Abstract
Background
Reports have demonstrated the prognostic function of long non-coding RNAS (lncRNAS) in patients with cancer. However, their prognostic functions in non small cell lung cancer (NSCLC) remain controversial. We therefore performed a meta-analysis on six lncRNAs (PVT1, AFAP1-AS1, LINC01133, ANRIL, MEG3 and UCA1) to clarify their prognostic roles in NSCLC.
Results
Thirty-six studies involving 6267 patients with NSCLC and 34 lncRNAs were included. Of the listed lncRNAs, 20 were shown to negatively affect patients' overall survival while the high expression of 13 lncRNAs indicated better survival outcomes.
Materials and Methods
The log-rank p value and Kaplan–Meier survival curves of survival outcomes were extracted for hazard ratio (HR) calculation. Survival outcomes were measured by overall survival (OS) and event free survival (EFS) which were then analyzed by calculating pooled hazard ratios. The heterogeneity was detected by Q statistic and I-squared statistic.
Conclusions
The abnormal expression of lncRNAs may significantly affect NSCLC patients' survival and may serve as a novel predictive factor for prognosis of NSCLC patients.
Keywords: lncRNAs, NSCLC, prognosis, meta-analysis
INTRODUCTION
Lung cancer is one of the most common causes of cancer-related deaths worldwide and non small cell lung cancer (NSCLC) accounts for 80% of all cases [1]. GLOBOCAN 2012 reported that there were approximately 14.1 million cancer patients in the world and 8.2 million of them died in 2012, most of which were population from less developed countries [2]. Patients with lung cancer are usually diagnosed at advanced stages with relatively poor prognosis. The estimated overall 5-year survival rate of advanced stage lung cancer is 0–14% [3, 4], while the 5-year survival rate of early stage NSCLC can be as high as 83%., which informs us that the early diagnosis and the finding of new molecular targets for NSCLC are the key to improve clinical strategies and outcomes of NSCLC [5]. Long non-coding RNAs (lncRNAs) are non-protein-coding RNA molecules with a length of more than 200 nucleotides and often expressed in a spatial, temporal and tissue-specific pattern [6, 7]. In the past, lncRNAs were merely viewed as transcriptional “noise” [8]. Recently, a growing number of genome-wide transcriptome studies have identified about 3000 lncRNASs and at the same time indicated their diverse biological functions in both normal and degenerated tissues, including cell growth, differentiation and disease progression [9]. lncRNAs may act as primary regulators of the molecular interaction with DNA-binding proteins and epigenetically regulate the expression of target genes [10].
So far, controversy about the prognostic role of lncRNAs in NSCLC still exists. Some studies drew statistically insignificant conclusions [11, 12], while some studies showed that lncRNAs could be important biomarkers for the assessment of overall survival and recurrence. Due to the limitation of sample size and research number, a single study may not be able to reflect the facts accurately. Therefore, we conducted a meta-analysis to identify the exact role of lncRNAs in NSCLC patients' prognosis. At the same time, we summarized in our study the relation of different lncRNAs to patients' prognosis. Kaplan–Meier survival analysis and log-rank tests were performed in our enrolled studies to further evaluate the correlation between lncRNA expression and the prognosis of NSCLC patients. Pooled results indicated that lncRNAs played an important role in NSCLC overall survival time, which provided us with new insights in the therapeutic strategies of NSCLC.
RESULTS
Study selection
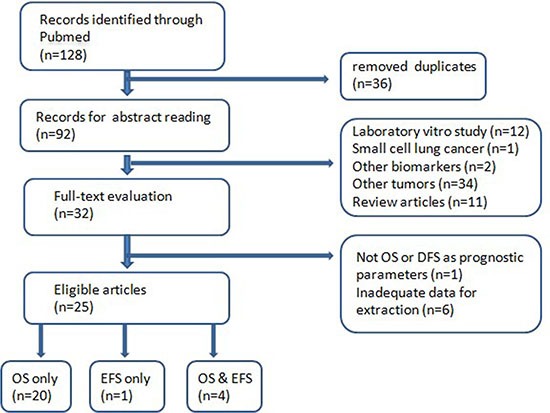
After full-text assessment of all included articles, we excluded studies that did not use EFS or OS as survival parameters. Studies that lacked information for calculation with methods developed by Parmar, Williamson , and Tierney (Parmar et al., 1998; Williamson et al., 2002; Tierney) were also excluded. The initial search returned 128 articles, from which 36 duplicated records were removed. Abstracts of the remaining 92 articles were carefully read by two authors independently and we excluded 60 unqualified literatures: laboratory studies (n = 12), review articles (n = 11), other biomarkers (n = 2) and other types of cancer (n = 35). Next we went through the full texts of the remaining 32 studies and 25 with adequate data for calculation were finally enrolled. The flow chart of selection process is shown in Figure 1. The supplementary search returned 46 articles, 11 of which contain useful information.
Figure 1. The flow chart of selection process.
Characteristics of included studies
Among the 36 studies, one article [11] used progression free survival instead of disease free survival, we therefore combined DFS and PFS together and use event free survival EFS as prognosis parameter of our study. 28 studies used overall survival OS as prognosis parameter, one study used event free survival EFS and four contained OS as well as EFS. All 36 studies used the quantitative real-time reverse transcription-PCR (qRTPCR) method to measure the expression of lncRNAs in tumor samples. All the included studies analyzed the prognosis of 6267 patients with NSCLC and the correlation between 34 lncRNAs levels and survival outcomes. All necessary data of included trials are listed in Table 1 and Table 2.
Table 1. Criteria for the inclusion of prognostic lncRNA studies.
| Study design | Prospective or retrospective cohort |
| Time of study | After 2006 |
| Tumor type | Non-small cell lung cancer (NSCLC) |
| Assay type | Tumor tissue or blood sample |
| RNA measurement | qRT-PCR or ISH |
| Outcome type | Overall Survival (OS) or Disease Free Survival (DFS) |
| Included results | Univariate and multivariate survival analysis (Cox proportional hazards regression model) including HRs, 95% CIs and P value / Kaplan Meier survival curves with enough data for calculation |
| Population size | ≥ 30 |
| Length of follow-up | ≥ 1 year |
| Source | Peer-reviewed journals |
| Language | English |
Abbreviations: OS, overall survival; DFS, disease-free survival; CI, confidence interval.
Table 2. Frequency of studies assessing prognostic value of lncRNAs in NSCLC.
| Name of lncRNA | Number of studies | Reference |
|---|---|---|
| PVT1 | 2 | Yan-Rong Yang, |
| Di Cui | ||
| CASC2 | 1 | Xuezhi He |
| PANDAR | 1 | L han |
| SPRY4-IT1 | 1 | Sun M |
| TUG1 | 1 | Eb Zhang |
| AFAP1-AS1 | 2 | Jun Deng |
| Zhaoyang Zeng | ||
| ANRIL | 2 | Ling Lin |
| Feng-qi Nie | ||
| BANCR | 1 | Ming Sun |
| CARLo-5 | 1 | Jie Luo |
| GAS6-AS1 | 1 | Liang Han |
| H19 | 1 | Erbao Zhang |
| HMlincRNA717 | 1 | Xiao Xie |
| HOTAIR | 2 | Xiang-hua Liu |
| Takayuki Nakagawa | ||
| LINC01133 | 2 | Jing Zhang |
| Chongshuang Zang | ||
| MALAT-1 | 2 | Lars Henning Schmidt |
| Liqin Shen | ||
| Sox2ot | 1 | Zhibo Hou |
| UCA1 | 3 | Huimin Wang |
| Ningning Cheng | ||
| Wei Nie | ||
| MEG3 | 2 | Lin Su |
| Kaihua Lu | ||
| BC087858 | 1 | Hui Pan |
| XIST | 1 | Jing Fang |
| NEAT1 | 1 | Chengcao Sun |
| HNF1A-AS1 | 1 | Ying Wu |
| MVIH | 1 | Feng-qi Nie |
| LINC00342 | 1 | Li Wang |
| LINC00473 | 1 | Zirong Chen |
| TUSC7 | 1 | Zhongwei Wang |
The number of patients enrolled in each study ranged from 20 to 1926, and the follow-up duration varied from 25 months to 200 months. Among them, 33 studies involved participants from China [11–43] and three studies involved patients respectively from Japan [44], Germany [45] and USA [46]. All studies investigated patients with NSCLC and qRT–PCR was used to detect lncRNAs expression in tumor tissues.
Overall Analyses
20 lncRNAs were shown to negatively affect patients' overall survival while 13 lncRNAs were associated with better survival outcomes. One study [11] on ANRIL (Nie et al: OS HR = 2.23 , 95% CI: 0.89–5.59, P = 0.09) showed no significant prognostic effect of lncRNAs expression on patients' overall survival. Wang et al. [12] observed no correlation between the expression of TUSC7 and patients' DFS, but significant correlation between TUSC7 expression and patients' OS. The BC087858 expression level was also associated with prognosis but it just reached the marginal statistical significance (P =0.083) [38]. All HRs, 95% CI and P values of included studies are listed in Table 3.
Table 3. Basic information of included studies.
| Author | LncRNA | Population | Sample | Total patients | Assay | Survival analysis | Follow up(month) |
|---|---|---|---|---|---|---|---|
| Takayuki N | HOTAIR | Japan | Tumor tissue | 77 | qRT-PCR | EFS | 40–50 |
| Xianghua Liu | HOTAIR | China | Tumor tissue | 42 | qRT-PCR | OS | 60 |
| YanRong Yang | PVT1 | China | Tumor tissue | 82 | qRT-PCR | OS | 60 |
| Di Cui | PVT1 | China | Tumor tissue | 108 | qRT-PCR | OS,EFS | 30–40 |
| Jun Deng | AFAP1-AS1 | China | Tumor tissue | 121 | qRT-PCR | OS | 60 |
| Zhaoyang Zeng | AFAP1-AS1 | China | Tumor tissue | 332 | qRT-PCR | OS | 96–168 |
| Jing Zhang | LINC01133 | China | Tumor tissue | 39 | qRT-PCR | OS | 60 |
| Chongshuang Zang | LINC01133 | China | Tumor tissue | 68 | qRT-PCR | OS | 30–40 |
| Ling Lin | ANRIL | China | Tumor tissue | 87 | qRT-PCR | OS | 60 |
| Fengqi Nie | ANRIL | China | Tumor tissue | 68 | qRT-PCR | OS,EFS | 36 |
| Xuezhi He | CASC2 | China | Tumor tissue | 76 | qRT-PCR | OS | 60 |
| L han | PANDAR | China | Tumor tissue | 140 | qRT-PCR | OS | 60 |
| Sun M | SPRY4-IT1 | China | Tumor tissue | 121 | qRT-PCR | OS,EFS | 30–40 |
| Eb Zhang | TUG1 | China | Tumor tissue | 192 | qRT-PCR | OS | 60 |
| Ming Sun1 | BANCR | China | Tumor tissue | 113 | qRT-PCR | OS | 36 |
| Jie Luo | CARLo-5 | China | Tumor tissue | 62 | qRT-PCR | OS | 60 |
| Liang Han | GAS6-AS1 | China | Tumor tissue | 50 | qRT-PCR | OS | 60 |
| Erbao Zhang | H19 | China | Tumor tissue | 70 | qRT-PCR | OS | 60 |
| Zirong Chen | LINC00473 | USA | Tumor tissue | 469 | qRT-PCR | OS | > 50 |
| Zhongwei Wang | TUSC7 | China | Tumor tissue | 112 | qRT-PCR | OS,EFS | > 60 |
| Xiao Xie | HMlincRNA717 | China | Tumor tissue | 118 | qRT-PCR | OS | 80 |
| Liqin Shen | MALAT-1 | China | Tumor tissue | 78 | qRT-PCR | EFS | 60 |
| Zhibo Hou | Sox2ot | China | Tumor tissue | 47 | qRT-PCR | OS | 60 |
| Wei Nie | UCA1 | China | Tumor tissue | 112 | qRT-PCR | OS | 80 |
| Fengqi Nie | MVIH | China | Tumor tissue | 42 | qRT-PCR | OS | 36 |
| Lars H Schmidt | MALAT-1 | Germany | Tumor tissue | 102 | qRT-PCR | OS | 100–140 |
| Huimin Wang | UCA1 | China | Tumor tissue | 60 | qRT-PCR | OS | 60–80 |
| Ningning Cheng | UCA1 | China | Tumor tissue | 52 | qRT-PCR | EFS | 20–25 |
| Lin Su | MEG3 | China | Tumor tissue | 20 | qRT-PCR | OS | 60 |
| Kaihua Lu | MEG3 | China | Tumor tissue | 42 | qRT-PCR | OS | 40–60 |
| Hui Pan | BC087858 | China | Tumor tissue | 38 | qRT-PCR | EFS | 30 |
| Jing Fang | XIST | China | Tumor tissue | 53 | qRT-PCR | OS | 150–200 |
| Chengcao Sun | NEAT1 | China | Tumor tissue | 96 | qRT-PCR | OS | 40 |
| Ying Wu | HNF1A-AS1 | China | Tumor tissue | 856 | qRT-PCR | OS | 200 |
| Li Wang | LINC00342 | China | Tumor tissue | 1926 | qRT-PCR | OS | 200 |
| Meng Zhou | CTD-2358C21.4 | China | Tumor tissue | 196 | qRT-PCR | OS | 60 |
| RP11-94L15.2 | |||||||
| KCNK15-AS1 | |||||||
| AC104134.2 | |||||||
| RP11-21L23.2 | |||||||
| GPR158-AS1 | |||||||
| RP11-701P16.5 | |||||||
| RP11-379F4.4 |
After careful reading of 92 literatures of the first search after duplicates were removed and all literatures returned from the second search, we summarized all lncRNAs up to date whose prognostic roles in NSCLC were investigated (Table 4). Of the 34 lncRNAs presented, eight( RP11-21L23.2, GPR158-AS1, RP11-701P16.5, RP11-379F4.4, CTD-2358C21.4, RP11-94L15.2, KCNK15-AS1 and AC104134.2) lacked information for calculation but their influences on prognosis were clearly demonstrated in the study [41].
Table 4. Summary of hazard ratios of lncRNA expression in NSCLC.
| LncRNA | Hazard ratio | CI | p value | Log (HR) | SE | Expression related to bad prognosis | ||
|---|---|---|---|---|---|---|---|---|
| OS | EFS | Lower | Upper | |||||
| HOTAIR | 3.10 | 1.05 | 9.10 | 0.04 | 1.13 | 0.55 | High | |
| HOTAIR | 2.69 | 1.30 | 5.56 | 0.007 | 0.99 | 0.37 | High | |
| PVT1 | 3.25 | 1.84 | 5.75 | < 0.0001 | 1.18 | 0.29 | High | |
| PVT1 | 1.72 | 1.01 | 2.91 | 0.05 | 0.54 | 0.27 | High | |
| 1.97 | 1.01 | 3.84 | 0.05 | 0.68 | 0.34 | High | ||
| AFAP1-AS1 | 8.94 | 3.10 | 25.75 | < 0.0001 | 2.19 | 0.54 | High | |
| AFAP1-AS1 | 1.90 | 1.17 | 3.08 | 0.009 | 0.64 | 0.25 | High | |
| 2.90 | 1.54 | 5.47 | 0.001 | 1.06 | 0.32 | High | ||
| LINC01133 | 2.39 | 1.03 | 5.54 | 0.04 | 0.87 | 0.43 | High | |
| LINC01133 | 2.25 | 1.25 | 4.05 | 0.007 | 0.81 | 0.30 | High | |
| ANRIL | 2.53 | 1.28 | 5.03 | 0.008 | 0.93 | 0.35 | High | |
| ANRIL | 2.23 | 0.89 | 5.59 | 0.09 | 0.80 | 0.47 | – | |
| 3.53 | 1.64 | 7.57 | 0.001 | 1.26 | 0.39 | High | ||
| H19 | 1.08 | 1.04 | 1.13 | < 0.0001 | 0.08 | 0.02 | High | |
| MALAT-1 | 1.79 | 1.09 | 2.92 | 0.02 | 0.58 | 0.25 | High | |
| MALAT-1 | 2.36 | 1.19 | 4.69 | 0.01 | 0.86 | 0.35 | High | |
| Sox2ot | 2.80 | 1.14 | 6.90 | 0.03 | 1.03 | 0.46 | High | |
| UCA1 | 1.94 | 1.06 | 3.26 | 0.029 | 0.66 | 0.29 | High | |
| UCA1 | 3.25 | 1.17 | 9.02 | 0.02 | 1.18 | 0.52 | High | |
| UCA1 | 1.40 | 1.07 | 1.85 | 0.02 | 0.34 | 0.14 | High | |
| MVIH | 2.01 | 1.08 | 3.77 | 0.03 | 0.70 | 0.32 | High | |
| CARLo-5 | 2.20 | 1.20 | 4.05 | 0.01 | 0.79 | 0.31 | High | |
| LINC00473 | 1.73 | 1.27 | 2.37 | 0.0006 | 0.55 | 0.16 | High | |
| XIST | 6.3 | 4.09 | 9.69 | < 0.0001 | 1.84 | 0.22 | High | |
| NEAT1 | 1.82 | 1.07 | 3.09 | 0.03 | 0.6 | 0.27 | High | |
| HNF1A-AS1 | 1.19 | 1.01 | 1.39 | 0.03 | 0.17 | 0.08 | High | |
| LINC00342 | 1.16 | 1.05 | 1.28 | 0.03 | 0.15 | 0.05 | High | |
| BC087858 | 2.51 | 0.89 | 7.10 | 0.083 | − | − | − | |
| TUSC7 | 0.26 | 0.10 | 0.66 | 0.005 | −1.35 | 0.48 | Low | |
| 0.7 | 0.42 | 1.16 | 0.17 | −0.36 | 0.26 | − | ||
| HMlincRNA717 | 0.40 | 0.21 | 0.75 | 0.004 | −0.91 | 0.32 | Low | |
| CASC2 | 0.28 | 0.10 | 0.76 | 0.01 | −1.29 | 0.52 | Low | |
| PANDAR | 0.65 | 0.46 | 0.93 | 0.02 | −0.43 | 0.18 | Low | |
| SPRY4-IT1 | 0.45 | 0.24 | 0.82 | 0.01 | −0.80 | 0.31 | Low | |
| 0.44 | 0.26 | 0.73 | 0.001 | −0.83 | 0.26 | Low | ||
| TUG1 | 0.78 | 0.69 | 0.88 | < 0.0001 | −0.25 | 0.06 | Low | |
| BANCR | 0.50 | 0.26 | 0.95 | 0.03 | −0.70 | 0.33 | Low | |
| GAS6-AS1 | 0.15 | 0.03 | 0.87 | 0.03 | −1.90 | 0.90 | Low | |
| MEG3 | 0.33 | 0.12 | 0.88 | 0.03 | −1.11 | 0.5 | Low | |
| MEG3 | 0.26 | 0.12 | 0.57 | 0.007 | −1.35 | 0.40 | Low | |
Subgroup analysis
Among the 20 listed lncRNASs, eight (HOTAIR, PVT1, AFAP1-AS1, LINC01133 and ANRIL, UCA1, MALAT-1, MEG3) have been studied by two or more articles. We then carried out meta analyses and obtained the combined HRs. While other studies have sufficient information for pooled analysis, studies on HOTAIR and MALAT-1 looked into OS and EFS separately and we were therefore unable to conduct relevant meta analysis.
PVT1
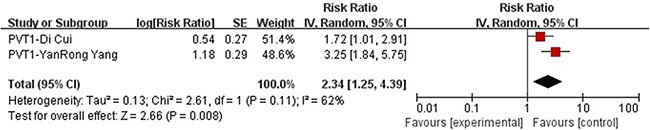
We performed meta-analysis on articles choosing lncRNA PVT1 as a prognostic marker. The two studies included in meta-analysis [14, 15], both conducted multivariate Cox regression analysis and the data such as HR is therefore directly extracted and put into pooled analysis. The median follow-up period is 41 months [14] and 32 months [15] respectively and the information of a total number of 190 patients were collected. There was evidence of considerable heterogeneity in these two groups (P = 0.11, I2 = 62%) so the random effect model was selected. A combined HR of 2.34 (95% CI: 1.25–4.39, P = 0.008) for those patients with high expression of PVT1 was found, from which we drew a conclusion that high expression of long non-coding RNAS PVT1 is a predictor of poorer overall survival (Figure 2).
Figure 2. Forrest plots of studies evaluating hazard ratios of high PVT1 expression as compared to low expression.
AFAP1-AS1
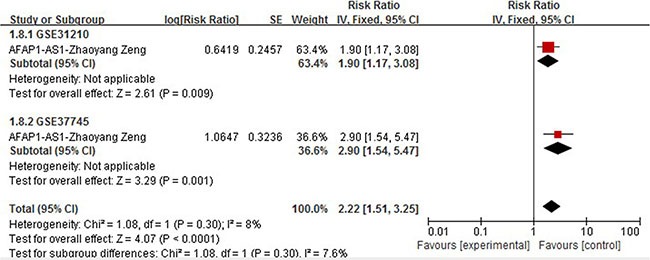
Two studies [16, 17] described the elevated expression of long non-coding RNAS AFAP1-AS1 as predictive of poor OS in NSCLC (n = 332). Deng et al enrolled 121 patients diagnosed with NSCLC who had never received any therapy before surgery. Multivariate Cox regression analysis was performed and HR for high AFAP1-AS1 expression was 8.947 (95% CI = 3.115–25.694, P = 0.000). Zang et al included two independent cohorts, GSE31210 (N = 226) [47] and GSE37745 (N = 106) [48] which had complete follow-up data. This article presented Kaplan-Meier curve with precise number of patients and the death ratio in high and low expression group, instead of Cox regression analysis result. The combined HR (HR = 2.22,95% CI: 1.51–3.25, P < 0.0001) from subsequent pooled analysis of these two cohorts is shown in Figure 3.
Figure 3. Forrest plots of studies evaluating hazard ratios of high AFAP1-AS1 expression as compared to low expression with 2 cohorts of one study.
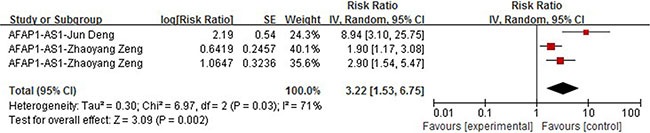
We then carried out meta analysis with these two articles containing three groups of data and the subsequent combined HR is shown in Figure 4. Significant heterogeneity among selected studies according to Q-test (chi2 = 6.97) and I-squared result (I2 = 71%, P = 0.03) was observed, so the random model was applied to calculate a pooled HR (HR = 3.22,95% CI: 1.53–6.75, P =0.002), which indicated that an elevated expression level of AFAP1-AS1 was a strong predictor of poorer OS.
Figure 4. Forrest plots of studies evaluating hazard ratios of high AFAP1-AS1 expression as compared to low expression.
LINC01133
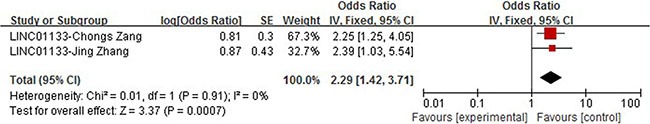
We included two studies investigating the correlation of LINC01133 expression with patients overall survival. Ling et al studied a cohort of 79 pairs of NSCLC tumor tissues, including 39 lung squamous cell cancer (LSCC) and 40 lung adenocarcinoma (LAD). Both studies conducted Kaplan-Meier survival analysis and no significant heterogeneity was observed (I2 = 0%, P = 0.91). Further meta analysis using the fixed effect model revealed that high expression of LINC01133 could develop as an independent factor for predicting the prognosis of NSCLC patients (HR = 2.29, 95% CI: 1.42–3.71, P = 0.0007) (Figure 5).
Figure 5. Forrest plots of studies evaluating hazard ratios of high LINC01133 expression as compared to low expression.
ANRIL
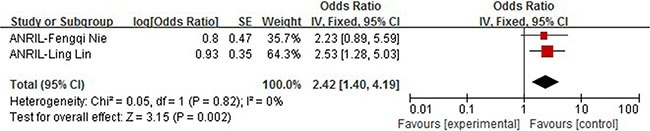
Two studies involved the multivariate Cox regression analysis of prognostic parameters including the expression of ANRIL in NSCLC patients. Ling et al (N = 87) and Nie et al (N = 68) had clinical follow-ups of 60 months and 36 months respectively. In Nie's study, the ANRIL over-expression did not show a significant influence on OS (HR = 2.23, 95% CI: 0.89–5.59, P = 0.09). In order to clarify the impact of ANRIL expression on patients' survival, we performed a pooled analysis. We observed no heterogeneity between studies (I2 = o%, P = 0.82) and therefore fixed effect model was applied to calculate the association between high tumoral ANRIL expression and OS (HR 2.42, 95% CI: 1.40–4.19, P = 0.002). These results suggest that high expression of ANRIL could predict worse prognosis of NSCLC patients regarding overall survival and may be an independent prognostic marker (Figure 6).
Figure 6. Forrest plots of studies evaluating hazard ratios of high ANRIL expression as compared to low expression.
UCA1
Two article about lncRNA UCA1 studied OS and were therefore included in meta-analysis [14, 15]. Both studies conducted multivariate Cox regression analysis and the data such as HR is therefore directly extracted and put into pooled analysis. We observed no heterogeneity between studies (I2 = 0%, P = 0.32) and therefore fixed effect model was applied. A combined HR of 1.49 (95% CI: 1.17–1.91, P = 0.001) for those patients with high expression of PVT1 was observed. We could then conclude that high expression of lncRNA UCA1 can be used as a predictor of poorer overall survival (Figure 7).
Figure 7. Forrest plots of studies evaluating hazard ratios of high UCA1 expression as compared to low expression.

MEG3
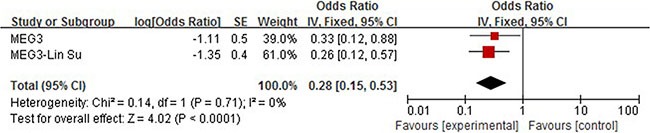
We carried out meta analysis with two articles describing the correlation between elevated expression of Meg3 and overall survival. The subsequent combined HR is shown in Figure 4. No heterogeneity among selected studies according to Q-test (chi2 = 0.14) and I-squared result (I2 = 0%, P = 071) was observed, so the fixed model was applied to calculate a pooled HR (HR = 0.28, 95% CI = 0.15–0.53, P < 0.0001), which indicated that elevated expression of MEG3 could positively affect patients' overall survival (Figure 8).
Figure 8. Forrest plots of studies evaluating hazard ratios of high MEG3 expression as compared to low expression.
DISCUSSION
The current meta-analysis investigating the correlation between lncRNAs and cancer prognosis, demonstrated that the over-expression of lncRNAs was an effective predictor of survival in a variety of cancers, in terms of both OS and EFS. For NSCLC, it is of great interest to identify its prognostic biomarkers, which can help cast light on the stratification of patients and make clinical decisions. In recent years, an increasing number of studies have proved the aberrant expression of lncRNAs in human cancer including NSCLC [49].
Our study included 36 recently published articles and a total number of 6267 patients, which is considered powerful enough to consolidate and perform the subgroup analyses. In this study, we listed 34 lncRNAs that were potential prognostic biomarkers for prognosis (Table 3). Our meta-analysis looked into six lncRNAs (PVT1, AFAP1-AS1, LINC01133 and ANRIL, UCA1, MALAT-1, MEG3) whose prognostic roles have been clearly demonstrated in two or more articles. The combined HRs suggested that elevated expressions of PVT1, AFAP1-AS1, LINC01133, ANRIL, UCA1, MALAT-1 and MEG3 were significantly correlated with patients' poor prognosis (Figures 2, 4, 5, 6, 7, 8). Although one study on ANRIL alone showed no statistical significance (HR = 2.23, 95% CI: 0.89–5.59, P = 0.09), the pooled outcome of two studies added convincing evidence that increased expression of ANRIL indicates shorter overall survival time (HR = 2.42, 95% CI: 1.40–4.19, P = 0.002). Due to the limitation of the study number, these conclusions need more clinical trials for verification. The heterogeneity of the population was probably due to the difference in source of population, the cut-off value of lncRNAs and the duration of follow-ups.
Distinct from earlier studies, this meta-analysis have summarized the prognostic role of all published lncRNAs in NSCLC and carried out pooled analysis on some certain lncRNAs with enough data. To the best of our knowledge, this is the first meta-analysis summarizing information about the prognostic value of all available lncRNAs in NSCLC patients. We strictly followed the literature inclusion criteria and all enrolled literatures were examined independently by two authors. Furthermore, we paid substantial attention to the details of study design and data reporting in quality assessment. We extracted data only of multivariate analysis to avoid the influence of heterogeneity among the included studies and to further explore the potential role of lncRNAs as prognostic biomarkers of NSCLC. As for Kaplan Meier survival curves, we carefully selected studies with valid information and strictly followed methods developed by Parmar, Williamson, and Tierney. Blurred curves were retouched with Microsoft Paint to make it precise for calculation. Furthermore, all data of extracted lncRNAs were based on frozen tissue samples of clear clinical origins. It was proven that the type of samples could influence the experimental outcomes in terms of RNASs detection [50]. All enrolled studies used qRT-PCR to measure lncRNAs which made pooled data from different studies more persuasive considering the consistent measurement background. Last but not least, all returned studies of our search strategy have been covered in this study which demonstrated the prognostic value of various lncRNA expression in NSCLC.
However, some details of our study need to be further refined. To start with, the number of eligible articles is relatively small, which lead to the relative insufficiency of studies in subgroup analyses. The possible cause for this was that studies reporting positive results were more likely to be published or that published literatures in other languages were missed during our search process. For the same reason, publication bias and sensitivity analyses were not performed, which might lead to the lack of statistical power. Second, the main ethnicities of the patients in our analysis were Asian. Thus, standardized analyses are expected in order to apply our results to other populations. Third, although all four sets of pooled outcomes of HR for OS in patients with high lncRNA expression were proven to be statistically significant (all HR > 2), some independent outcomes are not strong enough to have clinical value. Because empirically, a predictive HR value of more than 2.0 was considered to be statistically strong [51]. Although these results remain to be verified by larger numbers of clinical trials, they still possess statistic validity to reflex the general correlation of lncRNA expression with OS. The prognostic performance of lncRNAs in NSCLC has been proven. However, further clinical studies are warranted to Figure out the complicated molecular networks through which lncRNAs act to exert an influence on NSCLC patients.
MATERIALS AND METHODS
Search strategy
A comprehensive search was done via Pubmed database for literatures that analyzed the prognostic value of lncRNAs in NSCLC patients. Studies were selected using the varying combination of the following keywords: long non-coding RNAs, prognosis, lung cancer or NSCLC. The last search update was performed on May 19th , 2016. A second search was done on September 13th, 2016, using the following words: long non-coding RNAs, survival, lung cancer or NSCLC. Additional studies mentioned in those review articles were manually added to our evaluation list.
Inclusion criteria
We referred to the guidelines of Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) Statement issued in 2009 as well as the checklist of the Dutch Cochrane Centre represented by MOOSE [52]. We then came up with a criteria for studies that are considered eligible for our full-text evaluation: (i) studies about the relation between lncRNAs expression in tumor or blood samples and prognosis of patients with NSCLC; (ii) the survival outcomes were measured with overall survival (OS) or event free survival (EFS) including disease free survival (DFS) and progression free survival ( PFS). The inclusion criteria is shown in Table 5.
Table 5. Summary of lncRNAs in the prognosis of NSCLC and authors' attitudes.
| Lnc RNA | Attitude | Sample size | HR provided | Reference |
|---|---|---|---|---|
| HOTAIR | Negative | 119 | yes | [15, 46] |
| PVT1 | Negative | 190 | yes | [16, 17] |
| AFAP1-AS1 | Negative | 453 | yes | [18, 19] |
| LINC01133 | Negative | 107 | yes | [20, 21] |
| ANRIL | Negative | 155 | yes | [11, 22] |
| H19 | Negative | 70 | yes | [29] |
| MALA T-1 | Negative | 180 | yes | [32, 47] |
| Sox2ot | Negative | 47 | yes | [33] |
| UCA1 | Negative | 224 | yes | [34, 36, 37] |
| MVIH | Negative | 42 | yes | [35] |
| CARLo-5 | Negative | 62 | yes | [28] |
| LINC00473 | Negative | 469 | yes | [48] |
| XIST | Negative | 53 | yes | [45] |
| NEAT1 | Negative | 96 | yes | [44] |
| HNF1A-AS1 | Negative | 856 | yes | [42] |
| LINC00342 | Negative | 1926 | yes | [41] |
| RP11-21L23.2 | Negative | 196 | No | [43] |
| GPR158-AS1 | Negative | 196 | No | [43] |
| RP11-701P16.5 | Negative | 196 | No | [43] |
| RP11-379F4.4 | Negative | 196 | No | [43] |
| BC087858 | − | 38 | yes | [40] |
| TUSC7 | Positive | 112 | yes | [12] |
| HMlincRNA717 | Positive | 118 | yes | [31] |
| CASC2 | Positive | 76 | yes | [23] |
| PANDAR | Positive | 140 | yes | [24] |
| SPRY4-IT1 | Positive | 121 | yes | [25] |
| TUG1 | Positive | 192 | yes | [26] |
| BANCR | Positive | 113 | yes | [27] |
| GAS6-AS1 | Positive | 50 | yes | [18, 19] |
| MEG3 | Positive | 62 | yes | [38, 39] |
| CTD-2358C21.4 | Positive | 196 | No | [43] |
| RP11-94L15.2 | Positive | 196 | No | [43] |
| KCNK15-AS1 | Positive | 196 | No | [43] |
| AC104134.2 | Positive | 196 | No | [47] |
Negative= Higher expression of the lncRNAs indicates poor prognosis.
Positive= Higher expression of the lncRNAs indicates better prognosis.
Studies were excluded based on any of the following conditions: (i) review articles, laboratory articles or letters; (ii) articles about the prognosis of other tumors or other markers. When two articles involving the same medical center with similar data, the article with a larger sample size was selected. Two authors independently selected studies, and disagreements were resolved by consulting a third author.
Data extraction
All data were extracted independently by two authors and any disagreements were resolved by consensus and consultation with a third investigator. We extracted the results of multivariate Cox hazard regression analysis provided in the articles. However, if these data were not directly available, we extracted the log-rank p value and Kaplan–Meier survival curves of survival outcomes with the number of patients at risk in each expression group for further calculation. The following data were extracted: name of first author, investigated lncRNAs, number of patients, HR with 95% CI, P value, population, sample site, assay and survival outcome parameter.
Statistical methods
All HRs and 95% confidence interval(CI) were calculated with Tierney's method. The logHR and SE (logHR) (SE) were recorded for aggregation of the survival outcomes of different long non-coding RNAs. Pooled analysis of the survival outcomes of specific lncRNAs was then performed. A test of heterogeneity of combined HRs was carried out using Cochran's Q test and Higgins I-squared statistic. P value of < 0.05 or I2 > 50% was considered statistically significant. A random effect model (Der Simonian and Laird method) was applied if heterogeneity was observed (P < 0.05 or I2 > 50%), otherwise the fixed effect model was used [53]. All P values were two sided and a P value of less than 0.05 was considered to be statistically significant.
Footnotes
CONFLICTS OF INTEREST
None.
REFERENCES
- 1.Ramalingam S, Pawlish K, Gadgeel S, Demers R, Kalemkerian GP. Lung cancer in young patients: analysis of a Surveillance, Epidemiology, and End Results database. Journal of clinical oncology. 1998;16:651–657. doi: 10.1200/JCO.1998.16.2.651. [DOI] [PubMed] [Google Scholar]
- 2.Globocan 2012 IARC. http://globocaniarcfr/Pages/fact_sheets_canceraspx. Accessed July 18, 2014.
- 3.Chansky K, Sculier JP, Crowley JJ, Giroux D, Van Meerbeeck J, Goldstraw P. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. Journal of thoracic oncology. 2009;4:792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 4.van Rens MT, de la Riviere AB, Elbers HR, van Den Bosch JM. Prognostic assessment of 2,361 patients who underwent pulmonary resection for non-small cell lung cancer, stage I, II, and IIIA. Chest. 2000;117:374–379. doi: 10.1378/chest.117.2.374. [DOI] [PubMed] [Google Scholar]
- 5.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 6.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Molecular cancer therapeutics. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Jin Y, Ren H, Ma X, Wang B, Wang Y. Downregulation of the long non-coding RNA TUSC7 promotes NSCLC cell proliferation and correlates with poor prognosis. American journal of translational research. 2016;8:680–687. [PMC free article] [PubMed] [Google Scholar]
- 13.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. International journal of clinical and experimental pathology. 2014;7:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 15.Cui D, Yu CH, Liu M, Xia QQ, Zhang YF, Jiang WL. Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumour biology. 2015 doi: 10.1007/s13277-015-4261-x. [DOI] [PubMed] [Google Scholar]
- 16.Deng J, Liang Y, Liu C, He S, Wang S. The up-regulation of long non-coding RNA AFAP1-AS1 is associated with the poor prognosis of NSCLC patients. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2015;75:8–11. doi: 10.1016/j.biopha.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X, Zhang W, Deng H, Zhou M, Peng S, Li G, Xiong W. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour biology. 2015 doi: 10.1007/s13277-015-3860-x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Zhu N, Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour biology. 2015;36:7465–7471. doi: 10.1007/s13277-015-3460-9. [DOI] [PubMed] [Google Scholar]
- 19.Zang C, Nie FQ, Wang Q, Sun M, Li W, He J, Zhang M, Lu KH. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget. 2016;7:11696–707. doi: 10.18632/oncotarget.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin L, Gu ZT, Chen WH, Cao KJ. Increased expression of the long non-coding RNA ANRIL promotes lung cancer cell metastasis and correlates with poor prognosis. Diagnostic pathology. 2015;10:14. doi: 10.1186/s13000-015-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He X, Liu Z, Su J, Yang J, Yin D, Han L, De W, Guo R. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and regulates cell proliferation in non-small cell lung cancer. Tumour biology. 2016 doi: 10.1007/s13277-016-4787-6. [DOI] [PubMed] [Google Scholar]
- 22.Han L, Zhang EB, Yin DD, Kong R, Xu TP, Chen WM, Xia R, Shu YQ, De W. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell death & disease. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, Yang JS, Xu TP, Liu YW, Zou YF, Lu BB, Yin R, Zhang EB, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell death & disease. 2014;5:e1298. doi: 10.1038/cddis.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, De W, Shu YQ, Wang ZX. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell death & disease. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, Chen JF, Zhang EB, De W, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Molecular cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo J, Tang L, Zhang J, Ni J, Zhang HP, Zhang L, Xu JF, Zheng D. Long non-coding RNA CARLo-5 is a negative prognostic factor and exhibits tumor pro-oncogenic activity in non-small cell lung cancer. Tumour biology. 2014;35:11541–11549. doi: 10.1007/s13277-014-2442-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhang E, Li W, Yin D, De W, Sun S, Han L. c-Myc-regulated long non-coding RNA H19 indicates a poor prognosis and affects cell proliferation in non-small-cell lung cancer. Tumour biology. 2015 doi: 10.1007/s13277-015-4185-5. [DOI] [PubMed] [Google Scholar]
- 28.Han L, Kong R, Yin DD, Zhang EB, Xu TP, De W, Shu YQ. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Medical oncology (Northwood, London, England) 2013;30:694. doi: 10.1007/s12032-013-0694-5. [DOI] [PubMed] [Google Scholar]
- 29.Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu FQ, Hu R, Wang MS. LncRNA HMlincRNA717 is down-regulated in non-small cell lung cancer and associated with poor prognosis. International journal of clinical and experimental pathology. 2014;7:8881–8886. [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. Journal of neuro-oncology. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 31.Hou Z, Zhao W, Zhou J, Shen L, Zhan P, Xu C, Chang C, Bi H, Zou J, Yao X, Huang R, Yu L, Yan J. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. The international journal of biochemistry & cell biology. 2014;53:380–388. doi: 10.1016/j.biocel.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer letters. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 33.Nie FQ, Zhu Q, Xu TP, Zou YF, Xie M, Sun M, Xia R, Lu KH. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour biology. 2014;35:7587–7594. doi: 10.1007/s13277-014-2009-7. [DOI] [PubMed] [Google Scholar]
- 34.Cheng N, Cai W, Ren S, Li X, Wang Q, Pan H, Zhao M, Li J, Zhang Y, Zhao C, Chen X, Fei K, Zhou C, et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget. 2015;6:23582–23593. doi: 10.18632/oncotarget.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang HM, Lu JH, Chen WY, Gu AQ. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. International journal of clinical and experimental medicine. 2015;8:11824–11830. [PMC free article] [PubMed] [Google Scholar]
- 36.Su L, Han D, Wu J, Huo X. Skp2 regulates non-small cell lung cancer cell growth by Meg3 and miR-3163. Tumour biology. 2016;37:3925–3931. doi: 10.1007/s13277-015-4151-2. [DOI] [PubMed] [Google Scholar]
- 37.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan H, Jiang T, Cheng N, Wang Q, Ren S, Li X, Zhao C, Zhang L, Cai W, Zhou C. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget. 2016;7:49948–49960. doi: 10.18632/oncotarget.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Chen Z, An L, Wang Y, Zhang Z, Guo Y, Liu C. Analysis of Long Non-Coding RNA Expression Profiles in Non-Small Cell Lung Cancer. Cellular physiology and biochemistry. 2016;38:2389–2400. doi: 10.1159/000445591. [DOI] [PubMed] [Google Scholar]
- 40.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M, Guo M, He D, Wang X, Cui Y, Yang H, Hao D, Sun J. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. Journal of translational medicine. 2015;13:231. doi: 10.1186/s12967-015-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, Li D. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377–3p-E2F3 pathway. Oncotarget. 2016;7:51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochemical and biophysical research communications. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochemical and biophysical research communications. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. Journal of thoracic oncology. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, Li JL, Lin S, Cao C, Gimbrone NT, Yang R, Fu DA, Carper MB, Haura EB, Schabath MB, Lu J, Amelio AL, Cress WD, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. The Journal of clinical investigation. 2016 doi: 10.1172/JCI85250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer research. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 48.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Ponten F, Konig A, Fernandes O, Karlsson M, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clinical cancer research. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 49.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends in cell biology. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, Bowman ED, Mathe EA, Takenoshita S, Yokota J, Haugen A, Harris CC. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clinical cancer research. 2011;17:1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. Journal of mammary gland biology and neoplasia. 2001;6:375–392. doi: 10.1023/a:1014778713034. [DOI] [PubMed] [Google Scholar]
- 52.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. Jama. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 53.DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemporary clinical trials. 2015;45:139–145. doi: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


