Abstract
Background
Specific biomarkers for outcome prediction of lung squamous cell carcinoma (LUSC) are still lacking. This study assessed the prognostic value of differentially expressed miRNAs of LUSC patients.
Results
Twelve of the 133 most significantly altered miRNAs were associated with overall survival (OS) across different clinical subclasses of the Cancer Genome Atlas (TCGA) LUSC cohort. A linear prognostic model of seven miRNAs was developed to divide patients into high- and low-risk groups. Patients assigned to the high-risk group exhibited poor OS compared with patients in the low-risk group, which was further validated in the validation cohort and entire LUSC cohort.
Methods
MiRNA expression profiles with clinical information of 447 LUSC patients were obtained from TCGA. Most significantly altered miRNAs were identified between tumor and normal samples. Using survival analysis and supervised principal components method, a seven-miRNA signature for prediction of OS of LUSC patients was established. Survival receiver operating characteristic (ROC) analysis was used to assess the performance of survival prediction. The biological relevance of predicted miRNA targets was also analyzed using bioinformatics method.
Conclusions
The current study suggests that seven-miRNA signature may have clinical implications in the outcome prediction of LUSC.
Keywords: lung squamous cell carcinoma, microRNA, prognostic marker
INTRODUCTION
Lung cancer is the most common and fatal cancer in the world [1]. Non-small cell lung cancer (NSCLC) is the most frequent type of lung cancer and can be further subtyped mainly as adenocarcinomas (LAD) and squamous cell carcinomas (LUSC). Despite decades-long improvements in early detection and treatment, survival of advanced stage patients remains poor [2]. Effective biomarkers to identify patients with high recurrence and death risk are still lacking. Moreover, since the genetic and epigenetic alterations between LUSC and lung LAD are quite different, the targeted agents for LAD cannot be applied to LUSC. Thus, there is an urgent need to identify effective biomarkers for the prognosis of LUSC.
MicroRNAs (miRNAs) are small, non-coding RNAs which regulate gene expression at the posttranscriptional level [3]. By binding to the 3’ [4] or 5’ untranslated region (UTR) [5] of the target transcripts, miRNAs can modulate genes expression through translational repression or cleavage of mRNA. MiRNAs can function as either tumor suppressors or oncogenes by regulating genes involved in tumorigenesis. Moreover, miRNAs have been found in the blood samples and have been demonstrated to be remarkable stable even under harsh conditions such as treatment with RNase and DNase or multiple freeze-thaw cycles [6]. This suggests that miRNAs can be potential noninvasive biomarkers for cancer.
Although a number of miRNAs have been identified for predicting outcome for NSCLC, there is significant inconsistences among previous studies. This may have resulted from the small sample sizes as well as the heterogeneous histological subtypes and tumor stages. Thus, our study aims to identify and assess the prognostic value of candidate miRNA biomarkers for LUSC patients with large cohort.
RESULTS
TCGA dataset and patients characteristics
In this study, a total of 447 LUSC patients from The Cancer Genome Atlas (TCGA) were enrolled. 45 normal tissues (n = 45) were also included for differential expression analysis of miRNAs. The patients were separated into the training set (n=224) and testing set (n=223) randomly. No significant difference in clinical covariates was observed between the two sets (Table 1).
Table 1. Characteristics of the study population.
| Variable | Total (n=447) | Training Set (n=224) | Testing Set (n=223) | P |
|---|---|---|---|---|
| Age(yeras) | 0.104* | |||
| < 65 | 154 (34.5%) | 69 (30.8%) | 85 (38.1%) | |
| ≥ 65 | 293 (65.5%) | 155 (69.2%) | 138 (61.9%) | |
| Sex | 0.569 | |||
| Male | 332 (74.4%) | 169 (75.4%) | 163 (73.1%) | |
| Female | 115 (25.7%) | 55 (24.6%) | 60 (26.9%) | |
| Vital status | 0672† | |||
| Alive | 263 (58.8%) | 134 (59.8%) | 129 (57.8%) | |
| Dead | 184 (41.2%) | 90 (40.2%) | 94 (42.2%) | |
| Stage | 0.366 | |||
| I | 211 (47.2%) | 113 (50.4%) | 98 (43.9%) | |
| II | 153 (34.2%) | 75(33.5%) | 78 (35.0%) | |
| III | 78 (17.4) | 33 (14.6%) | 45 (20.2%) | |
| IV | 5 (1.1%) | 3 (1.3%) | 2 (0.9%) | |
| T stage | 0.896 | |||
| T1 | 179 (40.0%) | 87 (38.8%) | 92 (41.3%) | |
| T2 | 203 (45.5%) | 103 (46.0%) | 100 (44.8%) | |
| T3 | 55 (12.3%) | 28 (12.5%) | 27 (12.1%) | |
| T4 | 10 (2.2%) | 6 (2.7%) | 4 (1.8%) | |
| N stage | 0.903 | |||
| N0 | 279 (62.4%) | 142 (63.4%) | 137 (61.4%) | |
| N1 | 130 (29.1%) | 63 (28.1%) | 67 (30.0%) | |
| N2 | 35 (7.8%) | 18 (8.0%) | 17 (7.6%) | |
| N3 | 3 (0.7%) | 1 (0.4%) | 2 (0.9%) | |
| M stage | 1.000 | |||
| M0 | 442 (98.9%) | 221 (98.7%) | 221 (99.1%) | |
| M1 | 5 (1.1%) | 3 (1.3%) | 2 (0.9%) | |
| Smoke status | 0.337 | |||
| Smoker | 293 (65.5%) | 142 (63.4%) | 151 (67.7%) | |
| Nonsmoker | 154 (34.5%) | 82 (36.6%) | 72 (32.3) | |
| Adjuvant treatment | 0.279 | |||
| None | 296 (66.2%) | 154 (68.8%) | 142 (63.7%) | |
| Chemotherapy | 94 (21.0%) | 44 (19.6%) | 50 (22.4) | |
| Radiotherapy | 16 (3.6%) | 10 (4.5%) | 6 (2.7%) | |
| Chemoradiotherapy | 41 (9.2%) | 16 (7.1%) | 25 (11.2%) |
Identification of dysregulated miRNAs in LUSC
The miRNA expression profiles of 45 pairs of LUSC tumor tissues with normal lung tissues were analyzed. 133 miRNAs were identified differentially expressed miRNAs with p value less than 0.05 after FDR adjustment (Supplementary Table S1). Among these, 85 miRNAs were up-regulated and 58 miRNAs were down-regulated.
Association of miRNAs expression and clinical parameters with overall survival of LUSC patients
We conducted univariate Cox regression assays to identify common miRNAs correlated with overall survival (OS) within each subclass of the following clinical parameters: pathologic N stage, pathologic T stage, and pathologic M stage. MiRNAs were selected if they were significantly correlated with OS in at least two subclasses. Twelve miRNAs were identified in this analysis. The hazard ratios (HR) for the association of miRNAs with OS in each category were shown in Table 2.
Table 2. MiRNAs associated with prognosis in different clinical subclasses.
| miRNA | T1-2 HR(95%CI) | T3-4 HR(95%CI) | N0 HR(95%CI) | N1-3 HR(95%CI) | M0 HR(95%CI) | M1 HR(95%CI) |
|---|---|---|---|---|---|---|
| miR-101-2 | - | - | - | 0.70(0.51-0.96) | 0.81(0.68-0.98) | - |
| miR-1269 | - | 0.90(0.82-0.98) | - | 0.93(0.87-0.99) | - | - |
| miR-138-1 | - | - | 0.86(0.75-0.99) | 1.20(1.02-1.40) | - | - |
| miR-139 | 1.28(1.08-1.51) | - | 1.25(1.03-1.51) | -- | 1.17(1.01-1.35) | - |
| miR-144 | - | - | 0.87(0.77-0.98) | - | 0.89(0.81-0.98) | - |
| miR-182 | 0.82(0.71-0.96) | - | - | 0.75(0.62-0.92) | 0.85(0.74-0.97) | - |
| miR-183 | 0.86(0.74-0.99) | - | - | 0.77(0.62-0.94) | - | - |
| miR-190 | 0.82(0.70-0.97) | 0.80(0.64-0.99) | - | 0.85(0.73-0.98) | - | |
| miR-195 | - | 0.58(0.38-0.87) | - | 0.73(0.56-0.97) | - | - |
| miR-326 | 1.13(1.02-1.26) | - | - | 1.17(1.02-1.33) | 1.13(1.03-1.25) | - |
| miR-451 | - | - | 0.87(0.77-0.98) | - | 0.91(0.83-0.99) | - |
| miR-944 | - | - | - | 0.91(0.83-0.98) | 0.93(0.88-0.99) | - |
Establishment of miRNA prognostic model
By using the supervised principal component (SPC) method, seven of the 12 miRNAs identified above were selected within the training set. Of these 7 miRNAs, 2 (hsa-mir-139, hsa-mir-326) were negatively correlated with survival, while the other 5 (miR-101-2, miR-182, miR-183, miR-190, hsa-miR-944) were protective. Next, we developed a miRNA prognostic model using the miRNA expression levels based on the log2 reads per million of total aligned miRNA reads. Risk-score = (-0.226 × expression value of hsa-miR-101-2) + (0.147 × expression value of hsa-miR-139) + (-0.084 × expression value of hsa-miR-182) + (-0.026 × expression value of hsa-miR-183) + (-0.125 × expression value of hsa-miR-190) + (0.036 × expression value of hsa-miR-326) + (-0.032 × expression value of hsa-miR-944). The 447 patients were separated into high-risk group or low-risk group using the optimum cutoff point of miRNA scores according to receiver operating characteristic (ROC) curve for predicting 5-year survival in the training set. As shown in Figure 1, high-risk miRNAs tended to up-regulated in patients with high scores, whereas protective miRNAs tended to high expressed in patients with low scores (Figure 1A and 1B).
Figure 1. Heatmap and predictor-score of the seven-microRNA signature in LUSC cohort.
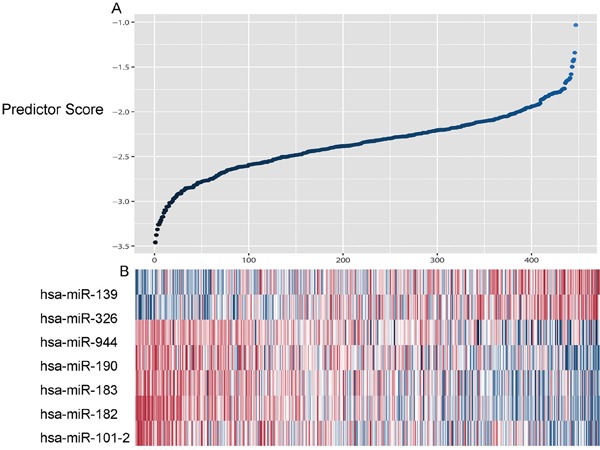
A. MicroRNA predictor-score distribution. B. Heatmap of the seven-miRNA expression profiles in LUSC patients.
Prognostic value of the seven- microRNA signature in LUSC
The ability of prognostic prediction of the seven-miRNA signature was then tested in the testing set and the entire LUSC cohort, respectively. Kaplan-Meier analysis revealed that patients in high-score group had poorer OS in both testing set (p=0.027, Figure 2A) and the entire LUSC cohort (p<0.001, Figure 3A).
Figure 2. Kaplan–Meier and ROC curves for the seven-miRNA signature in testing set.

A. The Kaplan–Meier curves for testing set (n = 223) divided by the optimum cutoff point. Patients with high scores had poorer outcome in terms of OS (Median OS: 629.6 days vs. 358.3 day, p=0.027. B. The ROC curve for predicting 60 months survival in the testing set.
Figure 3. Kaplan–Meier and ROC curves for the seven-miRNA signature in LUSC cohort.

A. The Kaplan–Meier curves for entire LUSC cohort divided by the optimum cutoff point. Patients with high scores had poorer outcome in terms of OS (Median OS: 2086 days vs. 947 day, p<0.001. B. The ROC curve for predicting 60 months survival in the LUSC cohort.
Time-dependent ROC curves were also applied to evaluate the prognostic value of the signature model. The area under ROC curve (AUC) of signature model for the testing set and the entire LUSC cohort was 0.604 (Figure 2B) and 0.610 (Figure 3B), respectively.
We also assessed the prognostic power of the seven-miRNA signature in early stage patients (stage I and II patients, n = 364). Kaplan-Meier analysis revealed that patients in high-risk group are associated with worse OS (p<0.001, log-rank test).
Finally, multivariate Cox regression analysis were used to investigate the independent prognostic value of the seven-miRNA signature. Age, gender, T stage, N stage and the miRNA signature were used as covariates. The miRNA signature (HR = 1.965, p < 0.001) and T stage (HR = 1.684, p =0.004) are showed to be as independent prognostic factors related with OS (Table 3).
Table 3. Multivariate analysis of overall survival of patients.
| Characteristic | HR(95%CI) | P value |
|---|---|---|
| Sex (male vs. female) | 0.800(0.566-1.133) | 0.209 |
| Age (< 65 vs. ≥ 65 years) | 1.262(0.908-1.755) | 0.166 |
| Smoking status | 0.995(0.735-1.347) | 0.995 |
| T stage (T1-2 vs T3-4) | 1.705(1.200-2.424) | 0.003 |
| N stage (N0 vs N1-3) | 0.773(0.421-1.419) | 0.406 |
| miRNA signature | 2.809(1.812-4.355) | <0.001 |
Correlation between miRNA signature and clinical characteristics
We examined the association of seven-miRNA signature with clinical factors in LUSC. No significant differences were observed when patients were stratified by gender, age and smoke status (Supplementary Table S2).
In silico analysis of target genes and pathways
The target genes of seven miRNAs was downloaded from miRecords. A total of 4242 target genes predicted by more than 4 data sets were selected for further analysis. Next, we performed a functional enrichment analysis to elucidate the biological function of target genes of seven miRNAs. A total of 39 Kyoto Encylopedia of Genes and Genomes (KEGG) pathways and 732 Gene Ontology (GO) pathways were enriched (Supplementary Table S3). The results showed that the predicted target genes involved in many important pathways associated with cancer development, e.g., adherens junction, Wnt, TGF-beta and MAPK signaling pathways (Table 4). Moreover, many target genes were enriched in cancer-related pathways for lung cancer.
Table 4. Results of over-representation analysis of the predicted target genes.
| Pathway | Target gene |
|---|---|
| Wnt signaling pathway | BTRC, FZD6, JUN, RAC1, WNT1, WNT2 |
| MAPK signaling pathway | FOS, TGFBR2, BDNF CACNA1C, MAPK9 |
| Adherens junction | ACTN4, CREBBP, CTNND1, TJP1, IQGAP1 |
| Apoptosis | FOXO1, IGF1R, MCL1, MITF, NOTCH1, NOTCH2, PTGS2, RARG |
| TGF-beta signaling pathway | ACVR2B, TGFBR1, TGFBR2 |
| VEGF signaling pathway | ITGA5, VEGFA, ITGB3 |
| Non-small cell lung cancer | CDK6, E2F3, FOXO3, GRB2, KRAS, MAPK1, PIK3R1, PRKCA, RARB, RASSF1, RXRA, RXRB, STK4, TGFA |
DISCUSSION
Accumulating evidence reveals that miRNAs could play a crucial role in tumorigenesis and progression of lung cancer. Many studies have reported the potential of miRNAs as biomarkers for early detection, molecular classification, prediction of outcome and treatment efficacy for lung cancer [7]. Although several studies had identified a number of miRNAs with prognostic value, most of these studies did not accurately analyze the expression of miRNAs in the different histotypes and stages. Since the miRNA expression patterns are quite different in different pathological types and tumor stages, the miRNAs identified in those studies may have less power in the clinical application for lung cancer outcome prediction.
In the current study, from the 133 differential expressed miRNAs, 12 miRNAs associate with OS of LUSC patients were identified in clinical subgroups. By using supervised principal components method, a seven-miRNA (miR-101-2, miR-139, miR-182, miR-183, miR-190, miR-326 and miR-944) signature was established and was validated to be an independent factor for outcome prediction for LUSC patient. This signature was demonstrated with high prognostic ability in both the entire LUSC set and the early stage patients.
Dysregulation of miR-139 has been observed in a variety of cancers [8]. Liu et al. reported that down-regulation of miR-139 was correlated with poor survival of colon cancer patients [9]. MiR-139 regulates diverse biological processes, such as proliferation, invasion and metastasis [8]. Low expression of miR-139 was significantly related with invasiveness and lymph node metastasis of NSCLC [10]. Wang et al. reported that downregulation of miR-101 accelerates the progression of lung cancer in RUNX1 dependent manner [11]. MiR-101 was also reported to suppress lung tumorigenesis through inhibition of DNMT3a [12]. By upregulating miR-101, Curcumin could inhibit the expression of EZH2, thus inhibiting lung cancer growth and metastasis [13]. MiR-190 could downregulate the expression of PH domain leucine-rich repeat protein phosphatase (PHLPP) and enhance the activation of Akt which leads to carcinogenesis [14]. Gennarino et al. reported that miR-190 could inhibit TGF-beta signaling and its effects on cell proliferation, morphology and scattering in NSCLC [15]. Recent studies reported that mir-326 was involved in carcinogenesis [16], metastasis, invasion of tumor[17, 18], and chemotherapy resistance [19]. MiR-326 was also identified as a biomarker for outcome prediction for prostate cancer and gastric cancer [20]. MiR-326 could regulate cell proliferation and migration of lung cancer by targeting phox2a [21], CCND1 [22] and NSBP [23]. Several reports of miR-182 highlighted its crucial roles in various cancers [24–26]. Stenvold et al. reported that up-regulation of miR-182 was associated with a good prognosis in LUSC [27]. By targeting different target genes, miR-182 could inhibit cell proliferation and invasion of LAD and LUSC [28–30]. Zhang et al. reported that serum miR-182 and miR-183 could be an effective biomarker for early diagnosis of NSCLC [31]. Powrozek et al. reported that plasma miR-944 could serve as biomarker for diagnosis of LUSC [32]. Through comprehensive analysis of characterization of LAD and LUSC, miR-944 was identified a potential drive-miRNA in classifying tumor histology [33]. Liu et al. reported that miR-944 affects cell growth by targeting EPHA7 [34] or SOCS4 [35] in NSCLC. Up-regulation of miR-944 is correlated with lymph node metastasis and advanced-stage of LUSC [35].
Based on the public data with large sample size, the prognostic value of miRNAs have been evaluated in other tumor types, including lung adenocarcinoma [36], gliomas [37], bladder cancer [38], pancreatic cancer [39], breast cancer [40], hepatocellular carcinoma [41], head and neck squamous cell carcinomas [42, 43], cervical cancer [44], melanoma [45]. Except miR-326 which have been reported as a member of a seven-miRNA signature predicting survival in hepatocellular carcinoma, the other six miRNAs identified in our study have not been recommended as biomarkers with best prognostic value in other tumor types. These results further supported the specificity of the seven-miRNA signature in predicting outcome for LUSC.
To gain a further insight into the functional role of the seven miRNAs, we retrieved their target genes and analyzed their related pathways. Bioinformatic analysis revealed that some important tumor-related genes are simultaneously by two or more miRNAs. Type 1 insulin-like growth factor receptor (IGF1R) is predicted to be the target gene of three miRNAs (miR-182, miR-326 and miR-944). The over-expression of IGF1R, which mediates tumor growth, adhesion, and protection from apoptosis, has been observed in various cancers [46]. ATRX is the predicted target of miR-101, miR-139 and miR-944. Misexpression of ATRX could modify the histone variant composition of chromatin by promoting genomic instability or gene expression changes related with tumorigenesis [47]. In addition, we also found that these miRNAs could regulate several crucial signaling pathways. Dysregulation of TGF-beta signaling pathway is important in cancer progression and cell invasion. Pajares et al. found that TGF-beta-induced protein expression was an independent outcome predictor for adjuvant-treated LUSC patients. Our informatics analysis identified several target genes that are involved in TGF-beta signaling pathway. Yang et al. found that high expression levels of VEGF-B is associated poor survival of LUSC patients. Three target genes related to VGEF signaling pathway have been identified. However, further molecular investigations are needed to confirm these predictions.
Nevertheless, some limitations may exist in the current study. Firstly, the censored rate of TCGA LUSC dataset was relatively high. This may have an impact on the reliability of the survival analysis. Secondly, the value of the miRNA signature should be validated in other independent cohort with long-term follow up.
In summary, our current study identified a seven-miRNA signature as potential outcome predictor for LUSC patients. Future studies using independent cohorts of large sample size from multiple institutions are needed to validate our findings for clinical practice. Future functional investigations are also required to explore the underlying mechanisms of these miRNAs in LUSC development.
MATERIALS AND METHODS
TCGA dataset
The LUSC dataset, including Level 3 miRNA expression data with clinical information, was downloaded from TCGA data portal (March 2016). To exclude unrelated causes of death, cases with less than 1-month follow-up less were excluded in the subsequent analysis.
Identification of dysregulated miRNAs in LUSC
The raw counts of miRNA expression data (Illumina HiSeq Systems) of 45 LUSC with their paired normal tissue were obtained from the TCGA dataset. MiRNA-expression data was normalized by using the R/Bioconductor package edgeR, which is designed for digital gene expression data [48]. MiRNAs with logFC (log2 fold change) < −1 or >1 (p less than 0.05, after FDR adjusted) were considered as differentially expressed miRNAs and were selected for further analysis.
Identification of miRNAs with prognostic value in LUSC
Semi-supervised method which combines the gene expression profile with clinical imformation was used to conduct univariate Cox regression analyses [49, 50]. Common miRNAs associated with OS were identified within each of the subgroups stratified by the TNM system. Common miRNAs identified in at least two independent subclasses were selected for the subsequent studies, using a HR>1 or HR<1 with p<0.05 as the cutoff.
Definition of prognostic risk model and ROC curve analysis
An importance score was calculated by the SPC method and was assigned to each miRNA [49]. Ten-fold cross validation was used to calculate the best threshold in SPC model and to select significant miRNAs. The TCGA dataset was separated into the training group and the testing group randomly. The linear signature prognostic model was developed based on the SPC method. Then, using the prognostic model, risk scores were compute for all the 447 patients. The best cutoff value of prognostic score was decided in the ROC curve analysis for predicting 5-year survival of the training set. The OS curves were evaluated using the Kaplan–Meier and log-rank method. Time-dependent ROC curves were also applied to assess the predict power of the prognostic model. All analyses were performed by the R/BioConductor (version 3.3.1).
Bioinformatic analysis of miRNA-target genes and pathways
Potential target genes of the candidate miRNAs were obtained from miRecords v4.0 (www.mirecords.biolead.org) database, which offers a comprehensive data of possible miRNA targets of 11 different data sets. The pathway enrichment analysis was conducted with the GeneTrail gene set enrichment tool. The results were considered significant when p value was less than 0.05 after FDR corrected [36] [51].
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Footnotes
CONFLICTS OF INTEREST
The authors declared that there is no conflicts of interest in this work.
REFERENCE
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Zhao H, Gao X, Wei F, Zhang X, Su Y, Wang C, Li H, Ren X. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget. 2016;7:26070–26086. doi: 10.18632/oncotarget.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 4.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Tsai NP, Lin YL, Wei LN. MicroRNA mir-346 targets the 5'-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem J. 2009;424:411–418. doi: 10.1042/BJ20090915. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 7.Munson R, Jr, Bailey C, Grass S. Diversity of the outer membrane protein P2 gene from major clones of Haemophilus influenzae type b. Mol Microbiol. 1989;3:1797–1803. doi: 10.1111/j.1365-2958.1989.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang HD, Jiang LH, Sun DW, Li J, Tang JH. MiR-139-5p: promising biomarker for cancer. Tumour Biol. 2015;36:1355–1365. doi: 10.1007/s13277-015-3199-3. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Duan B, Dong Y, He C, Zhou H, Sheng H, Gao H, Zhang X. MicroRNA-139-3p indicates a poor prognosis of colon cancer. Int J Clin Exp Pathol. 2014;7:8046–8052. [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe K, Amano Y, Ishikawa R, Sunohara M, Kage H, Ichinose J, Sano A, Nakajima J, Fukayama M, Yatomi Y, Nagase T, Ohishi N, Takai D. Histone methylation-mediated silencing of miR-139 enhances invasion of non-small-cell lung cancer. Cancer Med. 2015;4:1573–1582. doi: 10.1002/cam4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zhao Y, Qian H, Huang J, Cui F, Mao Z. The miR-101/RUNX1 feedback regulatory loop modulates chemo-sensitivity and invasion in human lung cancer. Int J Clin Exp Med. 2015;8:15030–15042. [PMC free article] [PubMed] [Google Scholar]
- 12.Yan F, Shen N, Pang J, Xie D, Deng B, Molina JR, Yang P, Liu S. Restoration of miR-101 suppresses lung tumorigenesis through inhibition of DNMT3a-dependent DNA methylation. Cell Death Dis. 2014;5:e1413. doi: 10.1038/cddis.2014.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu GQ, Chai KQ, Zhu XM, Jiang H, Wang X, Xue Q, Zheng AH, Zhou HY, Chen Y, Chen XC, Xiao JY, Ying XH, Wang FW, et al. Anti-cancer effects of curcumin on lung cancer through the inhibition of EZH2 and NOTCH1. Oncotarget. 2016;7:26535–26550. doi: 10.18632/oncotarget.8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beezhold K, Liu J, Kan H, Meighan T, Castranova V, Shi X, Chen F. miR-190-mediated downregulation of PHLPP contributes to arsenic-induced Akt activation and carcinogenesis. Toxicol Sci. 2011;123:411–420. doi: 10.1093/toxsci/kfr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gennarino VA, D'Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, Mutarelli M, Belcastro V, Ballabio A, Verde P, Sardiello M, Banfi S. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 2012;22:1163–1172. doi: 10.1101/gr.130435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kefas B, Comeau L, Floyd DH, Seleverstov O, Godlewski J, Schmittgen T, Jiang J, diPierro CG, Li Y, Chiocca EA, Lee J, Fine H, Abounader R, Lawler S, Purow B. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29:15161–15168. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valencia K, Martin-Fernandez M, Zandueta C, Ormazabal C, Martinez-Canarias S, Bandres E, de la Piedra C, Lecanda F. miR-326 associates with biochemical markers of bone turnover in lung cancer bone metastasis. Bone. 2013;52:532–539. doi: 10.1016/j.bone.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y, Kim H, Park H, Park D, Lee H, Lee YS, Choe J, Kim YM, Jeoung D. miR-326-histone deacetylase-3 feedback loop regulates the invasion and tumorigenic and angiogenic response to anti-cancer drugs. J Biol Chem. 2014;289:28019–28039. doi: 10.1074/jbc.M114.578229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, Wagar N, Yoon Y, Cho HT, Scala S, Shim H. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol. 2010;79:817–824. doi: 10.1016/j.bcp.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Kristensen H, Thomsen AR, Haldrup C, Dyrskjot L, Hoyer S, Borre M, Mouritzen P, Orntoft TF, Sorensen KD. Novel diagnostic and prognostic classifiers for prostate cancer identified by genome-wide microRNA profiling. Oncotarget. 2016;7:30760–30771. doi: 10.18632/oncotarget.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, Chen X, Xu T, Xia R, Han L, Chen W, De W, Shu Y. MiR-326 regulates cell proliferation and migration in lung cancer by targeting phox2a and is regulated by HOTAIR. Am J Cancer Res. 2016;6:173–186. [PMC free article] [PubMed] [Google Scholar]
- 22.Sun C, Huang C, Li S, Yang C, Xi Y, Wang L, Zhang F, Fu Y, Li D. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget. 2016;7:8341–8359. doi: 10.18632/oncotarget.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D, Du X, Liu A, Li P. Suppression of nucleosome-binding protein 1 by miR-326 impedes cell proliferation and invasion in non-small cell lung cancer cells. Oncol Rep. 2016;35:1117–1124. doi: 10.3892/or.2015.4403. [DOI] [PubMed] [Google Scholar]
- 24.Lei R, Tang J, Zhuang X, Deng R, Li G, Yu J, Liang Y, Xiao J, Wang HY, Yang Q, Hu G. Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis. Oncogene. 2014;33:1287–1296. doi: 10.1038/onc.2013.65. [DOI] [PubMed] [Google Scholar]
- 25.Yang MH, Yu J, Jiang DM, Li WL, Wang S, Ding YQ. microRNA-182 targets special AT-rich sequence-binding protein 2 to promote colorectal cancer proliferation and metastasis. J Transl Med. 2014;12:109. doi: 10.1186/1479-5876-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Ren R, Hu H, Tan C, Han M, Wang X, Zheng Y. MiR-182 is up-regulated and targeting Cebpa in hepatocellular carcinoma. Chin J Cancer Res. 2014;26:17–29. doi: 10.3978/j.issn.1000-9604.2014.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stenvold H, Donnem T, Andersen S, Al-Saad S, Busund LT, Bremnes RM. Stage and tissue-specific prognostic impact of miR-182 in NSCLC. BMC Cancer. 2014;14:138. doi: 10.1186/1471-2407-14-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, Wang Y, Zang W, Wang H, Chu H, Li P, Li M, Zhang G, Zhao G. Downregulation of microRNA-182 inhibits cell growth and invasion by targeting programmed cell death 4 in human lung adenocarcinoma cells. Tumour Biol. 2014;35:39–46. doi: 10.1007/s13277-013-1004-8. [DOI] [PubMed] [Google Scholar]
- 29.Zhu YJ, Xu B, Xia W. Hsa-mir-182 downregulates RASA1 and suppresses lung squamous cell carcinoma cell proliferation. Clin Lab. 2014;60:155–159. doi: 10.7754/clin.lab.2013.121131. [DOI] [PubMed] [Google Scholar]
- 30.Sun Y, Fang R, Li C, Li L, Li F, Ye X, Chen H. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem Biophys Res Commun. 2010;396:501–507. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- 31.Zhu W, Zhou K, Zha Y, Chen D, He J, Ma H, Liu X, Le H, Zhang Y. Diagnostic Value of Serum miR-182, miR-183, miR-210, and miR-126 Levels in Patients with Early-Stage Non-Small Cell Lung Cancer. PLoS One. 2016;11:e0153046. doi: 10.1371/journal.pone.0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powrozek T, Krawczyk P, Kowalski DM, Winiarczyk K, Olszyna-Serementa M, Milanowski J. Plasma circulating microRNA-944 and microRNA-3662 as potential histologic type-specific early lung cancer biomarkers. Transl Res. 2015;166:315–323. doi: 10.1016/j.trsl.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Lazar V, Suo C, Orear C, van den Oord J, Balogh Z, Guegan J, Job B, Meurice G, Ripoche H, Calza S, Hasmats J, Lundeberg J, Lacroix L, Vielh P, Dufour F, Lehtio J, et al. Integrated molecular portrait of non-small cell lung cancers. BMC Med Genomics. 2013;6:53. doi: 10.1186/1755-8794-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M, Zhou K, Cao Y. MicroRNA-944 Affects Cell Growth by Targeting EPHA7 in Non-Small Cell Lung Cancer. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17101493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J, Mannoor K, Gao L, Tan A, Guarnera MA, Zhan M, Shetty A, Stass SA, Xing L, Jiang F. Characterization of microRNA transcriptome in lung cancer by next-generation deep sequencing. Mol Oncol. 2014;8:1208–1219. doi: 10.1016/j.molonc.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Shi Y, Yin Z, Xue X, Zhou B. An eight-miRNA signature as a potential biomarker for predicting survival in lung adenocarcinoma. J Transl Med. 2014;12:159. doi: 10.1186/1479-5876-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan W, Li R, Liu Y, Yang P, Wang Z, Zhang C, Bao Z, Zhang W, You Y, Jiang T. MicroRNA expression patterns in the malignant progression of gliomas and a 5-microRNA signature for prognosis. Oncotarget. 2014;5:12908–12915. doi: 10.18632/oncotarget.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou H, Tang K, Xiao H, Zeng J, Guan W, Guo X, Xu H, Ye Z. A panel of eight-miRNA signature as a potential biomarker for predicting survival in bladder cancer. J Exp Clin Cancer Res. 2015;34:53. doi: 10.1186/s13046-015-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou X, Huang Z, Xu L, Zhu M, Zhang L, Zhang H, Wang X, Li H, Zhu W, Shu Y, Liu P. A panel of 13-miRNA signature as a potential biomarker for predicting survival in pancreatic cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X, Wang X, Huang Z, Xu L, Zhu W, Liu P. An ER-associated miRNA signature predicts prognosis in ER-positive breast cancer. J Exp Clin Cancer Res. 2014;33:94. doi: 10.1186/s13046-014-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Chong CC, Chen GG, Lai PB. A Seven-microRNA Expression Signature Predicts Survival in Hepatocellular Carcinoma. PLoS One. 2015;10:e0128628. doi: 10.1371/journal.pone.0128628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong N, Khwaja SS, Baker CM, Gay HA, Thorstad WL, Daly MD, Lewis JS, Jr, Wang X. Prognostic microRNA signatures derived from The Cancer Genome Atlas for head and neck squamous cell carcinomas. Cancer Med. 2016;5:1619–1628. doi: 10.1002/cam4.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi H, Chen J, Li Y, Li G, Zhong R, Du D, Meng R, Kong W, Lu M. Identification of a six microRNA signature as a novel potential prognostic biomarker in patients with head and neck squamous cell carcinoma. Oncotarget. 2016;7:21579–21590. doi: 10.18632/oncotarget.7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B, Ding JF, Luo J, Lu L, Yang F, Tan XD. Seven protective miRNA signatures for prognosis of cervical cancer. Oncotarget. 2016;7:56690–56698. doi: 10.18632/oncotarget.10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jayawardana K, Schramm SJ, Tembe V, Mueller S, Thompson JF, Scolyer RA, Mann GJ, Yang J. Identification, Review, and Systematic Cross-Validation of microRNA Prognostic Signatures in Metastatic Melanoma. J Invest Dermatol. 2016;136:245–254. doi: 10.1038/JID.2015.355. [DOI] [PubMed] [Google Scholar]
- 46.Ribeiro TC, Jorge AA, Montenegro LR, Almeida MQ, Ferraz-de-Souza B, Nishi MY, Mendonca BB, Latronico AC. Effects of Type 1 Insulin-Like Growth Factor Receptor Silencing in a Human Adrenocortical Cell Line. Horm Metab Res. 2016 doi: 10.1055/s-0042-108196. [DOI] [PubMed] [Google Scholar]
- 47.Watson LA, Goldberg H, Berube NG. Emerging roles of ATRX in cancer. Epigenomics. 2015;7:1365–1378. doi: 10.2217/epi.15.82. [DOI] [PubMed] [Google Scholar]
- 48.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi H, Chen J, Li Y, Li G, Zhong R, Du D, Meng R, Kong W, Lu M. Identification of a six microRNA signature as a novel potential prognostic biomarker in patients with head and neck squamous cell carcinoma. Oncotarget. 2016;7:21579–21590. doi: 10.18632/oncotarget.7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP. GeneTrail--advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


