Abstract
Recent evidence from a comprehensive genome analysis and functional studies have revealed that FOXM1 is a crucial metastatic regulator that drives cancer progression. However, the regulatory mechanism by which FOXM1 exerts its metastatic functions in cancer cells remains obscure. Here, we report that DLX1 acts as a FOXM1 downstream target, exerting pro-metastatic function in ovarian cancers. Both FOXM1 isoforms (FOXM1B or FOXM1C) could transcriptionally upregulate DLX1 through two conserved binding sites, located at +61 to +69bp downstream (TFBS1) and −675 to −667bp upstream (TFBS2) of the DLX1 promoter, respectively. This regulation was further accentuated by the significant correlation between the nuclear expression of FOXM1 and DLX1 in high-grade serous ovarian cancers. Functionally, the ectopic expression of DLX1 promoted ovarian cancer cell growth, cell migration/invasion and intraperitoneal dissemination of ovarian cancer in mice, whereas small interfering RNA-mediated DLX1 knockdown in FOXM1-overexpressing ovarian cancer cells abrogated these oncogenic capacities. In contrast, depletion of FOXM1 by shRNAi only partially attenuated tumor growth and exerted almost no effect on cell migration/invasion and the intraperitoneal dissemination of DLX1-overexpressing ovarian cancer cells. Furthermore, the mechanistic studies showed that DLX1 positively modulates transforming growth factor-β (TGF-β) signaling by upregulating PAI-1 and JUNB through direct interaction with SMAD4 in the nucleus upon TGF-β1 induction. Taken together, these data strongly suggest that DLX1 has a pivotal role in FOXM1 signaling to promote cancer aggressiveness through intensifying TGF-β/SMAD4 signaling in high-grade serous ovarian cancer cells.
Introduction
Forkhead box M1 (FOXM1) is a member of the Forkhead box family, with a conserved winged-helix DNA-binding domain.1 It is critically involved in embryogenesis and organ development.2, 3 Alternative splicing of FOXM1 generates three variants; FOXM1A contains alternative exons Va and VIIa, FOXM1C contains Va, and FOXM1B contains none of these exons. Both FOXM1B and FOXM1C are transcriptionally active, whereas FOXM1A is transcriptionally inactive, due to an insertion of exon VIIa in the transactivation domain (TBD).4 Emerging evidence has documented that aberrant upregulation of FOXM1 is frequently observed in various human cancers.5, 6, 7, 8 According The Cancer Genome Atlas (TCGA), activated FOXM1 is significantly associated with the majority of high-grade serous ovarian cancers, which is the most common and deadly subtype of epithelial ovarian cancer.9 FOXM1 exhibits potent oncogenic properties in promoting cell proliferation in human cancer cells, and acts as a major activator of cancer metastasis through enhancing the epithelial–mesenchymal transition, invasion, cell migration and angiogenesis.10, 11, 12 Indeed, we have previously reported a stepwise increase in FOXM1 expression from low- to high-grade ovarian cancer.13 We have also demonstrated that FOXM1B has a higher capacity to enhance cell migration and cell invasion, while FOXM1C is involved in not only cell migration and invasion of ovarian cancer cells but also cell proliferation.13 Given that FOXM1 acts as a crucial master regulator of tumorigenesis and metastasis in human cancers, it is of interest to understand the underlying molecular mechanism of FOXM1 in the transcriptional regulation of the diverse signaling pathways in each step of tumorigenesis. The identification of downstream targets of FOXM1 will provide reliable biomarkers and better therapeutic targets for the tailored treatment of ovarian cancers.
The DLX homeobox family is a group of transcription factors that show sequence homology to the Drosophila distal-less genes (Dll).14 DLX genes are essential in the development of appendages, craniofacial structures, sensory organs, brains, bones and blood, but their expression is variable in different developmental stages.15 Aberrant expression of homeobox genes has been found in a variety of human cancers. For examples, DLX4 is highly correlated with high-grade and metastatic stages of ovarian cancer.16 The oncogenic function of DLX4 is due to its capacity to inhibit the expression of p15lnk4B and p21WAF1/Cip1 by blocking Smad4 in the Transforming growth factor-β (TGF-β) signaling pathway.17 Moreover, DLX5 upregulation promotes ovarian cancer cell growth via the AKT signaling pathway.18 Moreover, the expression of DLX2 and DLX5/6 is associated with the metastatic potential of a variety of human cancer cells.15, 19 Within the DLX family, little is known about the oncogenic role of DLX1. However, recent reports have shown that DLX1 is important for controlling the proliferation and migration of GABAergic cortical interneuron.20, 21 Importantly, DLX1 has been found to be associated with the metastatic state in prostate cancer,22 indicating that DLX1 might have an oncogenic role in cancer progression.
In this study, we have identified DLX1 as a novel target of FOXM1 and showed that DLX1 is upregulated in high-grade ovarian cancer. In vitro and in vivo tumorigenic assays revealed that DLX1 could promote cell growth and migration/invasion, two common metastatic properties in high-grade ovarian cancer, by modulating the TGF-β1/SMAD4 signaling pathway. Taken together, these data highlight the possibility that DLX1 could be used as a biomarker and therapeutic target in combating ovarian cancer in the future.
Results
Prediction of the putative downstream targets of FOXM1
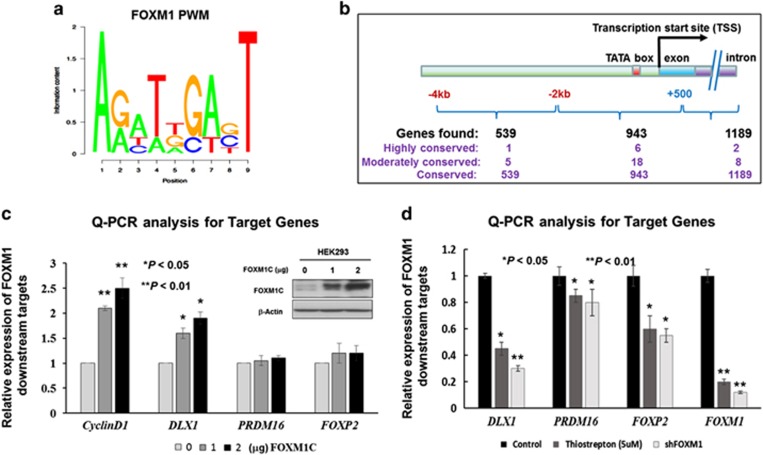
The position weight matrix (PWM) of FOXM1 (Figure 1a) was used to scan all human promoters for putative FoxM1-binding sites based on their predicted binding affinity according to our FastPval calculation.23 The search led to the identification of three categories of putative transcription factor binding sites (TFBSs) (highly conserved, moderately conserved, and conserved, with conservation P-value at 0.001, 0.005 and 0.05 respectively) (Figure 1b). To avoid the false positive targets, we only selected the predicted TFBSs with moderately conservation (P-value<0.005) between human and mouse for further screening (Supplementary Table S1). To verify that these putative genes are transcriptionally upregulated by FOXM1, a plasmid expressing the most common FOXM1 isoform, FOXM1C, was transiently transfected into HEK293 cells, and the quantitative real-time PCR (qRT-PCR) results showed that three out of the 18 putative targets (namely DLX1, PRDM16 and FOXP2) exhibited a progressive increase in mRNA expression that was concomitant with Cyclin D1 expression in dose-dependent manner, and the increased DLX1 mRNA was most remarkable (Figure 1c). On the other hand, other 15 genes just showed mild to moderate increases in their mRNA levels (data not shown). On the other hand, after inhibiting FOXM1 expression using Thiostrepton (FOXM1 inhibitor; 5 μm) (76.7% reduction) or an shRNAi approach (87% reduction) on A2780cp cells, we found that there were concomitant decreases in expression of DLX1 (50% to 65%), PRDM16 (15% to 18%) and FOXP2 (48–50%) compared with the untreated or scrambled control A2780cp cells (Figure 1d). In view of these findings, DLX1 was the most responsive candidate that best correlated with FOXM1 expression in ovarian cancer cells.
Figure 1.
Prediction of the putative downstream targets of FOXM1 by PWMSCAN. (a) PWM for FOXM1-binding motif sequence. (b) A schematic diagram summarizing the computational predictions of the putative targets of FOXM1, which are divided into three groups based on the positions of the FOXM1-binding sites. The genes are further classified into three subgroups according to degree of human–mouse conservation of their putative binding sites. (c) Western blot and qRT-PCR analyses showed concentration-dependent increases in DLX1, PRDM16 and FOXP2 expression following transient transfection of a FOXM1 expression plasmid. The expression of CyclinD1 was used as a positive control for FOXM1 expression. Both β-actin and GAPDH were used as internal controls. (d) The qRT-PCR analysis showed that the inhibition of FOXM1 by treatment with Thiostrepton inhibitor (5 μm, 24 h) or shRNAi-mediated FOXM1 knockdown was accompanied by a remarkable reduction of the three putative targets, DLX1, PRDM16 and FOXP2, compared with their respective untreated or scrambled controls of A2780cp (*P<0.01 and **P<0.05, Student's t-test).
DLX1 is a downstream target of FOXM1 in ovarian cancer cells
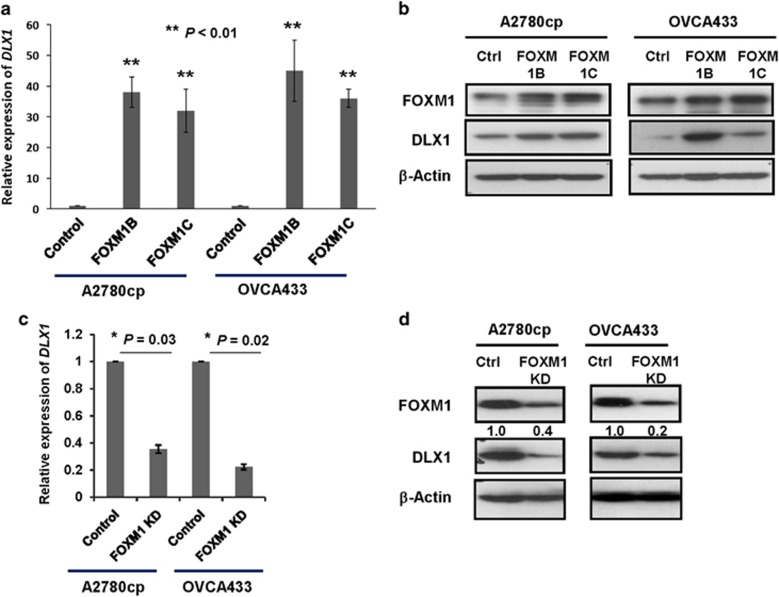
Previous findings have suggested that two FOXM1 active isoforms, FOXM1B and FOXM1C, have differential oncogenic capacities in ovarian cancer cells.13, 24 To confirm which FOXM1 isoform has a higher transcriptional ability to upregulate DLX1, both FOXM1B and FOXM1C expression plasmids were individually transiently transfected into A2780cp and OVCA433 cells. The qRT-PCR analysis showed that FOXM1B and FOXM1C upregulated the expression of DLX1 by 45-fold and 35-fold, respectively, in A2780cp cells, and by 50-fold and 40-fold, respectively, in OVCA433 cells (Figure 2a). Similarly, the western blot analysis confirmed that the transient transfection of FOXM1B or FOXM1C induced a concomitant upregulation of DLX1 in both A2780cp and OVCA433 cells (Figure 2b), and the induction of DLX1 by both isoforms varied in the different ovarian cancer cells, indicating that there was no significant difference in the ability of both isoforms to activate DLX1 transcription (Supplementary Figure S1). On the other hand, silencing of endogenous FOXM1 with an shRNA reduced the DLX1 mRNA expression by more than 50% in both the A2780cp and OVCA433 cells (P<0.05; Figure 2c). The western blots also showed a decrease in the levels of the DLX1 protein in A2780cp and OVCA433 cells when FOXM1 was depleted by a FOXM1 shRNA (Figure 2d). These findings suggest that DLX1 is the downstream target of FOXM1 in ovarian cancer cells.
Figure 2.
Identification of DLX1 as a downstream target of FOXM1. (a) The qRT-PCR analysis showed that DLX1 expression was upregulated by 40-fold and 35-fold in the A2780cp cells, and by 50-fold and 40-fold in OVCA433 cells (**P<0.01, Student's t-test) after transient transfection of FOXM1B or FOXM1C, respectively. (b) The western blot analysis showed that both FOXM1B and FOXM1C upregulate DLX1 expression in OVCA433 and A2780cp cells. (c) The qRT-PCR analysis demonstrated a 20 and 40% reduction of DLX1 expression in the A2780cp (*P=0.03, Student t-test) and OVCA433 cells (*P=0.02, Student's t-test) following the depletion of endogenous FOXM1 by shRNA, respectively. (d) The western blot analysis showed that FOXM1 knockdown reduces DLX1 expression in the A2780cp and OVCA433 cells. The 18S RNA was used as an internal control for all qRT-PCR analyses.
Transcriptional activation of DLX1 by FOXM1
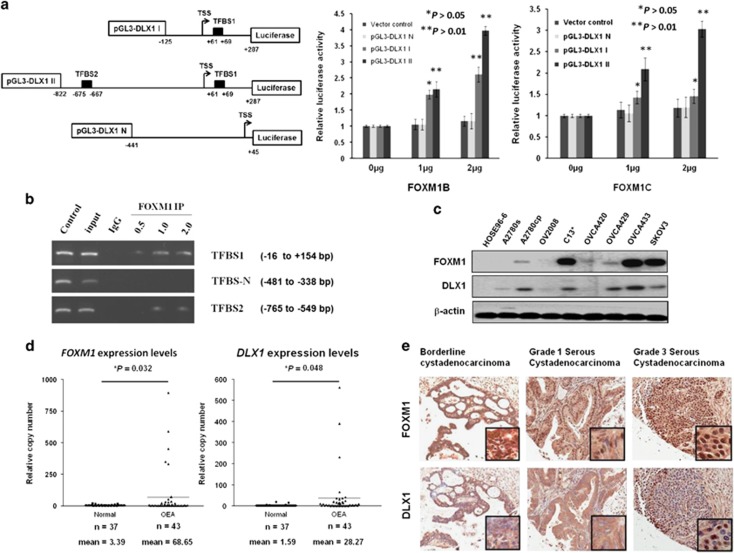
As DLX1 is a downstream target of FOXM1, we hypothesized that FOXM1 directly regulates DLX1 by transcriptional activation of the DLX1 promoter. To test this notion, we constructed three DLX1 promoter luciferase reporters and used them in transient transfections and reporter assays. According to the Transfac database using the PWM as a comparison, two putative FOXM1-binding sites in the DLX1 promoter located at +61 to +69bp and −675 to −667bp upstream and downstream of the transcription start site (TSS) were identified. The +61 to +69bp binding site 1 (TFBS1; ACTCCATTT, reverse strand AAATGGAGT) is the more conserved binding site, while the −675 to −667bp binding site 2 (TFBS2; ACTCCAGCT, reverse strand AGCTGGAGT) is less conserved. Three luciferase reporter constructs were generated; pGL3-DLX1 II includes both TFBS1 and TFBS2, whereas pGL3-DLX1 I includes only TFBS1. pGL3-DLX1 N, which was used as a negative control, does not contain any of the predicted binding sites (Figure 3a). We found that both pGL3-DLX1 I and pGL3-DLX1 II could be activated by either FOXM1B or FOXM1C overexpression (P<0.05; Figure 3a). Noticeably, pGL3-DLX1 II, which contains both the conserved TFBS1 and the less conserved TFBS2, showed stronger activity compared with pGL3-DLX1 I, which contains only the conserved TFBS1 (Figure 3a). On the other hand, pGL3-DLX1 N, which does not have any of the predicted binding sites, did not show an increase in activity upon FOXM1B or FOXM1C overexpression (Figure 3a). These data showed that without TFBS2, the luciferase activity of the DLX1 promoter with TFBS1 was still induced by twofold when transfected with FOXM1B (P<0.01) or by 1.5-fold when transfected with FOXM1C (P<0.05).
Figure 3.
FOXM1 and DLX1 are concomitantly expressed in high-grade serous ovarian cancers. (a) TFBS1 and TFBS2 are the two conserved FOXM1-binding sites on the DLX1 promoter. (left) A schematic diagram illustrating the DLX1 promoter luciferase reporter constructs: pGL3-DLX1 I with only one putative FOXM1-binding site, pGL3-DLX1 II with both putative FOXM1-binding sites, and pGL3-DLX1 II with none of the predicted binding sites. (right) The luciferase promoter assay demonstrated the ability of FOXM1 to transcriptionally activate DLX1 by binding to the two FOXM1-binding sites. Both FOMX1B and FOXM1C could increase the luciferase activity of the pGL3-DLX1 I and pGL3-DLX1 II constructs in a concentration-dependent manner (*P<0.05 and **P<0.01, Student's t-test). pGL3-DLX1 N, which does not have any of the predicted binding sites, shows no observable change in luciferase reporter activity. (b) The ChIP assay demonstrated that PCR fragments from the TFBS1 and TFBS2 sites were pulled down with the FOXM1 antibody (C-20) in a concentration-dependent manner. TFBSN was used as a negative control. (c) Western blot showing the FOXM1 and DLX1 levels in panels of ovarian cancer cell lines and a HOSE cell line. β-actin was used as a control. (d) The qRT-PCR analysis illustrated that FOXM1 (*P=0.032, Student's t-test) and DLX1 (*P=0.048, Student's t-test) expression were upregulated in ovarian cancer samples (n=43 cases) compared with the normal ovaries (n=37 cases). The 18S RNA was used as the internal control. (e) Representative photos showing the immunohistochemical images of FOXM1 and DLX1 staining on the ovarian cancer tissue array. Both FOXM1 and DLX1 expression were progressively increased from borderline cystadenocarcinoma to Grade 3 serous cystadenocarcinoma. (Magnification x100).
A chromatin immunoprecipitation (ChIP) assay was performed to demonstrate the direct binding of FOXM1 on the DLX1 promoter. Using the flanking primers at TFBS1 and TFBS2, our data clearly showed that FOXM1 could directly interact with TFBS1 or TFBS2 on the DLX1 promoter (Figure 3b).
Collectively, our findings strongly support that FOXM1 could transcriptionally activate DLX1 by interacting with two predicted binding sites, TFBS1 and TFBS2, in the DLX1 promoter.
Elevated DLX1 expression correlates with high-grade ovarian cancers
As DLX1 is a downstream target of FOXM1 and is capable of promoting FOXM1-mediated ovarian cancer cell growth and migration, we sought to evaluate the expression status of DLX1 and FOXM1 in human ovarian and cervical cancers. Out of the seven tested cancer cell lines, A2780cp, A2780s, C13*, SKOV3, OVCA429 and OVCA433 showed an increased DLX1 expression concomitant with FOXM1 expression compared with the immortalized ovarian surface epithelium cell line HOSE 96-6 (Figure 3c).
We next determined the expression levels of the DLX1 and FOXM1 mRNAs in normal ovarian clinical samples (n=37) and ovarian cancer samples (n=43) by qRT-PCR. FOXM1 was remarkably elevated in the ovarian cancer samples (20-fold) compared with the normal ovaries (P=0.032), while the expression of DLX1 was approximately 14-fold higher in ovarian cancer samples (P=0.048; Figure 3d). As expected, FOXM1 and DLX1 expression showed a strong positive correlation in the ovarian cancer samples, as tested by Spearman's non-parametric correlation test (coefficient value = 0.813, P<0.01; Supplementary Figure S2).
A commercially available ovarian cancer tissue array (OVC1021) was subjected to immunohistochemical analysis to further examine the expression of DLX1 and its clinical significance. As expected, both FOXM1 and DLX1 were frequently overexpressed in ovarian cancer samples. Using twofold and eightfold as the cutoff points for the expression of FOXM1 and DLX1, respectively, we found that that DLX1 overexpression was significantly associated with high-grade serous ovarian cancers (P<0.001) and FOXM1 overexpression (P=0.002) (Table 1). Intriguingly, the expression of both FOXM1 and DLX1 showed a progressive increase from low- to high-grade serous ovarian cancer, based on the semi-quantitative scoring system using the staining intensity and area of the positive immunohistochemical analysis staining (Figure 3e). Together, these data further support the notion that DLX1 is a downstream target of FOXM1 and that their overexpression is closely associated with the development of high-grade ovarian cancer that features increased cellular proliferation and migration.
Table 1. Immunohistochemical analysis of FOXM1 and DLX1 on an ovarian cancer tissue array (OVC1021, Pantomics, Inc.).
| Characteristics | Total |
DLX1 expression (fold change) |
P-value | |
|---|---|---|---|---|
| ⩽Eightfold | >Eightfold | |||
| All cases | 97 | 47 (48.5%) | 50 (51.5%) | |
| Stage | ||||
| Early (1) | 48 | 25 (52.1%) | 23 (47.9%) | |
| Late (2 and 3) | 49 | 22 (44.9%) | 27 (55.1%) | 0.545 |
| Grade | ||||
| Low (1 and 2) | 50 | 36 (72.0%) | 14 (28.0%) | |
| High (3) | 46 | 10 (21.7%) | 36 (78.3%) | <0.001* |
| Metastasis | ||||
| No | 73 | 36 (49.3%) | 37 (50.7%) | |
| Yes | 24 | 11 (45.8%) | 13 (54.2%) | 0.817 |
| FOXM1 (2xfold) | ||||
| Underexpression | 58 | 36 (62.1%) | 22 (37.9%) | 0.002* |
| Overexpression | 39 | 11 (28.2%) | 28 (71.8%) | |
Higher expression of DLX1 was found in higher tumor grading (*P<0.001, Fisher's exact test). No significant difference was found in tumor stages and metastatic status. DLX1 was positively correlated with FOXM1 expression in immunohistochemical analysis (*P=0.002, Fisher's exact test).
The role of DLX1 in ovarian cancer cell growth and cell migration
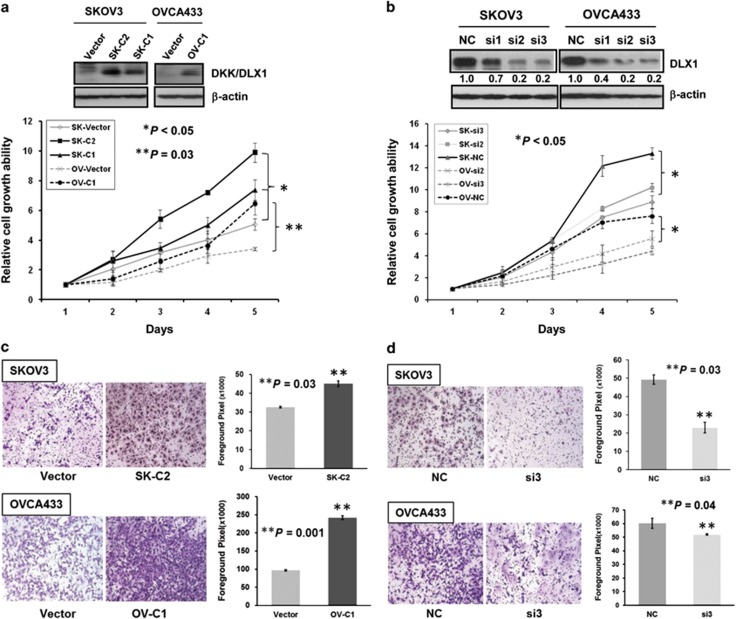
FOXM1 is a crucial metastatic regulator that governs a wide range of cellular behaviors, including uncontrolled cell proliferation, increased cell survival, increased DNA repair and an enhanced epithelial–mesenchymal transition in numerous human cancers.12 If DLX1 is indeed a physiological downstream target of FOXM1 in mediating cancer metastasis, we would expect that, like FOXM1, DLX1 would be critically required for cancer metastasis. Emerging evidence has shown that DLX1 is important for GABAergic neuron development,25 while Dlx1 knockout mice showed a substantial loss of GABAergic neurons.26 DLX1 may be similarly required for regulating ovarian cancer cell proliferation. To examine the oncogenic functions of DLX1 in ovarian cancer cells, transient transfection of DLX1 was initially performed in ovarian cancer cells. Results showed that enforced expression of DLX1 significantly enhanced cell proliferation (P<0.001) and cell migration rates of OVCA433 (P=0.04) and SKOV3 (P<0.0001) cells (Supplementary Figure S3). To further investigate the functional role of DLX1 in ovarian cancer cells, clones that stably express DLX1 were generated in the SKOV3 (SK-C1 and SK-C2) and OVCA433 (OV-C1) cells (Figure 4a). As expected, we found that DLX1 overexpression exhibited a 1.5–3-fold increase in the cell growth rate of the SKOV3 (SK-C1 and SK-C2; P<0.05) and OVCA433 (OV-C1) cells using the XTT cell viability assay (P=0.03; Figure 4a). In contrast, depletion of DLX1 in two FOXM1-overexpressing ovarian cancer cell lines (SKOV3 and OVCA433) by siRNA knockdown decreased the growth rate. Two out of the three siRNAs (si2 and si3) showed a significant reduction of 60–80% and 70–90% of endogenous DLX1 in the SKOV3 and OVCA433 cells, respectively (Figure 4b). The XTT cell viability assay showed that the depletion of DLX1 resulted in an approximately 1.5-fold reduction of the cell growth rate in the SKOV3 and OVCA433 cells (P<0.05; Figure 4b). Moreover, a focus formation assay confirmed that DLX1 overexpression in the SKOV3 (SK-C1 and SK-C2) and OVCA433 (OV-C1) cells increased both the colony number and colony size compared with the vector controls (Supplementary Figure S4).
Figure 4.
DLX1 is able to enhance cell proliferation and cell migration. (a) SKOV3 and OVCA433 (OV-C1) cells stably expressing DLX1. Vector represents the vector control. The XTT cell proliferation assay showed that cell proliferation was significantly increased in the SKOV3 cells by 1.5-fold (SK-C1 and SK-C2) (*P<0.05, Student's t-test) and in the OVCA433 cells (OV-C1) by twofold (**P=0.03, Student's t-test) following DLX1 overexpression. (b) Successful DLX1 knockdown in SKOV3 and OVCA433 cells with two siRNA oligonucleotides (si2 and si3). The XTT cell proliferation assay demonstrated that the cell growth rate of the DLX1 knockdown SKOV3 and OVCA433 cells was decreased by ~1.5-fold and by twofold, respectively, compared with their scrambled controls (*P<0.05, Student's t-test). (c) The cell migration rate of the SKOV3 (SK-C2) (*P=0.03, Student t-test) and OVCA433 (OV-C1) cells was increased by 1.5-fold and 2.5-fold, respectively (**P=0.001, Student's t-test), following DLX1 overexpression. (d) The migration of the SKOV3 and OVCA433 cells was reduced by twofold (**P=0.03, Student's t-test) and onefold (**P=0.04, Student's t-test), respectively, following DLX1 knockdown.
FOXM1 upregulation promotes cell proliferation, cell migration and invasion.11, 13 It is worth noting that DLX1 is able to modulate the migration of GABAergic interneurons from the proliferative zone to the differentiated zone of the cortex in previous studies,27, 28 but its role in cancer cell migration remains unclear. Therefore, it is worth testing whether DLX1 has a role in FOXM1-mediated ovarian cancer cell migration. Using a wound-healing assay, we showed that DLX1 overexpression in the SKOV3 (SK-C1 and SK-C2; P<0.05) and OVCA433 (OV-C1; P=0.002) cells increased the wound closure rate by twofold compared with the vector control (Supplementary Figure S5A). Conversely, DLX1 knockdown significantly reduced the cell migration rate by 50% (P<0.05) and 60% (P<0.01) in SKOV3 and OVCA433 cells, respectively (Supplementary Figure S5B). In addition, using the Transwell cell migration assay, we further confirmed that DLX1 overexpression could increase the cell migration rate by ~1.5-fold in the SKOV3 cells (SK-C2; P=0.03) and by 2.5-fold in the OVCA433 cells (OV-C1; P=0.001; Figure 4c). In contrast, DLX1 depletion by the siRNA markedly blocked the cell migration rate by 60% (P=0.03) and 13% (P=0.04) in the SKOV3 and OVCA433 cells, respectively (Figure 4d).
Taken together, these findings suggest that DLX1 is required for ovarian cancer cell proliferation and migration, and it may be an important effector of FOXM1-stimulated ovarian cancer metastasis.
DLX1 has a key role in FOXM1 signaling to promote cancer metastasis
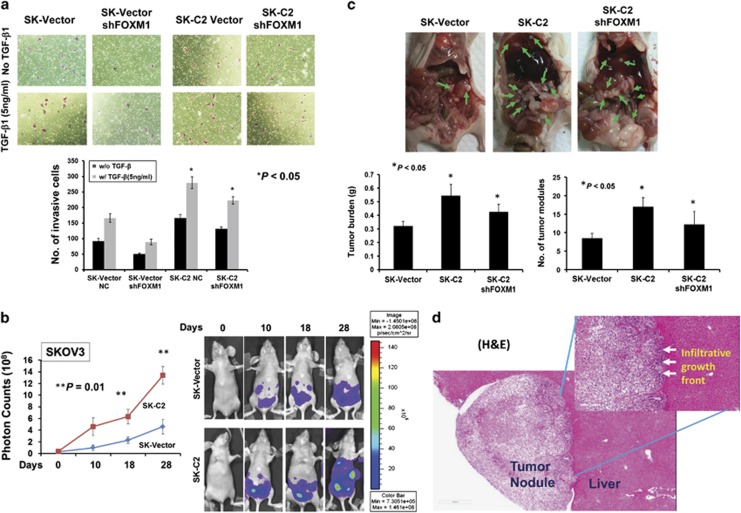
Previous studies have reported that TGF-β1 enhances the ovarian cancer cell epithelial–mesenchymal transition, migration/invasion and metastasis;29, 30, 31 however, it is not clear whether DLX1 synergistically increases the ability of TGF-β1 in cancer cell migration/invasion. Using the Transwell cell invasion assay, TGF-β1 treatment (5 ng/ml) increased the invasive capacity of both the vector control cells and DLX1 stably overexpressing clone (SK-C2) by ~45%, while the SK-C2 cells exhibited higher invasive capacity (Figure 5a). Moreover, knockdown of FOXM1 by shRNA strongly inhibited SKOV3 cell migration in the absence and presence of TGF-β1 (5 ng/ml) (Figure 5a), suggesting FOXM1 is a critically involved in ovarian cancer cell invasion. Of note, DLX1 overexpression in the FOXM1 knockdown SKOV3 cells (SK-C2) could still maintain higher cellular invasion rate with or without TGF-β1 (5 ng/ml) treatment (Figure 5a). This finding indicates that DLX1 is the dominant regulator of FOXM1-mediated ovarian cancer cell invasion.
Figure 5.
DLX1 dominantly regulates the tumor burden and cancer cell metastasis. (a) The Transwell cell invasion assay demonstrated a higher invasion rate of the SKOV3 cells upon treatment with TGF-β1 (5 ng/ml). The invasion rate was further enhanced by ~twofold in the DLX1-overexpressing SKOV3 cells (SK-C2) in the presence or absence of TGF-β1 (5 ng/ml). Stable depletion of FOXM1 by shRNA reduced the invasiveness of the SKOV3 cells (SK-vector) by ~twofold, but reduced the invasion rate of the DLX1-overexpressing cells (SK-C2) to a lesser extent (*P<0.05, Student's t-test). (b) SKOV3-luc cells with or without DLX1 expression were intraperitoneally injected into BALB/c nu/nu female mice. The DLX1-expressing SKOV3 (SK-C2) cells exhibited an ~threefold increase in tumor growth compared with the vector control (SK-vector) (*P<0.01, Student's t-test). The mice were imaged at different time points (0, 10, 18 and 28 days) using a Xenogen IVIS 100 system and Living Imaging, version 2.50.1. (c) The mice were killed on day 28; the gross images show the extensive dissemination of metastatic nodules (green arrows) of the SK-vector, SK-C2 and SK-C2 with stable knockdown of FOXM1 clones. The quantification of the intraperitoneal tumor burden and the number of intraperitoneal nodules are presented in bar charts (*P<0.05, Student's t-test). (d) Hemotoxylin and eosin analysis showed a tumor nodule colonized on the liver. The boundary of tumor nodule showed an infiltrative growth front was toward the liver with blood vessels penetrated from the liver to the tumor tissue.
Given that DLX1 enhances cell proliferation and cell migration/invasion in vitro, we next assessed the tumorigenic capacity of DLX1 in tumor growth and cancer cell dissemination using an orthotopic mouse model of ovarian cancer. Luc-labeled SKOV3 cells with or without stable DLX1 expression were injected intraperitoneally (i.p.) into 5-week-old female nude mice. The tumor burden was monitored by a Xenogen living animal system. As expected, the DLX-1-expressing SKOV3 cells (SK-C2) developed tumors 2.5-fold faster than the vector control (SK-vector) from 0 to 28 days (P=0.01, Fisher's exact test; Figure 5b). A post-mortem examination revealed that the DLX1-expressing SKOV3 (SK-C2) cells had higher capacity of peritoneal dissemination than the vector control (SK-vector) SKOV3 cells. All nude mice (n=5) that were i.p. injected with the SK-C2 cells showed ~48% more disseminated tumor nodules throughout the peritoneal cavity and a ~43% higher tumor burden than SK-vector cells (Figure 4c). Even after depletion of FOXM1, the DLX1-expressing cells (SK-C2) still exhibited ~38% more disseminated tumor nodules and a ~32% higher tumor burden than the vector control cells (Figure 5c). Histologically, the disseminated tumor showed that the blood vessels had infiltrated from disseminated site (liver) to the tumor nodule, whereas there were no invasive cancer cells present in liver tissue (Figure 5d). These data confirm that DLX1 has a critical role in enhancing the peritoneal dissemination of ovarian cancer cells in vivo.
DLX1 modulates TGF-β1 signaling in ovarian cancer cells by interacting with SMAD4
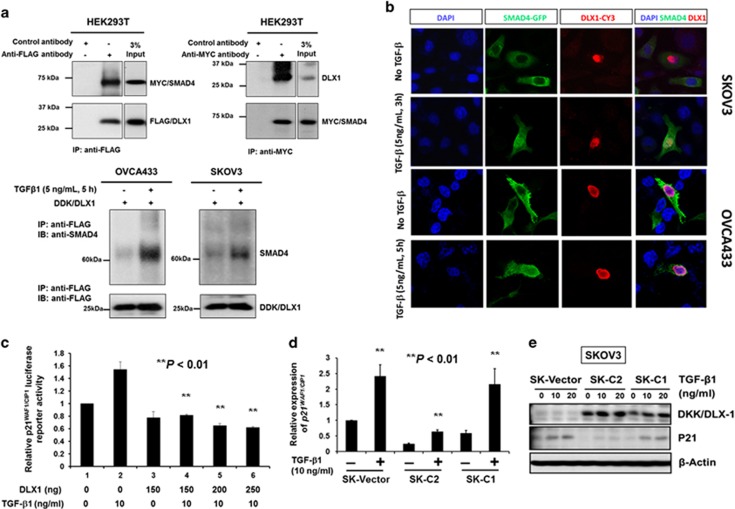
The above data indicate that DLX1 requires TGF-β1 to promote ovarian cancer cell proliferation and cell migration/invasion. Indeed, a recent study reported that DLX1 interacts with SMAD4 and blocks the action of TGF-β1 family members (for example, activin A, TGF-β1 and BMP-4),32 suggesting that DLX1 governs these ovarian cancer cell phenotypes by modulating TGF-β/SMAD4 signaling. To confirm DLX1 whether actually interacts with SMAD4 in ovarian cancer cells, the SKOV3 cells were transfected with the Myc-DDK-tagged HuDLX1 plasmid and treated with/without of TGF-β1 (5 ng/ml) for 5 h. Intriguingly, only the TGF-β1-treated SKOV3 cells exhibited an interaction between DDK-tagged DLX1 and endogenous SMAD4 (Figure 6a). This observation was further confirmed by immunofluorescent analysis showing that DDK/DLX1 was predominantly localized in the nucleus, and co-localized with GFP/SMAD4 when the SKOV3 and OVCA433 cells were treated with TGF-β1 (5 ng/ml, 3 h; Figure 6b). Our study also showed that DLX1 interacts with the majority of SMAD4 in the nucleus of ovarian cancer cells upon TGF-β1 stimulation (Figure 6b), and that DLX1 significantly inhibited TGF-β1 (10 ng/ml)-induced p21WAF1/Cip1 transcriptional activity in a concentration-dependent manner (Figure 6c). Both the qRT-PCR and western blot analyses confirmed that DLX1 reduced the mRNA and protein levels of p21WAF1/Cip1 in ovarian cancer cells (Figures 6d and e). To investigate whether the expression patterns of other cell cycle regulators affected by DLX1/SMAD4 upon TGF-β1 treatment, we employed western blot analysis to evaluate the common cell cycle regulators, for example, p15Ink4B, p18Ink4c and p27Kip1 in OVCA433 cells with transiently transfected with DDK/DLX1 and GFP/SMAD4. Results showed that p21 WAP1/Cip1 was significantly suppressed, while p15Ink4B was moderately reduced by co-expression of DLX1 and SMAD4 (Supplementary Figure S6A). But another two cell cycle regulators, p18Ink4c and p27Kip1, were slightly or no change when compared with their controls (Supplementary Figure S6A). These findings suggest that DLX1 promotes ovarian cancer cell growth by mainly suppression of TGF-β1-induced p21WAF1/Cip1 and partially by inhibition of p15Ink4B. To examine whether the interaction of DLX1 interferes the recruitment of SMAD4 to the promoter of p21WAF1/Cip1, we used SMAD4 motif from Jaspar database to search the SMAD-binding elements (SEBs) on the p21WAF1/Cip1 promoter (Supplementary Figure S6B). Taking the promoter region (±3k to TSS) of p21WAF1/Cip1: chr6:336641237-336647237, we found that two SEBs; SEB1 (hg19_ct_SBE_7386_SBE1: AGGTCTGGCCCCT, −1759~−1747 to TSS) and SEB2 (hg19_ct_SBE_7386_SBE2: CACTCTGTCACCC, located at −992~−980 to TSS), showing higher potential of SMAD4 binding and were supported by a recent study.33 ChIP results showed that SMAD4 occupancy on the above two SBEs of the p21WAF1/Cip1 promoter were apparently attenuated by overexpression of DLX1 in ovarian cancer cells (Supplementary Figure S6B), suggesting that DLX1 could retard the recruitment of SMAD4 to the p21WAF1/Cip1 promoter.
Figure 6.
DLX1 activates TGF-β1/SMAD4 signaling through a direct interaction with SMAD4 in the nuclei of ovarian cancer cells. (a) Reciprocal co-immunoprecipitation (co-IP) demonstrated that DLX1 interacts with SMAD4. (upper) Co-transfection of DDK/MYC/DLX1 and MYC/SMAD4 in HEK293T cells and co-IP using anti-Flag (DDK) showed that DDK/MYC/DLX1 could pull down MYC/SMAD4 confirmed by immunoblotting using anti-MYC. Conversely, transfection of MYC/SMAD4 in HEK293T cells showed that co-IP using anti-MYC could pull down endogenous DLX1 confirmed by immunoblotting using anti-DLX1. (lower) Co-IP assay using anti-Flag (DDK) confirmed that TGF-β1 (5 ng/ml, 5 h) induction could enhance DDK/MYC/DLX1 to pull down more MYC/SMAD4 in SKOV3 and OVCA433 cells, indicating that TGF-β1 induces more MYC/SMAD4 translocating into the nucleus and increases DLX1-SMAD4 interaction. (b) Immunofluorescent analysis demonstrating DLX1-SMAD4 co-localization in the nuclei of the SKOV3 and OVCA433 ovarian cancer cells after TGF-β1 (5 ng/ml, 3 h and 5 h for SKOV3 and OVCA433, respectively) stimulation. (c) DLX1 inhibits p21WAF1/CIP1 promoter activity upon TGF-β1 treatment. Various amounts of the Myc-DDK-tagged HuDLX1 vector were co-transfected with the pWWP-luciferase reporter construct and empty vector pCMV2 into HEK293 cells. TGF-β1 (10 ng/ml) was added 24 h after transfection and incubated for another 5 h before the luciferase activity was measured. The relative p21WAF1/CIP1 luciferase reporter activity of DLX1 was calculated by comparing the test samples with the empty vector control (**P<0.001, Student's t-test). (d) The qRT-PCR (**P<0.001, Student's t-test) and (e) Western blot analyses showed a significant reduction in the expression of p21WAF1/CIP1 in the SKOV3 cells that stably express DLX1 (C1 and C2) upon TGF-β1 treatment (10 and/or 20 ng/ml, 4 h).
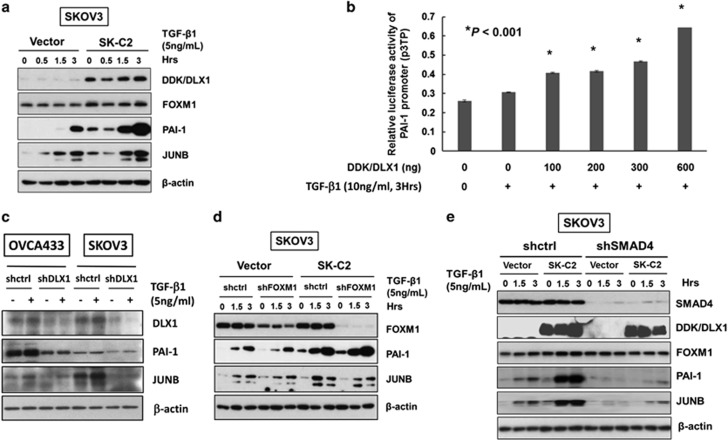
On the other hand, the expression of plasminogen activator inhibitor 1 (PAI-1) and JUNB, two TGF-β responsive genes involved in cancer cell migration and invasion,34 was significantly upregulated in the DLX1-overexpressing SKOV3 cells (SK-C2) compared with the vector control (Figure 7a). Intriguingly, the upregulation of both PAI-1 and JUNB were co-existed with the suppression of p21WAF1/Cip1 and p15Ink4B when co-expression of DLX1 and SMAD4 simultaneously in OVCA433 cells (Supplementary Figure S6A). Previous studies have shown that the expression of urokinase-type plasminogen activator (uPA) and PAI-1 are regulated by TGF-β/SMAD4 signaling in metastatic cancer cells.34, 35 We thus attempted to examine whether DLX1 enhances the TGF-β/SMAD4-mediated transcriptional activity of PAI-1, a PAI-1 promoter luciferase (p3TP) assay was performed. The results showed that the PAI-1 promoter activity was significantly upregulated by DLX1 and TGF-β1 treatment in a concentration-dependent manner (Figure 7b). However, depletion of SMAD4 by shRNA approach impaired DLX1 in upregulation of PAI-1 in SKOV3 cells (Figure 7e). These results indicate that PAI-1 is a direct target and can be transcriptionally activated by DLX1 through interaction of SMAD4.
Figure 7.
DLX1 is a critical downstream effector of FOXM1 that activates TGF-β1/SMAD4 signaling. (a) The western blot analysis showed increased expression of two TGF-β signaling targets, PAI-1 and JUNB, in SKOV3 cells stably expressing DLX1 (SK-C2) in response to TGF-β1 (5 ng/ml) treatment compared with the vector control (SK-vector). (b) DLX1 induced a concentration-dependent increase in PAI-1 luciferase activity upon TGF-β1 treatment (10 ng/ml, 3 h). (c) Depletion of DLX1 by lentiviral shRNA approach remarkably reduce the expressions of PAI-1 and JUNB in OVCA433 and SKOV3 cells upon treatment of TGF-β1 (5 ng/ml) for 5 h. (d) FOXM1 knockdown does not alter the ability of DLX1 to upregulate PAI-1 and JUNB expression upon TGF-β1 (5 ng/ml) treatment in a SKOV3 cell model stably expressing DLX1 (SK-C2). (e) The western blot analysis showed that SMAD4 knockdown markedly attenuated TGFβ1-induced expression of PAI-1 and JUNB, with or without stably overexpressing DDK/DLX1 in SKOV3 cells (SK-C2 and empty vector control).
FOXM1 is known to be a master transcription factor modulating the downstream transcriptional network.36 As DLX1 is a downstream target of FOXM1, it is of interest to examine the role of DLX1 in regulating FOXM1-induced oncogenic genes. We thus applied shRNA approach to knockdown DLX1 in OVCA433 and SKOV3 cells. The results showed that depletion of DLX1 significantly reduced the expression levels of PAI-1 and JUNB in ovarian cancer cells (Figure 7c). On the other hand, it is generally known that FOXM1 is frequently overexpressed in cancer cells, including ovarian cancer cells.13 To determine whether DLX1 is the critical downstream effector of FOXM1, we used FOXM1 knockdown in SKOV3 cells that stably express DLX1 (SK-C2). The results showed that FOXM1 could not alter the expression of PAI-1 and JUNB in these cells (Figure 7d), supporting our notion that DLX1 is a dominant downstream effector of FOXM1 in regulating the oncogenes associated with cell migration or invasion. Moreover, knockdown of SMAD4 (shSMAD4) in DLX1 stably overexpressing SKOV3 (SK-C2) and subsequently treated them with TGF-β1 (5 ng/ml), the expression of PAI-1 and JUNB were significantly attenuated compared with the scrambled control (shctrl) (Figure 7e), indicating that SMAD4 is required for DLX1-mediated regulation of PAI-1 and JUNB expression. Collectively, these findings suggest that DLX1 is a major downstream regulator controlling the expression of the TGF-β responsive genes associated with cell growth and migration/invasion through binding to nuclear SMAD4.
Discussion
Emerging evidence has demonstrated that FOXM1 is an oncogenic transcription factor and a master regulator of tumor progression and metastasis.5, 12, 37 However, the regulatory network of FOXM1, particularly the proteins that promote the metastatic potential of cancer cells, is still not fully understood. In this study, we identified DLX1 as the downstream target of FOXM1, and its expression was temporally associated with FOXM1. Functionally, DLX1 exhibited similar oncogenic functions in promoting cell proliferation and cell migration by activating TGF-β/SMAD4 signaling. Importantly, DLX1 overexpression was correlated with high-grade ovarian cancer. To the best of our knowledge, this is the first demonstration of the role of a direct downstream target of FOXM1 in governing these oncogenic capacities in high-grade ovarian cancer.
A recent comprehensive genomic analysis indicated that the FOXM1 and NOTCH signaling pathways are frequently deregulated and significantly associated with ovarian cancer oncogenesis (The Cancer Genome Atlas Research Network). Indeed, we and others have reported that aberrant activation of FOXM1 signaling is frequently involved in cancer progression by regulating the cell cycle, cell proliferation, the epithelial–mesenchymal-like transition, cell migration, invasion, angiogenesis and so on.12, 13, 38, 39, 40 However, the downstream targets or signaling pathways regulated by FOXM1 during ovarian cancer progression are still largely unknown. Therefore, investigations of this gray area may help to delineate the molecular mechanism underlying the role of FOXM1 in ovarian cancer oncogenesis. Our study also sheds light on biomarkers and therapeutic targets for the early diagnosis and treatment of this disease.
In this study, we identified DLX1 as a downstream target of FOXM1, based on bioinformatics predictions and the results from a series of biochemical and in vitro tumorigenic assays. On the basis of the genomic predictions, two possible FOXM1-binding sites, TFBS1 and TFBS2, are located near the TSS in the DLX1 promoter. TFBS1, located at +61 to +69 bp, is relatively more conserved, whereas TFBS2, located at −675 to −667 bp, is less conserved. These two binding sites, TFBS1 (ACTCCATTTT, reverse strand AAATGGAGT) and TFBS2 (ACTCCAGCT, reverse strand AGCTGGAGT), were identified using the Transfac database and ARATKGAST as the FOXM1 motif for the PWM analysis. However, this motif is different from the FOXM1 motif that was reported in a recent study, which showed that FOXM1 is involved in modulating the transcriptional activity of ERalpha in breast cancer.41 This difference in the FOXM1-binding sites may be due to a cell type- and context-specific sequences.42 For example, a recent study used MCF7 breast cancer cells to investigate FOXM1-binding, while we used ovarian cancer cells in this study. Moreover, the availability of co-factors or cellular stimulators, such as estrogen, in different cellular contexts in different tumor developing stages may also affect the binding sites for the same transcription factor.42 Therefore, our study found three out of 18 putative genes could be remarkably upregulated by enforced expression of FOXM1. Further biochemical and functional analyses revealed that DLX1 is a direct target of FOXM1 in mediating cell migration/invasion of high-grade serous ovarian cancer cells. In this study, we showed that FOXM1 could remarkably activate the DLX1 promoter, a response that was reduced by 50 and 100% when the less conserved binding site or both binding sites were deleted, respectively, suggesting that TFBS1 and TFBS2 are required for FOXM1-induced activation of DLX1 transcription. Furthermore, accumulating evidence shows that both FOXM1B and FOXM1C are predominantly upregulated in human cancers.5, 13, 43, 44, 45 Indeed, both FOXM1B and FOXM1C were able to increase the level of DLX1, with the former exhibiting a higher capacity than the latter in some ovarian cancer cells. This finding is consistent with our recent finding that FOXM1B has higher oncogenic potential than FOXM1C in human cancers.46 Numerous studies have consistently shown that FOXM1B is essential for tumor development and tumor metastasis.38, 43, 44, 45 However, this study showed that DLX1 is a common downstream target of both FOXM1B and FOXM1C, while the transcriptional activity of DLX1 may be higher in some FOXM1B-overexpressing cell lines to promote their migration and invasion.
DLX1 is a transcription factor from the DLX homeobox family, which shares sequence homology with the Drosophila distal-less genes (Dll).47 To date, only a few reports have analyzed the expression of the DLX family of transcription factors in human cancers, and the oncogenic functions of DLX1 remain unclear. For example, DLX4 and DLX5 have been reported to be upregulated in ovarian carcinoma,15, 18 and DLX4 enhances cancer metastasis by activating TWIST.48 On the other hand, there are several reports showing that DLX-1 and DLX-4 are able to interact with SMAD4 and block the autocrine TGF-β/BMP signaling cascade.17, 32 Indeed, the aberrant activation of the TGF-β signaling pathway frequently promotes cancer progression and metastasis.49 We and others have reported that the TGF-β-mediated cell proliferative and metastatic activities are significantly attenuated in high-grade ovarian cancer.50, 51 Moreover, emerging evidence has suggested that FOXM1 interacts with TGF-β signaling in governing cancer metastasis.52, 53 The critical feature for the activation of TGF-β signaling is the sustained accumulation of SMAD complexes in the nuclei of cancer cells.54 On the other hand, FOXM1 is frequently overexpressed in a variety of human cancers, including ovarian cancer.6, 55 A recent study reported that FOXM1 interacts with SMAD3 and retains the SMAD3/SMAD4 complex in the nucleus, thus promoting TGF-β-dependent cancer metastasis.53 Therefore, we proposed that other factors may crosstalk with and synergistically enhance the oncogenic capacities of these two pathways in cancer cells. Based on the findings in this study, DLX1 is a direct target of FOXM1 and promotes cancer cell proliferation and metastasis by interacting with SMAD4 and blocking TGF-β/BMP signaling. In addition, DLX1 has functions similar to DLX4 and could directly interact with nuclear SMAD4 upon TGF-β stimulation.17, 32 In contrast, SMAD4 depletion impaired DLX1-mediated activation of TGF-β/BMP signaling as well as two key factors, PAI-1 and JUNB, which control cancer cell migration and invasion.34 Importantly, although FOXM1 is an oncogenic TF that transcriptionally activates a number of oncogenes, FOXM1 knockdown failed to while DLX1 depletion could remarkably alter these responses, suggesting that DLX1 is a dominant downstream effector of FOXM1 in mediating ovarian cancer cell growth and metastasis, which involves the activation of TGF-β/SMAD4 signaling.
We have demonstrated that the TGF-β-mediated expression of PAI-1 and JUNB in high-grade ovarian cancer cells is enhanced by DLX1. These factors are required for cancer cell metastasis and have been shown to be transcriptionally upregulated by TGF-β/SMAD4 signaling, as SMAD4 binding sites are present on the PAI-1 and JUNB promoters.56, 57 Intriguingly, our results also showed that DLX1 is able to exert cell growth arrest by blocking TGF-β/SMAD4-induced p21WAF1/Cip1 and p15Ink4B. The co-activation of PAI-1 and JUNB in parallel with the suppression of p21WAF1/Cip1 and p15Ink4B appears paradoxical because these factors are downstream targets of TGF-β/SMAD4 signaling. It is known that TGF-β/SMAD4 signaling exerts its anti-proliferative activity by transcriptional activation of the cyclin-dependent kinase inhibitors p16INK4, p15Ink4B and p21WAF1/Cip1 for the maintenance of normal tissue homeostasis58 and the development of resistance to TGF-β-induced growth arrest, particularly in advanced stage tumors.50, 59 However, a recent report supports our finding that DLX4 attenuates the anti-proliferative action of TGF-β in cancer cells by preventing SMAD4 from forming complexes with SMAD2 and SMAD3, but not with Sp1.17 Indeed, our ChIP assays using anti-SMAD4 antibody has confirmed that DLX1 could interfere the recruitment of SMAD4 to the SMAD-binding elements (SBE1 and SBE2) on the p21WAF1/Cip1 promoter in ovarian cancer cells. Therefore, given that DLX1 and DLX4 are Homeobox proteins and function similarly in their interactions with SMAD4, we postulated that DLX1 inhibits not only p21WAF1/Cip1 but also p15Ink4B activation in ovarian cancer cells using the same mechanism (Supplementary Figure S7).
Furthermore, both the immunohistochemical and qRT-PCR analyses showed that the upregulation of DLX1 was significantly correlated with FOXM1 expression in ovarian cancer, particularly in high-grade tumors. High-grade ovarian tumors are well known to be the more aggressive and metastatic ovarian cancer subtype.60 Our in vitro tumorigenic assays on several FOXM1-overexpressing ovarian cancer cell models (for example, SKOV3 and OVCA433 cells) consistently indicated that DLX1 is a key effector of FOXM1 in promoting ovarian cancer cell proliferation and migration, as well as tumor growth and colonization. These in vitro findings are consistent with our clinicopathological analysis demonstrating that concomitant expression of DLX1 and FOXM1 promotes metastasis in high-grade ovarian cancer.
In conclusion, our data strongly support the hypothesis that DLX1 is a novel target of FOXM1 and is highly expressed in high-grade serous ovarian tumors. Our in vitro and in vivo functional studies clearly showed that DLX1 possesses tumorigenic capabilities, which may contribute to the aggressiveness of high-grade serous ovarian cancers. These findings indicate that DLX1 is a strong candidate biomarker and therapeutic target in high-grade ovarian tumors.
Materials and methods
Clinical samples and cell lines
The total RNA samples from 37 normal ovaries from patients with benign diseases and 43 ovarian cancer samples were selected for the qRT-PCR analysis. The histological subtypes and disease stages of the tumors were classified according to the criteria of the International Federation of Gynaecology and Obstetrics (FIGO). The use of the clinical specimens was approved by the local institutional ethics committee (IRB no. UW11-298). Three immortalized human ovarian surface epithelial cell lines (HOSE 6-3, HOSE 17-1 and HOSE 11–12), six ovarian cancer cell lines (A2780cp, A2780s, SKOV3, OVCA429, OVCA433 and ES2) (ATCC, Rockville, MD, USA), two cervical cancer cell lines (OV2008 and C13*) and HEK293 cells (ATCC) were used in this study. The cell line authentication was done by in-house STR DNA profiling analysis and checked for negative of mycoplasma contamination. The FOXM1 inhibitor Thiostrepton was purchased from Calbiochem (La Jolla, CA, USA).
Plasmids, siRNAs and cell transfection
The Myc-DDK-tagged HuDLX1 plasmid with the full length human DLX1 cDNA (OriGene Technologies, Rockville, MD, USA) was used to express DLX1. The pcDNA3-HA-FOXM1B and pcDNA3-HA-FOXM1C plasmids were used to express FOXM1B and FOXM1C, respectively.5, 13 The pTracer-CMV and pcDNA3 vectors were used as negative controls. For the luciferase reporter assays, three DLX1 promoter fragments were subcloned into the pGL3-basic vector (Promega Corporation, Madison, WI, USA) to prepare pGL3-DLX1 I, pGL3-DLX II and pGL3-DLX N. pGL3-DLX I carries −125 to +287 bp of the DLX1 promoter, which contains one (TFBS1) of the two predicted FOXM1-binding sites, whereas pGL3-DLX1 II carries −822 to +287 bp of the DLX1 promoter, which contains both (TFBS1 and TFBS2) of the two predicted FOXM1-binding sites. As a negative control, pGL3-DLX1 N carries +45 to −441 bp of the DLX1 promoter, which contains neither TFBS1 nor TFBS2. The full length p21WAF1/CIP1 promoter luciferase construct (pWWP) (a gift from Dr Mark Feitelson, Mercer Laboratory, Thomas Jefferson University, Philadelphia, PA, USA) and the p3TP-lux plasmid containing a portion of the plasminogen activator inhibitor 1 (PAI-1) promoter region (Addgene, Cambridge, MA, USA) were used for luciferase reporter assays to evaluate the transcriptional activities of p21WAF1/CIP1 and PAI-1, respectively. The TriFECTA Dicer-substrate RNAi kit (Integrated DNA Technologies, Inc., Singapore) includes three siRNA duplexes targeting the human DLX1 open reading frame at three different sites, while a scrambled siRNA duplex was used as a negative control. The FOXM1 shRNA plasmids (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) were used to target the human FOXM1B and FOXM1C open reading frames, and a copGFP Control Plasmid was used as a negative control (Santa Cruz). The CMV-GFP-T2A-Luciferase (Packaged Lentivirus) (System Biosciences, Mountain View, CA, USA) was used for the luc-labeled cells. The Myc-tagged SMAD4 expressing plasmid was kindly provided from Prof. Makoto Mark Taketo (Graduate School of Medicine, Kyoto University, Sakyo, Kyoto, Japan). Cell transfection was conducted using LipofectAMINE 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions.
RNA extraction and quantitative RT-PCR analysis
The total RNA was extracted by TRIzol as according to previously described procedure.50 The cDNA synthesis was performed using a Reverse transcription reagent kit (Applied Biosystems, Foster City, CA, USA). The qRT-PCR analysis of DLX1 and p21/WAF1 expression was performed using a Taqman Gene expression Assay with probes specific for human DLX1 (Assay ID: Hs00165626_m1), PRDM16 (Assay ID: Hs00922674_m1), FOXP2 (Assay ID: Hs00922674_m1), FOXM1 (Assay ID: Hs01073586_m1) and p21WAF1/Cip1 (Assay ID Hs00355782_m1) (Applied Biosystems). The PCR was performed in a 7500 Real Time PCR system (Applied Biosystems). The relative amount of DLX1 expression was determined by the comparative CT method using the 7500 system SDS software (Applied Biosystems, version 1.3.1), with the 18S RNA as the internal control for normalization.
Cell viability analysis
Cell viability was measured with the Cell Proliferation Kit II (XTT) (Roche, Indianapolis, IN, USA), and/or BrdU Cell Proliferation Assay Kit (Biovision, Inc., Milpitas, CA, USA), according to the manufacturer's instructions, Three independent experiments were performed.
Colorimetric cell migration assay
To quantify the cell migratory capacity of ovarian cancer cells, the Transwell cell migration assay kits (Chemicon International, Inc., Temecula, CA, USA) were used according to the manufacturer's instructions. The Transwell filters were visualized with a microscope (TE300, Nikon) and the number of cells was counted. This assay was repeated at least three times.
FOXM1-binding site prediction, chromatin immunoprecipitation (ChIP) and Luciferase reporter assays
PWMSCAN was used for the computational prediction of the FOXM1 downstream targets.61, 62 The known FOXM1-binding sites are represented in terms of the PWM, as annotated in the TRANSFAC database,63 with a consensus motif of 'ARATKGAST' (http://cistrome.org/~jian/motif_collection/databases/Transfac/raw_html/M00630?view=LocusReport&protein_acc=PR000007731). The transfac is a well-known database that is used to analyze the TFBSs identified by experiments, such as EMSA or SELEX. The human (hg18) and mouse (mm8) genomic sequences, pairwise (human and mouse) and multiple alignment files (human, mouse, rat, chicken, dog, chimp and pig) were obtained from the UCSC genome database (http://genome.ucsc.edu) and processed with an in-house pipeline.64 The RefSeq annotated TSS was used to determine the promoter region of the genes (2000 bp upstream to 500 bp downstream of the TSS). The PWM of FOXM1 was used to scan the all of the human promoters with a P-value cutoff of 10−4, which is the probability of finding the binding site in random sequences. The pairwise and multiple species conservations were then checked to reduce the false positives. The highly conserved and moderately conserved groups were defined as multiple species conservation P-values of <0.001 and 0.01, respectively, and the conserved binding sites were defined as a human–mouse conservation P-value<0.05 according to our FastPval calculation.23
The ChIP Assay Kit (Millipore, Billerica, MA, USA) was used to examine the interaction between FOXM1 and its predicted binding sites in the DLX1 promoter, as well as the recruitment of SMAD4 to the p21WAF1/Cip1 promoter in vector control and DLX1-overexpressing cells. The immunoprecipitated chromatin was analyzed by PCR (94 °C (8 min), 40 cycles of 94 °C (20 sec), 55 °C (20 s) FOXM1 on the DLX1 promoter) or 60 °C (SMAD4 on the p21WAF1/Cip1 promoter), and 72 °C (20 s), followed by an extension step at 72 °C (5 min)). The PCR products were separated on a 2% agarose gel. The ChIP primers targeted the DLX1 promoter at TFBS1 (S: 5′-GACCTTCGCTGAGTCAAAGC-3′ AS: 5′-TCCCTTTTGGGGGTCTCTAT-3′); TFBS2 (S: 5′-GGGAGGGAAAGAGGATGTGT-3′ AS: 5′-CTCCCTGCGAACTCACTCAC-3′); and at TFBSN (S: 5′-CGCTAGGCACAAGGCTCTTA-3′ AS: 5′-GGTCCCTAACACCGGACAC-3′). The primers targeted the SMAD-binding elements (SBEs) on the p21WAF1/Cip1 promoter are illustrated in Supplementary Figure S6B.
The luciferase reporter assay was used to investigate the interaction and transcriptional activity of FOXM1 on the DLX1 promoter. The Renilla luciferase reporter plasmid (pRL-CMV), each pGL3-DLX1 promoter luciferase construct, and pcDNA3-HA-FOXM1B or pcDNA3-HA-FOXM1C were co-transfected into the HEK293 cells at different concentrations, whereas the empty vector pcDNA3.1 was used for normalization. To analyze the effect of DLX1 on the induction of TGF-β1/p21/WAF1 signaling, various amounts of the Myc-DDK-tagged HuDLX1 vector with the pWWP-luciferase reporter construct were transiently transfected into HEK293 cells. To analyze the effect of DLX1 on PAI-1 induction, the p3TP-lux plasmid was co-transfected with various concentrations of the Myc-DDK-tagged HuDLX1 vector into HEK293 cells. The TGF-β1 treatment and luciferase activity analysis were the same as reported in a previous publication.50 The dual luciferase reporter assay system (Promega Corporation) was used to measure the firefly and Renilla luciferase activity, according to a previously described protocol.65 All experiments were repeated three separate times.
Western blot, immunopreciptation and immunohistochemical analyses
The cell lysates were prepared from the cell pellets using Cell Lysis Buffer (Cell Signaling Technology, Beverly, MA, USA) containing a Protease Inhibitor Cocktail (Roche) and PMSF (phenylmethylsulphonyl fluoride) (Sigma Chemical Co., St Louis, MO, USA). The samples were separated by SDS–polyacrylamide gel electrophoresis and electroblotted onto an Immobilon-P Transfer Membrane (Millipore Corporation). The blots were first blocked with 5% skim milk, followed by incubation with primary antibodies against FOXM1 (sc-502), p21WAF1/CIP1 (sc-397), p15Ink4B(sc-271791), p18Ink4c(sc-865), p27Kip1(sc-528), SMAD2 (sc-7960) and SMAD4 (sc-7966) (Santa Cruz Biotechnology, Inc.), DLX1 (PA5-28899, Thermo Scientific, Waltham, MA USA), DKK (EPR4759, OriGene Technologies, Rockville, MD, USA), PAI-1(#612024)(BD Transduction Laboratories, San Jose, CA, USA), JUNB(G53) (#3746) (Cell Signaling Technology), or β-actin (A5316, Sigma). The blots were then incubated with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse secondary antibodies (Amersham Pharmacia Biotech, Cleveland, OH, USA) and visualized by enhanced chemiluminescence (ECL) (Amersham).
The immunoprecipitation assay was performed to study the interaction of DLX1 and SMAD4 in ovarian cancer cells, as described by Chan et al.65 Briefly, the A2780cp cells were seeded onto 100 mm culture plates, transfected with either the Myc-DDK-tagged HuDLX1 plasmid or Myc-DKK-tagged control plasmid and treated with TGF-β1 (1.5 ng/ml for 3 h). The cells were then harvested in NET lysis buffer (Cell Signaling) with 1% NP40, pH 8.0, 0.1 mm PMSF, and 1 mm Complete TM protease inhibitor cocktail (Roche). One mg of cell lysate was incubated at 4°C with 1 μg of the mouse anti-IgG and anti-DKK antibodies. After the incubation, 40 μl of Protein A/G Plus-Agarose beads (Santa Cruz) were added and rotated overnight at 4 °C. After washing four times with NET lysis buffer, sample buffer (Cell Signaling) was added and the sample was heat denatured before electrophoresis.
Immunohistochemical staining for FOXM1 and DLX1 was performed on an ovarian cancer tissue array (OVC1021) (Pantomics Inc., San Francisco, CA, USA) using primary polyclonal anti-FOXM1 (Santa Cruz) and anti-DLX1 (Novus Biologicals, LLC, Littleton, CO, USA) antibodies. The immunoreactivity for each sample was calculated by multiplying the percentage of immunopositive cells (10 to 100%) by the intensity of the immunostaining which was scored as 0 (negative), 1 (faint), 2 (moderate), 3 (strong) and 4 (marked). The relative fold change of each sample was then obtained by normalizing the immunoreactive value of each sample with the mean value of the immunoreactivity from the normal-benign cases. The cutoff point (fold change) for each gene was determined according to the Receiver operating characteristic (ROC) curve. The quantification of the immunohistochemical staining was scored by two independent observers.
Tumor xenograft mouse model
SKOV3-luc cells with or without DLX1 overexpression (clone 2 or C2) (1.5 × 106 cells/100 μl) were intraperitoneally (i.p.) injected into 5-week BALB/c nu/nu female mice with 100 μl of Matrigel Matrix (BD Biosciences, San Jose, CA, USA). For statistical significance, 5 mice per group were used according to statistical calculation (36). The tumor size was monitored by the Xenogen IVIS 100 system and Living Imaging, version 2.50.1 (Xenogen, Alameda, CA, USA). All mice were killed on day 28 and the intraperitoneal tumor nodules were extracted and weighed. All of the animal experiments were approved by the University of Hong Kong Committee on the Use of Live Animals in Teaching and Research (CULATR No.2560-11).
Statistical analysis
The clinical parameters were analyzed by SPSS 13.0 software (SPSS, Chicago, IL, USA). Fisher's exact test (for parametric data) and the Mann-Whitney test (for non-parametric data) were used to compare the values between subgroups. The Student's t-test was used to analyze the cell viability and migration data. A P-value of <0.05 was considered statistically significant.
Acknowledgments
We thank Dr Shigeru Chiba (Division of Hematology, Institute of Clinical Medicine, University of Tsukuba, Japan) for providing pTracer-CMV-DLX1-Flag plasmid, Professor Makoto Mark Taketo (Graduate School of Medicine, Kyoto University, Sakyo, Kyoto, Japan) for providing Myc-tagged SMAD4 expressing plasmid, and Prof. George Tsao (Department of Anatomy, The University of Hong Kong) for providing HOSE 6-3, HOSE 17-1 and HOSE 11-12. This study was supported by Wong Check She Charitable Foundation.
Author contributions
DWC and WWYH designed the research. DWC, WWYH, JW, LMNH and MMHY performed the experiments. DWC, THYL, KKLC, YQ, JW and K-MY contributed new reagents-analytic tools. DWC, WWYH, JW, YQ, DX, K-MY, BKT and HYSN analyzed and interpreted the data. DWC wrote the manuscript. All authors were involved in editing the manuscript and had final approval of the submitted and published versions.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Myatt SS, Lam EW. Targeting FOXM1. Nat Rev Cancer 2008; 8: 242. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Holterman AX, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology 2003; 38: 1331–1347. [DOI] [PubMed] [Google Scholar]
- Kim IM, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem 2005; 280: 22278–22286. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta 2007; 1775: 92–102. [DOI] [PubMed] [Google Scholar]
- Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN et al. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol 2008; 215: 245–252. [DOI] [PubMed] [Google Scholar]
- Wen N, Wang Y, Wen L, Zhao SH, Ai ZH, Wang Y et al. Overexpression of FOXM1 predicts poor prognosis and promotes cancer cell proliferation, migration and invasion in epithelial ovarian cancer. J Transl Med 2014; 12: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia JT, Wang H, Liang LJ, Peng BG, Wu ZF, Chen LZ et al. Overexpression of FOXM1 is associated with poor prognosis and clinicopathologic stage of pancreatic ductal adenocarcinoma. Pancreas 2012; 41: 629–635. [DOI] [PubMed] [Google Scholar]
- Zeng J, Wang L, Li Q, Li W, Bjorkholm M, Jia J et al. FoxM1 is up-regulated in gastric cancer and its inhibition leads to cellular senescence, partially dependent on p27 kip1. J Pathol 2009; 218: 419–427. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao B, Wang Z, Ali S, Kong D, Banerjee S, Ahmad A et al. Over-expression of FoxM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J Cell Biochem 2011; 112: 2296–2306. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Huang C, Qiu Z, Wang L, Peng Z, Jia Z, Logsdon CD et al. A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res 2012; 72: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res 2011; 71: 4329–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok GT, Chan DW, Liu VW, Hui WW, Leung TH, Yao KM et al. Aberrant activation of ERK/FOXM1 signaling cascade triggers the cell migration/invasion in ovarian cancer cells. PloS One 2011; 6: e23790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo GR, Zerega B, Paleari L, Trombino S, Mantero S, Levi G. Multiple functions of Dlx genes. Int J Dev Biol 2000; 44: 619–626. [PubMed] [Google Scholar]
- Morini M, Astigiano S, Gitton Y, Emionite L, Mirisola V, Levi G et al. Mutually exclusive expression of DLX2 and DLX5/6 is associated with the metastatic potential of the human breast cancer cell line MDA-MB-231. BMC Cancer 2010; 10: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara F, Samuel S, Liu J, Rosen D, Langley RR, Naora H. A homeobox gene related to Drosophila distal-less promotes ovarian tumorigenicity by inducing expression of vascular endothelial growth factor and fibroblast growth factor-2. Am J Pathol 2007; 170: 1594–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh BQ, Barengo N, Naora H. Homeodomain protein DLX4 counteracts key transcriptional control mechanisms of the TGF-beta cytostatic program and blocks the antiproliferative effect of TGF-beta. Oncogene 2011; 30: 2718–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Cheung M, Pei J, Menges CW, Godwin AK, Testa JR. Upregulation of DLX5 promotes ovarian cancer cell proliferation by enhancing IRS-2-AKT signaling. Cancer Res 2010; 70: 9197–9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkova J, Gadgil S, Qiu YH, Zhang N, Hermanova I, Kornblau SM et al. Up-regulation of homeodomain genes, DLX1 and DLX2, by FLT3 signaling. Haematologica 2011; 96: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maira M, Long JE, Lee AY, Rubenstein JL, Stifani S. Role for TGF-beta superfamily signaling in telencephalic GABAergic neuron development. J Neurodev Disord 2010; 2: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH et al. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron 2000; 28: 727–740. [DOI] [PubMed] [Google Scholar]
- Cantile M, Kisslinger A, Cindolo L, Schiavo G, D'Anto V, Franco R et al. cAMP induced modifications of HOX D gene expression in prostate cells allow the identification of a chromosomal area involved in vivo with neuroendocrine differentiation of human advanced prostate cancers. J Cell Physiol 2005; 205: 202–210. [DOI] [PubMed] [Google Scholar]
- Li MJ, Sham PC, Wang J. FastPval: a fast and memory efficient program to calculate very low P-values from empirical distribution. Bioinformatics 2010; 26: 2897–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AK, Ngan AW, Leung MH, Kwok DC, Liu VW, Chan DW et al. FOXM1b, which is present at elevated levels in cancer cells, has a greater transforming potential than FOXM1c. Front Oncol 2013; 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JL. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Mol Cell Neurosci 2009; 40: 167–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC et al. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci 2005; 8: 1059–1068. [DOI] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron 2007; 54: 873–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TN, Du G, Fonseca M, Zhou QP, Wigle JT, Eisenstat DD. Dlx homeobox genes promote cortical interneuron migration from the basal forebrain by direct repression of the semaphorin receptor neuropilin-2. J Biol Chem 2007; 282: 19071–19081. [DOI] [PubMed] [Google Scholar]
- Lin SW, Lee MT, Ke FC, Lee PP, Huang CJ, Ip MM et al. TGFbeta1 stimulates the secretion of matrix metalloproteinase 2 (MMP2) and the invasive behavior in human ovarian cancer cells, which is suppressed by MMP inhibitor BB3103. Clin Exp Metastasis 2000; 18: 493–499. [DOI] [PubMed] [Google Scholar]
- Lin SW, Ke FC, Hsiao PW, Lee PP, Lee MT, Hwang JJ. Critical involvement of ILK in TGFbeta1-stimulated invasion/migration of human ovarian cancer cells is associated with urokinase plasminogen activator system. Exp Cell Res 2007; 313: 602–613. [DOI] [PubMed] [Google Scholar]
- Teng Y, Zhao L, Zhang Y, Chen W, Li X. Id-1, a protein repressed by miR-29b, facilitates the TGFbeta1-induced epithelial-mesenchymal transition in human ovarian cancer cells. Cell Physiol Biochem 2014; 33: 717–730. [DOI] [PubMed] [Google Scholar]
- Chiba S, Takeshita K, Imai Y, Kumano K, Kurokawa M, Masuda S et al. Homeoprotein DLX-1 interacts with Smad4 and blocks a signaling pathway from activin A in hematopoietic cells. Proc Natl Acad Sci USA 2003; 100: 15577–15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Zhao B, Rasul A, Qin H, Li J, Li X. PIAS1-modulated Smad2/4 complex activation is involved in zinc-induced cancer cell apoptosis. Cell Death Dis 2013; 4: e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A, Zieba A, Vasilaki E, Herrera Hidalgo C, Soderberg O, Koinuma D et al. Specific interactions between Smad proteins and AP-1 components determine TGFbeta-induced breast cancer cell invasion. Oncogene 2013; 32: 3606–3615. [DOI] [PubMed] [Google Scholar]
- Shiou SR, Datta PK, Dhawan P, Law BK, Yingling JM, Dixon DA et al. Smad4-dependent regulation of urokinase plasminogen activator secretion and RNA stability associated with invasiveness by autocrine and paracrine transforming growth factor-beta. J Biol Chem 2006; 281: 33971–33981. [DOI] [PubMed] [Google Scholar]
- Wierstra I. The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res 2013; 118: 97–398. [DOI] [PubMed] [Google Scholar]
- Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta 2012; 1819: 28–37. [DOI] [PubMed] [Google Scholar]
- Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene 2007; 26: 6212–6219. [DOI] [PubMed] [Google Scholar]
- Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V et al. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest 2007; 117: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res 2007; 67: 8293–8300. [DOI] [PubMed] [Google Scholar]
- Sanders DA, Ross-Innes CS, Beraldi D, Carroll JS, Balasubramanian S. Genome-wide mapping of FOXM1 binding reveals co-binding with estrogen receptor alpha in breast cancer cells. Genome Biol 2013; 14: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Metivier R, Salbert G. Defining specificity of transcription factor regulatory activities. J Cell Sci 2009; 122: 4027–4034. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev 2004; 18: 830–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Gusarova G, Wang Z, Carr JR, Li J, Kim KH et al. Deregulation of FoxM1b leads to tumour metastasis. EMBO Mol Med 2011; 3: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res 2008; 68: 8733–8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AY, Ngan AW, Leung MH, Kwok DC, Liu VW, Chan DW et al. FOXM1b, which is present at elevated levels in cancer cells, has a greater transforming potential than FOXM1c. Front Mol Cell Oncol 2013; 3: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezot F, Thomas B, Greene SR, Hotton D, Yuan ZA, Castaneda B et al. Physiological implications of DLX homeoproteins in enamel formation. J Cell Physiol 2008; 216: 688–697. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yang M, Gan L, He T, Xiao X, Stewart MD et al. DLX4 upregulates TWIST and enhances tumor migration, invasion and metastasis. Int J Biol Sci 2012; 8: 1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgis P. TGFbeta signaling in tumor initiation, epithelial-to-mesenchymal transition, and metastasis. J Oncol 2015; 2015: 587193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Liu VW, To RM, Chiu PM, Lee WY, Yao KM et al. Overexpression of FOXG1 contributes to TGF-beta resistance through inhibition of p21WAF1/CIP1 expression in ovarian cancer. Br J Cancer 2009; 101: 1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao ZY, Cai MY, Yang GF, He LR, Mai SJ, Hua WF et al. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-beta1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis 2010; 31: 1576–1583. [DOI] [PubMed] [Google Scholar]
- Li J, Wang Y, Luo J, Fu Z, Ying J, Yu Y et al. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells. FEBS Lett 2012; 586: 3761–3765. [DOI] [PubMed] [Google Scholar]
- Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. J Clin Invest 2014; 124: 564–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development 2009; 136: 3699–3714. [DOI] [PubMed] [Google Scholar]
- Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res 2013; 119: 191–419. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J 1998; 17: 3091–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem 1998; 273: 21145–21152. [DOI] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell 2006; 125: 929–941. [DOI] [PubMed] [Google Scholar]
- Jakowlew SB. Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev 2006; 25: 435–457. [DOI] [PubMed] [Google Scholar]
- Chan DW, Hui WW, Cai PC, Liu MX, Yung MM, Mak CS et al. Targeting GRB7/ERK/FOXM1 signaling pathway impairs aggressiveness of ovarian cancer cells. PloS One 2012; 7: e52578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Hannenhalli S. Identification of transcription factor binding sites in the human genome sequence. Mammalian Genome 2002; 13: 510–514. [DOI] [PubMed] [Google Scholar]
- Wang JW, Zhang SL, Schultz RM, Tseng H. Search for basonuclin target genes. Biochem Biophys Res Commun 2006; 348: 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 2006; 34: D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li MLJ, Wang PW, Zhang MQ, Wang JW. ChIP-Array: combinatory analysis of ChIP-seq/chip and microarray gene expression data to discover direct/indirect targets of a transcription factor. Nucleic Acids Res 2011; 39: W430–W436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Liu VW, Leung LY, Yao KM, Chan KK, Cheung AN et al. Zic2 synergistically enhances Hedgehog signalling through nuclear retention of Gli1 in cervical cancer cells. J Pathol 2011; 225: 525–534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.