Figure 1. In vivo and in vitro shRNA screening using a human breast cancer cell line (MCF10DCIS.com).
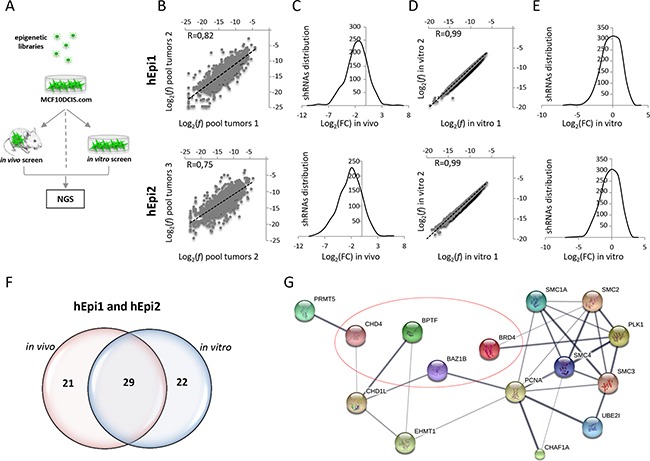
A. Graphical representation of the experimental procedure: MCF10DCIS.com cells were infected with hEpi1 and hEpi2 libraries and then orthotopically transplanted in the 4th mammary gland of NOD/SCID mice (in vivo screen), or cultured in vitro for 21 days (in vitro screen). Genomic DNAs (gDNAs) were extracted from transduced cells as reference, tumors and in vitro cultures and subsequently subjected to PCR amplification and Next Generation Sequencing (NGS) to quantify shRNAs representation. B. Scatter plot representation of the shRNAs log2 frequencies (f) of two different pools (pool tumors 1 and 2) composed of four tumors each of hEpi1 (upper panel) and hEpi2 (lower panel) transduced cells. Black dotted lines represent the axis bisectors. Pearson correlation coefficient (R) indicates the similarity between the two samples. C. Distribution of the shRNA reads expressed as log2 Fold Change (FC) in the three pooled samples (mean of replicates) in hEpi1 (upper panel) and hEpi2 (lower panel) samples. D. Scatter plot representation of the shRNAs log2(f) of in vitro duplicates of hEpi1 (upper panel) and hEpi2 (lower panel) libraries. Black dotted lines represent the axis bisectors. Pearson correlation coefficient (R) indicates the similarity between the two samples. E. Distribution of the shRNA reads expressed as log2(FC) of two in vitro samples (mean of replicates) of hEpi1 (upper panel) and hEpi2 (lower panel) transduced cells. F. Venn diagrams reporting the number of genes scoring as depleted targets in vivo, in vitro and in common between the two settings. G. Protein interaction network of 15 out of 29 genes scoring by Ingenuity Pathway Analysis and significantly enriched for “Cell cycle” regulation analyzed in STRING. Line thickness represents the strength of data confidence. Encircled genes were selected for in vivo and in vitro validation of the screen.
