Abstract
Background
Sputum smear microscopy for acid fast bacilli (AFB) is used by most public health programmes to detect tuberculosis. While most AFB in countries endemic for tuberculosis are Mycobacterium tuberculosis (MTB), some may also be non-tuberculous mycobacteria (NTM). The inability to differentiate NTM from MTB by sputum smear microscopy may lead to erroneous diagnoses of tuberculosis, leading in turn to inappropriate therapy.
Methods
This was a retrospective study of consecutive sputum samples received from November 2013 to March 2015 in the Department of Microbiology, Bhopal Memorial Hospital & Research Centre, Bhopal, India. Samples underwent smear microscopy, line probe assay (LPA) for MTB complex, culture, biochemical tests and LPA for NTM.
Results
Of 4095 sputum samples, 2886 were AFB smear positive (70.5%). Of these, MTB complex was detected in 2611 (90.5%) samples by LPA. Of the remaining 275 samples, 47 grew AFB on culture. Nine strains belonged to the MTB complex. The remaining 38 (1.3%) were NTM, and could be speciated in 26 strains; 14 (53.8 %) were M. abscessus; 10 (38.4%) M. intracellulare, one (3.8%) M. kansasii and one (3.8%) M. fortuitum. The remaining 12 NTM could not be speciated.
Conclusion
NTM were present in at least 1.3% of all smear positive samples. It is important for public health programs to recognize the avoidable burden on logistics, infrastructure and finances caused by this. Detection and quantification of this burden would help design an appropriate strategy for optimal tuberculosis control.
Keywords: Acid fast bacilli, microscopy, non-tuberculous mycobacteria, tuberculosis
Introduction
Sputum smear microscopy has been the long-standing primary method for diagnosis of pulmonary tuberculosis (TB) in low and middle income countries, where nearly 95% of TB cases occur. Hence, it remains an integral part of the global strategy for TB control, as well as public health programs for TB control in these countries.1 Diagnosis of pulmonary TB by sputum smear microscopy involves detection of acid fast bacilli (AFB) in the smear. While most of the AFB seen in smears in countries endemic for tuberculosis are usually Mycobacterium tuberculosis (MTB), there are other potentially pathogenic mycobacteria called non-tuberculous mycobacteria (NTM) which may be present in a proportion of sputa. These mycobacteria include Mycobacterium species that are not members of the MTB complex and that can cause atypical mycobacterial infections. The inability to differentiate this group of organisms from MTB by sputum smear microscopy may lead to an erroneous diagnosis of tuberculosis in such patients, leading in turn to inappropriate/unwarranted therapy. The present study aims to assess the proportion of NTM in AFB smear positive sputum samples. This information would provide some indicators on the number of patients who may be receiving inappropriate therapy due to an erroneous diagnosis of pulmonary tuberculosis based on AFB smear positivity. It may also unmask a hitherto unidentified reservoir of these emerging pathogens.
Methods
Study setting
This was a retrospective study carried out in the Department of Microbiology, Bhopal Memorial Hospital & Research Centre (BMHRC), Bhopal, India, on samples received from November 2013 to March 2015 as a part of routine, mandated diagnostic workup for tuberculosis.
Study samples
The sample size included all consecutive sputum samples from routine presumptive TB cases that were received at the Department of Microbiology, BMHRC, Bhopal, for TB diagnosis and drug sensitivity testing from November 2013 to March 2015. Ethics approval was obtained from the Institutional Ethics Committee (IEC/08/Micro/14 dated 28 March 2014) to carry out this study. The samples were received from all over the central state of Madhya Pradesh, India. This study analyzed the results of culture and molecular tests used for identification of AFB in the clinical samples. Samples were collected at peripheral districts in screw capped containers and were transported the same day at temperatures between 15°C to 20°C to our laboratory. The temperatures were checked on receipt of the samples in the laboratory. All patients from whom samples were collected were registered with the Revised National Tuberculosis Control Programme (RNTCP) of India for diagnosis and treatment of tuberculosis.
Inclusion criteria
The study applied the following inclusion criteria: 1) sputum samples with a Q score of at least 1+, and 2) AFB smear positive on microscopy. Sample quality rating was done by using the Q score. Numbers of neutrophils and squamous epithelial cells were assigned a value per low-powered field (10x) and the results were summed up. The potential range for neutrophils and squamous epithelial cells was 0 to +3 and -3 to 0 respectively which resulted in a potential range of values for the Q score from -3 to +3.2
Exclusion criteria
The study applied the following exclusion criteria: 1) mislabelled samples, and 2) evidence of leakage of sputum sample from its container on receipt of the sample.
Smear microscopy
Sputum samples were screened by fluorescent microscopy using Auramine O stain and smears found to be positive were confirmed by light microscopy using Ziehl-Neelsen’s stain as per the standard protocols of both staining methods.3,4 Sputa were graded for positivity of AFB as per RNTCP (India) guidelines,3,4 decontaminated according to standard RNTCP (India) guidelines5 and divided into two parts.
Line probe assay for MTB complex
Mycobacterial DNA was extracted from one part of the decontaminated smear positive sputum samples using GenoLyse®, VER1.0 (Hain Lifescience, GmBH, Nehren, Germany) according to the manufacturer’s guidelines6 and stored at 4°C for a maximum of two days in order to batch the samples. Line Probe Assay was carried out using GenoType® MTBDRplus,VER 2.0 (Hain Lifescience, GmBH, Nehren, Germany) to look for the presence of MTB complex as well as drug resistance to rifampicin and isoniazid, as per manufacturer’s instructions.6
Culture
Decontaminated samples of those sputum samples that were found to have no members of MTB complex were cultured on Löwenstein-Jensen (LJ) media as per standard RNTCP (India) protocol5 and incubated at 37°C for a maximum of 8 weeks. Any strain of AFB grown from these samples was put up for biochemical tests and an rRNA based DNA hybridization assay (Accuprobe® System; Gen-Probe Inc., San Diego, CA, USA) to detect the presence of MTB complex, if any, according to the manufacturer’s guidelines.7
Line probe assay for NTM
The strains negative for MTB complex were confirmed as NTM by negative niacin accumulation test, growth on paranitrobenzoic acid (PNB) incorporated LJ media, positive catalase test and a negative result of a ribosomal RNA based DNA hybridization assay for Mycobacterium tuberculosis complex (Accuprobe® System Gen-Probe Inc., San Diego, CA, USA). DNA was extracted from these NTM using GenoLyse®, VER1.0 (Hain Lifescience, GmBH, Nehren, Germany) according to the manufacturer’s instructions. Line probe assay for NTM was carried out using GenoType® Mycobacterium common mycobacteria (CM), VER 1.0 (Hain Lifescience, GmBH, Nehren, Germany) to identify the NTM as per the manufacturer’s guidelines.8
If NTM were detected in a sputum sample, a request was made to the treatment providers to organize to send three consecutive sputum samples from the patient in order to understand whether there was an NTM infection according to the established American Thoracic Society (ATS) criteria.9 Smear microscopy, culture and LPA were then again carried out as described above.
Treatment outcomes
Details of treatment outcomes (cured/defaulted/lost to follow up/dead/ongoing treatment) of those patients in whom NTM were detected, were collected from their treatment providers.
A defaulter was a patient whose TB treatment was interrupted for two or more consecutive months for any reason. A patient who was lost to follow up never returned to the programme.
Results
Study samples
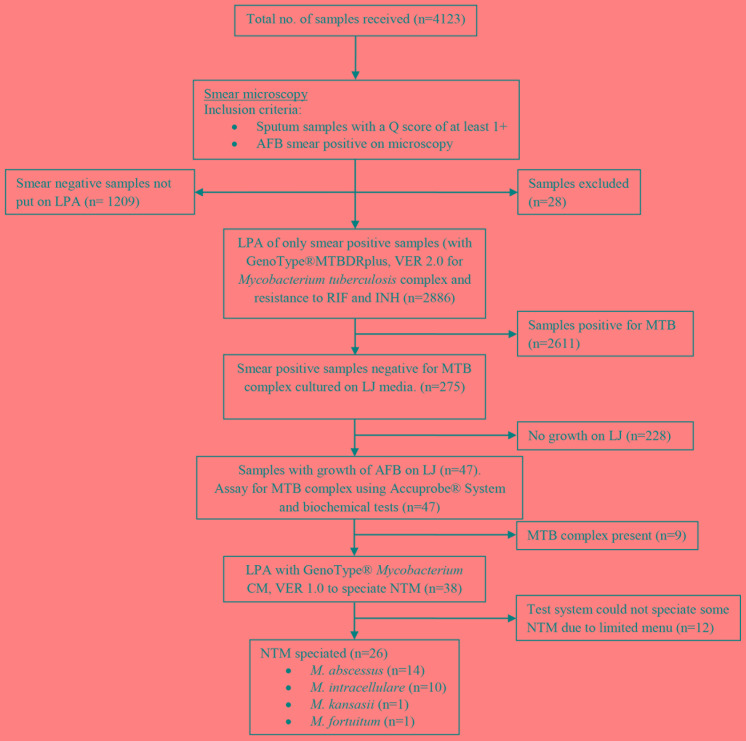
Of the 4123 samples received in the laboratory during the given time period, evidence for leakage of sputum was seen in 28 sample containers, and these samples were not processed (Figure 1). There were 4095 sputum samples processed for tuberculosis workup in the given time period, out of which 2886 (70.5%) were AFB smear positive while 1209 (29.5%) were AFB smear negative and excluded from our study.
Figure 1. Study flow diagram showing samples included in the analysis and test results.
Sample quality rating was done by using the Q score, which assessed the numbers of neutrophils (0 to +3) and squamous epithelial cells (-3 to 0) per low-powered field (10x).
AFB – acid fast bacilli; INH – isoniazid; LJ – Löwenstein-Jensen; LPA – line probe assay; MTB – Mycobacterium tuberculosis; NTM – non-tuberculous mycobacteria; RIF – rifampicin.
LPA for MTB complex
Out of all smear positive sputa, MTB complex was detected in 2611 (90.5%) samples by LPA using GenoType® MTBDRplus, VER 2.0.
Culture on LJ media
The remaining 275 (9.5%) samples out of 2886 smear positive samples were cultured on LJ media. Forty seven samples showed growth of strains of acid fast bacilli on LJ media. Ribosomal RNA based DNA hybridization assay for MTB complex (Accuprobe® System) of the strains from LJ media detected MTB complex in nine strains grown from nine samples respectively.
The remaining 38 were NTM, as confirmed by negative niacin accumulation test, growth on PNB incorporated LJ media, positive catalase test and a negative result of ribosomal RNA based DNA hybridization assay for MTB complex (Accuprobe® System).
LPA for NTM
DNA isolates from strains of NTM grown from the 38 samples were tested by LPA using GenoType® Mycobacterium CM, VER 1.0. NTM could be speciated from the DNA isolates of 26 strains. The remaining 12 NTM could not be speciated. Out of the 26 DNA isolates that could be speciated, 14 (53.8%) were identified as M. abscessus, 10 (38.4%) as M. intracellulare, one (3.8%) as M. kansasii and one (3.8%) as M. fortuitum by GenoType® Mycobacterium CM, VER 1.0 (Table 1).
Table 1. Frequency of NTM species speciated by GenoType® Mycobacterium CM assay.
| Non tuberculous mycobacteria species | Frequency, n (%) |
|---|---|
| M. abscessus | 14 (53.8%) |
| M. intracellulare | 10 (38.4%) |
| M. kansasii | 1 (3.8%) |
| M. fortuitum | 1 (3.8%) |
| Total | 26 |
We could also retrieve demographic data (gender and age) of all 38 patients whose sputum samples showed positive NTM growth. The demographic characteristics of the patients are given in Table 2. Out of the 38 patients from whom NTM were isolated, 28 (73.6%) were males and 10 (26.3%) were females. The prevalence of NTM isolation increased with age, ranging from 0% in patients younger than 20 years to 60.5% in patients aged 41-60 years.
Table 2. Demographic characteristics of NTM positive individuals (n=38).
| Characteristic | Number (%) of patients |
|---|---|
| Gender | |
| Males | 28 (73.6%) |
| Females | 10 (26.3%) |
| Age group | |
| <20 years | 0 (0%) |
| 21-40 years | 10 (26.3%) |
| 41-60 years | 23 (60.5%) |
| >60 years | 5 (13.1%) |
Outcomes
Since MTB complex was detected in 2611 (90.5%) of the AFB smear positive sputum samples, it is probable that NTM were present in the remaining 275 (9.5%) samples when all other variables were accounted for. However, our testing algorithm (with its inherent limitations) was able to conclusively determine that NTMs were present in 38 out of 2886 smear positive sputum samples (1.3%) received in the laboratory for diagnosis of TB.
All 38 patients with NTM in their sputa had received anti-TB therapy (ATT) as per national RNTCP (India) guidelines,3 based on the presence of AFB in their sputa. Of them, only 12 (31.6%) were cured as per RNTCP (India) guidelines.3 Six (15.8%) patients died; 13 (34.2%) were lost to follow up; one (2.6%) defaulted; and six (15.8%) were still on standard treatment (Table 3). During the same period in this region, among patients diagnosed with tuberculosis, 86% were cured, 4% died, 1% failed treatment and 5% defaulted.10
Table 3. Outcome details of patients whose samples were positive for NTM (n=38).
| Outcome details | Frequency, n (%) |
|---|---|
| Cured | 12 (31.6%) |
| Died | 6 (15.8%) |
| Lost to follow up | 13 (34.2%) |
| Defaulted | 1 (2.6%) |
| Still on treatment | 6 (15.8%) |
Of all the patients in whom NTM were detected, three consecutive sputum samples were received from only two patients. In one patient, NTM was not grown on culture in the three consecutive samples. The other patient had already been cured before the samples were sent to us.
Discussion
In our study, out of 2886 AFB smear positive sputum samples, MTB complex was detected in 2611 (90.5%) samples. Of the remaining 275 samples, 47 grew AFB on culture, of which 9 strains belonged to the MTB complex. The remaining 38 (1.3%) were NTM, and species identification could be done only in 26 strains. Out of 26 DNA isolates, 53.8% were M. abscessus, 38.4% M. intracellulare, 3.8% M. kansasii and 3.8% M. fortuitum.
The current scenario
In 2013, an estimated nine million people worldwide developed TB, of whom 1.5 million died from the disease.11 India is the second most populous country in the world, and one fourth of the global incident TB cases occur in India annually. There were 619,923 new smear positives in 2013, with a treatment failure rate of 2%.12 This would mean that 12,398 new smear positive patients failed treatment in 2013. The reasons for treatment failures are many, but one reason could be an erroneous diagnosis of TB due to dependence on only smear microscopy for initial diagnosis of the disease. This could be due, in part, to the presence of NTM in sputum samples, and the fact that smear microscopy is not an effective tool to differentiate between NTM and MTB.
Dependency on smear microscopy and the risk of inappropriate therapy
In many developing countries, particularly among those that have the largest burden of TB, sputum smear microscopy still remains the only bacteriologic basis for diagnosis of TB in primary health care centers. As a consequence, despite recent advances in rapid diagnostics, smear microscopy is possibly the only method by which diagnosis and treatment can be accessed in these areas.
The public health program in India also depends mainly on sputum AFB smear positivity for diagnosis and treatment of TB, and our results indicate that at least 1.3% of AFB smear positive patients are at risk of receiving inappropriate diagnosis and therapy. The latest available national figures state that the annual detection of AFB smear positive sputum samples in India is 928,190 (data for 2013).12 Assuming that 1.3% of 928,190 are falsely diagnosed as having infection with MTB while they have NTM instead, this would mean that there are approximately 12,000 such patients annually. Even though this proportion of patients may not be significant, the fact still remains that a large number of patients may be at risk of receiving inappropriate diagnosis and therapy.
NTM in clinical samples
There has been increased isolation of NTM causing mycobacterial diseases.13,14 In a previous study from India, NTM were obtained in 8.6% of sputum specimens.15 A recent study reported 1.9% culture confirmed cases of NTM in routine clinical samples, that were misdiagnosed and treated as TB. The authors stated that inefficient screening plus suboptimal diagnostic procedures in primary health care facilities may lead to unrealistic detection of TB cases.16 Active case finding activities may detect even more cases.
The most common species of NTM in our samples was M. abscessus. This is in contrast with results from a study in North India, which found that M. fortuitum and M. intracellulare were the most common isolates.17 This indicates that the distribution of NTM species varies by geographic area.18 M. intracellulare and M. kansasii are generally associated with pulmonary symptoms, lymphadenitis and disseminated infections. M. abscessus and M. fortuitum are usually associated with cutaneous infections but are also unusual causative agents for pulmonary disease and lymphadenitis.9
More than 160 NTM species have been identified worldwide.19 At least 42 species are known to be associated with infections.20 They have been detected in soil and water samples.21 Infection with NTM, when it occurs, is thought to be due to exposure to these environmental sources.22 In developing countries, pulmonary symptoms suggestive of mycobacterial disease are almost universally considered to be caused by MTB.23 As clinical and radiographic features of NTM infections are similar to MTB infection,24 NTM positive smears could also be misclassified as MTB and receive conventional ATT, which is inappropriate.
An increased number of NTM may be reported due to contamination of specimen during collection, contamination of reagents used in specimen processing,25,26 or contamination of medical tools.9 Therefore, patients testing positive for NTM should be closely assessed to ascertain the clinical significance of the laboratory result. Identification of true infection by NTM is very important, particularly because treatment of NTM disease is different and often complicated. ATS established a clear set of criteria to allow diagnosis of pulmonary NTM infection and to differentiate true infection from colonization or contamination.9,27
One study showed that the presence of NTM in patients with chronic TB substantially impacted clinical management.28 The study found NTM infection in 11/61 (18%) of chronic TB cases, and all were either on treatment for MDR-TB, or eligible for MDR-TB treatment. However, there is no information as to whether MDR was detected. Some TB patients had co-infection with NTM. The authors suggested that the presence of NTM in sputa may lead to misdiagnosis of MDR and, potentially, inappropriate treatment of the presumed MDR cases, while emphasizing the need to consider NTM when dealing with putative TB treatment failure. In our study, of the 38 patients who had NTM in their sputa, only 12 (31.6%) were cured with ATT and six (15.8%) were still on treatment. The remaining patients had defaulted or had died, none of which are favorable outcomes. The patients who were lost to follow up may have improved without treatment because smear positivity may have been due to colonization with NTM, and not infection.
The current need
While difficulties with active case finding, contact tracing problems and socioeconomic issues are problems that our TB control program needs to tackle, lack of access to appropriate diagnostic testing is also one obstacle to achieving optimal outcomes. The cost of providing this access is nominal, when compared to the cost of inappropriate treatment. The availability of appropriate diagnostic testing would reduce suboptimal management of patients. Public health programs require the resources to generate appropriate diagnostic algorithms based on their country’s needs.28
The limitation of our study algorithm was that it could only attempt to look for NTM in 47 strains of mycobacterial species which were isolated on culture out of 275 samples that did not have MTB complex. It is possible that there may have been non-viable NTM in many of the remaining samples that could have been detected by DNA sequencing. Also, we used GenoType® Mycobacterium common mycobacteria assay (Hain Lifescience, GmBH, Nehren, Germany) that could identify and speciate only 26 NTM strains of the 38 NTM that were isolated. We could not identify the remaining 12 NTM species. It is possible that we could have identified these 12 species had we used an additional line probe assay, for example the GenoType® Mycobacterium common mycobacteria/additional species (CM/AS) assay (Hain Lifescience, GmBH, Nehren, Germany). Financial constrains prevented us from carrying out additional line probe assay and DNA sequencing. Another limitation of our study was our failed attempt to obtain three consecutive sputum samples from patients whose samples came positive for NTM.
Conclusion
The presence of NTM in routine diagnostic smear positive sputum samples can interfere with appropriate diagnosis and control of TB. The public health program in India depends mainly on sputum AFB smear positivity for diagnosis and treatment of TB. Our results show that many patients may be at risk of receiving inappropriate therapy due to the ineffectiveness of the standard of care smear microscopy test in identifying NTM. This highlights the necessity of a cheaper, easier, more sensitive and specific TB screening algorithm for effective disease control. It is important for public health programs to recognize the avoidable burden on logistics, infrastructure and finances caused by this. Initiatives to detect and quantify this burden would go a long way towards designing an appropriate strategy for optimal TB control.
Acknowledgement
The authors gratefully acknowledge the role of the healthcare providers in providing information about patients whose samples were tested.
Footnotes
Authors’ contributions statement: PD, SA, MP designed and supervised the study and drafted the manuscript. KT and NP collected and analyzed the data and performed the background literature review for the manuscript. SK, MC, RV, SBM, AS carried out the laboratory work and conducted analyses. All authors reviewed and approved the final version of the manuscript.
Conflicts of interest: All authors – none to declare.
Funding: Mandated services funded through the National TB Control Programme of India.
References
- 1.Desikan P. Sputum smear microscopy in tuberculosis: is it still relevant? Indian J Med Res. 2013;137:442–4. [PMC free article] [PubMed] [Google Scholar]
- 2.Nair B, Stapp J, Stapp L, Bugni L, Van Dalfsen J, Burns JL. Utility of Gram staining for evaluation of the quality of cystic fibrosis sputum samples. J Clin Microbiol. 2002;40:2791–4. doi: 10.1128/JCM.40.8.2791-2794.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Central Tuberculosis Division, Directorate General of Health Services, Ministry of Health and Family Welfare, Nirman Bhavan, New Delhi Technical and Operational Guidelines for TB Control in India 2016. [Accessed on: 01 April 2016]. Available at http://tbcindia.gov.in/index1.php?lang=1&level=2&sublinkid=4573&lid=3177.
- 4.Revised National Tuberculosis Control Programme, Central TB Division, Directorate General of Health Services, Ministry of Health and Family Welfare, Nirman Bhavan, New Delhi Manual for fluorescent microscopy. [Accessed on: 01 April 2016]. Available at http://tbcindia.nic.in/WriteReadData/l892s/7890638455Flourescence_Microscopy%20Manual.pdf.
- 5.Revised National TB Control Programme Training Manual for Mycobacterium tuberculosis culture & drug susceptibility testing. [Accessed on: 01 April 2016]. Available at http://tbcindia.nic.in/WriteReadData/l892s/6995271860Training%20manual%20M%20tuberculosis%20C%20DST.pdf.
- 6.HAIN Lifescience Standard operating procedure for GenoType MTBDR plus, VER 2.0, Instructions for Use, IFU-304A-02 2/2012. http://www.hain-lifescience.de/ifu.html Accessed on: 01 April 2016. Available at.
- 7.AccuProbe Mycobacterium tuberculosis complex culture identification test. [Accessed on: 01 April 2016.]. Available at http://www.hologic.com/search/site/accuprobe.
- 8.HAIN Lifescience Genotype Mycobacterium common mycobacteria (CM) [Accessed on: 01 April 2016.]. VER 1.0. Available at http://mycobactoscana.it/Manuale/PDF/Alleg08.2.pdf.
- 9.Griffith DE, Aksamit T, Brown-Elliott BA, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 10.TB India 2016 Revised National TB Control Programme. Annual Status Report. [Accessed on: 27 August 2016.]. http://www.tbcindia.nic.in/showfile.php?lid=3180 Available at.
- 11.World Health Organization Global tuberculosis report 2014. [Accessed on: 02 April 2016.]. Available at http://www.who.int/tb/publications/global_report/en/
- 12.TB India 2013 Revised National TB Control Programme. Annual Status Report. [Accessed on: 01 April 2016.]. Available at http://www.tbcindia.nic.in/showfile.php?lid=3163.
- 13.Ding LW, Lai CC, Lee LN, Hsueh PR. Disease caused by non-tuberculous mycobacteria in a university hospital in Taiwan, 1997–2003. Epidemiol Infect. 2006;134:1060–7. doi: 10.1017/S0950268805005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai CC, Lee LN, Ding LW, Yu CJ, Hsueh PR, Yang PC. Emergence of disseminated infections due to nontuberculous mycobacteria in non-HIV-infected patients, including immunocompetent and immunocompromised patients in a university hospital in Taiwan. J Infect. 2006;53:77–84. doi: 10.1016/j.jinf.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Paramasivan CN, Govindan D, Prabhakar R, Somasundaram PR, Subbammal S, Tripathy SP. Species level identification of non-tuberculous mycobacteria from South Indian BCG trial area during 1981. Tubercle. 1985;66:9–15. doi: 10.1016/0041-3879(85)90048-0. [DOI] [PubMed] [Google Scholar]
- 16.Hoza AS, Lupindu AM, Mfinanga SGM, Moser I, König B. The role of non tuberculous mycobacteria in the diagnosis, management and quantifying risks of tuberculosis in Tanga, Tanzania. Tanzan J Health Res. 2016;18:1–9. [Google Scholar]
- 17.Maurya AK, Nag VL, Kant S, et al. Prevalence of non-tuberculous mycobacteria among extrapulmonary tuberculosis cases in tertiary care centers in Northern India. Biomed Res Int. 2015;2015:465403. doi: 10.1155/2015/465403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai CC, Tan CK, Chou CH, et al. Increasing incidence of non-tuberculous mycobacteria, Taiwan, 2000–2008. Emerg Infect Dis. 2010;16:294–6. doi: 10.3201/eid1602.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Anazi KA, Al-Jasser AM, Al-Anazi WK. Infections caused by non-tuberculous mycobacteria in recipients of hematopoietic stem cell transplantation. Front Oncol. 2014;4:311. doi: 10.3389/fonc.2014.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tortoli E. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin Microbiol Rev. 2003;16:319–54. doi: 10.1128/CMR.16.2.319-354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falkinham JO. Impact of human activities on the ecology of nontuberculous mycobacteria. Future Microbiol. 2010;5:951–60. doi: 10.2217/fmb.10.53. [DOI] [PubMed] [Google Scholar]
- 22.Mangione EJ, Huitt G, Lenaway D, et al. Nontuberculous mycobacterial disease following hot tub exposure. Emerg Infect Dis. 2001;7:1039–42. doi: 10.3201/eid0706.010623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verweij KE, Kamerik AR, van Ingen J, et al. Application of modern microbiological diagnostic methods for tuberculosis in Macha, Zambia. Int J Tuberc Lung Dis. 2010;14:1127–31. [PubMed] [Google Scholar]
- 24.Al Jarad N, Demertzis P, Jones DJ, et al. Comparison of characteristics of patients and treatment outcome for pulmonary non-tuberculous mycobacterial infection and pulmonary tuberculosis. Thorax. 1996;51:137–9. doi: 10.1136/thx.51.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnow PM, Bakir M, Thompson K, Bova JL. Endemic contamination of clinical specimens by Mycobacterium gordonae. Clin Infect Dis. 2000;31:472–6. doi: 10.1086/313940. [DOI] [PubMed] [Google Scholar]
- 26.Blossom DB, Alelis KA, Chang DC, et al. Pseudo-outbreak of Mycobacterium abscessus infection caused by laboratory contamination. Infect Control Hosp Epidemiol. 2008;29:57–62. doi: 10.1086/524328. [DOI] [PubMed] [Google Scholar]
- 27.Van Ingen J, Bendien SA, De Lange WC, et al. Clinical relevance of non-tuberculous mycobacteria isolated in the Nijmegen-Arnhem region, The Netherlands. Thorax. 2009;64:502–6. doi: 10.1136/thx.2008.110957. [DOI] [PubMed] [Google Scholar]
- 28.Maiga M, Siddiqui S, Diallo S, et al. Failure to recognize non-tuberculous mycobacteria leads to misdiagnosis of chronic pulmonary tuberculosis. PLoS One. 2012;7:e36902. doi: 10.1371/journal.pone.0036902. [DOI] [PMC free article] [PubMed] [Google Scholar]