Abstract
A simplified coupling of surface plasmon resonance (SPR) immuno-biosensing with ambient ionization mass spectrometry (MS) was developed. It combines two orthogonal analysis techniques: the biosensing capability of SPR and the chemical identification power of high resolution MS. As a proof-of-principle, deoxynivalenol (DON), an important mycotoxin, was captured using an SPR gold chip containing an antifouling layer and monoclonal antibodies against the toxin and, after washing, the chip could be taken out and analyzed by direct spray MS of the biosensor chip to confirm the identity of DON. Furthermore, cross-reacting conjugates of DON present in a naturally contaminated beer could be successfully identified, thus showing the potential of rapid identification of (un)expected cross-reacting molecules.
Surface plasmon resonance (SPR) is a powerful biosensing tool for the real-time detection of a wide range of molecules. Apart from that, SPR provides important information about the binding kinetics of a biorecognition event. However, SPR does not provide the chemical identity of the binding analyte.1 Monoclonal antibodies are used in SPR for the selective immuno-capturing and detection of the analyte. Although antibodies are highly specific, conjugates of the analyte can sometimes cross-react with the antibodies. Such cross-reacting conjugates cannot be distinguished from the targeted analyte by SPR and may lead to overestimation of the analyte concentration. Therefore, coupling of SPR with mass spectrometry (MS) would not only confirm the identity of the SPR-detected target analyte(s) but also might help to find any (un)expected cross-reacting analytes.2 As the bioreagents used for SPR are not MS-compatible, initial elution-based SPR-MS methods involved online collection of the desorbed analyte on a precolumn, followed by sample cleanup and off-line transfer of the precolumn with the sample to an electrospray ionization (ESI) tandem MS system.3 This leads to sample losses, which complicates the MS identification due to the minute amounts present. More sophisticated elution-based SPR-MS couplings allow real online MS analysis of analytes; however, the interfacing can be rather complex and expensive.4−6 Alternatively, coupling of SPR and MS based on matrix-assisted laser desorption ionization (MALDI) allows direct analysis of the biosensor chip7,8 but requires the addition of an excess of MALDI matrix. The abundant matrix (cluster) ions can easily obscure the ions of small molecules (<700 Da) present at subnanogram levels; hence, most of the SPR-MALDI MS studies focus on the identification of peptides and proteins.
Ambient ionization methods for mass spectrometry, such as direct analysis in real time (DART)9 and desorption electrospray ionization (DESI),10,11 have gained significant attention in the past decade as analyses can be performed at room temperature, under atmospheric conditions, often require minimal sample preparation, and are suitable for small molecules.12 Ionization methods based on direct spray,13 where the sample is loaded onto a solid substrate (paper, metal, wood, glass, etc.)14−16 followed by application of a high voltage (HV) to generate ions, have become popular due to their simplicity. These methods rely on extraction/desorption of the analytes from the surface of the substrate using an organic solvent and, consequently, the overall selectivity is entirely dependent on the resolution of the mass analyzer. Recently, in an attempt to get rid of interfering components, coated blade spray was demonstrated in which solid-phase micro extraction (SPME) was coupled with a desorption electrospray ionization (DESI) source.17 As the analyte of interest was captured by the SPME coating, the method offered both sample enrichment and removal of other components using a washing step prior to MS analysis.
The aim of the present paper was the development of a simplified SPR-MS coupling. An SPR biochip coated with antibodies was used to selectively capture the analyte in an SPR apparatus. The SPR chip was taken out, and following the application of a solvent and a high voltage, the analytes were desorbed and directly sprayed into a high-resolution MS (HRMS). In contrast to other direct MS approaches using affinity surfaces, such as, for example, surface-enhanced laser desorption/ionization (SELDI)18 and self-assembled monolayers laser desorption/ionization (SAMDI),19,20 the proposed concept combines two orthogonal analysis techniques. Moreover, the analysis of low molecular weight compounds was not obstructed by MALDI matrix ions.
Experimental Section
Carboxymethylated dextran (CMD) coated flat gold chips were purchased from Xantec (Düsseldorf, Germany). Nanostructured gold chips were purchased from Plasmore Srl. (Ispra, Italy) and were further modified with CMD by Xantec. SPR measurements on flat gold chips were performed using a Biacore 3000, and iSPR measurements on nanostructured gold chips were obtained using a prototype portable imaging nanoplasmonics instrument (Plasmore Srl., Italy).21,22 Ethanolamine hydrochloride, 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide hydrochloride (EDC), and N-hydroxysuccinimide (NHS) were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands). Methanol was purchased from VWR (Amsterdam, The Netherlands). HBS-EP buffer (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% v/v surfactant P20) was purchased from GE Healthcare (Diegem, Belgium). Milli-Q water (18.3 MΩ·cm resistivity) was obtained using a Merck (Amsterdam, The Netherlands) water purification system.
The immobilization of antibodies on the CMD-modified gold chips for SPR was performed manually on the bench: the entire chip was activated using 50 μL of a 1:1 mixture of 0.1 M NHS and 0.4 M EDC for 30 min, followed by washing with water and drying with nitrogen. Next, 50 μL of antibody solution (50 μg/mL in 10 mM acetate buffer, pH 4.5) was added to the activated chip. The chip was kept in an airtight container for 2 h to avoid evaporation and drying out of the solution. Following the incubation, the chips were washed with water and dried with nitrogen. Unreacted groups were blocked with ethanolamine for 30 min. Then, the chips were rinsed with Milli-Q water and dried with nitrogen. The chips were stored at 4–8 °C until use. The CMD chips (with or without antibodies) were mounted on the SPR flow cell and flushed with HBS-EP buffer for 5 min to allow swelling of the hydrogel. This was followed by injection of 50 μL of sample (blank or spiked buffer or beer). The chip was then flushed with HBS-EP (5 min) followed by water (5 min). Finally, the chip was unmounted from the SPR flow cell, dried with nitrogen, and mounted in the Biochip Spray MS setup.
The chip was clamped by an alligator clip, which was part of a modified DESI ion source (Prosolia, USA) equipped with a rotational and x–y–z positioner, and was directly connected to the HV supply of the ion source (Figure 1A). The square chip was positioned at an angle (2–4°) with one of the corners pointing downward toward the MS inlet and at a distance of 4–6 mm (Figure 1B). Five μL of spray solvent was added using an Eppendorf pipet (0.5–10 μL). After a waiting time of 30 s, a voltage of 5 kV was applied. All MS analyses were performed with a model Q-Exactive Focus quadrupole orbitrap high resolution MS (Thermo Fisher Scientific) to obtain full-scan positive ion measurements with a scan range of m/z 150.0–1000.0, a mass resolution of 70 000, a maximum injection time of 100 ms, and a scan rate of 1 Hz. The capillary temperature was 250 °C, and the S-lens RF level was 50. CAUTION: High voltages are involved! Prior to adding the spray solvent, always check that the voltage is actually 0 V and only switch it on when nobody is near the tip. The setup should not be touched during the experiment. Thermo Scientific Xcalibur software was used for data acquisition and processing. The intensity of the ions with m/z values within ±5 ppm of the theoretical m/z is shown in the extracted ion chronogram (EIC).
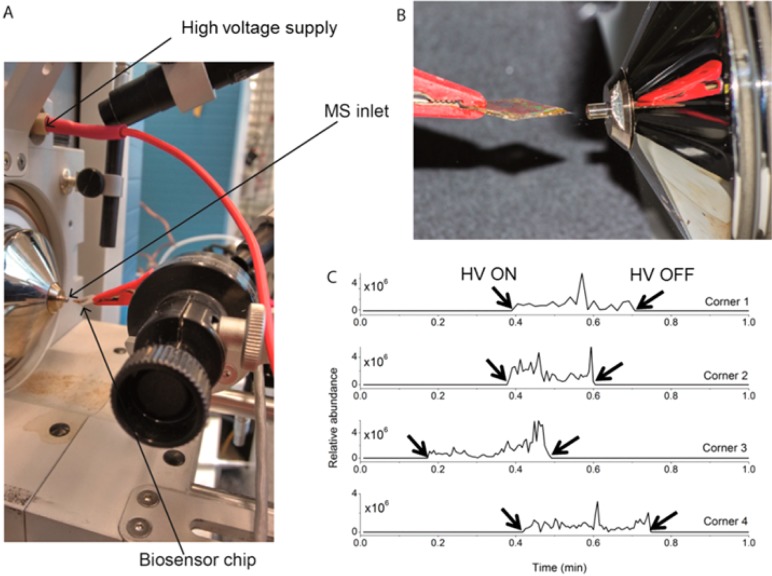
Figure 1.
(A) Setup used for Biochip Spray MS using a gold biosensor chip held in front of the MS inlet using an alligator clip and (B) spray obtained after adding 10 μL of methanol and applying a voltage of 5 kV. (C) Extracted ion chronogram for m/z 297.1333 ([DON + H]+) recorded in positive ion mode, as obtained from four different corners of a single 1 cm2 square carboxymethylated dextran (CMD) modified gold chip. Five μL of a 1 μg/mL DON solution in methanol was pipetted on each corner, allowed to dry, and afterward sprayed using methanol. The time points when the high voltage (HV) was turned on and off are indicated by arrows.
Results and Discussion
Although any analyte–antibody pair could have been selected to show the concept, this study uses a low molecular weight mycotoxin, deoxynivalenol (DON), and a corresponding monoclonal antibody (anti-DON), due to its societal relevance. Deoxynivalenol is a secondary metabolite of fungi and is commonly found in several foods such as nuts, cereals, coffee, oil seeds, and fruits, as well as in beverages and animal feed.23−25 Due to the thermal stability and relatively good water solubility, DON can be carried over from a contaminated starting ingredient like barley to malt and finally into beer.26,27 First, to demonstrate the feasibility of direct spray from a gold surface for analysis of DON, 5 μL of 1 μg/mL (5000 pg) DON was placed in one corner of a CMD-modified gold biosensor chip (without any biorecognition) after which the solvent was allowed to evaporate. Then, 10 μL of methanol was placed on the same corner of the chip followed by application of a high voltage to generate the spray. The voltage and distance between the chip and the MS inlet were optimized in order to obtain a stable spray without electric discharge. Optimal settings were 5 kV at a distance of 4–6 mm between the chip and the MS. Although shorter distances than 4 mm required a lower voltage for spraying, electric discharge was occasionally seen whereas larger distances (>6 mm) in combination with higher voltages led to either an unstable spray or no spray at all. Furthermore, the angle was adjusted such that the added methanol neither spread over the chip nor started to drip off from the chip but rather remained at one corner. Under these optimized conditions, a stable spray for about 10–20 s was generated. Ions for [DON + H]+ (m/z 297.1326), [DON + NH4]+ (m/z 314.1588), [DON + Na]+ (m/z 319.1150), and [DON + K]+ (m/z 335.0882) were observed in positive ion mode (see Figure S1A). Although the [DON + Na]+ ion showed the highest intensity, [DON + H]+ was chosen for identification as an unknown positive ion with m/z 319.1150 (Figure S1B) was also seen in blank measurements (methanol spraying solvent only). As the unidentified interferent ion had, within experimental mass accuracy error, the same exact mass as [DON + Na]+, using a higher resolution mass analyzer was not an option to resolve the [DON + Na]+ and the interfering agent ions. Measurements were performed in negative ion mode as well, but positive ionization was chosen because the intensities of the ions were at least an order of magnitude higher. When four corners of the 1 × 1 cm chips were subsequently used, quite reproducible average signal intensities (total area of the signal (in arbitrary units) divided by the time of signal duration (in min)) of 8.1 × 107, 2.7 × 108, 1.5 × 108, and 1.1 × 108, respectively, were obtained (Figure 1C). The comparable results obtained for the four different corners suggest that a single chip can be analyzed four times, for example, serving the purpose of replicate analyses or even multiplexing by immobilizing four different biorecognition elements in each corner of the square chip. No physical damage (scratching) was observed on the side used earlier for clamping as the CMD modified chips are quite scratch-resistant. Next, serial dilutions of DON were analyzed (5000, 500, 50, and 5 pg), yielding a decrease of an order of magnitude of average signal intensities at each dilution step (108 for 5000 pg, 107 for 500 pg, and 106 for 50 pg). No signal for DON could be observed when only 5 pg of the toxin was spiked onto the chip. Washing of the CMD gold chip (after spiking with DON and drying) with HBS-EP and water was performed to check if any DON could be nonspecifically adsorbed to the chip surface without antibodies. Even when spiked at the highest level (5000 pg), no ions for DON were observed following washing.
For the Biochip Spray measurements, anti-DON was covalently immobilized on gold chips coated with carboxymethylated dextran. This was followed by introduction of buffer or beer containing the toxins (10 μg/mL DON) and washing of the chips (HBSEP followed by water), both performed in the SPR instrument. Washing of the chip with buffer (HBS-EP) helped to get rid of any nonspecifically adsorbed mycotoxin or sample components while the washing with water removed buffer salts, making the chip suitable for direct spray MS experiments. The chip was then unmounted from the SPR flow cell, dried with nitrogen, and subsequently positioned in front of the MS inlet capillary. The choice of solvent is an additional parameter as it must not only be efficient for creating a stable electrospray but also should be able to disrupt the interaction of antibodies (anti-DON) with the mycotoxins (DON). Like in direct spray, 100% methanol gave a reproducible and stable spray. Using a lower concentration of methanol (90% methanol; 10% water) was not feasible as it often required higher voltages (>6 kV), and the spray was not reproducible. In earlier SPR studies, acidic (10 mM HCl) and basic conditions (20 mM NaOH) were found to be suitable for disrupting the interaction of antibody and antigen.28,29 However, these SPR chip regenerants are not compatible with MS experiments and must be replaced with, for example, formic acid or ammonium hydroxide. Thus, the addition of 1–2% formic acid or ammonium hydroxide to methanol was tested as well, but the best disruption and direct spray performance was still obtained with 100% methanol. As can be seen from Figure 2A,B, ions for DON could be observed in both spiked buffer and spiked beer. In the Biochip Spray experiment, 10 μg/mL toxins was used to ensure saturation of the available binding sites of the antibodies on the surface as observed in the calibration curves from the SPR experiments (Figure S2B). However, only a limited amount can be captured and thus be later detected by Biochip Spray MS. When comparing the obtained average signal intensity (106) with the spiked solution experiments described above, approximately ∼50 pg of DON, i.e., in the range expected on the basis of the amount of anti-DON present according to SPR, was immuno-captured and subsequently analyzed by Biochip Spray MS. To ensure that desorption was complete, the chip after having been used for the Biochip Spray MS experiment (no signals were observed in the second spray) was placed in a methanol solution (200 μL) for 5 min with occasional shaking. The resulting extract was evaporated to dryness, redissolved in 5 μL of methanol, and analyzed by ESI-MS. The absence of any ions from DON confirmed the complete desorption. Another parameter that has been suggested for optimization in the literature is the waiting time before starting the spray.17 Therefore, 0, 30, and 60 s of waiting time were tested to see if there was any effect on the amount of analyte desorbed. Insignificant differences were observed between the different waiting times, so 30 s of waiting time was chosen to ensure sufficient and controlled timing between adding the methanol solution and applying the voltage. To confirm the selectivity of the developed method, two negative control experiments were performed. The first negative control was a chip containing anti-DON but incubated in blank beer: the absence of DON ions was confirmed by Biochip Spray MS. Another control experiment was performed by immobilizing an antibody against another mycotoxin, fumonisin (anti-FB1), elsewhere on the same chip. On the basis of previous research,28 anti-FB1 does not cross-react with DON. After introduction of beer spiked with DON and the standard washing procedure, high voltage was applied to the chip. A change indicating the onset of the spray was seen in the total ion chronogram (Figure S3A), but no ions for DON were observed (zero signal in the extracted ion chronogram, Figure S3B). The above-mentioned experiments serve as true negative controls for two reasons. First, they could be performed on another corner of the same chip, the first half of which was used for immobilization of anti-DON. Second, the anti-FB1 used is also an IgG and would account for any nonspecific adsorption that could occur during the experiment. Reuse of the SPR sensor chip is still a big challenge in SPR-MS as the conditions used for desorption and ionization involve organic solvents that not only disrupt the antibody–antigen interaction but also unfold the immobilized antibodies.7 Indeed, we observed similar problems: the chips coated with antibodies could only be used for SPR biosensing once. Upon reincubation of the same chip in a solution containing the analyte, no DON signal could be obtained. However, the proposed four corner approach for replicate analysis, multiplexing, or negative control experiments offers a reasonable compromise between reliable results and economy of use.
Figure 2.
Extracted ion chronogram for m/z 297.1333 ([DON + H]+) recorded in positive mode obtained from a CMD-modified gold chip with immobilized anti-DON, flushed in the iSPR flow cell with (A) spiked buffer containing 10 μg/mL DON and (B) spiked beer containing 10 μg/mL DON, followed by washing with buffer and water and transfer to the Biochip Spray MS setup.
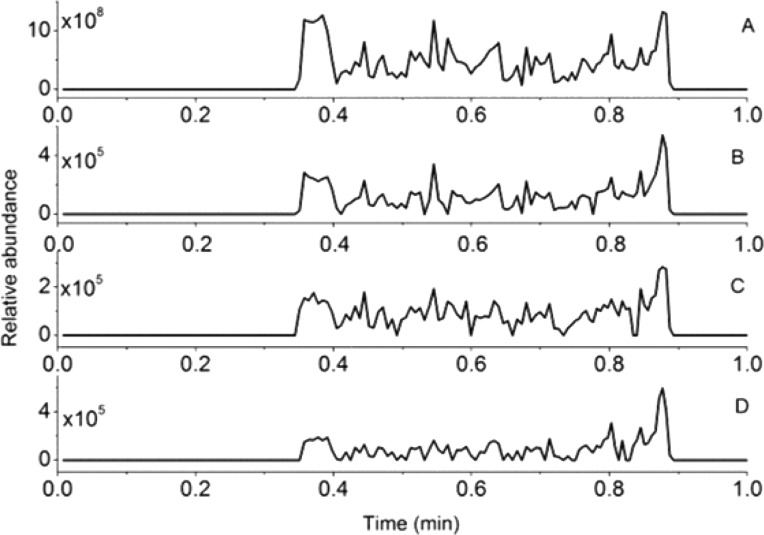
Finally, to demonstrate the real-life application of the newly developed method, a naturally contaminated beer sample containing 2760 ± 95 ng/mL DON plus 3883 ± 137 ng/mL deoxynivalenol-3-glucoside (D3G) according to LC-MS/MS30 or 5290 ± 370 ng/mL according to SPR (DON plus conjugates) was used.29 Large molecular weight analytes, such as proteins, yield high signals in SPR sensing with an antibody biochip and may be analyzed in a direct SPR assay (Figures S2A and S4A) without signal enhancer. Subsequently, the binding molecules may be identified in a direct manner by MS using MALDI7,8 or following elution3 using ESI MS or potentially by Biochip Spray MS (Figure S4A). However, low molecular weight analytes, such as DON, are much more challenging to detect in label-free biosensing approaches such as SPR: in a direct SPR mode with an antibody biochip, a low and noisy signal (Figure S2A, solid line, Biacore) or no signal (iSPR) will be obtained, unless the assay is changed into a competition format using a signal enhancer such as DON-OVA (Figure S2A, dashed line). Moreover, direct MS identification of low-molecular weight binding molecules would be challenging as well due to, e.g., matrix interference in MALDI MS and ion suppression and interference from residual assay reagents in ESI MS. Therefore, we performed SPR biosensing of DON in the contaminated beer sample in two different modes: (i) a competitive direct mode, using an anti-DON biochip and the addition of DON-OVA to the beer sample as a signal enhancer (Figures S2B and S4B) and (ii) a competitive indirect mode using an immobilized DON biochip and the beer sample mixed with anti-DON (Figures S2D and S4C). In both SPR modes, almost full signal inhibition was observed due to the presence of DON and conjugates in the contaminated beer (Figure S2C,E). Please note that, as for any large-scale routine SPR analysis of small molecules, a competitive indirect mode using an immobilized DON biochip (Figures S2D and S4C) would be the method of choice since such a chip will show extreme durability and, in the present case, could be used more than 400 times.29 Obviously, an immobilized DON chip would not directly trap the low molecular weight ligands for subsequent chemical identification by MS. As a way out, a so-called “recovery-chip” approach was applied as demonstrated previously for the coupling of inhibition SPR with nanoLC MS.4 Such a recovery chip is identical to the SPR screening chip, but the ligand binding interaction is reversed, i.e., immobilized antibodies on the chip to trap sufficient numbers of analyte molecules for chemical identification (Figure S4D). Thus, the contaminated beer sample was reinjected into the SPR apparatus but now onto a chip containing the anti-DON followed by washing with HBS-EP and water (Figure S4D). The chip was then taken out of the SPR instrument, dried with nitrogen, and analyzed using the Biochip Spray MS (Figure 3). In addition to the expected ions for DON, ions of D3G (m/z 481.1680, [D3G + Na]+) and acetyl DON (ADON, m/z 339.1438, [ADON + H]+) were observed. Both D3G and ADON are conjugates of DON known to cross-react with anti-DON.28 No ions from nivalenol (NIV) were visible, and this is in-line with our previous SPR research, in which no cross-reactivity was observed.28
Figure 3.
(A) Total ion chronogram along with the extracted ion chronogram for m/z (B) 297.1333 ([DON + H]+), (C) 481.1680 ([D3G + Na]+), and (D) 339.1438 ([ADON + H]+), recorded in positive ion mode obtained from a CMD-modified gold chip with immobilized anti-DON. The chip was loaded in an iSPR apparatus, and naturally contaminated beer was injected, followed by washing with buffer and water and transferring to the Biochip Spray MS setup.
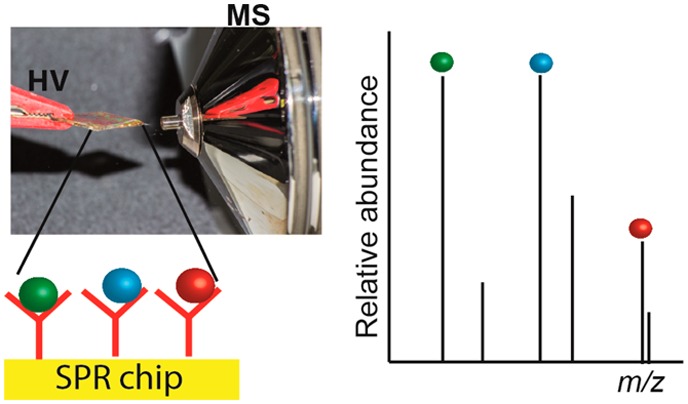
To conclude, a simplified coupling of SPR with MS through direct Biochip Spray was developed that allows chemical identification of low molecular weight analytes in SPR ligand binding assays. The technique is based on selective capturing of a target analyte on an SPR biosensor chip containing antibodies (or any other biorecognition element), followed by identification of the analyte as well as any (un)expected cross-reacting conjugates using ambient ionization MS directly from the gold SPR chip. The method may be applied for those samples, which give a response in either indirect or direct SPR biosensor screening modes (it is fair to say that not many low molecular weight analytes will yield a significant response in the direct SPR mode). The aforementioned method is, in principle, generic and could be applied to any MS-amenable analyte provided that antibodies are available and that they can be immobilized on an SPR chip. In this work, the antibodies were immobilized via the amine group and thus randomly oriented on the surface. Approaches for oriented immobilization of antibodies could be explored for better antigen binding and thus stronger signals in the MS. A 4-plex imaging SPR (iSPR) approach in which each corner of the biochip contains a different capturing antibody with the corresponding analyte that can be subsequently identified by Biochip Spray MS can be envisaged.
Acknowledgments
We would like to thank Dr. Milena Zachariasova (Institute of Chemical Technology, Prague, CZ) for the contaminated beer samples and Dr. Wilco Duvivier (RIKILT, Wageningen, NL) and Ing. Frank Claassen (Wageningen University, NL) for technical assistance. This research received funding from The Netherlands Organization for Scientific Research (NWO) in the framework of the Technology Area COAST (project no. 053.21.107) with WU, VU Amsterdam, RIKILT, Heineken, Synthon, Technex, EuroProxima, and Waterproef as partners and Plasmore and Bionavis as associated partners.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.6b04012.
Chip Spray mass spectra obtained by DON in methanol and blank methanol; different SPR assay modes, calibration curves, and sensorgrams of blank and contaminated beer; TIC and EIC for DON using a chip containing anti-FB1; schematic representation of the workflow options for large and small molecules by SPR-MS (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Homola J. Chem. Rev. 2008, 108, 462–493. 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- Stigter E. C. A.; de Jong G. J.; van Bennekom W. P. TrAC, Trends Anal. Chem. 2013, 45, 107–120. 10.1016/j.trac.2012.09.004. [DOI] [Google Scholar]
- Natsume T.; Nakayama H.; Jansson Ö.; Isobe T.; Takio K.; Mikoshiba K. Anal. Chem. 2000, 72, 4193–4198. 10.1021/ac000167a. [DOI] [PubMed] [Google Scholar]
- Marchesini G. R.; Buijs J.; Haasnoot W.; Hooijerink D.; Jansson Ö.; Nielen M. W. F. Anal. Chem. 2008, 80, 1159–1168. 10.1021/ac071564p. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Li X.; Nie H.; Yang L.; Li Z.; Bai Y.; Niu L.; Song D.; Liu H. Anal. Chem. 2015, 87, 6505–6509. 10.1021/acs.analchem.5b01272. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Xu S.; Wen L.; Bai Y.; Niu L.; Song D.; Liu H. Analyst 2016, 141, 3343–3348. 10.1039/C6AN00561F. [DOI] [PubMed] [Google Scholar]
- Krone J. R.; Nelson R. W.; Dogruel D.; Williams P.; Granzow R. Anal. Biochem. 1997, 244, 124–132. 10.1006/abio.1996.9871. [DOI] [PubMed] [Google Scholar]
- Urban P. L.; Amantonico A.; Zenobi R. Mass Spectrom. Rev. 2011, 30, 435–478. 10.1002/mas.20288. [DOI] [PubMed] [Google Scholar]
- Cody R. B.; Laramée J. A.; Durst H. D. Anal. Chem. 2005, 77, 2297–2302. 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- Takáts Z.; Wiseman J. M.; Gologan B.; Cooks R. G. Science 2004, 306, 471–473. 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- Nielen M. W. F.; Hooijerink H.; Zomer P.; Mol J. G. J. TrAC, Trends Anal. Chem. 2011, 30, 165–180. 10.1016/j.trac.2010.11.006. [DOI] [Google Scholar]
- Manova R. K.; Joshi S.; Debrassi A.; Bhairamadgi N. S.; Roeven E.; Gagnon J.; Tahir M. N.; Claassen F. W.; Scheres L. M. W.; Wennekes T.; Schroën K.; van Beek T. A.; Zuilhof H.; Nielen M. W. F. Anal. Chem. 2014, 86, 2403–2411. 10.1021/ac4031626. [DOI] [PubMed] [Google Scholar]
- Klampfl C. W.; Himmelsbach M. Anal. Chim. Acta 2015, 890, 44–59. 10.1016/j.aca.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Hu B.; So P.-K.; Yao Z.-P. Anal. Chim. Acta 2014, 817, 1–8. 10.1016/j.aca.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Jiang J.; Zhang H.; Li M.; Dulay M. T.; Ingram A. J.; Li N.; You H.; Zare R. N. Anal. Chem. 2015, 87, 8057–8062. 10.1021/acs.analchem.5b02390. [DOI] [PubMed] [Google Scholar]
- Paine M. R. L.; Barker P. J.; Blanksby S. J. Mass Spectrom. Lett. 2012, 3, 25–28. 10.5478/MSL.2012.3.1.025. [DOI] [Google Scholar]
- Gómez-Ríos G. A.; Pawliszyn J. Angew. Chem., Int. Ed. 2014, 53, 14503–14507. 10.1002/anie.201407057. [DOI] [PubMed] [Google Scholar]
- Seibert V.; Wiesner A.; Buschmann T.; Meuer J. Pathol., Res. Pract. 2004, 200, 83–94. 10.1016/j.prp.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Patrie S. M.; Mrksich M. Anal. Chem. 2007, 79, 5878–5887. 10.1021/ac0701738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrksich M. ACS Nano 2008, 2, 7–18. 10.1021/nn7004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsesia A.; Marabelli F.; Giudicatti S.; Marchesini G. R.; Rossi F.; Colpo P. World Patent, WO 2013/007448 A1, 2013.
- Bottazzi B.; Fornasari L.; Frangolho A.; Giudicatti S.; Mantovani A.; Marabelli F.; Marchesini G.; Pellacani P.; Therisod R.; Valsesia A. J. Biomed. Opt. 2014, 19, 017006–017010. 10.1117/1.JBO.19.1.017006. [DOI] [PubMed] [Google Scholar]
- Kostelanska M.; Hajslova J.; Zachariasova M.; Malachova A.; Kalachova K.; Poustka J.; Fiala J.; Scott P. M.; Berthiller F.; Krska R. J. Agric. Food Chem. 2009, 57, 3187–3194. 10.1021/jf803749u. [DOI] [PubMed] [Google Scholar]
- Papadopoulou-Bouraoui A.; Vrabcheva T.; Valzacchi S.; Stroka J.; Anklam E. Food Addit. Contam. 2004, 21, 607–617. 10.1080/02652030410001677745. [DOI] [PubMed] [Google Scholar]
- Zachariasova M.; Hajslova J.; Kostelanska M.; Poustka J.; Krplova A.; Cuhra P.; Hochel I. Anal. Chim. Acta 2008, 625, 77–86. 10.1016/j.aca.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Inoue T.; Nagatomi Y.; Uyama A.; Mochizuki N. Biosci., Biotechnol., Biochem. 2013, 77, 1410–1415. 10.1271/bbb.130027. [DOI] [PubMed] [Google Scholar]
- Lancova K.; Hajslova J.; Poustka J.; Krplova A.; Zachariasova M.; Dostalek P.; Sachambula L. Food Addit. Contam., Part A 2008, 25, 732–744. 10.1080/02652030701779625. [DOI] [PubMed] [Google Scholar]
- Joshi S.; Segarra-Fas A.; Peters J.; Zuilhof H.; van Beek T. A.; Nielen M. W. F. Analyst 2016, 141, 1307–1318. 10.1039/C5AN02512E. [DOI] [PubMed] [Google Scholar]
- Joshi S.; Annida R. M.; Zuilhof H.; Van Beek T. A.; Nielen M. W. F. J. Agric. Food Chem. 2016, 64, 8263–8271. 10.1021/acs.jafc.6b04106. [DOI] [PubMed] [Google Scholar]
- Zachariasova M.; Vaclavikova M.; Lacina O.; Vaclavik L.; Hajslova J. J. Agric. Food Chem. 2012, 60, 9280–9291. 10.1021/jf302069z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.