Abstract
Reverse vaccinology (RV) has been widely used for screening of surface-exposed proteins (PSEs) of important pathogens, including outer membrane proteins (OMPs), and extracellular proteins (ECPs) as potential vaccine candidates. In this study, we applied a novel RV negative strategy and a pan-genome analysis for screening of PSEs from 17 L. interrogans strains covering 11 predominately epidemic serovars and 17 multilocus typing (MLST) sequence types (STs) worldwide. Our results showed, for instance, out of a total of 633 predicted PSEs in strain 56601, 92.8% were OMPs or ECPs (588/633). Among the 17 strains, 190 core PSEs, 913 dispensable PSEs and 861 unique PSEs were identified. Of the 190 PSEs, 121 were further predicted to be highly antigenic and thus may serve as potential vaccine candidates against leptospirosis. With the exception of LipL45, OmpL1, and LigB, the majority of the 121 PSEs were newly identified antigens. For example, hypothetical proteins BatC, LipL71, and the OmpA family proteins sharing many common features, such as surface-exposed localization, universal conservation, and eliciting strong antibody responses in patients, are regarded as the most promising vaccine antigens. Additionally, a wide array of potential virulence factors among the predicted PSEs including TonB-dependent receptor, sphingomyelinase 2, leucine-rich repeat protein, and 4 neighboring hypothetical proteins were identified as potential antigenicity, and deserve further investigation. Our results can contribute to the prediction of suitable antigens as potential vaccine candidates against leptospirosis and also provide further insights into mechanisms of leptospiral pathogenicity. In addition, our novel negative-screening strategy combined with pan-genome analysis can be a routine RV method applied to numerous other pathogens.
Keywords: reverse vaccinology (RV), negative selection strategy, surface-exposed proteins, vaccine candidate, L. interrogans
Introduction
Leptospirosis, caused by pathogenic spirochete bacteria of the genus Leptospira, is one of the most common zoonotic diseases worldwide. Leptospirosis has been recognized as an emerging disease with more than half a million patients reported annually (Adler et al., 2011). Pathogenic Leptospira spp. are transmitted mainly by direct contact with infected animals or by exposure to water or soil contaminated by the urine of infected animals (Faine, 1994). To date, more than 250 serovars have been observed in pathogenic Leptospira (Zhang et al., 2012). At the present time, available leptospira vaccines are inactivated whole cell products that provide inadequate protection against most serovars and cannot provide cross-protection against a large number of serogroups of pathogenic leptospires (Faisal et al., 2008). Therefore, there is an urgent need to develop a long-term and cross-protective vaccine set against leptospirosis.
A revolutionary vaccine research strategy, reverse vaccinology (RV), was able to identify five suitable serogroup B meningococcal vaccine candidates (Pizza et al., 2000). Subsequently, RV has been widely applied to a wide range of bacterial pathogens, including Streptococcus pneumoniae, S. agalactiae, Staphylococcus aureus, Porphyromonas gingivalis, Chlamydia pneumonia, and L. interrogans (Paton and Giammarinaro, 2001; Wizemann et al., 2001; Hava and Camilli, 2002; Gamberini et al., 2005; Maione et al., 2005; Mora et al., 2005; Tettelin et al., 2005; Falugi et al., 2008; Seib et al., 2012). Generally, Gram-negative bacteria have five subcellular location sites including cytoplasm, inner membrane, outer membrane, periplasm, and extracellular space. According to RV theory, except for cytoplasmic and inner membrane proteins, proteins located in the other sites can be regarded as PSEs, and are the most suitable vaccine candidates due to their high susceptibility to antibody recognition and eliciting protective immune responses. The in silico approach of RV is a novel and integrative method that uses available bioinformatic tools in the first step of vaccine development. The currently used in silico strategy of RV is to focus only on OMPs and ECPs positively predicted by several bioinformatic tools, such as PSORTb, Cello, and P-classifier. This approach may overlook numerous unknown proteins as potential vaccine candidates because a relatively high proportion of proteins are not covered by these bioinformatic tools. For instance, the most frequently used tool, PSORTb, achieved the greatest degree of precision, but as many as 30.8% (1,140) of str.56601 proteins were not selected as potential vaccine candidates for further screening, simply due to the fact that the localization sites of these proteins were unknown. This is illustrated by the extracellular virulence factor of Bordetella pertussis-pertussis toxin, the only indispensable component of acellular pertussis vaccines, which was predicted as an “unknown” protein by PSORTB. Furthermore, OMPs, ECPs, and Periplasmic proteins (PMs) were predicted far less accurately and reliably than cytoplasmic proteins (CYTs) and inner membrane proteins (IMPs) by these frequently used bioinformatic tools, including PSORTb, Cello, Proteome Analysis, Subloc, and LOCtree (Gardy and Brinkman, 2006). The usage of these popular bioinformatic tools remains a matter for further investigation, as they may miss or exclude highly antigenic vaccine candidates. Here, in this study, a novel RV prediction method employing a negative selection strategy was developed to reliably identify potential vaccine candidates by removing CYTs and IMPs. Based on our novel RV strategy, these “unknown” proteins that are further predicted as CYTs or IMPs by multiple tools according to our criteria are excluded, and the remaining “unknown” proteins, which might be surface-exposed, are retained in the final vaccine candidates list for further screening. Thus, we can greatly reduce the risk of missing potential vaccine candidates among these “unknown” proteins predicted by one of these computational methods.
Early RV efforts were focused mainly on a single genome of a pathogenic strain or species. This limited focus renders it impossible to develop a universal vaccine comprising biologically cross-protective antigens against multiple serovars, strains, or pathovars of one pathogen. To alleviate this shortcoming, pan-genome strategies have been developed to identify potential cross-protective antigens using multiple genomes of the same species, such as group B Streptococcus spp. (Maione et al., 2005).
In this study, we have applied a new in silico RV negative selection strategy combining a pan-genome analysis to screen PSEs as vaccine candidates to provide a framework for future vaccine development against leptospirosis. In addition, potential virulence factors of leptospira were also further analyzed in this study. Future efforts will be targeted toward the experimental characterization of these identified PSEs in our study, as well as screening their potential as vaccine candidates in an animal model.
Materials and methods
Selection of leptospiral genome sequences
Information for leptospiral serovars and multilocus sequence typing were combined to select suitable strains of L. interrogans. Finally, the 17 representative L. interrogans strains covering 11 dominantly epidemic serovars and 17 MLST sequence types (STs) worldwide were selected. For instance, more than 90% of Chinese epidemic or outbreak strains belonged to the 11 dominant serovars (Zhang et al., 2012). The proteomes of all strains were downloaded from the Pathosystems Resource Integration Center (PATRIC) website (www.patricbrc.org) and detailed information about the selected strains is presented in Table 1.
Table 1.
All information of the 17 representative strains of pathogenic L. interrogans used in this study.
| Strains | MLST | Serovar | Isolated location | Host | Contig |
|---|---|---|---|---|---|
| Leptospirainterrogansserovar Australia str. 2002000624 | ST51 | Australia | Hawaii, USA | Human | 146 |
| LeptospirainterrogansserovarBataviae str. 2006006976 | ST50 | Bataviae | Egypt | Human | 314 |
| LeptospirainterrogansserovarBataviae str. L1111 | ST42 | Bataviae | Thailand | Human | 157 |
| LeptospirainterrogansserovarBataviae str. UI 08561 | ST79 | Bataviae | Laos | Human | 300 |
| LeptospirainterrogansserovarBulgarica str. Mallika | ST112 | Bulgarica | India | Human | 335 |
| LeptospirainterrogansserovarCopenhageni str. Fiocruz L1-130 | ST17 | Copenhageni | Brazil | Human | 2 |
| LeptospirainterrogansserovarGrippotyphosa str. UI 08368 | ST77 | Grippotyphosa | Laos | Human | 369 |
| LeptospirainterrogansserovarGrippotyphosa str. UI 08434 | ST82 | Grippotyphosa | Laos | Human | 237 |
| LeptospirainterrogansserovarGrippotyphosa str. UI 12764 | ST85 | Grippotyphosa | Laos | Human | 147 |
| Leptospirainterrogansserovar Lai str. 56601 | ST1 | Lai | China | Human | 2 |
| LeptospirainterrogansserovarManilae str. M001_Tn_Mutant_Parent | ST57 | Manilae | Philippinnes | Human | 271 |
| LeptospirainterrogansserovarMedanensis str. UT053 | ST46 | Medanensis | Thailand | Human | 188 |
| LeptospirainterrogansserovarMuenchen str. Brem 129 | ST24 | Muenchen | Germany | Horse | 322 |
| Leptospirainterrogansserovar Pomona str. Pomona | ST37 | Pomona | Australia | Human | 118 |
| LeptospirainterrogansserovarPyrogenes str. 2006006956 | ST88 | Pyrogenes | Egypt | Human | 344 |
| LeptospirainterrogansserovarPyrogenes str. L0374 | ST49 | Pyrogenes | Thailand | Human | 169 |
| LeptospirainterrogansserovarPyrogenes str. SriLanka1 | ST75 | Pyrogenes | Tanzania | Rodent, Mastomyssp | 667 |
Predicting strategy for PSEs of L. interrogans
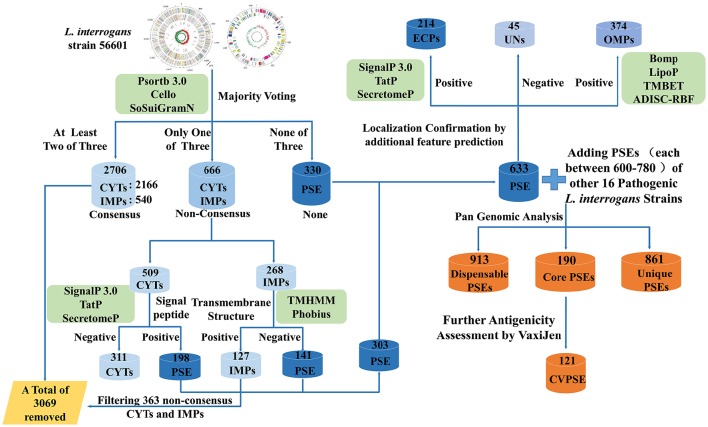
A novel RV approach employing a negative selection strategy was used in this work (Figure 1). At first, the three currently used bioinformatic tools, PSORTb3.0 (Yu et al., 2010), CELLO (Yu et al., 2004), and SOSUI-GramN (Imai et al., 2008), were used to predict subcellular localization of these proteins by a majority voting strategy. Proteins predicted as CYTs by at least two of the three bioinformatic tools were defined as consensus CYTs. Similarly, proteins predicted as IMPs by at least two of the three tools were defined as consensus IMPs. Proteins predicted as CYTs or IMPs by only one of the three tools were labeled as non-consensus CYTs or IMPs, respectively. The remaining proteins were labeled as PSEs. Thus, the predicted results were preliminarily divided into three groups: consensus CYTs/IMPs, non-consensus CYTs/IMPs, and PSEs. The consensus CYTs and IMPs as non-PSEs were directly removed from further study. Non-consensus CYTs and IMPs were further analyzed by combination of additional bioinformatic tools. If these non-consensus CYTs were predicted to be negative by SignalP3.0 (Bendtsen et al., 2004b), TatP (Juncker et al., 2003), and SecretomeP (Bendtsen et al., 2004a), they were removed from further analysis. Non-consensus CYTs with positive signal peptide results were retained as PSEs. Non-consensus IMPs with transmembrane structures predicted by TMHMM (Krogh et al., 2001) or Phobius (Kall et al., 2004) were also removed for further study. Non-consensus IMPs with no transmembrane structures predicted by TMHMM and Phobius were retained as PSEs. Thus, the remaining proteins classified as PSEs were categorized as follows: (1) ECPs or periplasmic proteins predicted by SignalP3.0, Tat and SecretomeP; (2) OMPs predicted by BOMP (Berven et al., 2004), TMBETADISC-RBF (Ou et al., 2008) and LipoP (Juncker et al., 2003); and (3) proteins with unknown localization. Finally, based on amino acid sequences, the antigenicity value of each PSE was predicted using the VaxiJen server with default parameter “bacteria” and the threshold of 0.5 (Doytchinova and Flower, 2007).
Figure 1.
Schematic representation of the novel strategy of reverse vaccinology applied to Pathogenic L.interrogans. In Figure 1, L. interrogans str.56601 was selected as a representative example for elucidating the step-by-step process and predicted results of PSEs using the novel RV negative strategy. Similarly, PSEs of the other 16 representative strains of pathogenic L. interrogans were predicted following same strategy as str.56601. First of all, PSORTb3.0, CELLO, and SOSUI-GramN were used to predict subcellular localization of these proteins by majority voting strategy. Proteins predicted as CYTs and IMPs by at least two of the three bioinformatic tools were defined as consensus CYTs and IMPs and were directly removed from further study. Proteins predicted as CYTs or IMPs by only one of the three tools were labeled as non-consensus CYTs or IMPs, respectively. The remaining proteins were labeled as PSEs. Then, both the non-consensus CYTs with no signal peptides predicted by all of SignalP3.0, TatP and SecretomeP and the non-consensus IMPs with positive transmembrane structures predicted by TMHMM or Phobius were defined as Non-PSEs and removed from further study, whereas the remaining non-consensus CYTs with positive signal peptide and non-consensus IMPs with no transmembrane structure were added into the predicted PSEs. In addition, SignalP3.0, Tat and SecretomeP as well as BOMP, TMBETADISC-RBF, and LipoP were utilized to further investigate extracellular features of these PSEs. Finally, pan-genome analysis of the predicted PSEs among the 17 pathogenic L. interrogans strains identified the core, dispensable, and unique PSEs. And the core PSEs with high antigenicity values predicted by the VaxiJen server were determined as final vaccine antigen candidates. PSE, potential surface-exposed proteins; CVPSE, Conserved Vaxijen antigenicity predicted PSE.
Bioinformatic tools used in reverse vaccinology
Subcellular localization of L. interrogans proteins was predicted by PSORTb, CELLO and SOSUI-GramN. These were classified into CYTs, IMPs, periplasmic proteins (PMs), OMPs, or ECPs. SignalP3.0, TatP, SecretomeP, LipoP, TMBETADISC-RBF, and BOMP were used for further extracellular feature prediction. String database was used for analyzing protein–protein interactions (PPI) of L. interrogans PSEs (Franceschini et al., 2013).
Pangenomic analysis of predicted PSEs among 17 leptospiral strains
Reciprocal blast with bidirectional best hit (BBH) and e-values of 10−10 were used for ortholog clustering of L. interrogans in a pan-genome analysis. Additionally, in order to avoid homologous mismatches, both the coverage and identity percent of cut-offs were set to at least 50%. The concepts of core, dispensable, and unique PSEs were used in this study according to the pan-genome classification. Core PSEs were highly conserved among all 17 strains. Dispensable PSEs and unique PSEs existed in less than 16 strains and exclusively in only one strain, respectively. Finally, these core PSEs with high antigenicity values predicted by the VaxiJen server were determined as the final vaccine antigens candidates against leptospirosis.
Results
General information of selected L. interrogans strains
A total of 17 leptospiral strains covering 11 different serovars and 17 STs were selected for analysis (Table 1). Among these strains, serovars Bataviae, Grippotyphosa, and Pyrogenes consisted of three different STs. The present study was focused mainly on those selected strains that are the most common serovars in China; further, the STs associated with evolutionary information were taken into account (Varni et al., 2013).
Prediction schema of PSEs by the negative selection method
The new combined RV strategy is illustrated by Figure 1. We chose L. interrogans str.56601 as an example. A total of 3,702 proteins were analyzed using our novel RV strategy; 2,706 consensus CYTs and IMPs, 666 non-consensus proteins, and 330 PSEs were predicted. Among these 2,706 proteins, 2,166 proteins were predicted as CYTs and 540 as IMPs by at least two of the three software (PSORTb3.0, CELLO and SOSUI-GramN). Moreover, these 666 non-consensus proteins predicted as CYT or IMP by only one of the three software were further assessed according to the following rules: For example, LA_0012 was predicted to be unknown in PSORT, OMP in Cello and CYT in SoSui-GramN, respectively; And LA_0009 was predicted to be unknown in PSORT, OMP in Cello, IMP in SoSui-GramN. A total of 398 non-consensus proteins like LA_0012 and 157 proteins like LA_0009 were subdivided as non-consensus CYTs and non-consensus IMPs, respectively. In addition, the remaining 111 non-consensus proteins like LA_0293 with unknown location in PSORTb, CYT in Cello and IMPs in SoSui-GramN, were defined as both non-consensus CYTs and IMPs. Therefore, the 666 non-consensus proteins were divided into 509 non-consensus CYTs (398 plus 111) and 268 non-consensus IMPs (157 plus 111). Among the 509 non-consensus CYTs, 311 were predicted negative using the three programs (SignalP3.0, TatP, and SecretomeP) and were removed from further analysis. There were 198 non-consensus CYTs with positive signal peptide results; these were retained as PSEs. Another 268 non-consensus IMPs were further analyzed by TMHMM (Krogh et al., 2001) or Phobius (Kall et al., 2004). One hundred and twenty-seven of these were predicted to have transmembrane structures and eliminated from further study. The remaining 141 with no transmembrane structure were retained and classified as PSEs. Finally, 303 were also predicted to be PSEs out of the 666 non-consensus proteins. Altogether, in addition to the 330 PSEs mentioned above, we predicted a total of 633 PSEs from 3,702 proteins in this study. Among them, the subcellular localization of 45 proteins was unknown and the remaining proteins were almost all predicted as OMPs or ECPs. The predicted PSEs were as high as 92.8% (588/633). The detailed information of PSEs in the remaining strains identified was shown in Figure 2.
Figure 2.
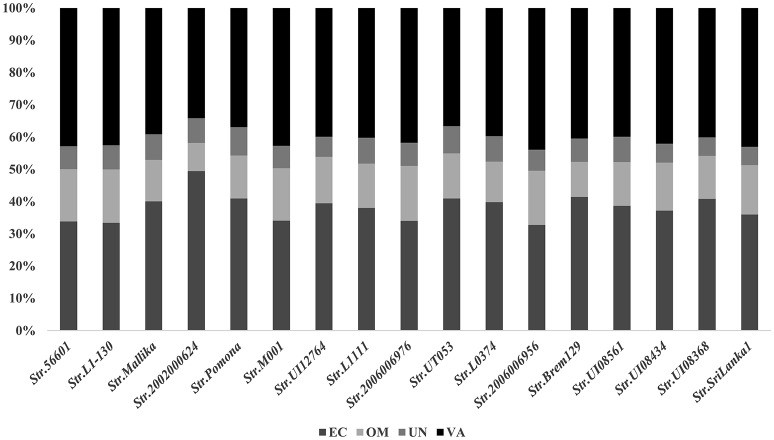
Subcellular localizations of these PSEs among the 17 representative strains of Pathogenic L. interrogans. EC, extracellular; OM, outer membrane; UN, unknown; VA, variable (proteins with multiple locations-EC or OM).
Pan-genome analysis of predicted PSEs among 17 leptospiral strains
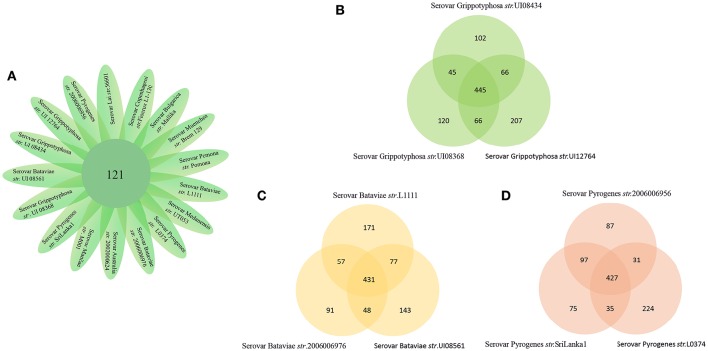
The number of predicted PSEs in the various strains of L. interrogans ranged from 600 to 780 (Figure 2). Gene accumulation curves showed that core genome size fits an exponential decay curve that reached a plateau at 11,043 proteins, whereas the pan PSE grouping fits a power law curve, suggesting the 17 leptospiral strains selected are sufficient to characterize pan core PSEs (Figure 3). Among the 1,103 leptospiral ortholog clusters, 190 core PSEs (17.2%) and 913 dispensable PSEs (82.8%) were shared by all 17 of L. interrogans strains and partly conserved among 2–16 strains, respectively. Furthermore, the pan PSEs included 861 unique PSEs that were found only in one strain. The numbers of unique PSEs in each strain range from 17 (serovar Manilae str.M001) to 103 (serovar Medanensis str.UT053). The dispensable and unique PSEs might be related to different serotypes. The detailed information of all strains and those three dependent serovars was shown in Figure 4. In the present study, our main goal was to predict potential novel protective antigens for the development of universal vaccines against leptospirosis; special attention was given to the 121 high antigenic PSEs from 190 core PSEs, including 37 ECPs, 83 OMPs, and 1 unknown protein localization (see Table 2). As more than 40% of L. interrogans proteins have been annotated as hypothetical proteins, further study of these proteins' functions is needed. Among them, only 55 were categorized into the following COG groups, including main cell wall/membrane/envelope biogenesis (9); function unknown (9); cell motility (7); inorganic ion transport and metabolism (5); general function prediction only (4); Posttranslational modification, protein turnover, chaperones (4); Carbohydrate transport and metabolism (3); Energy production and conversion (2), etc. 16 PSEs were predicted as being involved in(a) cell wall/membrane/envelope biogenesis or (b) cell motility, which are related to the classical function of PSEs (Table S1). In addition, we predicted dispensable and unique PSEs in our pan-genome analysis. For instance, there were 28 unique PSEs in str.56601 and 27 in str. Fiocruz L1-130 (Table S2).
Figure 3.

Calculation of core- and pan-genome sizes of Pathogenic L. interrogans including exponential law models.
Figure 4.
Pan-genome representation for the 17 representative strains of Pathogenic L. interrogans. (A) Core genes of the 17 L.interrogans strains. In our L.interrogans candidates, nine strains belong to three different serovars (B–D). Show pan-genome of the three serovars themselves.
Table 2.
The detailed information of the final 121 PSEs with predicted high antigenicity among the 17 representative strains of pathogenic L.interrogans.
| Str. 56601 Locus | Str. L1-130 Locus | Osmolarity in vivo/in vitro) (Matsunaga et al., 2007) | Low iron (Lo et al., 2010) | Serum (Patarakul et al., 2010) | Temperature P-E (Lo et al., 2006) | Temperature upshift (Lo et al., 2006) | Lai/IPAV (Zhong et al., 2011) | COG | Product | Localization | Vaxijen antigenicity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LA_0022 | LIC10021 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.7022 |
| LA_0071 | LIC10064 | – | – | – | – | – | – | COG4731S | Hypothetical protein | ECPs | 0.7164 |
| LA_0074 | LIC10067 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.6286 |
| LA_0075 | LIC10068 | – | – | ↓ | – | – | – | – | Hypothetical protein | ECPs | 0.5617 |
| LA_0136 | LIC10123 | – | – | – | – | – | ↓ | COG4254S | Hypothetical protein | OMPs | 0.6461 |
| LA_0303 | LIC10260 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.6756 |
| LA_0322 | LIC10280 | – | – | – | – | – | – | – | fibronectin binding protein | OMPs | 0.7026 |
| LA_0333 | LIC10288 | – | – | – | – | – | – | COG0603R | PP-loop superfamily ATPase | OMPs | 0.6805 |
| LA_0346 | LIC10298 | – | – | – | – | – | – | COG1558N | Flagellar basal body rod protein FlgC | ECPs | 0.633 |
| LA_0357 | LIC10307 | – | – | – | – | – | – | COG0412Q | Dienelactone hydrolase family protein | OMPs | 0.6134 |
| LA_0364 | LIC10313 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 1.094 |
| LA_0410 | LIC10359 | – | – | – | – | – | – | COG2834M | Outer membrane lipoprotein-sorting protein | OMPs | 0.6661 |
| LA_0419 | LIC10368 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.7143 |
| LA_0430 | LIC10377 | – | – | – | – | ↑ | – | – | Hypothetical protein | OMPs | 0.8101 |
| LA_0505 | LIC13050* | – | – | – | – | – | ↓ | – | Hypothetical protein | OMPs | 0.8807 |
| LA_0568 | LIC13002 | – | – | – | – | ↓ | – | COG2067I | Fatty acid transport protein | OMPs | 0.6682 |
| LA_0589 | LIC12986 | – | ↑ | – | – | – | – | – | hypothetical protein | OMPs | 0.5822 |
| LA_0591 | LIC12985 | – | – | – | ↑ | ↑ | – | – | Hypothetical protein | ECPs | 0.5631 |
| LA_0663 | LIC12930 | – | – | ↑ | – | – | – | – | Hypothetical protein | ECPs | 0.5964 |
| LA_0862 | LIC12765 | – | – | ↓ | – | ↓ | ↓ | COG2077O | Thiol peroxidase | ECPs | 0.5274 |
| LA_1122 | LIC12558 | – | – | – | ↑ | ↑ | – | – | Hypothetical protein | OMPs | 0.7834 |
| LA_1159 | LIC12525 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.5873 |
| LA_1167 | LIC12519 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.765 |
| LA_1168 | LIC12518 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.6311 |
| LA_1180 | LIC12509 | – | – | – | ↑ | – | – | – | Hypothetical protein | ECPs | 0.6433 |
| LA_1192 | LIC12499 | – | – | – | ↑ | ↑ | – | – | Hypothetical protein | OMPs | 0.564 |
| LA_1356 | LIC12374 | – | – | – | – | – | – | COG4206H | TonB-dependent outer membrane receptor | OMPs | 0.6274 |
| LA_1404 | LIC12337 | – | – | – | – | – | ↑ | – | hypothetical protein | OMPs | 0.5464 |
| LA_1458 | LIC12295 | – | – | – | – | – | – | COG1134GM | ABC transporter ATP-binding protein | OMPs | 0.5095 |
| LA_1499 | LIC12259 | – | – | – | – | – | – | COG0026F | Phosphoribosylaminoimidazole carboxylase ATPase subunit | OMPs | 0.6708 |
| LA_1507 | LIC12254 | – | – | – | – | – | – | COG4775M | Hypothetical protein | OMPs | 0.5181 |
| LA_1508 | LIC12253 | – | – | – | ↑ | ↑ | ↓ | COG4775M | Hypothetical protein | ECPs | 0.599 |
| LA_1897 | LIC12002 | – | – | ↓ | – | – | – | COG0753P | Catalase | ECPs | 0.5618 |
| LA_1931 | LIC11975 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.802 |
| LA_1968 | LIC11935 | – | – | – | – | – | ↓ | – | Hypothetical protein | OMPs | 0.5621 |
| LA_2069 | LIC11846 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 1.0393 |
| LA_2105 | LIC11813 | – | – | – | – | ↑ | – | COG1886NU | Flagellar motor switch protein | OMPs | 0.5388 |
| LA_2186 | LIC11739 | – | – | – | – | – | – | COG0405E | gamma-glutamyltranspeptidase | OMPs | 0.5669 |
| LA_2272 | LIC11665 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.6501 |
| LA_2316 | LIC11625 | – | – | – | – | – | – | COG2013S | Hypothetical protein | ECPs | 0.8212 |
| LA_2377 | LIC11568 | – | – | – | – | – | – | COG2133G | Glucose/sorbosone dehydrogenase | OMPs | 0.6489 |
| LA_2498 | LIC11467 | – | – | – | – | – | – | COG0739M | M23 family metalloendopeptidase | OMPs | 0.6045 |
| LA_2538 | LIC11435 | – | – | – | – | – | – | COG5184DZ | Regulator of chromosome condensation | OMPs | 0.6316 |
| LA_2550 | LIC11424 | – | – | – | – | – | ↑ | COG0596R | Esterase/lipase | ECPs | 0.7281 |
| LA_2595 | LIC11388 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.863 |
| LA_2601 | LIC11382 | – | – | – | – | ↑ | – | – | Hypothetical protein | OMPs | 0.5082 |
| LA_2613 | LIC11370 | – | – | – | – | – | – | COG0545O | FKBP-type peptidylprolyl isomerase | OMPs | 0.6189 |
| LA_2617 | LIC11366 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.7008 |
| LA_2641 | LIC11345 | – | – | – | – | – | – | COG1886NU | Endoflagellar motor switch protein | OMPs | 0.6637 |
| LA_2672 | LIC11320 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.5604 |
| LA_2741 | LIC11271* | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.715 |
| LA_2742 | LIC11270 | – | – | – | – | – | – | COG1629P | Ferrichrome-iron receptor | ECPs | 0.5877 |
| LA_2746 | LIC11268 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.7177 |
| LA_2757 | LIC11259 | – | – | – | – | ↓ | – | – | Hypothetical protein | OMPs | 0.517 |
| LA_2764 | LIC11254 | – | – | – | – | ↑ | – | COG1613P | Sulfate ABC transporter substrate-binding protein | OMPs | 0.5849 |
| LA_2796 | LIC11228 | – | – | ↓ | – | – | – | COG0768M | Transpeptidase/penicillin binding protein | OMPs | 0.6239 |
| LA_2815 | LIC11213 | – | – | – | – | – | – | COG1792M | Rod shape-determining protein MreC | OMPs | 0.6909 |
| LA_2823 | LIC11207 | – | – | – | ↑ | ↑ | – | – | Hypothetical protein | ECPs | 0.7306 |
| LA_2848 | LIC11188 | – | – | – | – | ↓ | – | – | Hypothetical protein | OMPs | 0.7231 |
| LA_2849 | LIC11187 | – | – | – | – | ↓ | – | – | Hypothetical protein | ECPs | 0.5677 |
| LA_2850 | LIC11186 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.6522 |
| LA_2854 | LIC11184 | – | – | ↓ | – | – | – | COG1749N | Flagellar hook protein FlgE | OMPs | 0.6733 |
| LA_2949 | LIC11112 | – | – | – | ↓ | ↓ | – | COG1843N | Flagellar hook assembly scaffolding protein | OMPs | 0.5626 |
| LA_2958 | LIC11103 | – | – | – | ↑ | ↑ | – | COG3144N | Flagellar protein | OMPs | 0.5731 |
| LA_2975 | LIC11087 | – | – | – | – | – | – | COG0265O | Serine protease | OMPs | 0.5597 |
| LA_2992 | LIC11074 | – | – | – | – | – | – | COG2267I | Hypothetical protein | ECPs | 0.7857 |
| LA_2993 | LIC11073 | – | – | – | – | – | – | COG1858P | Cytochrome c peroxidase | OMPs | 0.6704 |
| LA_2998 | LIC11067 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.8504 |
| LA_3026 | LIC11052* | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.6387 |
| LA_3050 | LIC11040 | – | – | – | – | ↑ | – | – | Hypothetical protein | OMPs | 0.5837 |
| LA_3064 | LIC11030 | – | – | – | – | – | – | COG1664M | Cell shape determination protein | OMPs | 0.7079 |
| LA_3097 | LIC11003 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.7837 |
| LA_3138 | LIC10973 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.6481 |
| LA_3145 | LIC10968 | ↑ | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.6892 |
| LA_3150 | LIC10963 | – | – | – | – | – | – | COG1652S | Hypothetical protein | ECPs | 0.6863 |
| LA_3210 | LIC10920 | – | – | – | – | ↓ | – | – | OmpL1 | OMPs | 0.9344 |
| LA_3268 | LIC10873 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.5264 |
| LA_3303 | LIC10845 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.6679 |
| LA_3319 | LIC10833 | – | – | – | – | – | – | COG2010C | Hypothetical protein | ECPs | 0.6143 |
| LA_3410 | LIC10760 | – | – | – | – | ↑ | – | – | Hypothetical protein | ECPs | 0.9564 |
| LA_3454 | LIC10723 | – | – | – | – | – | – | COG3865S | Hypothetical protein | OMPs | 0.5181 |
| LA_3468 | LIC10714 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.5529 |
| LA_3469 | LIC10713 | – | – | – | – | – | – | COG2353S | Hypothetical protein | OMPs | 0.6583 |
| LA_3470 | LIC10712 | – | – | – | – | – | – | COG1345N | Flagellar hook-associated protein FliD | ECPs | 0.5272 |
| LA_3501 | LIC10686 | – | – | – | – | – | – | COG3487P | Hypothetical protein | OMPs | 0.57 |
| LA_3508 | LIC10683 | – | – | – | – | – | – | COG3489R | Hypothetical protein | ECPs | 0.7949 |
| LA_3571 | LIC10628 | – | – | – | – | – | – | COG3794C | Methylamine utilization protein | OMPs | 0.6392 |
| LA_3711 | LIC10520 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.7514 |
| LA_3778 | LIC10464 | ↑ | – | ↑ | – | ↑ | – | COG2885M | OmpA family protein | OMPs | 0.5249 |
| LA_3834 | LIC13066 | ↑ | – | – | – | – | – | COG3607R | Glyoxalase/bleomycin resistance protein/dioxygenase | OMPs | 0.6016 |
| LA_3838 | LIC13070 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.5302 |
| LA_3849 | LIC13076 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.6748 |
| LA_3853 | LIC13078 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.5234 |
| LA_3867 | LIC13086 | ↑ | ↑ | – | – | ↑ | – | – | Hypothetical protein | OMPs | 0.5336 |
| LA_3870 | LIC13089 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.5439 |
| LA_3881 | LIC13101 | – | – | – | – | ↑ | – | – | Hypothetical protein | OMPs | 0.7749 |
| LA_4059 | LIC13238 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.7565 |
| LA_4083 | LIC13255 | – | – | – | ↓ | ↓ | – | COG2303E | Cholesterol oxidase precursor | OMPs | 0.667 |
| LA_4144 | LIC13306 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.5584 |
| LA_4178 | LIC13334 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.5203 |
| LA_4202 | LIC13354 | – | – | – | ↓ | ↓ | – | COG4642S | Hypothetical protein | OMPs | 0.6064 |
| LA_4203 | LIC13355 | – | – | – | – | – | ↓ | COG0331I | Fatty acid synthase subunit beta | OMPs | 0.5784 |
| LA_4272 | LIC13418 | – | – | – | – | – | ↓ | – | Hypothetical protein | OMPs | 0.7046 |
| LA_4291 | LIC13434 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.5418 |
| LA_4293 | LIC13436 | – | – | – | – | – | ↓ | – | polysaccharide deacetylase | OMPs | 0.6614 |
| LA_4335 | LIC13477 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.6873 |
| LB_001 | LIC20001 | – | – | – | – | – | ↓ | – | Hypothetical protein | OMPs | 0.6864 |
| LB_056 | LIC20042* | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.8468 |
| LB_072 | LIC20056 | – | – | – | – | – | – | – | Hypothetical protein | ECPs | 0.5987 |
| LB_098 | LIC20077 | – | – | – | – | – | – | – | Hypothetical Protein | ECPs | 0.5732 |
| LB_110 | LIC20087* | – | – | – | – | – | – | COG5010U | TPR repeat-containing protein | OMPs | 0.5788 |
| LB_191 | LIC20151 | – | – | – | – | – | – | – | Hypothetical protein | OMPs | 0.7302 |
| LB_192 | LIC20152 | – | – | – | – | – | – | COG1022I | Long-chain-fatty-acid–CoA ligase | OMPs | 0.6367 |
| LB_194 | LIC20153 | – | – | – | – | – | - | COG0726G | Xylanase/chitin deacetilase | OMPs | 0.6861 |
| LB_242 | LIC20185 | – | – | ↓ | – | – | – | – | Putative outermembrane protein | OMPs | 0.5456 |
| LB_258 | LIC20197 | – | – | – | – | – | – | COG4206H | Putative TonB-dependent outer membrane receptor protein | OMPs | 0.5271 |
| LB_268 | LIC20205 | – | – | – | – | – | ↓ | – | Hypothetical protein | ECPs | 0.5724 |
| LB_277 | LIC20212 | – | – | – | – | ↓ | – | COG4254S | Hypothetical protein | ECPs | 0.6918 |
| LB_279 | LIC20214 | – | – | – | – | – | ↑ | COG4870O | Cysteine protease | OMPs | 0.6034 |
| LB_280 | LIC20215 | – | – | – | – | – | – | COG2267I | Hypothetical protein | ECPs | 0.5028 |
| LA_1462 | LIC12293 | – | – | – | – | – | ↑ | COG1633S | Hypothetical protein | Unknown | 0.5146 |
Proteins were confirmed to be reactive with human serum (Lessa-Aquino et al., 2013). Osmolarity (in vivo/in vitro), physiologic osmolarity compared with those at low osmolarity; Low iron, genes differentially expressed under low-iron conditions; Serum, gene changed in serum; Temperature P-E, genes changes in physiological vs. environmental temperatures; Temperature upshift, genes changes in overnight 37° upshift vs. 30° long term; “↓,” down –regulated; “↑,” up-regulated; “–”, unknown or without changes.
Discussion
PSEs of pathogens are potential immune targets for the host immune system (Solis and Cordwell, 2011). In this study, we analyzed the PSEs of 17 leptospiral representative strains covering 11 main serovars and 17 STs, and identified potential vaccine candidates or virulence factors.
Recently, we identified a total of 33 highly reliable ECPs in serovar Lai str.56601 using a newly modified protein-free medium, and 26 of them were found in predicted PSEs of str.56601 in the current study, including LipL32, LipL36, LipL48, LenC, LenE, TonB receptor, OmpA family protein, and 8 putative lipoproteins and 6 hypothetical proteins (Zeng et al., 2013). In addition, a novel L. interrogans OMP microarray was developed and contained a total of 366 predicted lipoproteins and transmembrane OMPs (Pinne et al., 2012). About 70% (239/346) of these OMPs or lipoproteins in the protein array were found in our predicted PSEs of str. Fiocruz L1-130. It has been reported that 1,026 proteins in the TX-114 OMP-enriched fraction were found from the transcriptional and translational responses to temperature shift by high-throughput liquid chromatography tandem mass spectrometry (LC/MS-MS); however, only 154 of the 1026 proteins were found in our predicted PSEs of str.56601. The significant discrepancies could be due to lower coverage of OMPs or lipoproteins within the 1,026 proteins, which comprised no more than 80 predicted or known OMPs or lipoproteins (Lo et al., 2009). In order to comprehensively evaluate the advantages and disadvantages of our negative-screening RV strategy, we further compared another three different data sets including experimentally identified 78 surface-exposed antigens or virulence factors (see Table S3). 499 PSEs of L. interrogans were identified by a positive-selection RV strategy as previously described by Yang et al. (2006) and 346 OMPs/lipoproteins of L. interrogans in the L. interrogans OMP array (Pinne et al., 2012), with our negative-screening results (See Figure 5 and Table S3). Among all 78 known surface-exposed antigens, 63, 55, and 43 were identified in the OMP array (Pinne et al., 2012), in this study and Yang's studies (Yang et al., 2006), respectively. Actually, the highest consistency between protein array result and the known surface-exposed antigens might mainly be due to more than 90% (70/78) of known antigens being located in the outer membrane. Moreover, there are 95 common OMPs/Lipoproteins among Yang's, Pinne's and our study's antigen inventory. There were 84 common OMPs/Lipoproteins between Pinne's and our study while there were only 40 proteins between Pinne's and Yang's study. Thus, for OMP/Lipoprotein, our negative RV strategy predicted more proteins than that of Yang's positive RV strategy. However, the information of extracellular proteins in pathogenic Leptospira spp. is still limited. Further, studies to identify more ECPs and to assess the prediction precision of the two different RV strategies are needed.
Figure 5.
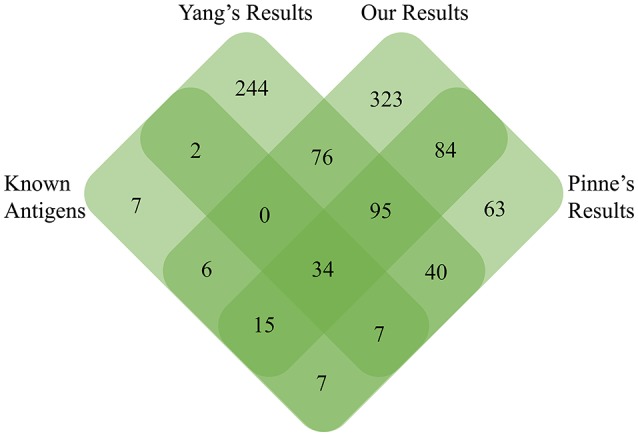
Venn diagram detailed the unique and common PSEs among our negative-screening, Yang's positive-screening, and Pinne's OMP array results with known experimentally identified surface-exposed antigens.
In this study, pan-genome analysis showed 121 highly antigenic PSEs conserved completely among all 17 strains. Except for several known proteins, including LipL45, OmpL1, and LigB, the majority of these candidates are identified in Leptospira for the first time (Pinne et al., 2012). Among the 121 PSEs, the most promising new vaccine antigens appear to be hypothetical proteins (LA_2741), BatC (LB_056), and lipL71/LruA (LA_3097). LA_2741 and BatC were recognized in leptospirosis patients and identified as differentially reactive antigens between acute- or convalescent-phase leptospirosis patients and healthy individuals (Lessa-Aquino et al., 2013). The lipoprotein LruA, present in pathogenic L. interrogans but not in non-pathogenic L. biflexa, could induce high levels of humoral antibody responses in equine uveitis eyes and in sera of humans with leptospiral uveitis (Verma et al., 2005). Thus, these three PSEs could be worthy of further investigation as novel vaccine candidates and/or diagnostic markers for leptospirosis because of common features, including surface-exposed localization, universal conservation, and eliciting strong antibody production in patients (Verma et al., 2005).
Surface-exposed proteins generally comprise a wide array of virulence factors involved in pathogen–host interactions and are responsible for causing disease. Comparing our predicted results to the previous leptospiral OMP microarray data (Pinne et al., 2012), 11 of 15 fibronectin-binding proteins were found in the predicted PSEs of str. Fiocruz L1-130, which were subdivided into four core PSEs (hypothetical protein, TonB-dependent receptor, iron-regulated lipoprotein, and OmpA family proteins) and seven dispensable PSEs (lipoprotein, Lsa66, leucine-rich repeat protein, sphingomyelinases 2 and 3; Pinne et al., 2012). All four core PSEs are involved in adherence to fibronectin during the initial attachment stage of infection and have significant potential to exhibit key roles in the pathogenesis of leptospirosis. For example, TonB-dependent receptor (LA_3468), and iron-regulated lipoprotein (LA_3469) are related to iron uptake, which is essential for pathogenic leptospires (Murray et al., 2008). In our study, iron-regulated lipoprotein (LA_3469) was confirmed to be up-regulated at 37°C as compared to 28°C and could activate the host's immune system to produce a high-level antibody response (our unpublished data), indicating this protein might have an indispensable function in the pathogenesis of L. interrogans. The dispensable PSEs sphingomyelinases Sph2 and Sph3 (LA_1029 and LA_4004) showed distinctly different conservation. It has been confirmed that Sph2 secreted as sphingomyelinase hemolysin has strong hemolytic activity against sheep erythrocytes as well as cytotoxic activity against mouse lymphocytes and macrophages (Zhang et al., 2005, 2008). Thus, Sph2 might be important as a novel virulence factor involved in leptospiral pathogenesis and might be associated with virulence differences among different leptospirosis serovars. Another dispensable PSE is the leucine-rich repeat protein (LA_3028) found exclusively in the highly pathogenic strains: str.56601 and str. Fiocruz L1-130. The leucine-rich repeat protein (LRR) has been reported frequently as a virulence factor in numerous pathogens involved in cell adhesion, invasion, and stimulation of host defense mechanisms (Kobe and Kajava, 2001; Brinster et al., 2007). The leucine-rich repeat protein was identified as a fibronectin-binding protein and it should be, at least partly, related to the high virulence of str.56601 and str. Fiocruz L1-130. The other core PSE like hypothetical protein LA_0505 predicted as a secretion protein through non-classical pathway, has been shown to bind some host extracellular matrices (such as laminin, plasma fibronectin, fibrinogen, etc.) and play an important role in adhesion of L. interrogans (Pinne et al., 2012). Interestingly, LA_0505 was found in the supernatant of L. interrogansstr. 56601 and up-regulated in vivo in our recent study (Zeng et al., 2013). Moreover, LA_0505 has a BIG domain as Ca2+-binding modules during the process of leptospirosis (Raman et al., 2010). The potential virulence factors in predicted PSEs are the four hypothetical proteins LA_1761–1764 identified here. These four PSEs are located in the 54 kb separate circular prophage of str.56601, which was inserted into the larger chromosome at the same time; however, the 54 kb prophage was absent from the genome of str. Fiocruz L1-130 (Bourhy et al., 2007). Until now, there was no experimental evidence suggesting these four proteins might be associated with the virulence of Leptospira; however, PPI analysis in the string database suggested that the four proteins interact mostly with other hypothetical proteins in the PPI network (Figure 6). LA_1762 interacts with lipoproteins LA_3730 and LA_3867, both of which were identified as putative extracellular proteins and thus were recommended as novel candidates for the development of leptospirosis vaccines (Viratyosin et al., 2008). LA_3867 was identified as one of the most strongly up-regulated genes of pathogenic L. interrogans at physiologic osmolarity as compared to low osmolarity, indicating over-expression of LA_3867 in pathogenic leptospires might be associated with transition from survival in the outside environment to infection of mammalian hosts (Matsunaga et al., 2007). Therefore, as an interacting partner of LA_3867, LA_1762 could have a crucial role in successful establishment of host infection.
Figure 6.
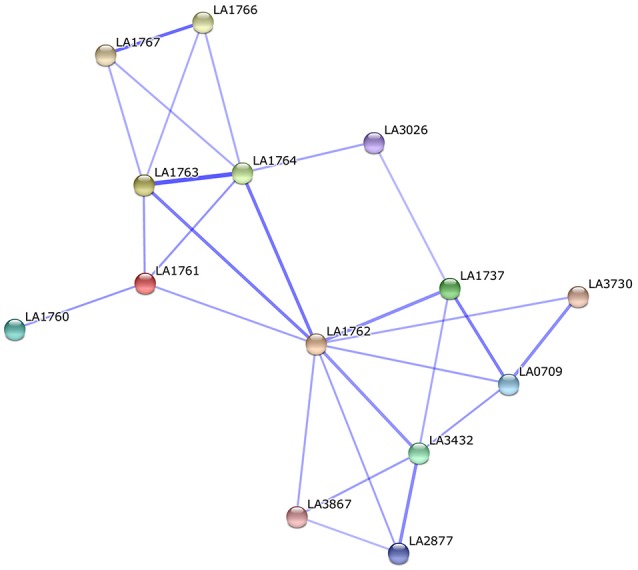
Protein-protein interaction of the potential virulence factors (LA_1761 to LA_1764) located in the 54 kb circular prophage of str.56601.
Conclusions
A new RV negative-screening strategy combined with pan-PSE analysis was used to screen PSEs among 17 L. interrogans strains. We identified 190 core PSEs, 913 dispensable PSEs, and 861 unique PSEs. Further, antigenicity analysis finally identified 121 highly antigenic PSEs as potential vaccine candidates from the 190 core PSEs, which include several known antigens, including LipL45, OmpL1, and LigB, and the vast majority of newly identified potential vaccine candidates against leptospirosis. At the same time, we also characterized many potential virulence factors in our inventory of predicted PSEs. Our prediction results may accelerate vaccine development against leptospirosis and deepen our understanding of leptospiral virulence mechanisms. Moreover, this in silico strategy combined with pan-genome analysis could be a routine method of reverse vaccinology applied widely to similar pathogens. Further, cloning, expression, and purification of these proteins and screening of these potential vaccine candidates are needed.
Author contributions
Conceived and design the experiment: YZZ, XG, Y-FC, and YY; compartive genomic analysis: LZ and DW; predicting subcellular localization: LZ, NH, QZ, and KC; screening known surface-exposed antigens: KD and YZ; write the manuscript: LZ, XG, YZZ, Y-FC, and YY.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from Health and Family Planning Commission of Jiangxi Province (20155090), Jiangxi Provincial Department of Science and Technology (20151BAB205059) and National Natural Science Foundation of China (31660035, 81271793, and 81460300). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00396/full#supplementary-material
All of core, dispensable and specific PSEs among the 17 representative strains of pathogenic L. interrogans.
Experimentally confirmed surface-exposed antigens or virulence factors of pathogenic Leptospira.
The detailed known antigens among Hakke's result, our result and Yang's result.
References
- Adler B., Lo M., Seemann T., Murray G. L. (2011). Pathogenesis of leptospirosis: the influence of genomics. Vet. Microbiol. 153, 73–81. 10.1016/j.vetmic.2011.02.055 [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D., Jensen L. J., Blom N., Von Heijne G., Brunak S. (2004a). Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356. 10.1093/protein/gzh037 [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D., Nielsen H., Von Heijne G., Brunak S. (2004b). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Berven F. S., Flikka K., Jensen H. B., Eidhammer I. (2004). BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 32, W394–W399. 10.1093/nar/gkh351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy P., Salaun L., Lajus A., Medigue C., Boursaux-Eude C., Picardeau M. (2007). A genomic island of the pathogen Leptospira interrogans serovar Lai can excise from its chromosome. Infect. Immun. 75, 677–683. 10.1128/IAI.01067-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster S., Posteraro B., Bierne H., Alberti A., Makhzami S., Sanguinetti M., et al. (2007). Enterococcal leucine-rich repeat-containing protein involved in virulence and host inflammatory response. Infect. Immun. 75, 4463–4471. 10.1128/IAI.00279-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I. A., Flower D. R. (2007). VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 8:4. 10.1186/1471-2105-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faine S. (1994). Leptospira and Leptospirosis. Boca Raton, MA: CRC Press. [Google Scholar]
- Faisal S. M., Yan W., Chen C. S., Palaniappan R. U., Mcdonough S. P., Chang Y. F. (2008). Evaluation of protective immunity of Leptospira immunoglobulin like protein A (LigA) DNA vaccine against challenge in hamsters. Vaccine 26, 277–287. 10.1016/j.vaccine.2007.10.029 [DOI] [PubMed] [Google Scholar]
- Falugi F., Zingaretti C., Pinto V., Mariani M., Amodeo L., Manetti A. G., et al. (2008). Sequence variation in group A Streptococcus pili and association of pilus backbone types with lancefield T serotypes. J. Infect. Dis. 198, 1834–1841. 10.1086/593176 [DOI] [PubMed] [Google Scholar]
- Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., et al. (2013). STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815. 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberini M., Gomez R. M., Atzingen M. V., Martins E. A., Vasconcellos S. A., Romero E. C., et al. (2005). Whole-genome analysis of Leptospira interrogans to identify potential vaccine candidates against leptospirosis. FEMS Microbiol. Lett. 244, 305–313. 10.1016/j.femsle.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Gardy J. L., Brinkman F. S. (2006). Methods for predicting bacterial protein subcellular localization. Nat. Rev. Microbiol. 4, 741–751. 10.1038/nrmicro1494 [DOI] [PubMed] [Google Scholar]
- Hava D. L., Camilli A. (2002). Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406. 10.1046/j.1365-2958.2002.03106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K., Asakawa N., Tsuji T., Akazawa F., Ino A., Sonoyama M., et al. (2008). SOSUI-GramN: high performance prediction for sub-cellular localization of proteins in gram-negative bacteria. Bioinformation 2, 417–421. 10.6026/97320630002417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juncker A. S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., Krogh A. (2003). Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662. 10.1110/ps.0303703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L., Krogh A., Sonnhammer E. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036. 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Kobe B., Kajava A. V. (2001). The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732. 10.1016/S0959-440X(01)00266-4 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Lessa-Aquino C., Borges Rodrigues C., Pablo J., Sasaki R., Jasinskas A., Liang L., et al. (2013). Identification of seroreactive proteins of Leptospira interrogans serovar copenhageni using a high-density protein microarray approach. PLoS Negl. Trop. Dis. 7:e2499. 10.1371/journal.pntd.0002499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M., Bulach D. M., Powell D. R., Haake D. A., Matsunaga J., Paustian M. L., et al. (2006). Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 74, 5848–5859. 10.1128/IAI.00755-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M., Cordwell S. J., Bulach D. M., Adler B. (2009). Comparative transcriptional and translational analysis of leptospiral outer membrane protein expression in response to temperature. PLoS Negl. Trop. Dis. 3:e560. 10.1371/journal.pntd.0000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M., Murray G. L., Khoo C. A., Haake D. A., Zuerner R. L., Adler B. (2010). Transcriptional response of Leptospira interrogans to iron limitation and characterization of a PerR homolog. Infect. Immun. 78, 4850–4859. 10.1128/IAI.00435-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maione D., Margarit I., Rinaudo C. D., Masignani V., Mora M., Scarselli M., et al. (2005). Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science 309, 148–150. 10.1126/science.1109869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J., Lo M., Bulach D. M., Zuerner R. L., Adler B., Haake D. A. (2007). Response of Leptospira interrogans to physiologic osmolarity: relevance in signaling the environment-to-host transition. Infect. Immun. 75, 2864–2874. 10.1128/IAI.01619-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M., Bensi G., Capo S., Falugi F., Zingaretti C., Manetti A. G., et al. (2005). Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U.S.A. 102, 15641–15646. 10.1073/pnas.0507808102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray G. L., Ellis K. M., Lo M., Adler B. (2008). Leptospira interrogans requires a functional heme oxygenase to scavenge iron from hemoglobin. Microbes Infect. 10, 791–797. 10.1016/j.micinf.2008.04.010 [DOI] [PubMed] [Google Scholar]
- Ou Y. Y., Gromiha M. M., Chen S. A., Suwa M. (2008). TMBETADISC-RBF: Discrimination of beta-barrel membrane proteins using RBF networks and PSSM profiles. Comput. Biol. Chem. 32, 227–231. 10.1016/j.compbiolchem.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Patarakul K., Lo M., Adler B. (2010). Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum. BMC Microbiol. 10:31. 10.1186/1471-2180-10-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton J. C., Giammarinaro P. (2001). Genome-based analysis of pneumococcal virulence factors: the quest for novel vaccine antigens and drug targets. Trends Microbiol. 9, 515–518. 10.1016/S0966-842X(01)02207-7 [DOI] [PubMed] [Google Scholar]
- Pinne M., Matsunaga J., Haake D. A. (2012). Leptospiral outer membrane protein microarray, a novel approach to identification of host ligand-binding proteins. J. Bacteriol. 194, 6074–6087. 10.1128/JB.01119-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizza M., Scarlato V., Masignani V., Giuliani M. M., Arico B., Comanducci M., et al. (2000). Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287, 1816–1820. 10.1126/science.287.5459.1816 [DOI] [PubMed] [Google Scholar]
- Raman R., Rajanikanth V., Palaniappan R. U., Lin Y. P., He H., Mcdonough S. P., et al. (2010). Big domains are novel Ca(2)+-binding modules: evidences from big domains of Leptospira immunoglobulin-like (Lig) proteins. PLoS ONE 5:e14377. 10.1371/journal.pone.0014377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib K. L., Zhao X., Rappuoli R. (2012). Developing vaccines in the era of genomics: a decade of reverse vaccinology. Clin. Microbiol. Infect. 18(Suppl. 5), 109–116. 10.1111/j.1469-0691.2012.03939.x [DOI] [PubMed] [Google Scholar]
- Solis N., Cordwell S. J. (2011). Current methodologies for proteomics of bacterial surface-exposed and cell envelope proteins. Proteomics 11, 3169–3189. 10.1002/pmic.201000808 [DOI] [PubMed] [Google Scholar]
- Tettelin H., Masignani V., Cieslewicz M. J., Donati C., Medini D., Ward N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U.S.A. 102, 13950–13955. 10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varni V., Ruybal P., Lauthier J. J., Tomasini N., Brihuega B., Koval A., et al. (2013). Reassessment of MLST schemes for Leptospira spp. typing worldwide. Infect. Genet. Evol. 22, 216–222. 10.1016/j.meegid.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Verma A., Artiushin S., Matsunaga J., Haake D. A., Timoney J. F. (2005). LruA and LruB, novel lipoproteins of pathogenic Leptospira interrogans associated with equine recurrent uveitis. Infect. Immun. 73, 7259–7266. 10.1128/IAI.73.11.7259-7266.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viratyosin W., Ingsriswang S., Pacharawongsakda E., Palittapongarnpim P. (2008). Genome-wide subcellular localization of putative outer membrane and extracellular proteins in Leptospira interrogans serovar Lai genome using bioinformatics approaches. BMC Genomics 9:181. 10.1186/1471-2164-9-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizemann T. M., Heinrichs J. H., Adamou J. E., Erwin A. L., Kunsch C., Choi G. H., et al. (2001). Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69, 1593–1598. 10.1128/IAI.69.3.1593-1598.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. L., Zhu Y. Z., Qin J. H., He P., Jiang X. C., Zhao G. P., et al. (2006). In silico and microarray-based genomic approaches to identifying potential vaccine candidates against Leptospira interrogans. BMC Genomics 7:293. 10.1186/1471-2164-7-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. S., Lin C. J., Hwang J. K. (2004). Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. 10.1110/ps.03479604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Zhang Y., Zhu Y., Yin H., Zhuang X., Zhu W., et al. (2013). Extracellular proteome analysis of Leptospira interrogans serovar Lai. OMICS 17, 527–535. 10.1089/omi.2013.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wang H., Yan J. (2012). Leptospirosis prevalence in Chinese populations in the last two decades. Microbes Infect. 14, 317–323. 10.1016/j.micinf.2011.11.007 [DOI] [PubMed] [Google Scholar]
- Zhang Y. X., Geng Y., Bi B., He J. Y., Wu C. F., Guo X. K., et al. (2005). Identification and classification of all potential hemolysin encoding genes and their products from Leptospira interrogans serogroup Icterohae-morrhagiae serovar Lai. Acta Pharmacol. Sin. 26, 453–461. 10.1111/j.1745-7254.2005.00075.x [DOI] [PubMed] [Google Scholar]
- Zhang Y. X., Geng Y., Yang J. W., Guo X. K., Zhao G. P. (2008). Cytotoxic activity and probable apoptotic effect of Sph2, a sphigomyelinase hemolysin from Leptospira interrogans strain Lai. BMB Rep. 41, 119–125. 10.5483/BMBRep.2008.41.2.119 [DOI] [PubMed] [Google Scholar]
- Zhong Y., Chang X., Cao X. J., Zhang Y., Zheng H., Zhu Y., et al. (2011). Comparative proteogenomic analysis of the Leptospira interrogans virulence-attenuated strain IPAV against the pathogenic strain 56601. Cell Res. 21, 1210–1229. 10.1038/cr.2011.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All of core, dispensable and specific PSEs among the 17 representative strains of pathogenic L. interrogans.
Experimentally confirmed surface-exposed antigens or virulence factors of pathogenic Leptospira.
The detailed known antigens among Hakke's result, our result and Yang's result.