Abstract
The stromal antigen 2 (STAG2) gene encodes a component of the cohesin complex that participates in the regulation of sister chromatid separation during mitosis. When activated, STAG2 may act as a ‘caretaker’ tumor suppressor gene. As it is unknown whether STAG2 gene is responsible for the occurrence and associated with the prognosis of acute myeloid leukemia (AML), the present study analyzed the relative expression levels of STAG2 in 127 de novo AML patients and 17 healthy volunteers using reverse transcription-quantitative polymerase chain reaction. In addition, AML patients were divided into three risk groups using cytogenetic and molecular genetic abnormalities to define their risk status. STAG2 gene expression was found to be significantly downregulated in de novo AML patients, when compared with the healthy controls; however, the expression was not significantly different in the various gender and age subgroups. Furthermore, no significant difference between risk groups was detected in AML patients. Thus, the STAG2 gene may serve an important role in AML development, but is not associated with prognosis in AML.
Keywords: stromal antigen 2, acute myeloid leukemia, prognosis, mutation, deletion
Introduction
The stromal antigen 2 (STAG2) gene encodes a cohesion complex component that regulates sister chromatid separation during mitosis (1–3). It is recognized as a tumor suppressor gene, which disrupts the mitotic sister chromatid separation when inactivated. Kim et al (4) reported the loss of STAG2 expression in certain gastric, colorectal and prostate cancer cells when compared with the expression in their normal cell counterparts, implying the involvement of the gene in tumorigenesis. In addition, Solomon et al (5) identified a series of somatic STAG2 mutations in glioblastomas, melanomas and Ewing's sarcomas, including missense, nonsense and splice site mutations, as well as intragenic deletions, and the remaining wild-type alleles were found to be on the inactivated X chromosome. Loss of STAG2 expression was also observed in malignant hematopoietic cells, including certain leukemia and lymphoma cell lines (5). However, whether the dysregulated expression of the STAG2 gene is responsible for the occurrence and prognosis of acute myeloid leukemia (AML) remains unclear.
In the present study, the alterations in STAG2 expression were investigated in de novo AML patients.
Patients and methods
Patients and healthy volunteers
Between January 2009 and December 2012, bone marrow samples were obtained from healthy volunteers (n=17) and from AML patients upon diagnosis, all of which had been referred to the Jiangsu Institute of Hematology (The First Affiliated Hospital of Soochow University, Suzhou, China). A total of 127 patients were diagnosed with de novo AML according to symptoms, blood tests and bown barrow aspirate examinations, classified according to the French-American-British (FAB) classification system (6), were selected for analysis. All patients were Han Chinese (median age, 36 years; range, 8–61 years) and predominantly male (59.8%). The healthy volunteers were also Han Chinese (median age, 32 years; range, 11–56 years) and predominantly male (58.8%). Their ages and genders were not significantly different from the patient group. The main patient characteristics are summarized in Table I. Sample preservation and genetic analysis were performed following written informed consent from the patients and approval by the Ethics Committee of the First Affiliated Hospital of Soochow University, and the study experiments were in accordance with the Declaration of Helsinki.
Table I.
Demographic and clinical characteristics of de novo acute myeloid leukemia patients (n=127).
| Parameter | Value |
|---|---|
| Median age (range), years | 36 (8–61) |
| Age group, n | |
| <35 years | 47 |
| 35–50 years | 43 |
| >50 years | 37 |
| Gender, n | |
| Male | 76 |
| Female | 51 |
| French-American-British classification | |
| M0 | 3 |
| M1 | 17 |
| M2 | 38 |
| M3 | 5 |
| M4 | 10 |
| M4EO | 17 |
| M5 | 32 |
| M6 | 4 |
| M7 | 1 |
| Karyotype | |
| Normal | 59 |
| t(8;21) | 13 |
| t(15;17) | 5 |
| inv(16), t(16;16) | 17 |
| 11q23/MLL | 9 |
| Complex abnormalities | 13 |
| Others | 11 |
| Risk status (based on cytogenetics and molecular abnormalities) | |
| Favorable risk | 31 |
| Intermediate risk | 66 |
| Poor risk | 30 |
Cytogenetic analysis of AML samples
Cytogenetic analysis of bone marrow cells was performed to diagnosis AML. Samples were processed using standard 24-h unstimulated cultures. Conventional R-banding assay was used for karyotype analysis. Where possible, no fewer than 20 metaphases per sample were analyzed. Abnormal clonal karyotypes were described using the International System for Human Cytogenetic Nomenclature (ISCN, 2005).
Molecular characterization of AML samples
The 29 common fusion genes identified in AML, including AML1-ETO, CBFβ/MYH11, PML/RARα, BCR/ABL, MLL/AF6, MLL/ELL and TLS/ERG, were assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) using previously described primers and PCR cycling conditions (7–9). Briefly, mononuclear cells were separated from the bone marrow samples of 127 de novo AML patients by Ficoll gradient centrifugation (400 × g for 30 min), and then subjected to total RNA extraction by TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA). The RNA concentrations were measured using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Inc.), according to the manufacturers instructions. Subsequently, cDNA was synthesized with a SuperScript II reverse transcriptase kit (Invitrogen; Thermo Fisher Scientific, Inc.) and random hexamers. A single PCR reaction was performed in a total volume of 25 µl, containing 1 µl of cDNA. The PCR amplification was conducted as follows: Initial denaturation at 95°C for 10 min, followed by 30 denaturation cycles at 95°C for 60 sec, primer annealing at 55°C for 30 sec and primer extension at 72°C for 30 sec. Separation of the PCR products was performed on 2% agarose, followed by visualization with GelRed (Biotium, Inc., Fremont, CA, USA). The quality of RNA was examined by PCR amplification and agarose gel electrophoresis analysis of ABL gene transcripts.
Gene mutation detection of AML samples
FLT3 internal tandem duplication, NPM1 and C-KIT mutations were detected as reported previously (10,11). All positive samples were confirmed by direct Sanger sequencing using an ABI 3730 DNA analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc., Foster City, CA, USA), according to the manufacturers instructions. Furthermore, CEBPA mutations were detected by direct DNA sequencing. The mutations were used to identify risk stasus according to the NCCN Clinical Practice Guidelines in Oncology (2015).
RT-qPCR for the detection of STAG2 expression
Using the appropriate primers and PCR conditions, the expression of STAG2 was assessed by RT-qPCR according to the manufacturer's protocol. Briefly, total RNA was extracted from bone marrow mononuclear cells prepared with Ficoll gradient centrifugation, and then RNA concentrations were measured by fluorometry. Subsequently, cDNA synthesis was performed using an M-MLV reverse transcriptase kit (Promega Corp., Madison, WI, USA). For qPCR, the cDNA in samples was amplified using TaqMan Universal PCR Master Mix and a probe of STAG2 (Hs00198227_m1) in an Applied Biosystems 7500 Real-Time PCR system according to the manufacturer's protocol (all from Thermo Fisher Scientific, Inc.). Porphobilinogen deaminase (PBGD) was used as the internal control for relative quantification of gene expression in qPCR. The oligonucleotide sequences of the primers and probe are shown in Table II. STAG2 expression was quantified using the comparative 2−ΔCq method.
Table II.
Oligonucleotide primer and probe sequences used for quantitative polymerase chain reaction.
| Primer | Oligonucleotide sequence (5′-3′) |
|---|---|
| PBGD forward | GGCAATGCGGCTGCAG |
| PBGD reverse | GGGTACCCACGCGAATCAC |
| PBGD probe | CATCTTTGGGCTGTTTTCTTCCGCC |
PBGD, porphobilinogen deaminase.
Statistical analysis
The Mann-Whitney U test was used to analyze differences in STAG2 expression levels between the following groups: Patients and healthy controls; male and female AML patients; and abnormal and normal karyotype groups of AML patients. Kruskal-Wallis H test was used to compare the favorable, intermediate and poor risk subgroups of AML patients, as well as the age subgroups. Differences were deemed statistically significant for P-values of <0.05. Statistical analysis was performed using the SPSS software package (version 17.0; SPSS, Inc., Chicago, IL, USA).
Results
STAG2 expression in de novo AML
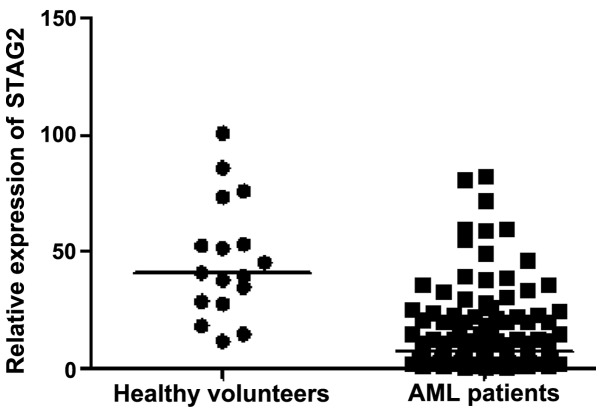
As shown in Fig. 1, the relative expression levels of STAG2 were non-normally distributed in 127 de novo AML patients (median value, 7.33; range, 0.60–82.44) and normally distributed in 17 healthy volunteers (median value, 40.91; range, 11.67–100.69). STAG2 expression was found to be significantly lower in AML patients when compared with that in the healthy volunteers (P<0.001). However, there was no statistically significant between-group difference in the gender distribution (P=0.890), or between the age subgroups in AML patients (P=0.288; data not shown).
Figure 1.
Comparison of STAG2 expression in acute myeloid leukemia patients and healthy volunteers. STAG2, stromal antigen 2.
As shown in Table I, the included AML patients were classified in the following FAB types: M0, M1, M2, M3, M4, M4EO, M5, M6 and M7. The median expression levels of STAG2 according to the FAB types are presented in Table III.
Table III.
STAG2 expression in de novo acute myeloid leukemia based on FAB type.
| FAB classification | Cases (n) | Median level of STAG2 expression (minimum, maximum) |
|---|---|---|
| M0 | 3 | 4.04 (2.60, 4.80) |
| M1 | 17 | 3.67 (0.63, 29.90) |
| M2 | 38 | 6.19 (0.60, 80.98) |
| M3 | 5 | 9.37 (5.55, 15.35) |
| M4 | 10 | 19.71 (2.39, 46.55) |
| M4EO | 17 | 8.64 (1.50, 59.90) |
| M5 | 32 | 8.36 (1.92, 82.44) |
| M6 | 4 | 9.91 (1.41, 22.69) |
| M7 | 1 | 36.26 |
| Total AML cases | 127 | 7.33 (0.60, 82.44) |
STAG2, stromal antigen 2; FAB, French-American-British.
Correlation of STAG2 expression with cytogenetic features
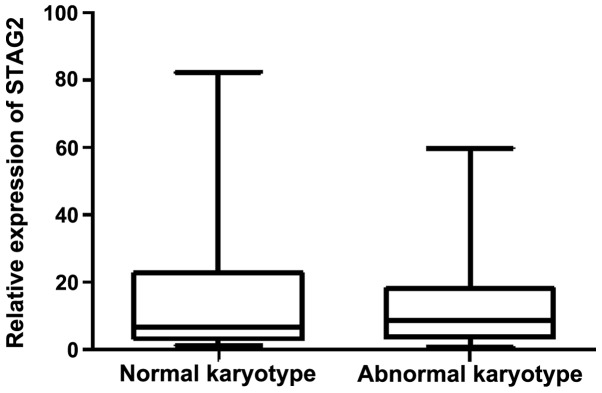
Among the 127 de novo AML patients, abnormal karyotypes were detected in 68 cases (53.5%), including t(8;21), t(15;17), inv(16), t(16;16), 11q23/MLL, complex abnormalities and other karyotypes (Table I). The median relative STAG2 expression levels in patients with abnormal and normal karyotypes were 8.59 (range, 0.60–59.95) and 6.71 (range, 1.41–82.44), respectively (Fig. 2). The results indicated that there was no significant difference in the STAG2 expression between the abnormal and normal karyotypes (P=0.896).
Figure 2.
Correlation of STAG2 expression with cytogenetic features. STAG2, stromal antigen 2.
Association of STAG2 expression with risk status determined based on cytogenetics and molecular abnormalities
The 127 de novo AML patients were divided into three risk groups: Favorable (24.4%; 31/127), intermediate (52.0%; 66/127) and poor 23.6% (30/127) risk. As shown in Fig. 3, the median relative STAG2 expression levels in these three groups were 9.37 (range, 1.50–59.90), 6.00 (range, 0.60–82.44) and 7.62 (range, 0.63–59.95), respectively. Between-group differences in STAG2 expression were not found to be statistically significant (P=0.503).
Figure 3.
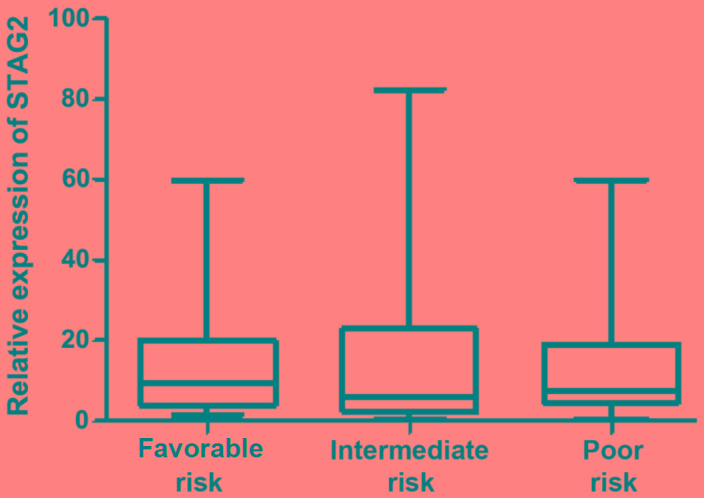
Association of STAG2 expression with risk status based on cytogenetics and molecular abnormalities. STAG2, stromal antigen 2.
Discussion
It has been suggested that STAG2 may be a ‘caretaker’ tumor suppressor gene that may lead to chromosomal instability when inactivated (5). In order to understand whether the expression of STAG2 is abnormal in AML, the present study determined the relative expression levels of STAG2 in 127 de novo AML patients and 17 controls (healthy volunteers) using RT-qPCR analysis.
The current results demonstrated a downregulated level of STAG2 gene expression in de novo AML when compared with that in the control subjects (P<0.001), suggesting that STAG2 gene may serve an important role in AML development. The downregulation in STAG2 expression may be attributed to genetic mutation or deletion. Deletions of chromosome Xq25, where STAG2 is located, are occasionally observed in AML and myelodysplasia (12,13), while STAG2 mutational hot spot regions (such as at exon 9, 11, 12 and 20) have been identified in acute leukemia (14). Mutations in the cohesin complex including STAG2 have recently been reported in a number of AML and myelodysplastic syndrome cases (15–20). In addition, Kon et al (21) reported the occurrence of multiple and recurrent mutations and deletions of cohesin complex genes, such as STAG2, RAD21, SMC1A and SMC3, in different myeloid neoplasms. Furthermore, a more recent study published in 2014 identified STAG2 as one of the most commonly mutated genes in bladder cancer (22). Furthermore, STAG2 may operate as a tumor suppressor through mechanisms that are distinct from those used to prevent aneuploidy. Previous observations have suggested that the recurrent inactivation of STAG2 (which is evident in bladder cancer) is not associated with aneuploidy (23).
The present study did not detected statistically significant differences in the gender distribution (P=0.890) and age subgroups (P=0.288) of the de novo AML patients. Furthermore, no association was observed between STAG2 expression and cytogenetic abnormalities, while no significant difference was identified in the levels of STAG2 expression between the favorable, intermediate and poor risk groups (P=0.503). These findings suggest that STAG2 gene expression is not associated with prognosis in AML patients.
In conclusion, the present study demonstrated the downregulation of STAG2 expression in de novo AML patients when compared with healthy controls; however, no significant difference in STAG2 expression was detected between the various risk groups. These results indicate that the downregulated STAG2 gene expression may contribute to AML development without being associated with AML prognosis. In order to investigate the underlying mechanism of the dysregulated STAG2 expression, the STAG2 gene in AML patients should be sequenced in future studies.
Acknowledgements
This study was supported in part by grants from the Priority Academic Program Development Project of Jiangsu Higher Education Institutions, Administration of Traditional Chinese Medicine of Jiangsu Province (LZ13207) and the Foundation of Jiangsu Province Health Department (Y201320).
References
- 1.Carramolino L, Lee BC, Zaballos A, Peled A, Barthelemy I, Shav-Tal Y, Prieto I, Carmi P, Gothelf Y, González de Buitrago G, et al. SA-1, a nuclear protein encoded by one member of a novel gene family: Molecular cloning and detection in hemopoietic organs. Gene. 1997;195:151–159. doi: 10.1016/S0378-1119(97)00121-2. [DOI] [PubMed] [Google Scholar]
- 2.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 4.Kim MS, Kim SS, Je EM, Yoo NJ, Lee SH. Mutational and expressional analyses of STAG2 gene in solid cancers. Neoplasma. 2012;59:524–529. doi: 10.4149/neo_2012_067. [DOI] [PubMed] [Google Scholar]
- 5.Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med. 1985;103:620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 7.Lower KM, Turner G, Kerr BA, Mathews KD, Shaw MA, Gedeon AK, Schelley S, Hoyme HE, White SM, Delatycki MB, et al. Mutations in PHF6 are associated with Börjeson-Forssman-Lehmann syndrome. Nat Genet. 2002;32:661–665. doi: 10.1038/ng1040. [DOI] [PubMed] [Google Scholar]
- 8.Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, De Moerloose B, Philippé J, González-García S, Toribio ML, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, Nicolas C, Payer AR, Fernandez HF, Tallman MS, et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011;25:130–134. doi: 10.1038/leu.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rocquain J, Carbuccia N, Trouplin V, Raynaud S, Murati A, Nezri M, Tadrist Z, Olschwang S, Vey N, Birnbaum D, et al. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer. 2010;10:401. doi: 10.1186/1471-2407-10-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boissel N, Leroy H, Brethon B, Philippe N, de Botton S, Auvrignon A, Raffoux E, Leblanc T, Thomas X, Hermine O, et al. Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML) Leukemia. 2006;20:965–970. doi: 10.1038/sj.leu.2404188. [DOI] [PubMed] [Google Scholar]
- 12.Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, Baty J, Heath S, Westervelt P, Watson MA, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA. 2009;106:12950–12955. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rocquain J, Gelsi-Boyer V, Adélaïde J, Murati A, Carbuccia N, Vey N, Birnbaum D, Mozziconacci MJ, Chaffanet M. Alteration of cohesin genes in myeloid diseases. Am J Hematol. 2010;85:717–719. doi: 10.1002/ajh.21798. [DOI] [PubMed] [Google Scholar]
- 14.Chung NG, Kim MS, Yoo NJ, Lee SH. Somatic mutation of STAG2, an aneuploidy-related gene, is rare in acute leukemias. Leuk Lymphoma. 2012;53:1234–1235. doi: 10.3109/10428194.2011.645819. [DOI] [PubMed] [Google Scholar]
- 15.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, Larson DE, McLellan MD, Dooling D, Abbott R, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network, corp-author. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter MJ, Shen D, Shao J, Ding L, White BS, Kandoth C, Miller CA, Niu B, McLellan MD, Dees ND, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013;27:1275–1282. doi: 10.1038/leu.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cazzola M, Porta MG Della, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122:4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 22.Black P. Frequent truncating mutations of STAG2 in bladder cancer. Urology. 2014;83:691–692. doi: 10.1016/j.urology.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 23.Balbás-Martínez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Márquez M, Vazquez M, Lapi E, Castro-Giner F, Beltran S, Bayés M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45:1464–1469. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]