Abstract
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-associated mortality in China and the third leading cause worldwide. A number of microRNAs (miRNAs) have been implicated in cell cycle progression, growth, apoptosis, angiogenesis and metastasis in HCC. In the present study, reverse transcription-quantitative polymerase chain reaction analysis was used to detect the levels of miR-302d expression in the tissues of 30 patients with HCC. Cell cycle, growth, apoptosis and migration were analyzed using a cell counting kit, flow cytometry and a Transwell migration assay. Dual-luciferase reporter assays and western blotting were also used to analyze the expression levels of transforming growth factor beta type II receptor (TGFBR2) in HCC cells. The present study evaluated the role of miR-302d in the development and progression of HCC. Abnormally high expression of miR-302d was observed in 80% of HCC specimens. Moreover, patients with lower levels of miR-302d expression experienced a longer survival time than those with higher levels of miR-302d expression. It was demonstrated that miR-302d promoted HCC cell growth and migration, suppressed cell apoptosis and affected cell cycle distribution in vitro, and augmented tumorigenicity in vivo. Furthermore, TGFBR2, which is a tumor suppressor, was confirmed as a target of miR-302d in HCC cells. Dual-luciferase reporter assays indicated that TGFBR2 expression was negatively regulated by miR-302d. Taken together, the results of the present study suggest that miR-302d may serve as a valuable tool for predicting the prognosis of patients with HCC.
Keywords: microRNA-302d, hepatocellular carcinoma, tumorigenicity, tumor promoter, transforming growth factor beta type II receptor
Introduction
Hepatocellular carcinoma (HCC) has been ranked as the second leading cause of cancer-associated mortality in China and the third leading cause of cancer mortality worldwide (1,2). Incidence and mortality rates of HCC continue to increase in various countries across the world, including the United States (3). Primary causes of HCC include alcohol consumption, dietary exposure to aflatoxin B1 and infection with hepatitis virus (4). Although numerous risk factors have been identified, the pathogenetic mechanisms underlying HCC development and progression remain unclear. Investigating the molecular mechanisms that contribute to HCC may enable the development of novel therapeutic strategies and improve patient prognosis.
MicroRNAs (miRNAs) are endogenous small noncoding RNAs and act as oncogenes or tumor suppressors in cancer (5). Previous studies investigating HCC have demonstrated that miRNA expression profiles are signatures of HCC development and progression (6–8). Various miRNAs, including miR-302d, have been implicated in the onset and development of HCC (9). The identification of novel functions of specific miRNAs may help elucidate the molecular mechanisms that underlie HCC development and progression.
It has been reported that miR-302d is highly expressed in human embryonic stem cells and, as such, it is recognized as a human embryonic stem cell-specific miRNA (10). miR-302d directly contributes to the regulation of p21 expression and governs the G1/S transition checkpoint in human embryonic stem cells (11). By inhibiting p21, miR-302d rescues Ras-induced senescence in human mammary epithelial cells (12). Additionally, miR-302d increases cell proliferation and inhibits cell death induced by oxidizing agents in human adipose tissue-derived mesenchymal stem cells (13). Increasing evidence suggests that embryogenesis and tumorigenesis involve the activation of the same pathways; therefore, miR-302d may perform the same functions during tumorigenesis as occurs during embryogenesis (14). However, the role of miR-302d in HCC remains unclear. The microarray expression profile of HCC indicates that miR-302d is highly expressed in HCC in the present study (data not shown). In the present study, in vitro and in vivo studies were performed to determine whether miR-302d promotes the proliferation and migration of HCC and enhances tumor growth, by investigating the association between miR-302d and transforming growth factor beta type II receptor (TGFBR2).
Materials and methods
Patients and tissue samples
A total of 140 patients with stage I–IV HCC admitted to the Department of Infectious Diseases, First Affiliated Hospital of Zhejiang University (Hangzhou, China) between February 2011 and October 2014 were enrolled in the present study. The inclusion criteria of the current study were that no patients had received radiotherapy or chemotherapy. Complete clinical and pathological follow-up data was available for 110 patients, who were grouped into two groups according to their median levels of miR-302d expression [high expression of miR-302d (n=55) vs. low expression of miR-302d (n=55)] and the association between miR-302d expression and the survival of patients following surgery was examined. The survival of HCC patients was defined as 0 and mortality of HCC patients was defined as 1, and the survival time of patients was analyzed using GraphPad Prism software ver. 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Specimens of HCC and paired non-cancerous tissues from surrounding adjacent areas were obtained from 30 patients via surgical resection. All tissues were snap frozen in liquid nitrogen immediately after resection to be used in subsequent experiments to measuremiR-302d expression. Ethical approval for the present study was provided by the Independent Ethics Committee of The First Affiliated Hospital of Zhejiang University. Informed and written consent was obtained from all patients or their family members in accordance with the Ethics Committee guidelines.
Cell culture and transfection
Cells from the SMMC-7721 human hepatocellular carcinoma cell line were purchased from JRDUN Biotechnology Co., Ltd., (Shanghai, China) and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified chamber containing 5% CO2. miR-302d mimics and inhibitors were purchased from Shanghai GenePharma Co., Ltd., (Shanghai, China). For transient transfection of cells in 6-well plates, 100 µM mimics or inhibitors were added with Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in OPTI-MEM media (Gibco; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from normal and HCC tissue using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA was synthesized from RNA using a MMLV RT reagent kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A specific primer (5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCACACTC-3′) was used to synthesize miR-302d cDNA. cDNA were amplified using a SYBRGreen PCR kit (Thermo Fisher Scientific, Inc.). PCR primers for miR-302d were as follows: Forward, 5′-ACAGTGTCGTTCACGGAGGT-3′ and reverse, 5′-AACTGGTGTCGTGGAGTCGGC-3′. PCR amplification was performed on an Applied Biosystems 7500 real-time PCR platform (Thermo Fisher Scientific, Inc.). The PCR cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 45 sec, and a final extension step of 95°C for 15 sec, 60°C for 1 min, 95°C for 15 sec and 60°C for 15 sec. The experiment was repeated three times. To normalize miR-302d, 5S rRNA was used as an internal standard. PCR primers for 5S rRNA were as follows: Forward, 5′-CCATACCACCCTGGAAACGC-3′ and reverse, 5′-TACTAACCGAGCCCGACCCT-3′. Relative quantification of miR-302d expression levels was determined using the 2−ΔΔCq method (15).
Cell growth analysis
Cells were seeded in a 96-well plate (1×105 cells/well) and cultured at 37°C for 24 h. Cells were subsequently transfected with miRNA mimics, antisense or negative controls (NC; untreated cells) for 72 h. Cell growth was assessed every 24 h using a cell counting kit (CCK)-8 assay (Beyotime Institute of Biotechnology, Shanghai, China). All samples were evaluated in triplicate for each group and the experiment was repeated at least twice.
Cell cycle analysis
To determine cell cycle properties, cells were transfected with miRNA mimics, antisense or NC. After 36 h, transfected cells were harvested by trypsinization and incubated with 0.5 µg/ml RNase A (Thermo Fisher Scientific, Inc.) and 100 µg/ml propidium iodide (PI) for 30 min at room temperature prior to fluorescence-activated cell sorting with a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Apoptosis assay
For the analysis of apoptosis, cells were seeded into 6-well plates and transfected with miRNA mimics, antisense or NC. At 48 h after transfection, cells were collected, washed and stained using the Annexin V fluorescein isothiocyanate/PI double staining kit (BD Biosciences) according to the manufacturer's instructions. A flow cytometer was used to analyze apoptotic cells (BD Biosciences).
In vitro migration assays
Cell migration was assessed using Transwell insert chambers (Corning, Inc., Corning, NY, USA). Cells transfected with miRNA mimics, antisense or NC were re-suspended in 100 µl RPMI1640 and placed into the upper chamber without Matrigel. RPMI1640 supplemented with 10% FBS was added to the lower chamber and functioned as the chemoattractant. Cells were incubated for 24 h. Migrated cells were fixed with 4% polyoxymethylene, stained with 0.2% crystal violet and imaged under an inverted microscope (Olympus Corp., Tokyo, Japan) at a magnification of ×200.
Tumor growth assay
A total of 12 male BALB/c nude mice (15–18 g), aged 4–5 weeks old, were purchased from Shanghai SLAC Laboratory Animal Co., Ltd., (Shanghai, China). Mice were equally divided into two groups (n=6) and received a subcutaneous injection in their right flank of either 1×107 SMMC-7721 cells stably expressing miR-302d inhibitor or SMMC-7721 cells without treatment (the NC). Mice were housed in the animal facility at a temperature of 25°C and humidity of 60–70% and subjected to a 12-h light-dark cycle with ad libitum access to food and water. Mice were observed over 6 weeks for tumor formation. Mice were subsequently sacrificed by intraperitoneal injection of 3% sodium pentobarbital (40 mg/kg; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and cervical dislocation was performed. The tumors were subsequently resected and the wet weights of each tumor were measured.
Western blot analysis
SMMC-7721 cells transfected with miRNA mimics, antisense or NC were lysed using radioimmunoprecipitation assay buffer supplemented with protease inhibitor (Beyotime Institute of Biotechnology). Protein concentration was estimated using the BCA Protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of protein (20 µg) were separated by 12% SDS-PAGE and transferred to nitrocellulose membranes (EMD Millipore, Billerica, MA, USA). Following blocking with fat-free milk for 1 h at 25°C, membranes were immunoblotted overnight at 4°C with primary antibodies against GAPDH (cat. no. 2251-1; 1:1,500; Fermentas; Thermo Fisher Scientific, Inc.) and TGFBR2 (cat. no. ab61213; 1:800; Abcam, Cambridge, MA, USA). Following washing, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (Ig) G (cat. no. A0208; 1,000; Beyotime Institute of Biotechnology) and goat anti-mouse IgG (cat. no. A0216; 1:1,000; Beyotime Institute of Biotechnology) secondary antibodies at 37°C for 1 h. Signals were detected using an enhanced chemiluminescence Western Blotting substrate (Pierce; Thermo Fisher Scientific, Inc.).
Luciferase reporter assays
TGFBR2 was predicted to interact with miR-302d by bioinformatics analysis using TargetScan (http://www.targetscan.org), which predicts biological targets of miRNAs by searching for the presence of 8 oligomer, 7 oligomer, and 6 oligomer sites that match the seed region of each miRNA (16). The 3′-untranslated region (UTR) of the human TGFBR2 gene predicted to interact with miR-302d was synthesized and inserted in the pGL3 vector (Promega Corp., Madison, WI, USA), downstream of the firefly luciferase gene to form pGL3-TGFBR2. SMMC-7721 cells (2×105/well) were seeded in 96-well plates, reporter plasmids were co-transfected with miR-302d mimics or NC using Lipofectamine 2000. Following 24 h, cells were lysed and the activities of Renilla and firefly luciferase were examined using the Dual-Luciferase Reporter assay system (Promega Corp.). Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software ver. 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Overall survival in relation to miR-302d expression was evaluated by Kaplan-Meyer survival curves and the log-rank nonparametric test. The results from different groups were compared using two-tailed Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Upregulated miR-302d expression is associated with poorer patient survival rates
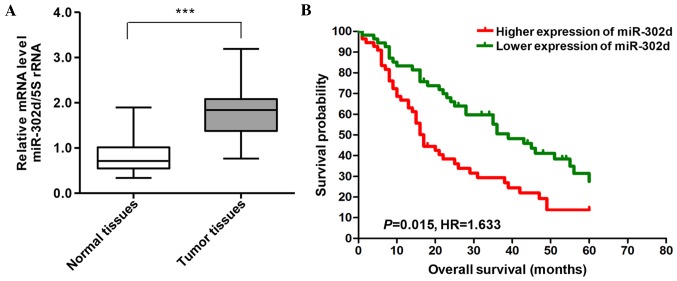
The microarray expression profile of HCC indicated that miR-302d is highly expressed in HCC (data not shown). To further verify this finding, RT-qPCR was performed on 30 pairs of HCC tissue and adjacent healthy tissue samples. It was detected that levels of miR-302d expression were significantly higher in the HCC tissues compared with those in adjacent noncancerous tissue (P<0.001; Fig. 1A). High expression of miR-302d (>1.50-fold) was more frequently observed in HCC tissue (24/30; 80%) than in noncancerous adjacent tissue (Fig. 1A). The HCC patients were divided into two groups according to the median level of miR-302d. Subsequently, the association between miR-302d expression and the survival of patients following surgery was examined. The results demonstrated that patients with lower levels of miR-302d expression experienced longer survival times than those with higher levels of miR-302d expression (P=0.015; Fig. 1B).
Figure 1.
Association between miR-302d expression and survival time of patients with HCC. (A) Analysis of miR-302d expression levels in 30 pairs of HCC samples and adjacent healthy tissue by reverse transcription-quantitative polymerase chain reaction. miR-302d expression was normalized to 5S. (B) Effect of levels of miR-302d expression on the overall survival of patients with HCC. The cut-off level was set at the median value of the miR-302d expression levels in 110 HCC patients. The group of patients with high miR-302d expression experienced poorer prognoses than that of the group with low miR-302d expression. ***P<0.001. miR, microRNA; HCC, hepatocellular carcinoma; HR, hazard ratio.
miR-302d increases HCC cell growth in vitro
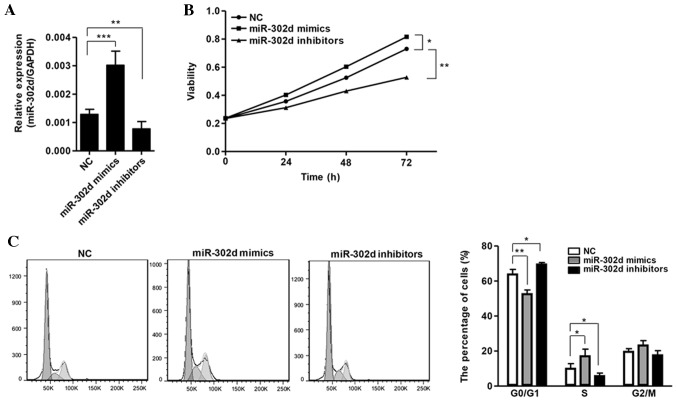
To determine the role of miR-302d in HCC progression, miR-302d mimics, inhibitors or NCs were transfected into SMMC-7721 cells (Fig. 2A). CCK-8 assays indicated that the viability of the miR-302d mimics group was significantly improved compared with the NC group (P<0.05), while the viability of the miR-302d inhibitor group was significantly reduced compared with the NC group (P<0.01; Fig. 2B). Cell cycle distribution analysis demonstrated that miR-302d overexpressing SMMC-7721 cells exhibited a significantly decreased percentage of cells in the G0/G1 phase (P<0.01) and a significant increase in the percentage of cells in the S-phase, compared with NC-transfected cells (P<0.05; Fig. 2C). By contrast, in the miR-302d inhibitor group, there was a significant increase in the percentage of cells in the G0/G1 phase (P<0.05) and a significant decrease in the percentage of cells in the S-phase, compared with the NC group (P<0.05; Fig. 2C).
Figure 2.
miR-302d promoted hepatocellular carcinoma cell growth and migration. (A) Relative expression levels of miR-302d were measured using reverse transcription-quantitative polymerase chain reaction in SMMC-7721 cells following transfection of miR-302d mimics, inhibitors or NC. (B) Cell viability was determined in SMMC-7721 cells transfected with miR-302d mimics, inhibitors or NC by CCK-8 assays. (C) The proportion of cells in each phase of the cell cycle was determined in SMMC-7721 cells following transfection of miR-302d mimics, inhibitors or NC by FACS. Histogram indicates the percentage of cells at the G0/G1, S and G2/M phases. *P<0.05, **P<0.01, ***P<0.001. NC, negative control; FACS, fluorescence-activated cell sorting; miR, microRNA; CCK-8, cell counting kit-8.
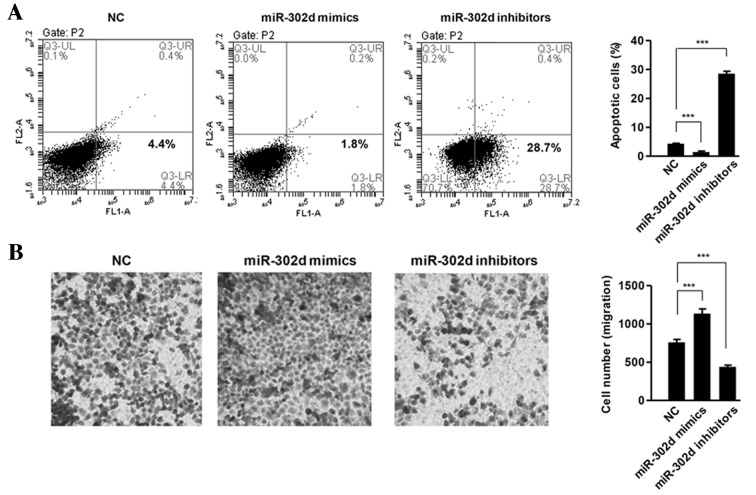
miR-302d inhibits apoptosis and promotes migration in HCC cells
The effect of miR-302d on apoptosis was assessed using fluorescence-activated cell sorting. The results demonstrated that cells overexpressing miR-302d exhibited a significant decrease in the percentage of cells undergoing apoptosis compared with NC (P<0.001), whereas cells expressing miR-302d inhibitors exhibited a significant increase in the percentage of apoptotic cells (P<0.001; Fig. 3A). In vitro migration assays indicated that miR-302d mimics significantly promoted the motility of SMMC-7721 cells (P<0.001), whereas inhibition of miR-302d significantly suppressed the motility of SMMC-7721 cells (P<0.001; Fig. 3B).
Figure 3.
miR-302d inhibited apoptosis. (A) Apoptosis was determined in SMMC-7721 cells following transfection of miR-302d mimics, inhibitors or NC by FACS. Histogram indicates the percentage of cells at apoptosis. (B) Cell migration abilities were determined in SMMC-7721 cells transfected with miR-302d mimics, inhibitors or NC by Transwell assay. Magnification, ×200. ***P<0.001. NC, negative control; FACS, fluorescence-activated cell sorting; miR, microRNA; CCK-8, cell counting kit-8.
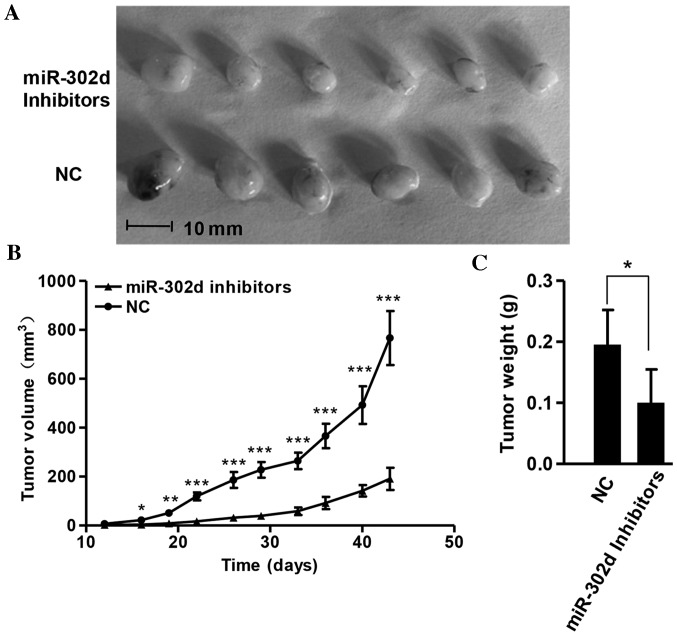
miR-302d promotes xenograft tumor growth in vivo
To further determine the function of miR-302d, its effect on tumorigenicity was examined using a mouse model. Mice injected with SMMC-7721 cells stably expressing miR-302d inhibitors exhibited tumors that grew more slowly than those of mice injected with NC. These differences were significantly increased until the end of the evaluation period at 6 weeks (P<0.05; Fig. 4A and B). Tumor weights were consistently decreased in miR-302d inhibitor expressing tumors compared with the controls (P<0.05; Fig. 4C). These data indicate that miR-302d may promote tumor growth in vivo.
Figure 4.
miR-302d inhibitors suppressed tumor growth in vivo. (A) Xenograft tumors from nude mice injected with SMMC-7721 cells stably expressing miR-302d inhibitors or NC. (B) Growth curve of tumor volumes. (C) Tumor weights. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; NC, negative control.
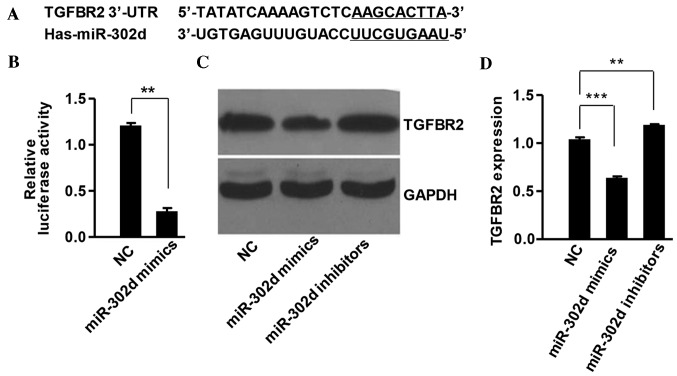
miR-302d directly targets TGFBR2 in HCC cells
To explore the regulation mechanism of miR-302d, TargetScan bioinformatics analysis (www.targetscan.org) was employed. TargetScan identified that TGFBR2 mRNA contained potential binding sites of miR-302d (Fig. 5A). To confirm that TGFBR2 was a target of miR-302d and regulated by it in HCC cells, the TGFBR2 3′-UTR was cloned and inserted into a luciferase reporter vector. The luciferase assay indicated that miR-302d significantly suppressed luciferase activity in the vector containing the TGFBR2 3′-UTR (P<0.01; Fig. 5B). Western blot analysis demonstrated that miR-302d overexpression significantly suppressed endogenous TGFBR2 expression (P<0.001), whereas inhibition of miR-302d significantly increased levels of TGFBR2 protein in SMMC-7721 cells (P<0.01; Fig. 5C and D). These results suggest that TGFBR2 is a target of miR-302d in HCC cells.
Figure 5.
miR-302d directly targeted TGFBR2 in hepatocellular carcinoma cells. (A) Base-pairing interaction between miR-302d seed sequences and TGFBR2, as predicted by TargetScan. (B) Luciferase assay. SMMC-7721 cells were co-transfected with pGL3-TGFBR2 and miR-302d mimics, inhibitors or NC. Firefly luciferase activity was normalized to Renilla luciferase activity. (C and D) TGFBR2 protein levels were examined in SMMC-7721 cells transfected with miR-302d mimics, inhibitors or NC. **P<0.01 and ***P<0.001. miR, microRNA; TGFBR2, transforming growth factor beta type II receptor; NC, negative control; 3′UTR, 3′ untranslated region.
Discussion
HCC is the third leading cause of cancer-associated mortality worldwide (1). However, the molecular mechanisms responsible for the development and progression of HCC remain largely unknown. It has been reported that miRNAs are associated with HCC development and progression and, according to their function in tumorigenesis, they may act as oncogenes or tumor suppressors (5). Various miRNAs are oncogenic in HCC, including miR-21, miR-221, miR-222 and miR-151; however it has been suggested that a number of miRNAs, including miR-29, miR-122, miR-125b, miR-101 and miR-139, may be tumor suppressive in HCC (17). It has previously been demonstrated that in HCC, miRNAs are involved in the regulation of cell survival and proliferation, angiogenesis and metastasis (9,17).
In the present study, results from RT-qPCR indicated that miR-302d is highly expressed in HCC, and a correlation was observed between increased miR-302d expression and decreased patient survival time. The data collected suggest that miR-302d promotes cell proliferation and migration, suppresses cell apoptosis, affects the cell cycle distribution of HCC cells in vitro, enhances tumorigenicity in vivo and may be used as a prognostic indicator of survival in patients with HCC. Therefore, miR-302d may be a novel oncogene in HCC.
miR-302d belongs to the miR-302 family, which consists of four highly homologous miRNA members, including miR-302c, miR-302b, miR-302d, mi-302a and miR-367 (18). It has been suggested that the expression of different forms of miR-302 may be associated with tumor progression (19,20). Furthermore, it has been identified that miR-302b suppresses the proliferation of HCC SMMC-7721 cells (21). However, it has also been demonstrated that miR-302d increases cell proliferation and inhibits cell death in human adipose tissue-derived mesenchymal stem cells (13). Therefore, miR-302d may promote tumor growth. This is consistent with results from the present study that implicate miR-302d as a novel oncogene in HCC.
Furthermore, the mechanism by which miR-302d influences the development of HCC was investigated. TGFBR2 was predicted to be the potential target gene of miR-302d by bioinformatic algorithms. A luciferase activity assay indicated that miR-302d directly targeted the 3′-UTR of TGFBR2 mRNA. In addition, miR-302d overexpression significantly downregulated the expression of TGFBR2. These results suggest that miR-302d may function as a novel oncogene in HCC and contribute to tumor progression in HCC.
TGF-β signaling is tumor suppressive in numerous different cell types (22). Reduced expression or loss of the TGFβ type I and type II receptors (TGFBR1 and TGFBR2) has been reported in various types of cancer, including HCC (23–25). TGFBR2 has been identified as the direct target of numerous miRNAs, including miR-655, miR-520c, miR-373 and miR-211 (26). The results of the present study suggest that TGFBR2 is a novel target of miR-302d in HCC and may partially explain the mechanism by which miR-302d functions in HCC, and why TGFBR2 expression is reduced in HCC.
In conclusion, the results of the present study provide novel evidence that miR-302d negatively regulates TGFBR2 expression, promotes HCC cell growth and migration, suppresses cell apoptosis and affects cell cycle distribution in vitro, and promotes tumorigenicity in vivo. Therefore, miR-302d may be a novel tumor promoter in HCC and may be developed as a novel therapeutic strategy to treat patients with HCC.
References
- 1.He J, Gu D, Wu X, Reynolds K, Duan X, Yao C, Wang J, Chen CS, Chen J, Wildman RP, et al. Major causes of death among men and women in China. N Engl J Med. 2005;353:1124–1134. doi: 10.1056/NEJMsa050467. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 3.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen VT, Law MG, Dore GJ. Hepatitis B-related hepatocellular carcinoma: Epidemiological characteristics and disease burden. J Viral Hepat. 2009;16:453–463. doi: 10.1111/j.1365-2893.2009.01117.x. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Varnholt H, Drebber U, Schulze F, Reynolds K, Duan X, Yao C, Wang J, Chen CS, Chen J, Wildman RP. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 7.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei R, Huang GL, Zhang MY, Li BK, Zhang HZ, Shi M, Chen XQ, Huang L, Zhou QM, Jia WH, et al. Clinical significance and prognostic value of microRNA expression signatures in hepatocellular carcinoma. Clin Cancer Res. 2013;19:4780–4791. doi: 10.1158/1078-0432.CCR-12-2728. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, Lu H, Wang X, Jin H. MicroRNAs in hepatocellular carcinoma: Regulation, function, and clinical implications. ScientificWorldJournal. 2013;2013:924206. doi: 10.1155/2013/924206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li SS, Yu SL, Kao LP, Tsai ZY, Singh S, Chen BZ, Ho BC, Liu YH, Yang PC. Target identification of microRNAs expressed highly in human embryonic stem cells. J Cell Biochem. 2009;106:1020–1030. doi: 10.1002/jcb.22084. [DOI] [PubMed] [Google Scholar]
- 11.Dolezalova D, Mraz M, Barta T, Plevova K, Vinarsky V, Holubcova Z, Jaros J, Dvorak P, Pospisilova S, Hampl A. MicroRNAs regulate p21(Waf1/Cip1) protein expression and the DNA damage response in human embryonic stem cells. Stem Cells. 2012;30:1362–1372. doi: 10.1002/stem.1108. [DOI] [PubMed] [Google Scholar]
- 12.Borgdorff V, Lleonart ME, Bishop CL, Fessart D, Bergin AH, Overhoff MG, Beach DH. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1) Oncogene. 2010;29:2262–2271. doi: 10.1038/onc.2009.497. [DOI] [PubMed] [Google Scholar]
- 13.Kim JY, Shin KK, Lee AL, Kim YS, Park HJ, Park YK, Bae YC, Jung JS. MicroRNA-302 induces proliferation and inhibits oxidant-induced cell death in human adipose tissue-derived mesenchymal stem cells. Cell Death Dis. 2014;5:e1385. doi: 10.1038/cddis.2014.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Zhao L, Xiao Q, Jiang L, He M, Bai X, Ma M, Jiao X, Wei M. miR-302a/b/c/d cooperatively inhibit BCRP expression to increase drug sensitivity in breast cancer cells. Gynecol Oncol. 2016;141:592–601. doi: 10.1016/j.ygyno.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Wong CM, Kai AK, Tsang FH, Ng IO. Regulation of hepatocarcinogenesis by microRNAs. Front Biosci (Elite Ed) 2013;5:49–60. doi: 10.2741/E595. [DOI] [PubMed] [Google Scholar]
- 18.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barroso-delJesus A, Romero-López C, Lucena-Aguilar G, Melen GJ, Sanchez L, Ligero G, Berzal-Herranz A, Menendez P. Embryonic stem cell-specific miR302-367 cluster: Human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Yao J, Shi X, Hu L, Li Z, Song T, Huang C. MicroRNA-302b suppresses cell proliferation by targeting EGFR in human hepatocellular carcinoma SMMC-7721 cells. BMC Cancer. 2013;13:448. doi: 10.1186/1471-2407-13-448. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 23.Ikushima H, Miyazono K. TGFbeta signalling: A complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 24.Bjerke GA, Pietrzak K, Melhuish TA, Frierson HF, Jr, Paschal BM, Wotton D. Prostate cancer induced by loss of Apc is restrained by TGFβ signaling. PLoS One. 2014;9:e92800. doi: 10.1371/journal.pone.0092800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mamiya T, Yamazaki K, Masugi Y, Mori T, Effendi K, Du W, Hibi T, Tanabe M, Ueda M, Takayama T, Sakamoto M. Reduced transforming growth factor-beta receptor II expression in hepatocellular carcinoma correlates with intrahepatic metastasis. Lab Invest. 2010;90:1339–1345. doi: 10.1038/labinvest.2010.105. [DOI] [PubMed] [Google Scholar]
- 26.Harazono Y, Muramatsu T, Endo H, Uzawa N, Kawano T, Harada K, Inazawa J, Kozaki K. miR-655 is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One. 2013;8:e62757. doi: 10.1371/journal.pone.0062757. [DOI] [PMC free article] [PubMed] [Google Scholar]