultured on control medium had only a small amount of KRT5 at the outflow end of the main duct (the main duct being identifiable owing to larger diameter and lack of branches) (Fig. 4 E, G). No KRT5 was observed within secondary ducts or endbuds in any control specimens. In contrast, ER cultured on medium containing BMS 493 exhibited dramatically upregulated levels of KRT5 in all ducts and endbuds (Fig. 4 F, H). Measuring the amount of fluorescence signal by confocal microscopy demonstrated that the amount of KRT5 protein per specimen was increased ≥ 4-fold in BMS 493-treated ER relative to control ER, (N=3 ER/condition × 2 experiments, p ≤ 0.002) (Fig. 4 J). Ectopic high level KRT5 expression was restricted to cells of the basal epithelial layer (Fig. 4 K). These data demonstrate that inhibition of RA signaling elevates KRT5 protein ectopically in the basal epithelial layer of all ducts and endbuds of cultured ER. The elevated levels of KRT5 in BMS 493-treated specimens indicate that KRT5 is negatively regulated by RA in specimens cultured on control medium.
Block of RA signaling coordinately upregulates expression of Krt5 and Krt14
In order to accurately quantify the level of Krt5 upregulation when RA signaling was blocked, and to determine if the upregulation occurred at the level of mRNA, we performed qPCR on ER cultured on BMS 493 or control medium. ER were cultured for 48 hours followed by RNA purification and qPCR analysis to assess the relative expression level of Krt5 and other genes. Analysis of mRNA levels by qPCR revealed Krt5 was ≥ 24-fold higher in BMS 493-treated ER relative to controls, N = 3 independent culture experiments with 6-8 ER/condition × 6 technical qPCR replicates (p=0.000002) (Fig. 5). These data demonstrate that the elevated level of KRT5 protein in BMS 493-treated specimens relative to controls (Fig. 4 E,F,J) results from a dramatic and significant upregulation of Krt5 mRNA.
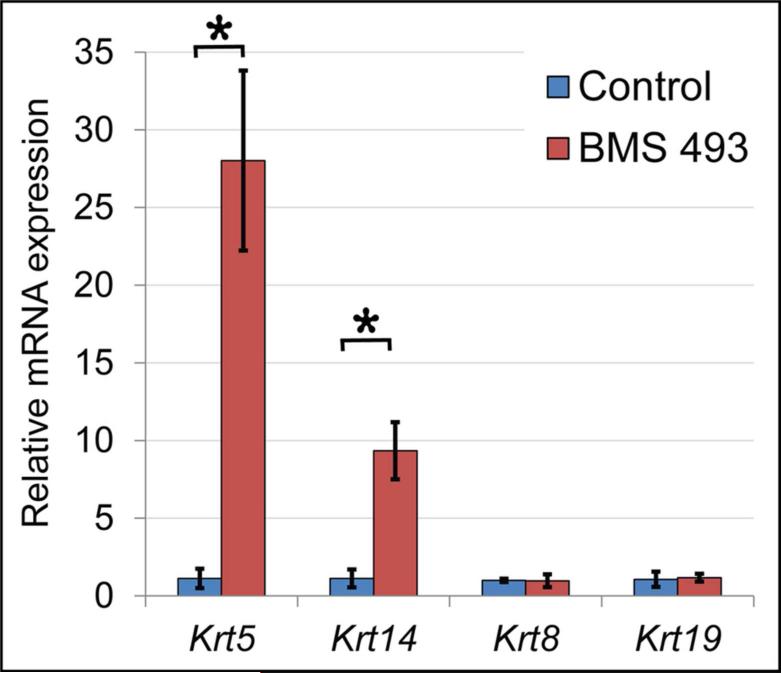
Figure 5. Inhibition of RA signaling upregulates expression of Krt5 and Krt14 mRNA.
Quantitation of gene expression by qPCR demonstrates that Krt5 and its dimerization partner Krt14 are upregulated by inhibition of RA signaling with BMS 493. Krt5 is upregulated ≥ 24 fold (p = 0.000002) and Krt14 is upregulated ≥ 8 fold (p ≤ 0.03) in ER grown in BMS 493 relative to controls. No significant change in expression is observed for keratins Krt8 or Krt19. Data represent averages for 3 independent culture experiments with 6-8 ER/condition. For Krt5 each cDNA sample was run as 6 technical qPCR replicates, for all others each sample was run as 3 technical replicates. Error bars represent standard deviations.
We investigated also whether blocking RA signal with BMS 493 altered expression of Krt5 specifically, or if expression of other members of the keratin family were likewise impacted. Keratins function as heterodimers that polymerize to form intermediate filaments. Krt5 heterodimerizes with Krt14 (Coulombe and Fuchs, 1990; Lee and Coulombe, 2009; Lee et al., 2012), and the two keratin genes are co-expressed together in basal epithelial cells of epidermis and airway epithelium (Rock et al., 2009). In developing salivary glands Krt5 and Krt14 are co-expressed together in some cells, and expressed separately in other cells (Lombaert et al., 2013). Krt8 and Krt19 are also expressed in developing salivary epithelium. To address whether inhibition of RA signaling is required for expression of specific keratins or keratins in general, RNA from cultured ER were subjected to qPCR analysis for Krt14, expressed in basal epithelium, and Krt8 and Krt19, which are present in luminal epithelial layers. The qPCR analysis revealed that Krt14 was upregulated ≥ 8-fold in BMS 493-treated ER specimens relative to controls, N = 3 independent culture experiments with 6-8 ER/condition × 3 technical qPCR replicates (p ≤ 0.03). No significant change in expression of Krt8 or Krt19 was observed. These data demonstrate that inhibition of RA signaling coordinately upregulates expression of the two basal epithelial keratins Krt5 and Krt14, but does not generally alter expression of other keratin family genes. The upregulation of Krt5 and Krt14 when RA signaling is inhibited demonstrates that RA signaling negatively regulates expression of these two basal epithelial keratins.
Stem cell marker Kit is regulated independently of Krt5/Krt14 by RA signaling
Because Krt5 and Krt14 have been implicated as markers of stem cells or progenitor cells in developing salivary gland epithelia (Knox et al., 2010; Lombaert et al., 2013), we investigated whether inhibition of RA signaling coordinately regulated stem cell genes in conjunction with Krt5 and Krt14. Expression of Sox2, cMyc, Klf4, and Kit were assessed by qPCR for control and BMS-treated ER. No significant difference was observed for Sox2, cMyc or Klf4 between control and BMS 493 treated specimens (Fig. 6). Thus, the stem cell markers Sox2, cMyc and Klf4 are not coordinately upregulated with Krt5/Krt14 by inhibition of RA signaling.
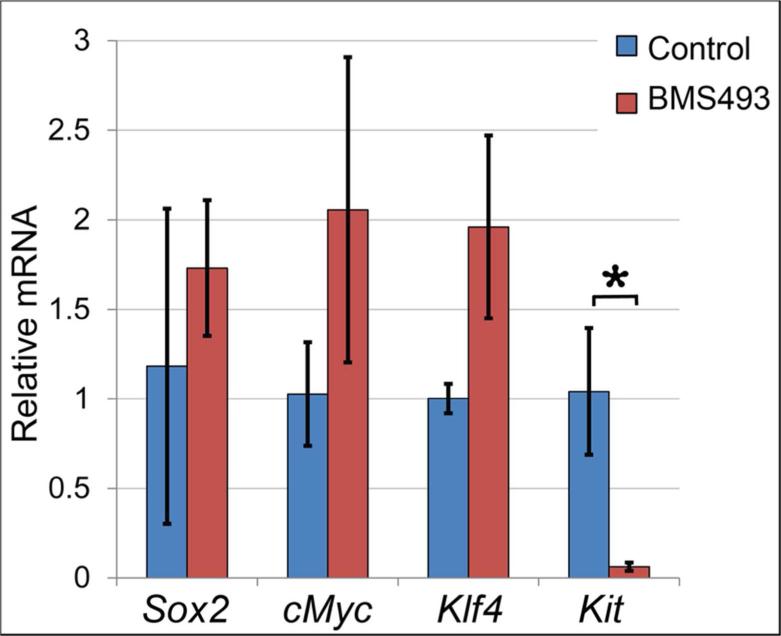
Figure 6. Inhibition of RA signaling downregulates expression of stem cell marker Kit.
Expression of genes associated with stem cell or progenitor cell character was assessed by qPCR for ER cultured on BMS 493 or control medium. Sox2, cMYC, and Klf4 were not significantly different between control or treated specimens. Kit was ≥16-fold downregulated in ER cultured on BMS 493 relative to ER cultured on control medium (p=0.0007). Data represent averages for 3 independent culture experiments with 6-8 ER/condition, each sample was run as 3 technical replicates. Error bars represent standard deviations.
While expression of Sox2, cMyc or Klf4, were not significantly changed by BMS 493 treatment, expression of the stem cell marker Kit was significantly altered by inhibition of RA signaling. Expression of Kit was reduced ≥16-fold in BMS 493-treated ER relative to ER grown on control medium, N = 3 independent culture experiments with 6-8 ER/condition × 3 technical qPCR replicates, p ≤ 0.0007, (Fig. 6). These data demonstrate that treatment with the RA signal inhibitor BMS 493 represses Kit expression in salivary epithelial cells, indicating that RA signaling positively regulates Kit expression. Moreover, because inhibition of RA signaling downregulates Kit expression, while upregulating expression of Krt5 and Krt14, the data show that the stem cell gene Kit is regulated inversely to Krt5 and Krt14 in this context.
DISCUSSION
We have previously demonstrated that Vitamin A metabolism and RA signaling is important for developmental growth and branching morphogenesis of SMG epithelium (Wright et al., 2015). For this study we extend our initial analysis, demonstrating that canonical RA signaling occurs in multiple tissues of the developing SMG, including epithelium, neurons, and non-neuronal mesenchyme (Fig. 1 A-D, Fig. 2A). We show that RA signaling occurs in isolated ER in culture (Fig. 2B), and demonstrate that RA signaling regulates epithelial branching morphogenesis by direct action in epithelial cells (Fig.3 B-D). We identify that RA signaling works in concordance with FGF10 signaling in SMG epithelium, stimulating Etv5 expression and cell proliferation (Fig. 3 F-H). Our data reveal that the two cytokeratins Krt5 and Krt14 are significantly upregulated by inhibition of RA signaling in culture (Fig. 4 E-H,J and Fig. 5). Importantly, we show inhibition of RA signaling in cultured embryonic salivary epithelium has little impact on expression of Sox2, Klf4, or Myc, but causes significant downregulation of the stem cell gene Kit (Fig.6). Thus, our data reveal that Krt5 and Krt14 are regulated independently of genes associated with stem cell or progenitor cell character in this context.
Using immunostain analysis of sectioned embryos we identify that RA signaling occurs within multiple tissues of the developing SMG, being present in epithelium, mesenchyme and neurons. Within the epithelium of E13.5 pseudoglandular SMG RA signaling is active ~10% of cells in both the basal and supra-basal compartments of the epithelium, in endbuds and ducts (Fig. 1 A-B, Fig. 2 A). Such analyses do not reveal whether the pattern of RA signaling is static or dynamic within E13.5 SMG epithelium. If static, then the 10% RA signal positive cells may have a unique identity from their neighboring epithelial cells. If, on the other hand, RA signaling is dynamic in this tissue, as it is in other developmental contexts (Bok et al., 2011; Schilling et al., 2012), it is possible that all E13.5 SMG epithelial cells are transiently positive for RA signaling, being different only in the timing of RA signal activity.
We show here, by inhibiting RA signaling in cultured isolated ER with the pan-RAR inhibitor BMS 493, that RA signaling is required directly within SMG epithelium to promote branching morphogenesis (Fig. 3 B-D). These data raise the possibility that modulation of RA signaling could be utilized in the context of in vitro methods aimed at generating salivary epithelial tissues for transplantation. In addition to its direct action in epithelium, RA signaling may also influence epithelium indirectly via action in nerve or mesenchyme.
It is well established that signaling by the growth factor FGF10 promotes growth and branching of salivary epithelial tissues in vivo and in vitro (De Moerlooze et al., 2000; Ohuchi et al., 2000; Entesarian et al., 2005; Jaskoll et al., 2005; Milunsky et al., 2006; Rohmann et al., 2006; Wells et al., 2013). One downstream target of FGF signaling is Etv5. RA signaling has been shown to stimulate expression of the Etv5 homolog Etv5a during morphogenesis of kidney epithelium in zebrafish embryos (Marra and Wingert, 2016). Here we report that inhibition of RA signal is associated with a reduction in expression of Etv5 in culture salivary ER (Fig. 3 F). Thus, RA signaling, like FGF10 signaling, increases expression of Etv5. Interactions between the RA signaling pathway and FGF signaling pathways occur in many tissues during embryonic morphogenesis (Diez del Corral et al., 2003; Dubrulle and Pourquie, 2004; Moreno and Kintner, 2004; Sirbu and Duester, 2006; Shen et al., 2007; Wilson et al., 2009; Sorrell and Waxman, 2011). In some cases the RA and FGF pathways work together in the same direction, in other cases RA and FGF pathways antagonize each other. One site where RA signaling and FGF10 signaling work together is the developing lung (Desai et al., 2004; Chen et al., 2007; Chen et al., 2010). The data reported here identify a new developmental context in which RA signaling and FGF10 signaling function in the same direction to promote developmental morphogenesis of an organ.
FGF10 promotes proliferation in cultured salivary epithelium (Steinberg et al., 2005). In a variety of epithelial tissues the mitogenic action of FGF10 is enhanced by heparin or heparan sulfate (Igarashi et al., 1998; Izvolsky et al., 2003; Patel et al., 2008), the latter of which is included in the culture medium for these experiments. We show here that the FGF10-dependent cell proliferation of cultured ER is significantly reduced when RA signaling is blocked (Fig. 3 G, H). Thus, for cell proliferation as for Etv5 expression, RA signaling functions in the same direction as FGF10 in cultured salivary epithelium.
The basal epithelial cytokeratin KRT5 has been the subject of much interest in studies of salivary epithelium and has been interpreted as a marker of stem cells or progenitor cells (Knox et al., 2010; Lombaert et al., 2011; Lombaert et al., 2013; Knosp et al., 2015). Here we show Krt5 expression is repressed by RA signaling in cultured ER (Fig. 4 E-H, J, Fig. 5). Inhibition of RA signaling results in elevated KRT5 in basal epithelial cells of all ducts and endbuds (Fig. 4 E-H, K). The observation that KRT5 is upregulated in nearly all basal epithelial cells of the ER when RA signaling is blocked is somewhat surprising given that RA signaling is normally active in only a fraction of the cells (Fig. 2 B, Fig. 4 C). The widespread KRT5 elevation may suggest that the subset of cells that lose their active RA signaling influence their neighbors. Alternatively, if RA signaling is dynamic and occurs in all epithelial cells over time, then inhibition of RA signaling could impact all cells of the epithelium by direct action in each cell.
By qPCR analysis of ER specimens we demonstrated that RA repression of Krt5 occurs at the mRNA level. The increase in Krt5 expression following inhibition of RA signal is consistent with previous studies demonstrating negative regulation of Krt5 by liganded RAR in epithelium from epidermis and other organs (Ohtsuki et al., 1992; Tomic-Canic et al., 1996; Radoja et al., 1997). Here we identify embryonic salivary epithelium as a new biological context for repression of Krt5 gene expression by RA. In whole SMG culture, expression of Krt5 has been shown to be downregulated after 24 hours of treatment with a combination of FGF10 and FGF7 (Knosp et al., 2015). Here we have examined the impact of RA inhibition on gene expression in ER at 48 hours. Although the whole gland study of FGF regulation of Krt5 at 24 hours is not directly comparable to our study of ER gene expression at 48 hours, it is possible that negative regulation of Krt5 expression represents an additional mechanism whereby FGF10 and RA signaling work in the same direction in salivary gland development.
BMS 493 is defined as a pan-RAR inverse agonist because it enhances interactions of RAR with the nuclear receptor corepressor NcoR (Germain et al., 2009). In that context BMS 493 inhibits expression of RA regulated genes. Here we identify a novel action of BMS 493 showing that this RAR inverse agonist can cause activation of a gene. We speculate that Krt5 gene activation by BMS 493 could possibly result from context dependent enhancement of interactions between RAR and coactivators, or disruption of interactions with ligand dependent corepressors such as TNIP, RIF1, Trim24, PRAME, LCoR, or RIP140 (Hu et al., 2004; White et al., 2004; Epping et al., 2005; Heim et al., 2007; Khetchoumian et al., 2007; Li et al., 2007; Gurevich and Aneskievich, 2009).
KRT5 functions as a component of intermediate filaments in combination with its hetero-dimerization partner KRT14 (Coulombe and Fuchs, 1990; Lee and Coulombe, 2009; Lee et al., 2012). Together, KRT5 and KRT14 have been implicated as marking stem cells in embryonic salivary glands and other epithelial tissues (Rock et al., 2009; Lombaert and Hoffman, 2010). We show here that Krt14 gene expression, like Krt5, is upregulated when RA signaling is inhibited (Fig. 5). The two keratins are co-expressed in the basal layer of many epithelial tissues (Moll et al., 1982; Purkis et al., 1990). In developing salivary epithelium they are co-expressed together in some cells, and expressed independently of each other in other cells (Lombaert et al., 2013). In some contexts expression of the two keratin genes is co-dependent. Reduction of Krt14 by RNA interference in cultured epithelial cells results in downregulation of Krt5 (Alam et al., 2011). Our data reported here indicate that RA signaling is one mechanism whereby Krt5, Krt14 are co-regulated.
Importantly, we show here that RA signaling does not coordinately regulate expression of stem cell and progenitor cell markers in conjunction with Krt5 and Krt14. Treatment with the RA signaling inhibitor BMS 493, which upregulates Krt5 and Krt14, causes no significant change in expression of Sox2, cMyc or Klf4, and results in significant downregulation of Kit (Fig. 6). These data demonstrate that Krt5 and Krt14 are regulated independently of progenitor cell character by RA signaling. From qPCR analysis of cultured ER specimens it is not possible to discern whether upregulation of Krt5 and Krt14 and downregulation of Kit occurs within the same cells or different cells, nor does it distinguish if one change is a consequence of another. Further experiments will be required to resolve these questions. Nonetheless the data presented here demonstrate that expression of Krt5 or Krt14 on their own are not reliable markers of salivary epithelial progenitor identity.
In summary, RA signaling regulates cell proliferation and branching morphogenesis of salivary epithelium. Thus, RA, or inhibitors of RA signaling, may have potential utility as pharmacologic agents for directing differentiation of salivary epithelial stem cells.
Experimental Procedures
Mice
FVB/NJ mice were obtained from Jackson laboratories. FVB/NJ embryos were used for ER culture experiments and for qPCR quantitation of gene expression (Fig. 3, Fig. 5, Fig. 6). RARE-lacZ reporter mice (Rossant et al., 1991), were obtained from Jackson laboratories (official name, Tg(RARE-Hspa1b/lacZ)12Jrt). RARE lacZ embryos were used for immunostain analysis of RA signaling (Fig. 1, Fig. 2, Fig. 4). The day of the vaginal plug was considered E0.5. All experiments involving mice were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee at the University of Louisville.
Culture of ER
Medium used for dissection and washes was DMEM/F12 containing HEPES (HyClone SH30126.01). Medium used for culture was DMEM/F12 without HEPES (HyClone SH30271.01) plus 50 μg/ml transferrin, 150 μg/ml ascorbic acid, Penicillin/Streptomycin, 500 ng/ml recombinant mouse FGF10 (RD Systems 6224-FG-025), and 500 ng/ml Heparan sulfate proteoglycan (Sigma Aldrich H4777).
ER culture methods were based on previously described protocol (Steinberg et al., 2005). E13.5 SMG with 3-6 endbuds were dissected under a stereomicroscope in dissection medium. Once isolated, SMG were treated with dispase I (Sigma-Aldrich D4818), 1.6 U/ml in phosphate buffered saline (PBS), to loosen epithelium from mesenchyme. Dispase I incubation was performed in a well of a glass staining plate for 17 min in a humidified chamber 37°C, 5% CO2. After dispase I treatment, dispase was inactivated by washing specimens 3 times in 7.5% BSA in dissection medium. ER were then separated from surrounding mesenchyme using fine tip forceps and were washed in dissection medium to remove BSA. Specimens were cultured inside a 15 μl drop of growth factor-reduced Matrigel (Corning catalog number 356230). Prior to each experiment an aliquot of Matrigel was thawed at 4°C overnight, then diluted 1:1 in dissection medium (4 mg/ml final). At the time of plating a 15μl drop of Matrigel was placed on top of a 13 mm diameter Nuclepore Track-Etch membrane filter, pore size 0.1 μm (Whatman, 110405). Filters were placed over 100 μl of culture medium supported by a silicone culturewell gasket (Grace Biolabs CW-4R-1.0) in a plastic petri dish, with a small petri lid filled with H20 to ensure humidity within the culture dish. Specimens were cultured at 3-8 ER/filter. ER were cultured 48 hours in a humidified incubator at 37 °C with 5%CO2/95% air. Cultures were fed daily by adding fresh culture medium to an empty well of the culturewell gasket and moving filter.
For inhibition of canonical RA signaling the pan-RAR inverse agonist BMS 493 (Tocris, #3509) was used. For each experiment, a fresh BMS 493 stock solution 5 mM in DMSO was prepared. The BMS 493 stock solution was then diluted in culture medium for final concentration of 5 μM. For control samples, an equivalent volume of DMSO was added to culture medium.
Specimens were imaged with transmitted light at beginning and end of the culture period on using a Leica M165 stereomicroscope with Leica imaging software. Branching morphogenesis and growth of ER specimens was assessed by counting of endbuds and by quantification of visible area using ImageJ software. The significance of difference in number of endbuds was calculated using Student's T-test.
Whole mount immunostain
Whole mount cultured ER specimens were fixed on filters with 4% formaldehyde at room temperature for 1 hr, then made permeable by incubation in 0.1% Triton in PBS. After permeablization, specimens were blocked in 0.1M Tris pH7.5, 0.15M NaCl with blocking reagent (Perkin Elmer FP1020). Primary antibody hybridization was performed in blocking solution overnight 4°C. Following primary antibody incubation, specimens were washed 5 × 1 hr in PBS at room temperature. Hybridization with fluorescent secondary antibodies was performed in blocking solution overnight at 4°C. After secondary antibody hybridization specimens were washed 3 × 20 min in PBS. To aid in finding tissues during confocal microscopy specimens were stained with DAPI (10nM, 10 min) and washed in PBS. After staining, ER specimens were post-fixed in 4% formaldehyde for 45 min at room temperature. All steps were performed with gentle rocking. For confocal imaging, stained ER specimens on filters were placed in a depression slide in PBS, covered with a coverslip affixed with vacuum grease, and imaged with an Olympus MPE FV1000 confocal microscope.
Frozen section immunostain
Embryonic heads were collected at E13.5 and E14.5 stages of development and fixed overnight in 4% formaldehyde at 4°C. Following fixation, samples were equilabrated in 30% sucrose overnight at 4°C. Samples were embedded in OCT compound and stored at −80°C. Tissues were cut at 12 μm thickness, vacuum dried 1 hr, and stored at −80°C. For staining, sections were enclosed with a hydrophobic barrier using a PAP pen and washed for 5 min in 0.1% Tween 20 in PBS (PBT). Blocking was performed in blocking solution (as described for whole mount immunostain) for 1 h at room temperature. Primary antibodies were hybridized overnight at 4 °C in a humidified chamber in blocking solution. Following incubation with primary antibodies, slides were washed 3 ×10 min in PBT. Secondary antibody hybridization was performed for 1 hr at room temperature in blocking solution. Unbound secondary antibody was removed by washing 3 ×10 min in PBT. Nuclei were stained with DAPI (10nM, 10 min), followed by a final was in PBT. Stained slides were mounted with Prolong Gold mounting medium (ThermoFisher P36930). Stained frozen sections were imaged on a Zeiss AxioImager.A1.
Quantification of mRNA by qPCR
For gene expression analysis ER were cultured 6-8 specimens per filter. Following culture, ER specimens were recovered from Matrigel by incubating with Corning Cell Recovery solution (Corning 354253) for 1 hr on wet ice at 4°C with gentle shaking. ER were then washed twice with ice cold PBS by spinning at 200G for 1 min. Total RNA was extracted with RNeasy Micro Kit (Qiagen 74004). DNA was removed by on-column DNase I digestion (Qiagen 79254). RNA was reverse transcribed to cDNA using random hexamers and the SuperScript III First-Strand Synthesis System (Invitrogen 18080-051). SYBRgreen qPCR was performed using SYBR Select Master Mix (Applied Biosystems 4472908). For each qPCR reaction 100 ng of cDNA was used as template. Data represent the average of 3 independent culture experiments with 6-8 ER/condition. Each sample was run as 3 technical replicates, except Krt5, which was run as 6 technical replicates. To identify a suitable control gene for normalization of mRNA levels in this experimental context expression of Gapdh and Actb was assessed in control and BMS-treated samples to determine if either gene varied relative to the other. No difference was observed between Gapdh and Actb expression patterns, demonstrating that either gene could be used as an appropriate normalization control for ER cultured on control or BMS 493 medium. Gapdh was therefore used for normalization of gene expression. All primers were validated for efficiency between 90% - 110%. Data was evaluated by the 2-ΔΔCT method (Livak and Schmittgen, 2001). Significance was evaluated by two tailed Student's T-test assuming unequal variance.
Antibodies
Primary antibodies used were:
anti-β-glactosidase (Abcam ab9361) 1:500,
anti-E-cadherin (BD Biosciences #610182) 1:50,
anti-Neuronal Class III β-Tubulin (Covance, PRB0435-P) 1:1000,
anti-Cytokeratin 5, (Abcam ab24647)1:1000.
anti-Cytokeratin-8 (DSHB TROMA-I)1:50.
Fluorescently conjugated secondary antibodies, each used at 1:300 were: Alexafluor 488, AlexaFluor 546, AlexaFluor 660 (Invitrogen), or Dynalight 488 (Abcam).
Primers
Many primers were identified using Primer Bank Database (Wang et al., 2012).
Table 1.
List of primers used for qPCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Actb | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| Etv5 | TCAGTCTGATAACTTGGTGCTTC | GGCTTCCTATCGTAGGCACAA |
| Gapdh | ACAGTCCATGCCATCACTGCC | GCCTGCTTCACCACCTTCTTG |
| Kit | TCATCGAGTGTGATGGGAAA | GGTGACTTGTTTCAGGCACA |
| Klf4 | GTGCCCCGACTAACCGTTG | GTCGTTGAACTCCTCGGTCT |
| Krt5 | TCCAGTGTGTCCTTCCGAAGT | TGCCTCCGCCAGAACTGTA |
| Krt8 | TCCATCAGGGTGACTCAGAAA | CCAGCTTCAAGGGGCTCAA |
| Krt14 | AGCGGCAAGAGTGAGATTTCT | CCTCCAGGTTATTCTCCAGGG |
| Krt19 | GGGGGTTCAGTACGCATTGG | GAGGACGAGGTCACGAAGC |
| Mki67 | ATCATTGACCGCTCCTTTAGGT | GCTCGCCTTGATGGTTCCT |
| Myc | ATGCCCCTCAACGTGAACTTC | CGCAACATAGGATGGAGAGCA |
| Sox2 | GCGGAGTGGAAACTTTTGTCC | CGGGAAGCGTGTACTTATCCTT |
| Top2a | CAACTGGAACATATACTGCTCCG | GGGTCCCTTTGTTTGTTATCAGC |
Acknowledgments
Funding:
Research in the Sandell laboratory is supported by R15 DE025960 and a Competitive Enhancement Grant from University of Louisville EVPRI. MAM was supported by F31 DE022679. The Microscopy Suite at the Cardiovascular Innovation Institute in Louisville, KY is supported by GM103507.
REFERENCES
- Alam H, Sehgal L, Kundu ST, Dalal SN, Vaidya MM. Novel function of keratins 5 and 14 in proliferation and differentiation of stratified epithelial cells. Mol Biol Cell. 2011;22:4068–4078. doi: 10.1091/mbc.E10-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aure MH, Konieczny SF, Ovitt CE. Salivary gland homeostasis is maintained through acinar cell self-duplication. Dev Cell. 2015;33:231–237. doi: 10.1016/j.devcel.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J, Raft S, Kong KA, Koo SK, Drager UC, Wu DK. Transient retinoic acid signaling confers anterior-posterior polarity to the inner ear. Proc Natl Acad Sci U S A. 2011;108:161–166. doi: 10.1073/pnas.1010547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broudy VC. Stem cell factor and hematopoiesis. Blood. 1997;90:1345–1364. [PubMed] [Google Scholar]
- Chen F, Cao Y, Qian J, Shao F, Niederreither K, Cardoso WV. A retinoic acid-dependent network in the foregut controls formation of the mouse lung primordium. J Clin Invest. 2010;120:2040–2048. doi: 10.1172/JCI40253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Desai TJ, Qian J, Niederreither K, Lu J, Cardoso WV. Inhibition of Tgf beta signaling by endogenous retinoic acid is essential for primary lung bud induction. Development. 2007;134:2969–2979. doi: 10.1242/dev.006221. [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M, Knutson D. Vitamin a in reproduction and development. Nutrients. 2011;3:385–428. doi: 10.3390/nu3040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colopy SA, Bjorling DE, Mulligan WA, Bushman W. A Population of Progenitor Cells in the Basal and Intermediate Layers of the Murine Bladder Urothelium Contributes to Urothelial Development and Regeneration. Developmental dynamics : an official publication of the American Association of Anatomists. 2014;243:988–998. doi: 10.1002/dvdy.24143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe PA, Fuchs E. Elucidating the early stages of keratin filament assembly. J Cell Biol. 1990;111:153–169. doi: 10.1083/jcb.111.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Desai TJ, Malpel S, Flentke GR, Smith SM, Cardoso WV. Retinoic acid selectively regulates Fgf10 expression and maintains cell identity in the prospective lung field of the developing foregut. Dev Biol. 2004;273:402–415. doi: 10.1016/j.ydbio.2004.04.039. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, Pourquie O. Coupling segmentation to axis formation. Development. 2004;131:5783–5793. doi: 10.1242/dev.01519. [DOI] [PubMed] [Google Scholar]
- Entesarian M, Matsson H, Klar J, Bergendal B, Olson L, Arakaki R, Hayashi Y, Ohuchi H, Falahat B, Bolstad AI, Jonsson R, Wahren-Herlenius M, Dahl N. Mutations in the gene encoding fibroblast growth factor 10 are associated with aplasia of lacrimal and salivary glands. Nat Genet. 2005;37:125–128. doi: 10.1038/ng1507. [DOI] [PubMed] [Google Scholar]
- Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Firnberg N, Neubuser A. FGF signaling regulates expression of Tbx2, Erm, Pea3, and Pax3 in the early nasal region. Dev Biol. 2002;247:237–250. doi: 10.1006/dbio.2002.0696. [DOI] [PubMed] [Google Scholar]
- Germain P, Gaudon C, Pogenberg V, Sanglier S, Van Dorsselaer A, Royer CA, Lazar MA, Bourguet W, Gronemeyer H. Differential action on coregulator interaction defines inverse retinoid agonists and neutral antagonists. Chem Biol. 2009;16:479–489. doi: 10.1016/j.chembiol.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Goswami PC, Roti Roti JL, Hunt CR. The cell cycle-coupled expression of topoisomerase IIalpha during S phase is regulated by mRNA stability and is disrupted by heat shock or ionizing radiation. Mol Cell Biol. 1996;16:1500–1508. doi: 10.1128/mcb.16.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I, Aneskievich BJ. Liganded RARα and RARγ interact with but are repressed by TNIP1. Biochemical and Biophysical Research Communications. 2009;389:409–414. doi: 10.1016/j.bbrc.2009.08.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim KC, White KA, Deng D, Tomlinson CR, Moore JH, Freemantle SJ, Spinella MJ. Selective repression of retinoic acid target genes by RIP140 during induced tumor cell differentiation of pluripotent human embryonal carcinoma cells. Mol Cancer. 2007;6:57. doi: 10.1186/1476-4598-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Chen Y, Farooqui M, Thomas MC, Chiang CM, Wei LN. Suppressive effect of receptor-interacting protein 140 on coregulator binding to retinoic acid receptor complexes, histone-modifying enzyme activity, and gene activation. J Biol Chem. 2004;279:319–325. doi: 10.1074/jbc.M307621200. [DOI] [PubMed] [Google Scholar]
- Hudson DL, Guy AT, Fry P, O'Hare MJ, Watt FM, Masters JRW. Epithelial Cell Differentiation Pathways in the Human Prostate: Identification of Intermediate Phenotypes by Keratin Expression. J Histochem Cytochem. 2001;49:271–278. doi: 10.1177/002215540104900214. [DOI] [PubMed] [Google Scholar]
- Igarashi M, Finch PW, Aaronson SA. Characterization of recombinant human fibroblast growth factor (FGF)-10 reveals functional similarities with keratinocyte growth factor (FGF-7). J Biol Chem. 1998;273:13230–13235. doi: 10.1074/jbc.273.21.13230. [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izvolsky KI, Shoykhet D, Yang Y, Yu Q, Nugent MA, Cardoso WV. Heparan sulfate- FGF10 interactions during lung morphogenesis. Dev Biol. 2003;258:185–200. doi: 10.1016/s0012-1606(03)00114-3. [DOI] [PubMed] [Google Scholar]
- Jaskoll T, Abichaker G, Witcher D, Sala F, Bellusci S, Hajihosseini M, Melnick M. FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC developmental biology. 2005;5:11. doi: 10.1186/1471-213X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho SH, Radoja N, Im MJ, Tomic-Canic M. Negative response elements in keratin genes mediate transcriptional repression and the cross-talk among nuclear receptors. J Biol Chem. 2001;276:45914–45920. doi: 10.1074/jbc.M103144200. [DOI] [PubMed] [Google Scholar]
- Khetchoumian K, Teletin M, Tisserand J, Mark M, Herquel B, Ignat M, Zucman-Rossi J, Cammas F, Lerouge T, Thibault C, Metzger D, Chambon P, Losson R. Loss of Trim24 (Tif1[alpha]) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39:1500–1506. doi: 10.1038/ng.2007.15. [DOI] [PubMed] [Google Scholar]
- Knosp WM, Knox SM, Hoffman MP. Salivary gland organogenesis. Wiley Interdisciplinary Reviews: Developmental Biology. 2012;1:69–82. doi: 10.1002/wdev.4. [DOI] [PubMed] [Google Scholar]
- Knosp WM, Knox SM, Lombaert IM, Haddox CL, Patel VN, Hoffman MP. Submandibular parasympathetic gangliogenesis requires sprouty-dependent Wnt signals from epithelial progenitors. Dev Cell. 2015;32:667–677. doi: 10.1016/j.devcel.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox SM, Lombaert IMA, Reed X, Vitale-Cross L, Gutkind JS, Hoffman MP. Parasympathetic Innervation Maintains Epithelial Progenitor Cells During Salivary Organogenesis. Science. 2010;329:1645–1647. doi: 10.1126/science.1192046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil K. Organ specificity in mesenchymal induction demonstrated in the embryonic development of the mammary gland of the mouse. Developmental Biology. 1969;20:46–71. doi: 10.1016/0012-1606(69)90004-9. [DOI] [PubMed] [Google Scholar]
- Lee CH, Coulombe PA. Self-organization of keratin intermediate filaments into cross- linked networks. J Cell Biol. 2009;186:409–421. doi: 10.1083/jcb.200810196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Kim MS, Chung BM, Leahy DJ, Coulombe PA. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat Struct Mol Biol. 2012;19:707–715. doi: 10.1038/nsmb.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Haque ZK, Chen A, Mendelsohn M. RIF-1, a novel nuclear receptor corepressor that associates with the nuclear matrix. J Cell Biochem. 2007;102:1021–1035. doi: 10.1002/jcb.21340. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jiang H, Crawford HC, Hogan BL. Role for ETS domain transcription factors Pea3/Erm in mouse lung development. Dev Biol. 2003;261:10–24. doi: 10.1016/s0012-1606(03)00359-2. [DOI] [PubMed] [Google Scholar]
- Liu Z, Hu Q, Rosenfeld MG. Complexity of the RAR-mediated transcriptional regulatory programs. Subcell Biochem. 2014;70:203–225. doi: 10.1007/978-94-017-9050-5_10. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lombaert IM, Abrams SR, Li L, Eswarakumar VP, Sethi AJ, Witt RL, Hoffman MP. Combined KIT and FGFR2b signaling regulates epithelial progenitor expansion during organogenesis. Stem Cell Reports. 2013;1:604–619. doi: 10.1016/j.stemcr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert IMA, Brunsting JF, Wierenga PK, Faber H, Stokman MA, Kok T, Visser WH, Kampinga HH, de Haan G, Coppes RP. Rescue of Salivary Gland Function after Stem Cell Transplantation in Irradiated Glands. PLoS ONE. 2008;3:e2063. doi: 10.1371/journal.pone.0002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert IMA, Hoffman MP. Epithelial Stem/Progenitor Cells in the Embryonic Mouse Submandibular Gland. Frontiers of Oral Biology. 2010;14:90–106. doi: 10.1159/000313709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert IMA, Knox SM, Hoffman MP. Salivary gland progenitor cell biology provides a rationale for therapeutic salivary gland regeneration. Oral Dis. 2011;17:445–449. doi: 10.1111/j.1601-0825.2010.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra AN, Wingert RA. Epithelial cell fate in the nephron tubule is mediated by the ETS transcription factors etv5a and etv4 during zebrafish kidney development. Dev Biol. 2016;411:231–245. doi: 10.1016/j.ydbio.2016.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D'Agati V, Licht JD, Martin GR, Costantini F. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:e1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunsky JM, Zhao G, Maher TA, Colby R, Everman DB. LADD syndrome is caused by FGF10 mutations. Clin Genet. 2006;69:349–354. doi: 10.1111/j.1399-0004.2006.00597.x. [DOI] [PubMed] [Google Scholar]
- Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Moreno TA, Kintner C. Regulation of segmental patterning by retinoic acid signaling during Xenopus somitogenesis. Dev Cell. 2004;6:205–218. doi: 10.1016/s1534-5807(04)00026-7. [DOI] [PubMed] [Google Scholar]
- Nanduri LS, Lombaert IM, van der Zwaag M, Faber H, Brunsting JF, van Os RP, Coppes RP. Salisphere derived c-Kit+ cell transplantation restores tissue homeostasis in irradiated salivary gland. Radiother Oncol. 2013;108:458–463. doi: 10.1016/j.radonc.2013.05.020. [DOI] [PubMed] [Google Scholar]
- Nedvetsky PI, Emmerson E, Finley JK, Ettinger A, Cruz-Pacheco N, Prochazka J, Haddox CL, Northrup E, Hodges C, Mostov KE, Hoffman MP, Knox SM. Parasympathetic innervation regulates tubulogenesis in the developing salivary gland. Dev Cell. 2014;30:449–462. doi: 10.1016/j.devcel.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Matsuzaki Y, Nishikawa S, Hayashi S, Kunisada T, Sudo T, Kina T, Nakauchi H, Nishikawa S. Expression and function of c-kit in hemopoietic progenitor cells. J Exp Med. 1991;174:63–71. doi: 10.1084/jem.174.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki M, Tomic-Canic M, Freedberg IM, Blumenberg M. Regulation of Epidermal Keratin Expression by Retinoic Acid and Thyroid Hormone. The Journal of Dermatology. 1992;19:774–780. doi: 10.1111/j.1346-8138.1992.tb03779.x. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N. FGF10 Acts as a Major Ligand for FGF Receptor 2 IIIb in Mouse Multi-Organ Development. Biochemical and Biophysical Research Communications. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- Patel VN, Likar KM, Zisman-Rozen S, Cowherd SN, Lassiter KS, Sher I, Yates EA, Turnbull JE, Ron D, Hoffman MP. Specific heparan sulfate structures modulate FGF10- mediated submandibular gland epithelial morphogenesis and differentiation. J Biol Chem. 2008;283:9308–9317. doi: 10.1074/jbc.M709995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkis PE, Steel JB, Mackenzie IC, Nathrath WB, Leigh IM, Lane EB. Antibody markers of basal cells in complex epithelia. J Cell Sci. 1990;97(Pt 1):39–50. doi: 10.1242/jcs.97.1.39. [DOI] [PubMed] [Google Scholar]
- Radoja N, Diaz DV, Minars TJ, Freedberg IM, Blumenberg M, Tomic-Canic M. Specific Organization of the Negative Response Elements for Retinoic Acid and Thyroid Hormone Receptors in Keratin Gene Family. J Invest Dermatol. 1997;109:566–572. doi: 10.1111/1523-1747.ep12337483. [DOI] [PubMed] [Google Scholar]
- Rebustini IT, Patel VN, Stewart JS, Layvey A, Georges-Labouesse E, Miner JH, Hoffman MP. Laminin alpha5 is necessary for submandibular gland epithelial morphogenesis and influences FGFR expression through beta1 integrin signaling. Dev Biol. 2007;308:15–29. doi: 10.1016/j.ydbio.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmann E, Brunner HG, Kayserili H, Uyguner O, Nurnberg G, Lew ED, Dobbie A, Eswarakumar VP, Uzumcu A, Ulubil-Emeroglu M, Leroy JG, Li Y, Becker C, Lehnerdt K, Cremers CWRJ, Yuksel-Apak M, Nurnberg P, Kubisch C, Schlessinger J, van Bokhoven H, Wollnik B. Mutations in different components of FGF signaling in LADD syndrome. Nat Genet. 2006;38:414–417. doi: 10.1038/ng1757. [DOI] [PubMed] [Google Scholar]
- Rossant J, Zirngibl R, Cado D, Shago M, Goguere V. Expression of retinoic acid response element-hsp lacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Nie Q, Lander AD. Dynamics and precision in retinoic acid morphogen gradients. Current Opinion in Genetics & Development. 2012;22:562–569. doi: 10.1016/j.gde.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CN, Marguerie A, Chien CY, Dickson C, Slack JM, Tosh D. All-trans retinoic acid suppresses exocrine differentiation and branching morphogenesis in the embryonic pancreas. Differentiation. 2007;75 doi: 10.1111/j.1432-0436.2006.00116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu IO, Duester G. Retinoic-acid signalling in node ectoderm and posterior neural plate directs left-right patterning of somitic mesoderm. Nat Cell Biol. 2006;8:271–277. doi: 10.1038/ncb1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell MR, Waxman JS. Restraint of Fgf8 signaling by retinoic acid signaling is required for proper heart and forelimb formation. Dev Biol. 2011;358:44–55. doi: 10.1016/j.ydbio.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg Z, Myers C, Heim VM, Lathrop CA, Rebustini IT, Stewart JS, Larsen M, Hoffman MP. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development. 2005;132:1223–1234. doi: 10.1242/dev.01690. [DOI] [PubMed] [Google Scholar]
- Tomic-Canic M, Day D, Samuels HH, Freedberg IM, Blumenberg M. Novel regulation of keratin gene expression by thyroid hormone and retinoid receptors. J Biol Chem. 1996;271:1416–1423. doi: 10.1074/jbc.271.3.1416. [DOI] [PubMed] [Google Scholar]
- Tomic M, Jiang CK, Epstein HS, Freedberg IM, Samuels HH, Blumenberg M. Nuclear receptors for retinoic acid and thyroid hormone regulate transcription of keratin genes. Cell Regul. 1990;1:965–973. doi: 10.1091/mbc.1.12.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS. Salivary gland development. Seminars in Cell & Developmental Biology. 2007;18:237–244. doi: 10.1016/j.semcdb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Research. 2012;40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells KL, Gaete M, Matalova E, Deutsch D, Rice D, Tucker AS. Dynamic relationship of the epithelium and mesenchyme during salivary gland initiation: the role of Fgf10. Biology Open. 2013 doi: 10.1242/bio.20135306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JH, Fernandes I, Mader S, Yang XJ. Corepressor recruitment by agonist-bound nuclear receptors. Vitam Horm. 2004;68:123–143. doi: 10.1016/S0083-6729(04)68004-6. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V, Olivera-Martinez I, Storey KG. Stem cells, signals and vertebrate body axis extension. Development. 2009;136:1591–1604. doi: 10.1242/dev.021246. [DOI] [PubMed] [Google Scholar]
- Wright DM, Buenger DE, Abashev TM, Lindeman RP, Ding J, Sandell LL. Retinoic acid regulates embryonic development of mammalian submandibular salivary glands. Dev Biol. 2015;407:57–67. doi: 10.1016/j.ydbio.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZA, Guan SP, Liew A-A, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, Crum CP, Xian W, McKeon F. p63+Krt5+ distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]