Abstract
Salmonella spp. is an important zoonotic pathogen related to foodborne diseases. Despite that quinolones/fluoroquinolones are considered a relevant therapeutic strategy against resistant isolates, the increase in antimicrobial resistance is an additional difficulty in controlling bacterial infections caused by Salmonella spp. Thus, the acquisition of resistance to quinolones in Salmonella spp. is worrisome to the scientific community along with the possibility of transmission of resistance through plasmids. This study investigated the prevalence of plasmid-mediated quinolone resistance (PMQR) in Salmonella spp. and its association with fluoroquinolone susceptibility in Brazil. We evaluated 129 isolates, 39 originated from food of animal sources, and 14 from environmental samples and including 9 from animals and 67 from humans, which were referred to the National Reference Laboratory of Enteric Diseases (NRLEB/IOC/RJ) between 2009 and 2013. These samples showed a profile of resistance for the tested quinolones/fluoroquinolones. A total of 33 serotypes were identified; S. Typhimurium (63) was the most prevalent followed by S. Enteritidis (25). The disk diffusion test showed 48.8% resistance to enrofloxacin, 42.6% to ciprofloxacin, 39.53% to ofloxacin, and 30.2% to levofloxacin. According to the broth microdilution test, the resistance percentages were: 96.1% to nalidixic acid, 64.3% to enrofloxacin, 56.6% to ciprofloxacin, 34.1% to ofloxacin, and 30.2% to levofloxacin. Qnr genes were found in 15 isolates (8 qnrS, 6 qnrB, and 1 qnrD), and the aac(6′)-Ib gene in 23. The integron gene was detected in 67 isolates with the variable region between ±600 and 1000 bp. The increased detection of PMQR in Salmonella spp. is a serious problem in Public Health and must constantly be monitored. Pulsed-field gel electrophoresis was performed to evaluated clonal profile among the most prevalent serovars resistant to different classes of quinolones. A total of 33 pulsotypes of S. Typhimurium were identified with a low percentage of genetic similarity (≤65%). This result demonstrates the presence of high diversity in the resistant clones evaluated in this study.
Keywords: foodborne diseases, Salmonella spp., quinolone resistance, plasmid mediated quinolone resistance, clonal profile
Introduction
Foodborne diseases caused by Salmonella spp. are a serious public health problem in many parts of the world. The variety of food sources, particularly foods of animal origin, and routes of transmission can lead to human infection (Scallan et al., 2011).
In addition, the progressive increase of antimicrobial resistance in foodborne Salmonella isolates is observed as due to the uncontrolled use of these drugs for therapeutic and prophylactic purposes in foods of animal origin such as poultry, pigs, and cattle. These events have reinforced the need for epidemiological studies describing the prevalence and patterns of resistance in these bacteria (Yang et al., 2010; Tamang et al., 2011). Antimicrobial-resistant bacteria emerge from the use of antimicrobial drugs to treat and prevent diseases and promote growth in large-scale animal production.
Quinolones, particularly fluoroquinolones, are commonly used for the treatment of multi-drug resistant salmonellosis “in human and veterinary medicine” because of their broad spectrum antimicrobial activity (Dalhoff, 2012).
Point mutations in DNA gyrase and topoisomerase IV genes are directly related to quinolone resistance in Enterobacteriaceae “by changes in the action target site called quinolone resistance-determining regions (QRDR).” “In Salmonella spp., these mutations are related to resistance to nalidixic acid (NAL) and reduced susceptibility to FQs such as that of ciprofloxacin (Cip) (Cavaco and Aarestrup, 2009).” It is believed that the resistance to quinolones is mediated only by this mechanism. However, the situation changed with the discovery of a variety of determinants of plasmid-mediated quinolone resistance (PMQR).
Currently three mechanisms are recognized as PMQRs. The qnr genes with five different qnr families, each with different numbers of alleles “(qnrA1–7, qnrS1–4, qnrB1–31, qnrC, and qnrD) (Jacoby et al., 2009)”; “a modified aminoglycoside acetyl-transferase gene [aac(6′)-1b-cr] (Robicsek et al., 2005)”; and a specific quinolone efflux pump (qepA) (Yamane et al., 2007) and multidrug resistance pumps such as oqxAB (Zhao et al., 2010). PMQR-positive isolates present a low-level of resistance to quinolones (only a small reduction in susceptibility to nalidixic acid). However, the ability to highlight pre-existing resistance mechanisms, such as chromosomal mutations in target regions of quinolones that still allow the selection of resistant mutants to quinolone concentrations (therapeutic doses), emphasize the importance of studying these genes (Cui et al., 2014).
The present study identified the occurrence of some PMQR in Salmonella spp. isolated between 2009 and 2013 from the food chain in Brazil, and characterized the genetic similarity profile of serovars of greatest importance in the dispersion of resistance to quinolones in Brazil.
Materials and methods
Bacterial isolates
129 Salmonella spp. strains with resistance to quinolone and/or fluoroquinolone were evaluated. Of these, 51.9% (67/129) were from human clinical isolates, 30.2% (39/129) from food products for human consumption (beef, eggs, milk), 7.1% (9/129) from food of animal origin for human consumption (poultry, swine, cattle), and 10.8% (14/129) from environmental samples (water and drag swabs); all samples were selected from a database. The studied strains were sent to the National Reference Laboratory of Enteric Diseases (NRLEB/IOC/RJ) between 2009 and 2013 and stored in phosphate-buffered agar at room temperature and/or in BHI/glycerol broth −70°C. The isolates were inoculated in Nutrient Broth (DIFCO) and incubated at 37°C for 12–18 h for subsequent tests, as confirmation of the biochemical, serological and antimicrobial resistance profile.
Antigenic characterization
The serological determination of Salmonella serotypes was determined according to the Kauffmann-White scheme using slide agglutination with O and H antisera prepared in the LRNEB/IOC/RJ.
Antimicrobial susceptibility
The obtained resistance profiles were confirmed by the disk diffusion test according to Clinical and Laboratory Standards Institute (2013, 2014). According to Pribul et al. (2016), this test was performed using representatives of the quinolone class (OXOID) for human and veterinary therapeutic use, such as Nalidixic Acid (NAL), Ciprofloxacin (CIP), Enrofloxacin (ENO), Ofloxacin (OFL), and Levofloxacin (LVX).
MIC determinations for Nalidixic Acid (SIGMA), Ciprofloxacin (SIGMA), Enrofloxacin (SIGMA), Levofloxacin (SIGMA), and Ofloxacin (SIGMA) were performed in 96-well-microplates and according to the Clinical and Laboratory Standards Institute (2013) broth microdilution assay.
Detection of PMQR
Total DNA was extracted using the DNEASY Tissue Qiagen® kit. The studied genes were detected by PCR amplification using primer sequences reported in Pribul et al. (2016). The qnrA, qnrB, and qnrS genes were amplified through multiplex PCR reactions; the rrs gene was used as the reaction control. The qnrC, qnrD, aac(6′)-Ib, integrase, and variable integron region genes were amplified by simplex PCR.
PFGE
The isolates from serovars S. Typhimurium, S. Muenchen, S. Infantis, and S. Heidelberg were subjected to molecular typing by pulsed-field gel electrophoresis, which clonally evaluates isolates. The PulseNet protocol was used in this study including DNA preparation according to Heir et al. (2000) and digestion with XbaI restriction enzyme, according to Pfaller et al. (1992), Tenover et al. (1997), and Cooper et al. (2006). The definition of clones was based on the recommendations of Tenover et al. (1997) and Barrett et al. (2006). S. Braenderup H9812, which is considered the universal strain for PulseNet (Hunter et al., 2005), was used as the standard. The restriction patterns were analyzed in the BioNumerics software IV (Applied Maths).
Results
Serovar identified
Altogether, 26 different Salmonella serovars were identified. Salmonella Typhimurium (48.8%, 63/129) was the predominant serovar followed by Salmonella Enteritidis (19.4%, 25/129). The prevalent serovars associated with resistance to quinolones are presented in Table 1.
Table 1.
Distribution of quinolone-resistant Salmonella spp. serovars isolated from food chain diseases.
| Salmonella spp. Serotype | Number of NTSa from | ||||
|---|---|---|---|---|---|
| Human | Food | Environment | animal | Total | |
| S. Typhimurium | 35 | 22 | 4 | 2 | 63 |
| S. Enteritidis | 24 | 1 | – | – | 25 |
| S. Muenchen | 2 | 2 | – | – | 4 |
| S. Infantis | 1 | 1 | – | – | 3 |
| S. Heidelberg | 2 | – | – | 1 | 3 |
| Others | 3 | 11 | 8 | 5 | 26 |
| Total | 67 | 37 | 12 | 8 | |
Non-typhoidal Salmonella.
Most of the studied samples were isolated in 2012 (88 of 129).
Among these 129 isolates that were previously resistant to Nalidixic Acid, five were sensitive to all tested quinolones (including Nalidixic Acid), 55 (42.6%) were resistant to Ciprofloxacin, 63 (48.8%) to Enrofloxacin, 51 (39.53%) to Ofloxacin, and 48 (37.2%) to Levofloxacin in the disc diffusion test.
The broth microdilution test identified 36.4% (47/129) isolates with decreased susceptibility to Ciprofloxacin (MICs between 0.125 and 0.5 mg/ml), 20.1% (26/129) to Enrofloxacin, 9.3% (12/129) to Ofloxacin, and 6.2% (8/129) to Levofloxacin (MICs between 0.5 and 1 mg/ml). The decreased susceptibility breakpoint to Nalidixic Acid is not reported by Clinical and Laboratory Standards Institute (2015). Seventy-three (56.6%) isolates were resistant to Ciprofloxacin, 83 (64.3%) to Enrofloxacin, 44 (34.1%) to Ofloxacin, and 39 (30.2%) to Levofloxacin. A total of 124 isolates (96.1%) were resistant to Nalidixic Acid.
The resistance profile obtained with the microdilution test showed that 37 (28.7%) isolates were resistant to all tested quinolones, 30 (23.2%) to Ciprofloxacin, Enrofloxacin, and Nalidixic Acid, 16 (12.4%) to Enrofloxacin and Nalidixic Acid, 2 (1.5%) to Ciprofloxacin and Nalidixic Acid, and 39 (30.2%) to Nalidixic Acid only.
The detection of resistance genes showed six isolates carrying the qnrB gene, eight the qnrS gene, and one the qnrD gene. Among these 15 positive isolates, 10 strains were recovered from human samples, 3 from food of animal origin, 1 from environmental samples, and 1 from animal samples. The most qnr-positive prevalent serovar was S. Typhimurium followed by S. Saintpaul and S. Livingstone. None of the isolates presented the qnrA or qnrC genes.
A total of 23 isolates showed the aac(6′)-Ib gene, which is prevalent in S. Typhimurium (14/23). The most prevalent source of isolation was human (10/23), followed by foodborne (7/23), animal (3/23), and amibental (3/23). Thirteen isolates aac(6′)-Ib positive were resistant to all tested quinolones.
Three qnr-positive isolates presented the aac(6′)-Ib gene in association: two S. Typhimurium and one S. Saintpaul. These two Salmonella ser. Typhimurium were resistant to all tested quinolones/fluoroquinolones in the broth microdilution assay at the highest concentration.
Sixty-seven isolates showed the presence of integrase gene within 600 to 1000 bp variable region range and were mainly identified in human samples (38/67) followed by food samples (15/67), ambiental (8/67), and animal (6/67). The S. Typhimurium serovar was the most frequent (39/67) isolate with the conserved region of class 1 integron and variable regions between ±900 and >1000 bp. Nine isolates of serovar S. Typhimurium carrying the aac(6′)-Ib gene were positive for the integron with ±900 bp; two of these were also positive for qnr.
Figure 1 shows the clonal profile comparison between the quinolones/fluoroquinolones resistant strains and other sensitive strains.
Figure 1.
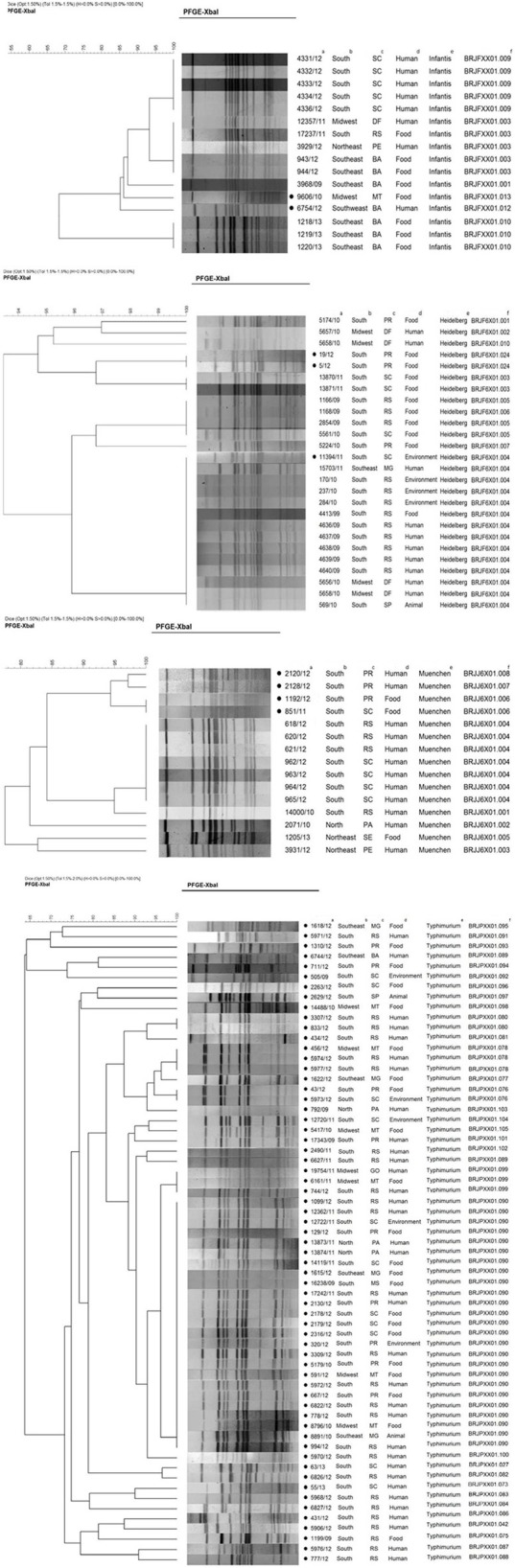
Distribution of phylogenetic groups of Salmonella ser. Infantis, Heidelberg, Muenchen, and Typhimurium according to PFGE. Isolates with resistance to quinolones; aID of IOC/year of isolation; bregion; cstate; dsource of isolation; eserovar; fpulsotype.
The genetic similarity among isolates with resistance to quinolones was approximated 84% in S. Infantis, 92% in S. Heidelberg, 88% in S. Muenchen, and 63% in S. Typhimurium despite their isolation in different periods, regions, and sources.
Thirty-three distinct pulsotypes were identified among strains with low percentage genetic similarity in serovar S. Typhimurium (≤65%), representing the highest diversity among resistant clones.
The Table 2 presents resistance profiles obtained with the microdilution test, detection of PMQR, size of variable integron region, and the pulsotype identified by the pfge technique.
Table 2.
Resistance profile and resistance genes in isolates evaluated by PFGE.
| Serovar | Source | IOC ID/Yeard | Pulsotype | PMQRc | Integron (bp) | Resistance profile (MICb) |
|---|---|---|---|---|---|---|
| Infantis | F | 6754/12 | BRJFXX01.13 | qnrD | – | CIP NAL ENOa |
| Infantis | H | 9606/10 | BRJFXX01.012 | – | – | NAL |
| Heidelberg | F | 19/12 | BRJF0X01.024 | – | 700 | CIP NAL ENO |
| Heidelberg | F | 5/12 | BRJF0X01.024 | – | – | CIP NAL ENO |
| Heidelberg | H | 11394/11 | BRJF0X01.004 | – | 900 | CIP NAL ENO |
| Muenchen | H | 2120/12 | BRJJ6X01.008 | qnrS | – | NAL ENO |
| Muenchen | H | 2128/12 | BRJJ6X01.007 | qnrS | – | NAL ENO |
| Muenchen | F | 1192/12 | BRJJ6X01.006 | aac(6′)-Ib | 600 | NAL ENO |
| Muenchen | F | 851/11 | BRJJ6X01.006 | - | 700 | CIP NAL ENO OFL |
| Typhimurium | F | 1618/12 | BRJPXX01.095 | aac(6′)-Ib | – | CIP NAL ENO LVX OFL |
| Typhimurium | H | 5971/12 | BRJPXX01.091 | aac(6′)-Ib | 1000 | NAL |
| Typhimurium | F | 1310/12 | BRJPXX01.093 | qnrS | – | NAL ENO |
| Typhimurium | H | 6744/12 | BRJPXX01.089 | qnrB | 1000 | NAL ENO |
| Typhimurium | F | 711/12 | BRJPXX01.094 | qnrD | – | NAL |
| Typhimurium | E | 505/09 | BRJPXX01.092 | – | 900 | CIP NAL ENO |
| Typhimurium | F | 2263/12 | BRJPXX01.096 | – | – | CIP NAL ENO LVX OFL |
| Typhimurium | A | 2629/12 | BRJPXX01.097 | aac(6′)-Ib | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | F | 14488/10 | BRJPXX01.098 | – | 900 | CIP NAL ENO |
| Typhimurium | H | 3307/12 | BRJPXX01.080 | – | 1000 | CIP NAL ENO |
| Typhimurium | H | 833/12 | BRJPXX01.080 | – | – | CIP NAL ENO |
| Typhimurium | H | 434/12 | BRJPXX01.081 | – | – | CIP NAL ENO |
| Typhimurium | F | 456/12 | BRJPXX01.078 | – | 1000 | NAL |
| Typhimurium | H | 5974/12 | BRJPXX01.078 | – | 1000 | NAL OFL |
| Typhimurium | H | 5977/12 | BRJPXX01.078 | – | 900 | CIP NAL ENO OFL |
| Typhimurium | F | 1622/12 | BRJPXX01.077 | – | – | NAL ENO |
| Typhimurium | F | 43/12 | BRJPXX01.076 | – | – | NAL |
| Typhimurium | E | 5973/12 | BRJPXX01.076 | – | 1000 | CIP NAL ENO |
| Typhimurium | H | 792/09 | BRJPXX01.103 | – | 900 | CIP NAL |
| Typhimurium | E | 12720/11 | BRJPXX01.104 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | F | 5417/10 | BRJPXX01.105 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 17343/09 | BRJPXX01.101 | – | 1000 | CIP NAL ENO |
| Typhimurium | H | 2490/11 | BRJPXX01.102 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 6627/11 | BRJPXX01.089 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 19754/11 | BRJPXX01.099 | – | – | CIP NAL ENO LVX OFL |
| Typhimurium | F | 6161/11 | BRJPXX01.099 | – | – | CIP NAL ENO LVX OFL |
| Typhimurium | H | 744/12 | BRJPXX01.099 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 1099/12 | BRJPXX01.090 | – | 1000 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 12362/11 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | E | 12722/11 | BRJPXX01.090 | – | – | CIP NAL ENO LVX OFL |
| Typhimurium | F | 129/12 | BRJPXX01.090 | aac(6′)-Ib | – | CIP NAL ENO LVX OFL |
| Typhimurium | H | 13873/11 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 13874/11 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | F | 14119/11 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | F | 1615/12 | BRJPXX01.090 | – | – | CIP NAL ENO LVX OFL |
| Typhimurium | F | 16238/09 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 17242/11 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 2130/12 | BRJPXX01.090 | aac(6′)-Ib | – | CIP NAL ENO LVX OFL |
| Typhimurium | F | 2178/12 | BRJPXX01.090 | aac(6′)-Ib | – | CIP NAL ENO LVX OFL |
| Typhimurium | F | 2179/12 | BRJPXX01.090 | aac(6′)-Ib | – | CIP NAL ENO LVX OFL |
| Typhimurium | F | 2316/12 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | E | 320/12 | BRJPXX01.090 | aac(6′)-Ib | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 3309/12 | BRJPXX01.090 | qnrB/aac(6′)-Ib | 900 | CIP NAL ENO |
| Typhimurium | F | 5179/10 | BRJPXX01.090 | – | – | CIP NAL ENO |
| Typhimurium | F | 591/12 | BRJPXX01.090 | – | 900 | NAL |
| Typhimurium | H | 5972/12 | BRJPXX01.090 | aac(6′)-Ib | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | F | 667/12 | BRJPXX01.090 | – | – | CIP NAL ENO |
| Typhimurium | H | 6822/12 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 778/12 | BRJPXX01.090 | qnrB | – | CIP NAL ENO LVX OFL |
| Typhimurium | F | 8796/10 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | A | 8891/10 | BRJPXX01.090 | – | 900 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 994/12 | BRJPXX01.090 | – | 900 | CIP NAL ENO OFL |
| Typhimurium | H | 5970/12 | BRJPXX01.100 | aac(6′)-Ib | – | CIP NAL ENO LVX OFL |
| Typhimurium | H | 63/13 | BRJPXX01.027 | – | 1000 | CIP NAL ENO |
| Typhimurium | H | 6826/12 | BRJPXX01.082 | – | 1000 | NAL |
| Typhimurium | H | 55/13 | BRJPXX01.073 | qnrD | – | CIP NAL ENO LVX OFL |
| Typhimurium | H | 5968/12 | BRJPXX01.083 | – | 1000 | NAL |
| Typhimurium | H | 6827/12 | BRJPXX01.084 | – | 1000 | NAL |
| Typhimurium | H | 431/12 | BRJPXX01.086 | – | – | – |
| Typhimurium | H | 5906/12 | BRJPXX01.042 | – | – | NAL |
| Typhimurium | F | 1199/09 | BRJPXX01.075 | – | 600 | CIP NAL ENO |
| Typhimurium | H | 5976/12 | BRJPXX01.087 | aac(6′)-Ib | 700 | CIP NAL ENO LVX OFL |
| Typhimurium | H | 777/12 | BRJPXX01.088 | qnrB/aac(6′)-Ib | – | CIP NAL ENO LVX OFL |
CIP, Ciprofloxacin; ENO, Enrofloxacin; NAL, Nalidixic Ácid; LVX, Levofloxacina; OFL, Ofloxacin;
MIC, Minimum Inhibitory Concentration;
PMQR, Plasmid-Mediated Quinolone Resistance;
IOC ID/Year, Institut Oswaldo Cruz Identification by Year;
eF, Food; A, Animal; E, Environment; H, Human.
Discussion
The variation in resistance to the different tested quinolones can be explained by the mechanism of resistance when the resistance level depends on the affected target enzyme, the number of accumulated mutations, and presence of PMQRs. Furthermore, there is a relationship between the level of specific resistance and potency of each drug, especially in the newest quinolones (Sanders, 2001; Ruiz et al., 2012).
Chong et al. (2010) reported that an increased resistance to fluoroquinolones based on the acquisition of qnr genes could be related with reduction in the clinical efficacy “of this class” of antimicrobial. “However, Jacoby et al. (2009)” argue that the genes involved in plasmid-level resistance to fluoroquinolones are still poorly understood when compared to other resistance mechanisms.
A high prevalence of isolates carrying PRQM genes is reported in the present study (27%, 35/129). The most prevalent serovar associated with the presence of PMQR genes was Salmonella ser. Typhimurium (18/35). A high level of detection of S. Typhimurium was expected because this serovar is directly related to detected genotypic and phenotypic profiles of antimicrobial resistance (Herrero et al., 2008; Kingsley et al., 2009). The presence of PMQR genes is related “to decreased susceptibility to fluoroquinolones,” accelerating the selection of fluoroquinolone-resistant mutants (Rodríguez-Martínez et al., 2011).
Three isolates presented association between the qnr and aac(6′)-Ib genes. A similar association has been reported by Park et al. (2006) in the United States, Xiong et al. (2011) while investigating the aac(6′)-Ib and qnr genes in Enterobacter cloacae “in China, and” Kim et al. (2013) in enterobacteria isolated from clinical samples in Korea. Not sequencing the aac(6′)-Ib gene to determine the cr variant was one limitation in the present study.
Regardless that some authors recognize the location of the aac(6′)-Ib gene mostly in class 1 integrons, our results show the absence of this gene in all analyzed strains [12 aac(6′)-Ib positive isolates without the integron region] (Rodríguez-Martínez et al., 2011; Kim et al., 2013).
The Enteritidis serovar was not assessed by PFGE because, according to the literature, these isolates have low clonal diversity (Spiliopoulou et al., 2007).
Six distinct pulsotypes were detected in S. Infantis serovar isolates. Those with resistance to quinolones are placed in two separate pulsotypes (BRJFXX01.13 and BRJFXX01.12) with a genetic similarity of ~85%. The quinolone resistance isolates were obtained from different sources, regions, and periods, and the resistance to quinolones showed variations. The 6754/12 isolate showed resistance to ciprofloxacin, nalidixic acid, and enrofloxacin and carried the qnrD gene; the 9606/10 isolate showed resistance to nalidixic acid only and did not carry resistant genes.
The S. Heidelberg serovar presented eight distinct pulsotypes. The resistant isolates showed a clonal ratio of 100% similarity between the isolates 5/12 and 19/12, and ~94% between them and isolate 11394/11. The 11394/11 isolate (environmental source from the Southern region) was detected in the BRJF6X01.004 pulsotype (pulsotype with 13 susceptible isolates). Isolates 5/12 and 19/12, within the same pulsotype, were foodborne and originated in the Southern region.
Three pulsotypes were identified in serovar Muenchen isolates with quinolone resistance profiles (pulsotypes BRJJ6X01008, BRJJ6X01007, and BRJJ6X01006), showing 88% of genetic similarity. Among the isolates resistant, the isolates of human origin presenting a profile similarity of ~97%. The isolates of foodborne origin presenting the same origin clonal being from different periods and states. The 2128/12, 2120/12, and 1192/12 isolates show similar quinolone resistance profiles. However, isolate 851/11 show a resistance profile to ciprofloxacin and ofloxacin. The 2120/12 and 2128/12 isolates show similar resistance profile and are carriers of the qnrS gene.
The detection of 33 different pulsotypes of S. Typhimurium indicates that different clones with resistance to quinolones are circulating in Brazil. The most prevalent pulsotype (BRJPXX01.090) is mainly represented in samples from the Southern region and are related to food and human sources. Most isolates of this pulsotype show the same resistance profile to quinolones/fluoroquinolones (except isolates 3309/11, 5179/10, 591/12, and 667/12) demonstrating a resistance profile to all tested quinolones/fluoroquinolones. Out of the 25 isolates showing resistance to quinolones, 4 did not carry resistance genes. Twelve isolates show an integron with a variable region of 900 bp and one with >1000 bp. One isolate shows the qnrB gene and, four show the aac(6′)-Ib gene. Two isolates show the 900 bp integron and the aac(6′)-Ib gene; one isolate shows the 900 bp integron and the qnrB, acc(6′)-Ib gene.
The profiles identified in the PFGE analysis show relatively high diversity in all serovars and, indicate that cases of resistance to quinolones are probably sporadic. This interpretation is in accordance with other results reported in the literature (Feasey et al., 2012).
Author contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Barrett T. J., Gerner-Smidt P., Swaminathan B. (2006). Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog. Dis. 3, 20–31. 10.1089/fpd.2006.3.20 [DOI] [PubMed] [Google Scholar]
- Cavaco L. M., Aarestrup F. M. (2009). Evaluation of quinolones for use in detection of determinants of acquired quinolone resistance, including the new transmissible resistance mechanisms qnrA, qnrB, qnrS, and aac(6′)Ib-cr, in Escherichia coli and Salmonella enterica and determinations of wild-type distributions. J. Clin. Microbiol. 47, 2751–2758. 10.1128/JCM.00456-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Y. P., Choi S. H., Kim E. S. (2010). Bloodstream infections caused by qnr-positive Enterobacteriaceae: clinical and microbiologic characteristics and outcomes. Diagn. Microbiol. Infect. Dis. 67, 70–77. 10.1016/j.diagmicrobio.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2013). Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standards. CLSI document M2-A3. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Clinical and Laboratory Standards Institute (2014). Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standards. CLSI document M2-A3. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Clinical and Laboratory Standards Institute (2015). Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standards. CLSI document M2-A3. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cooper K. L., Luey C. K., Bird M., Terajima J., Nair G. B., Kam K. M., et al. (2006). Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog. Dis. 3, 51–58. 10.1089/fpd.2006.3.51 [DOI] [PubMed] [Google Scholar]
- Cui S. F., Peng L. P., Zhang H. Z., Rasheed S., Vijaya K., Kumar K., et al. (2014). Novel hybrids of metronidazole and quinolones: synthesis, bioactive evaluation, cytotoxicity, preliminary antimicrobial mechanism and effect of metal ions on their transportation by human serum albumin. Eur. J. Med. Chem. 30, 318–334. 10.1016/j.ejmech.2014.08.063 [DOI] [PubMed] [Google Scholar]
- Dalhoff A. (2012). Resistance surveillance studies: a multifaceted problem - the fluoroquinolone example. Infection 40, 239–262. 10.1007/s15010-012-0257-2 [DOI] [PubMed] [Google Scholar]
- Feasey N. A., Dougan G., Kingsley R. A., Heyderman R. S., Gordon M. A. (2012). Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379, 2489–2499. 10.1016/S0140-6736(11)61752-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heir E., Linstedt B. A., Vardund T., Wasteson Y., Kapperud G. (2000). Genomic fingerprinting of shigatoxin-producing Escherichia coli (STEC) strains: comparison of pulsed-field gel electrophoresis (PFGE) and fluorescent amplified-fragment-length polymorphism (FAFLP). Epidemiol. Infect. 125, 537–548. 10.1017/S0950268800004908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A., Mendoza M. C., Rodicio R., Rodicio M. R. (2008). Characterization of pUO-StVR2, a virulence-resistance plasmid evolved from the pSLT virulence plasmid of Salmonella enterica serovar typhimurium. Antimicrob. Agents Chemother. 52, 4514–4517. 10.1128/AAC.00563-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S. B., Vauterin P., Lambert-Fair M. A., Van Duyne M. S., Kubota K., Graves L., et al. (2005). Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43, 1045–1050. 10.1128/JCM.43.3.1045-1050.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Gacharna N., Black T. A., Miller G. H., Hooper D. C. (2009). Temporal appearance of plasmid-mediated quinolone resistance genes. Antimicrob. Agents Chemother. 53, 1665–1666. 10.1128/AAC.01447-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Cho J. K., Kim K. S. (2013). Prevalence and characterization of plasmid-mediated quinolone resistance genes in Salmonella isolated from poultry in Korea. Avian Pathol. 42, 221–229. 10.1080/03079457.2013.779636 [DOI] [PubMed] [Google Scholar]
- Kingsley R. A., Msefula C. L., Thomson N. R., Kariuki S., Holt K. E., Gordon M. A., et al. (2009). Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19, 2279–2287. 10.1101/gr.091017.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. H., Robicsek A., Jacoby G. A., Sahm D., Hooper D. C. (2006). Prevalence of aac(6′)-Ib-cr enconding a ciprofloxacin-modifying enzyme in the United States. Antimicrob. Agents Chemother. 50, 3953–3955. 10.1128/AAC.00915-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Hollis R. J., Sader H. S. (1992). Chromosomal restriction fragment analysis by pulsed-field gel electrophoresis, in Clinical Microbiology Procedures Handbook, ed Isenberg H. D.(Washington, DC: American Society for Microbiology; ), 10.5.c.1.–10.5.c.12. [Google Scholar]
- Pribul B. P., Festivo M. L., de Souza M. M., Rodrigues Ddos P. (2016). Characterization of quinolone resistance in Salmonella spp. isolates from food products and human samples in Brazil. Braz. J. Microbiol. 47, 196–201. 10.1016/j.bjm.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek A., Sahm D. F., Strahilevitz J., Jacoby G. A., Hooper D. C. (2005). Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob. Agents Chemother. 49, 3001–3003. 10.1128/AAC.49.7.3001-3003.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Martínez J. M., Cano M. E., Velasco C., Martínez-Martínez L., Pascual A. (2011). Plasmid-mediated quinolone resistance: an update. J. Infect. Chemother. 17, 149–182. 10.1007/s10156-010-0120-2 [DOI] [PubMed] [Google Scholar]
- Ruiz J., Pons M. J., Gomes C. (2012). Transferable mechanisms of quinolone resistance. Int. J. Antimicrob. Ag. 40, 196–203. 10.1016/j.ijantimicag.2012.02.011 [DOI] [PubMed] [Google Scholar]
- Sanders C. C. (2001). Mechanisms responsible for cross-resistance and dichotomous resistance among the quinolones. Clin. Infect. Dis. 32, S1–S8. 10.1086/319369 [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States: major pathogens. Emer. Infect. Dis. 17, 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliopoulou I., Zografou S., Goula A., Dimitracopoulos G., Christofidou M. (2007). Molecular epidemiology and antibiotic resistance patterns of Salmonella enterica from outhwestern Greece. Chemotheraphy 53, 392–396. 10.1159/000109768 [DOI] [PubMed] [Google Scholar]
- Tamang M. D., Nam H. M., Kim T. S., Jang G. C., Jung S. C., Lim S. K. (2011). Emergence of extended-spectrum beta-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J. Clin. Microbiol. 49, 2671–2675. 10.1128/JCM.00754-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V. (1997). How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. molecular typing working group of the society for healthcare epidemiology of America. Infect. Control Hosp. Epidemiol. 18, 426–439. 10.2307/30141252 [DOI] [PubMed] [Google Scholar]
- Xiong X., Bromley E. H., Oelschlaeger P., Woolfson D. N., Spencer J. (2011). Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a Gram-negative bacterium. Nucleic Acids Res. 39, 3917–3927. 10.1093/nar/gkq1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Wachino J., Suzuki S., Kimura K., Shibata N., Kato H., et al. (2007). New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51, 3354–3360. 10.1128/AAC.00339-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Qu D., Zhang X., Shen J., Cui S., Shi Y., et al. (2010). Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int. J. Food Microbiol. 141, 63–72. 10.1016/j.ijfoodmicro.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Zhao X., Xu X., Zhu D., Ye X., Wang M. (2010). Decreased quinolone susceptibility in high percentage of Enterobacter cloacae clinical isolates caused only by Qnr determinants. Diagn. Microbiol. Infect. Dis. 67, 110–113. 10.1016/j.diagmicrobio.2009.12.018 [DOI] [PubMed] [Google Scholar]


