Abstract
Type 1 diabetes mellitus (T1DM) is caused by progressive autoimmune-mediated loss of pancreatic β-cell mass via apoptosis. The onset of T1DM depends on environmental factors that interact with predisposing genes to induce an autoimmune assault against β cells. Epidemiological, clinical and pathology studies in humans support viral infection — particularly by enteroviruses (for example, coxsackievirus) — as an environmental trigger for the development of T1DM. Many candidate genes for T1DM, such as MDA5, PTPN2 and TYK2, regulate antiviral responses in both β cells and the immune system. Cellular permissiveness to viral infection is modulated by innate antiviral responses that vary among different tissues or cell types. Some data indicate that pancreatic islet α cells trigger a more efficient antiviral response to infection with diabetogenic viruses than do β cells, and so are able to eradicate viral infections without undergoing apoptosis. This difference could account for the varying ability of islet-cell subtypes to clear viral infections and explain why chronically infected pancreatic β cells, but not α cells, are targeted by an autoimmune response and killed during the development of T1DM. These issues and attempts to target viral infection as a preventive therapy for T1DM are discussed in the present Review.
Type 1 diabetes mellitus (T1DM) arises when the pancreatic β cells undergo long-term autoimmune attack, killing the majority of the β-cell population while the neighbouring α cells and δ cells are spared1. Destruction of the β cells manifests as a failure to produce insulin; consequently, patients with T1DM remain insulin-dependent for their lifespan.
In most cases, T1DM is characterized by pancreatic islet inflammation (insulitis) and progressive β-cell loss by apoptosis1,2. Histological analysis has demonstrated the presence of increased β-cell apoptosis among both patients with new-onset T1DM and those with T1DM of long duration3,4. The defective insulin release characteristic of T1DM reflects progressive β-cell destruction in the range of 60–100%, depending on disease duration4–7, as well as functional defects (for example, defective glucose-induced insulin release and delayed conversion of proinsulin to insulin2,8) that are probably caused by local release of proinflammatory mediators by infiltrating immune cells2,9,10. Pancreatic pathology among patients with T1DM is heterogeneous, with varying degrees of insulitis and β-cell loss observed in different lobes of the pancreas1,10.
The genetic basis of T1DM is well established, with >50 candidate genes identified to date that explain ~80% of disease heritability11,12. Nevertheless, the estimated 1.5% annual rise in the incidence of T1DM in high-income countries1,13, the observation that migration changes the risk of T1DM according to the country of residence14, and differences in the penetrance rate between genetically similar populations, including monozygotic twins15, all point to the contribution of nongenetic variables in the pathogenesis of this disease. The triggering of T1DM, therefore, probably depends on environmental factors that interact with predisposing genes to induce an auto-immune assault against the pancreatic β cells2,12. Among the potential environmental factors, epidemiological, clinical and pathology studies in humans support a role for viral infections, particularly by enteroviruses (for example, coxsackievirus), as triggers for the development of T1DM16 (BOX 1).
In this Review, we will discuss potential mechanisms by which enteroviruses could contribute to the specific destruction of pancreatic β cells in T1DM, focusing on data obtained in clinical studies and human samples. Emphasis is given to the role of enteroviruses in the induction of insulitis and T1DM; how candidate genes for T1DM modulate the host response to the viral infection; and why pancreatic β cells are particularly susceptible to these infections. This issue is timely given that major advances in this area have occurred in the past 5 years, providing support for the role of viruses in the pathogenesis of T1DM.
Viruses as environmental triggers
The growing evidence for the role of viruses in T1DM has been examined in detail elsewhere16–20; therefore, this aspect will be only briefly discussed here.
The most striking evidence comes from cases of fulminant T1DM, which are particularly prevalent in Japan. In contrast to the slow progression and auto-antibody positivity usually observed in classic T1DM, fulminant T1DM is characterized by an abrupt onset of insulin-deficient hyperglycaemia and ketoacidosis among individuals without detectable autoantibodies21. The high prevalence (>70%) of preceding common-cold-like and gastrointestinal symptoms strongly suggests an infectious origin for fulminant T1DM21. Indeed, the onset of fulminant T1DM has been reported during acute infection with mumps, parainfluenza, human herpes virus 6 and enteroviruses21. The intense inflammation in the injured pancreas in response to the presence of viruses that is observed among patients with fulminant T1DM strongly suggests that pancreatic viral replication is linked to a devastating immune response, destroying not only β cells but also the surrounding exocrine pancreatic tissue22.
A frequent association has been observed between classic T1DM and enterovirus infections. Epidemiological studies identified an increased incidence of T1DM following enterovirus epidemics23. Furthermore, enteroviral RNA has been detected in the blood of patients with newly diagnosed T1DM24 and serological analysis confirmed a link between enteroviral infection and T1DM25, particularly for the coxsackievirus B1 (CVB1) serotype26. The presence of anti-CVB1 antibodies was also associated with an increased risk of β-cell auto-immunity27. A meta-analysis of 24 studies that included a total of 4,448 participants reported a clinically significant association between the presence of molecular markers of enteroviral infections, autoimmunity related to diabetes mellitus and T1DM28. Of particular relevance, enteroviral RNA was detected by in situ hybridization in the islets of four patients with new-onset T1DM29 and expression of the viral capsid protein VP1 was detected by immunohistochemistry in the islets of >60% of brain-dead organ donors with T1DM versus 8% among individuals without T1DM30–33. Only insulin-containing islets were positive for VP1, which indicates that coxsackievirus is able to infect β cells and to persist in the pancreatic islets of patients with T1DM. Coxsackieviruses isolated from pancreatic biopsy samples taken from six living patients with newly diagnosed T1DM failed to efficiently amplify in vitro, which suggests that despite persistent infection, the virus was poorly replicative33.
A naturally occurring deletion at the 5′ terminus of the coxsackievirus genome enables chronic infection in mouse and human myocardium and in the pancreata of nonobese diabetic (NOD) mice, a model of spontaneous autoimmune diabetes mellitus34,35. Viruses harbouring this deletion are less replicative than wild-type viruses and are noncytopathic35. Whether this deletion is present in the VP1-positive β cells of patients with T1DM remains unclear. This 5′ terminus deletion could explain the long-term noncytopathic persistence of coxsackie-virus in the pancreatic islets, contributing to the β-cell autoimmunity characteristic of T1DM.
Finally, the absence of insulitis in 60% of insulin-containing VP1-positive islets36, and in pancreatic tissue from an autoantibody-positive nondiabetic child37, suggests that viral infection precedes the onset of insulitis. The link between coxsackievirus infection and T1DM is thus more likely to be causal than opportunistic.
Autoimmunity and β-cell death
Loss of β cells and triggering of insulitis after viral infection could result from several nonexclusive mechanisms. During the acute phase of infection, these effects might be a direct consequence of viral amplification and an excessive antiviral immune response to destroy infected cells among genetically susceptible individuals. This putative cell destruction is particularly deleterious because of the limited capacity of human β cells to proliferate38 and thus compensate for their loss. Later on, progressive β-cell loss might be secondary to the activation of autoreactive39,40 and bystander CD8+ T cells41,42, leading to progressive destruction of insulin-producing cells in the course of a chronic autoimmune assault. Conversely, viruses such as lymphocytic choriomeningitis virus or the coxsackievirus B3 (CVB3) serotype seem to protect against T1DM in animal models by either reducing or abrogating the autoimmune process43,44. Lymphocytic choriomeningitis virus and CVB3 do not inflict any damage on β cells and protect against T1DM by triggering immunoregulatory mechanisms by at least two pathways. The first pathway involves upregulation of programmed cell death-1 ligand 1 on lymphoid cells, which prevents the expansion of diabetogenic CD8+ T cells expressing programmed cell death-1. The second pathway enhances the number of CD4+CD25+Foxp3+ regulatory T cells that produce TGF-β, which is crucial for the maintenance of immune tolerance in the periphery. The mechanisms leading to virus-induced autoimmunity and β-cell death are discussed in detail in the following sections.
Epitope spreading
During the acute phase of infection, viruses that specifically target pancreatic β cells replicate in these cells, directly leading to β-cell destruction and induction of a cytotoxic immune response45,46. With the exception of fulminant T1DM, the degree of β-cell loss is probably moderate during this phase, ending when the immune response arrests viral amplification30. Data obtained in mouse models suggest that β-cell loss might be alleviated to some extent (probably on a transitory basis) by conversion of α cells or precursor islet cells into β cells47. This effect might be secondary to de novo expression of key transcription factors for β-cell function, such as PDX1, PAX4 and NKX6.1 (REF. 47). During infection, the release of sequestered islet antigens could lead to presentation of β-cell antigens in the draining lymph nodes. If peripheral regulatory mechanisms fail, such antigen presentation will lead to epitope spreading48. In line with the relevance of virally induced islet-cell damage and consequent epitope spreading and development of autoimmunity, an epidemio logical analysis found a correlation between the pathogenicity of viral strains, the extent of the antiviral response and the incidence of autoimmunity49.
The presence of viral markers in the islets of patients with T1DM, up to several years after disease onset, indicates that coxsackieviruses establish a persistent infection in the β cells30–33. This chronic infection is associated with low levels of viral replication, as indicated by the observations that only 5% of the endocrine cells per islet are VP1-positive31; the percentage of VP1-positive islets ranges from 2%33 to 28%31; and the viral load obtained from pancreatic biopsy samples grown in culture is low33. Despite low levels of viral production, overexpression of the major histocompatability complex (MHC) class I protein is detected in both infected and noninfected β cells31, which suggests that the presence of the virus affects all of the β cells within the infected islets. This overexpression of MHC class I is probably a consequence of local production of type I interferons50 and consequent activation of the kinase TYK2 (REF. 51) (the product of a candidate gene for T1DM) and other downstream signals. MHC class 1 expression and presentation of β-cell derived peptides have a key role in islet-specific homing of CD8+ T cells, as demonstrated in NOD mice41. Long-term overexpression of MHC class I proteins could lead to continuous presentation of β-cell epitopes to the immune system, increasing the risk of autoimmunity. Interestingly, several candidate genes for T1DM regulate key steps of this process (outlined in subsequent sections).
Bystander damage
Viral infection promotes the recruitment of natural killer cells and T cells to the islets30 and the local production of inflammatory cytokines, particularly INF-α, INF-β, IFN-γ, tumour necrosis factor (TNF) and IL-1β52. The essential role of these cytokines in β-cell destruction has been demonstrated in NOD mice and rat models of diabetes mellitus53–57. The molecular mechanisms have been extensively studied and involve the induction of endoplasmic reticulum stress and activation of the intrinsic pathway of apoptosis in islets obtained from both patients with T1DM and rodent models of the disease2,58. Local production of cytokines thus contributes to β-cell destruction and the spreading of β-cell epitopes.
Molecular mimicry
The molecular mimicry hypothesis reflects potential crossreactivity between viral protein epitopes that share homology at the amino acid sequence level with host islet proteins targeted by autoimmune T lymphocytes. Homologies have been predicted between pancreatic autoantigens and viral proteins, including those expressed by coxsackievirus59–61. Nevertheless, attempts to detect crossreactivity between autoimmune antibodies or T-cell clones and coxsackievirus epitopes have failed48,62,63, arguing against epitope mimicry as a crucial factor in coxsackievirus-induced T1DM. By contrast, crossreactivity is observed with cytomegalo virus61, but no strong epidemiological evidence exists to support a role for cytomegalovirus infection in T1DM. Interestingly, crossreactivity between viral epitopes and self-epitopes can augment (but not initiate) autoimmune disease in the context of repeated viral infections in a transgenic mouse model that expresses a viral antigen in the pancreatic β cells and thymus64. This finding suggests that epitope mimicry induced by recurrent viral infections might contribute to late events that lead to T1DM; namely, acceleration of the disease once autoimmunity (as evaluated by the development of islet autoantibodies) is already present. However, it remains to be clarified whether this mechanism is relevant for humans.
Bystander activation
Bystander activation is characterized by T-cell activation that does not involve specific recognition of a peptide presented to the T-cell receptor. During infection of cells adjacent to the β cells, secretion of proinflamma-tory cytokines by dendritic cells could initiate bystander activation among circulating naive islet-specific T cells in pancreatic islets or lymph nodes, thus accelerating β-cell destruction48,65–67. Potential adjacent cells include exocrine, endothelial, neuronal and islet α cells68. In line with this possible role of immune and adjacent cells, infection of the islets of NOD mice with the CVB1, CVB3 or CVB4 serotypes of coxsackievirus accelerates development of diabetes mellitus in this model45,69,70; however, this effect depends on the presence of a threshold number of autoreactive T cells in the islets71.
The important role of IFN-α production has been underlined among patients infected with hepatitis C virus who were treated with IFN-α for up to 1 year. This long-term treatment with IFN-α increases the risk of developing T1DM by 10-fold to 18-fold72–74. Onset of T1DM is abrupt and irreversible for most patients (98%), indicating a rapid and complete loss of β cells. Whether systemic treatment with IFN-α stimulates epitope presentation in islets and/or bystander activation of β-cell-specific T cells or alternative deleterious mechanisms remains to be determined.
Candidate genes
In the course of an infection, damage to the host can be a consequence of the invading microorganism, the host response to infection, or both. This observation suggests that the host–microorganism interaction must be examined to fully understand diseases with an infectious component. For example, pulmonary tuberculosis will develop in only 10% of all individuals infected by Mycobacterium tuberculosis; in these individuals, an excessive inflammatory response damages the lung tissue75.
This situation is particularly relevant when considering an autoimmune disease such as T1DM. In most cases, the pathogenic role of the virus during the development of T1DM does not require massive lytic replication in the islet cells. Instead, T1DM occurs in the presence of a persistent low-grade infection that triggers different degrees of inflammatory response and consequently different degrees of β-cell damage and antigen release. Not understanding this nuanced context has led, in our view, to a misguided focus on identifying specific viruses that are present among patients with T1DM but not normoglycaemic individuals. One characteristic that can modulate an individual's response to viral infection is the genetic background. Here, we discuss candidate genes for T1DM.
Genome-wide association studies
Genome-wide association studies (GWAS) have identified >50 naturally occurring genetic variants (risk-conferring single-nucleotide polymorphisms (SNPs)) that are linked to T1DM susceptibility and explain ~80% of the heritability11,76.
The MHC complex displays the highest odds ratio (>6.5) for T1DM; however, most of the genetic loci identified by GWAS confer only a modest risk of developing this condition (odds ratio <2.0)76. MHC-related genes are predominantly associated with the risk of developing autoimmunity. By contrast, the other T1DM risk alleles probably regulate the anatomical location of the autoimmune attack (for example, targeting the β cells in T1DM or the joints in rheumatoid arthritis), as well as evolution from autoimmunity to the disease state and the speed of this process (the time between the appearance of islet autoantibodies and the onset of symptomatic T1DM)77. Of note, families predisposed to T1DM exhibit an increased innate inflammatory state, which might reflect genetic variants that potentiate immune pathways independent of autoantibodies or HLA status78. Many of these polymorphisms are shared with other autoimmune diseases, particularly those associated with the development of autoantibodies79, but not with type 2 diabetes mellitus.
The general assumption is that candidate genes for T1DM modify disease risk by acting at the level of the immune system80. However, data published in the past 4 years suggest that human pancreatic β cells express mRNA for >80% of the T1DM candidate genes12,51,81,82, which suggests a role for these genes in both the immune system and the target β cells. In line with this hypothesis, comparison of SNP locations against chromatin maps for different cell types indicates a primary signature of T1DM-related SNPs in the promoter regions of candidate genes in both T cells and pancreatic islets83.
Functions of candidate genes
Candidate genes for T1DM might contribute to disease by regulating antiviral responses, innate immunity, activation of apoptosis or the β-cell phenotype12,51,84–89 (FIG. 1). The potential roles of T1DM candidate genes at the levels of the immune system and the β cell have been reviewed elsewhere12,80. Here, we focus on emerging information on genetically regulated pathways that modulate antiviral responses in pancreatic β cells.
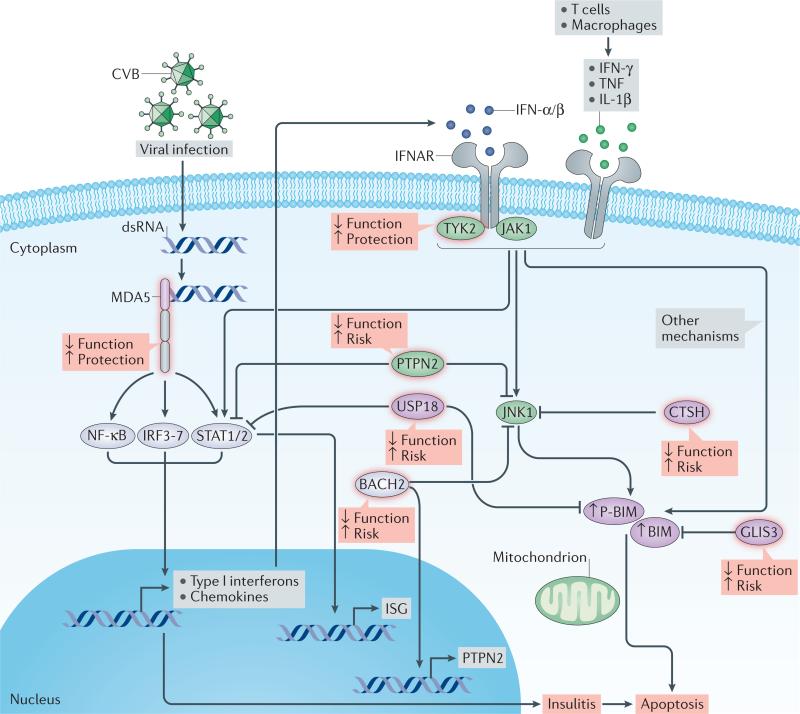
Figure 1. Regulation of key antiviral responses in pancreatic β cells.
Following infection, replicating coxsackievirus subtype B (CVB) produce cytosolic double-stranded RNA (dsRNA), a nonphysiological form of mRNA recognized by the cytoplasmic receptor MDA5. Binding of MDA5 to the dsRNA activates the transcription factors NF-κB, IRFs and STATs, triggering production of type I interferons and chemokines, thus contributing to local inflammation (insulitis). Type I interferons (IFN-α and IFN-β), type II interferon (IFN-γ) and the cytokines TNF and IL-1β contribute to β-cell destruction among genetically susceptible individuals. Type I interferons bound to the IFN-α/β receptor (IFNAR) signal via TYK2 and JAK1 and induce activation of STATs and expression of interferon-stimulated genes (ISGs) with antiviral properties. Proinflammatory cytokines promote the activation of JNK1, which induces the intrinsic (mitochondrial) apoptotic pathway through the proapoptotic protein BIM and its phosphorylated form (P-BIM). PTPN2 modulates β-cell death induced by interferons by regulating activation of P-BIM via JNK1. BIM and/or JNK1 are downregulated by BACH2, GLIS3 and CTSH. PTPN2 also functions as a negative regulator of the STAT signalling pathway, whereas USP18 exerts negative feedback on interferon-induced STAT signalling and mitochondrial apoptotic pathways in β cells. Candidate-gene regulated factors implicated in type 1 diabetes mellitus are framed in red and the consequences of their modulated expression and/or activity on biological function or type 1 diabetes mellitus risk indicated. Kinases and phosphatases are indicated by green ovals and transcription factors by grey ovals.
As shown in FIG. 1, the cellular response to viral infection starts by recognition of the whole virus particle or some of its components (for example, double-stranded RNA (dsRNA)) by pattern recognition receptors, such as TLR3, RIG-I or MDA5, followed by local release of type I interferons. Secreted interferons bind to cell-surface receptors (IFNAR) in an autocrine and paracrine manner and activate specific kinases (TYK2 and JAK). These enzymes phosphorylate and activate key transcription factors (STATs), thereby inducing signal transduction pathways that establish an antiviral response in target cells via activation of interferon-stimulated genes90.
Type I interferons
A role for type I interferons in human T1DM is evidenced by the fact that these cytokines and their downstream genes are expressed in pancreatic islets isolated from patients with T1DM16,91, and that a type I interferon signature is present in the peripheral blood of both children genetically at risk of T1DM92 and individuals with T1DM93. Transient blockade of either the type I interferon receptor or expression of Ifn-α by dendritic cells before the onset of insulitis markedly decreases the incidence of diabetes mellitus in NOD mice94,95. By contrast, NOD mice lacking a functional type I interferon receptor develop diabetes mellitus at the same rate as wild-type NOD mice96. A possible interpretation of these apparently contradictory findings is that viral infection and the subsequent production of type I interferons favours autoimmunity (and eventually T1DM) if the expression of interferons is induced at early and critical points during the development of this condition.
MDA5
A clear association exists between risk alleles for T1DM, viral recognition and interferon signalling pathways12 (FIG. 1). SNPs leading to decreased expression of IFIH1 (or MDA5) protect against T1DM97,98. Conversely, risk alleles in MDA5 promote rapid progression to T1DM compared with protective MDA5 alleles (31% and 11% within 5 years, respectively)99. MDA5 risk alleles are also associated with the development of autoantibodies targeting β cells100.
MDA5 is a cytoplasmic receptor for viral dsRNA. This receptor is expressed in human pancreatic islets and its mRNA expression levels are upregulated by enterovirus infection or exposure to a synthetic viral dsRNA molecule85. Inhibition of MDA5 mRNA expression in human and rodent pancreatic β cells by specific small interfering RNAs (siRNAs) decreases the chemokine expression and release that is usually induced in response to synthetic viral dsRNA85, potentially decreasing the homing of immune cells during insulitis. Mice with reduced levels of Mda5 expression exhibit a unique antiviral profile of type I interferons and induce a regulatory, rather than an effector, T-cell response during viral infection, thereby preventing immune-mediated diabetes mellitus101.
TYK2
SNPs in TYK2 — the gene that encodes the kinase that acts downstream of type I interferon signalling — are associated with systemic lupus erythematosus, multi ple sclerosis, rheumatoid arthritis and T1DM102,103. The SNP rs2304256:C>A is located in exon 8 of TYK2 on chromosome 19p13.2 and is thought to protect against T1DM. This variant causes a missense mutation in the TYK2 protein that decreases its interaction with IFNAR1 (REF. 104), as well as downstream signalling51. Inhibition of TYK2 by specific siRNAs in human β cells exposed to synthetic viral dsRNA decreases activation of the type I interferon pathway, lowering production of IFN-α and the chemokine CXCL10. These cells also exhibit decreased expression of MHC class I proteins, a hallmark of early β-cell inflammation in T1DM, and are less susceptible to apoptosis induced by synthetic viral dsRNA than are β cells treated with a control (inactive) siRNA51. By contrast, a spontaneous mutation that reduces Tyk2 expression in mice increases their susceptibility to virus-induced diabetes mellitus105, and a TYK2 promoter variant that reduces kinase activity in the protein increases the risk of fulminant T1DM among Japanese patients106.
Taken together, the available information on MDA5 and TYK2 suggests that SNPs that decrease biological function protect against autoimmune T1DM, which indicates that an excessive inflammatory response to viral infections contributes to autoimmunity and eventual T1DM among susceptible individuals. In this scenario, triggering of autoimmunity is secondary to the long-lasting presence of a noncytopathic virus in the β cells and the consequent protracted and/or excessive innate immune or inflammatory response. Conversely, in situations where the β cells are directly destroyed by the viral infection, as seems to be the case for fulminant T1DM21,22, decreased activity of these early antiviral responses might be deleterious and so accelerate disease106.
PTPN2
The gene encoding a phosphatase, PTPN2, is another candidate gene for T1DM107,108. The rs45450798 SNP accelerates progression to T1DM after the appearance of β-cell autoantibodies100. PTPN2 has an important role in the modulation of interferon signalling in β cells84–86. Inhibition of PTPN2 expression in β cells increases activation of STATs and augments apoptosis induced by IFN-α, IFN-β and IFN-γ via activation of the BH3-only protein BIM, particularly in its phosphorylated form (P-BIM), and subsequent triggering of the mitochondrial cell death pathway84–86. These observations suggest that SNPs that decrease PTPN2 expression sensitize β cells to apoptosis secondary to local inter-feron production in response to viral infection, and that the T1DM-associated risk allele of PTPN2 reduces the levels of its mRNA expression109.
Candidate gene networks
Some evidence indicates that BACH2, another candidate gene for T1DM, regulates expression of PTPN2 in β cells89. This finding reinforces the hypothesis that the risk of triggering T1DM during a viral infection depends on the presence of susceptibility variants in multiple genes interacting in pathways that leave individuals over-responsive to β-cell viral infections or to other danger signals12,85,110. In line with this possibility, an integrated analysis of gene networks and DNA sequence variations in T1DM identified the inter-feron regulatory factor 7 (IRF7)-regulated gene network (also known as IDIN, for ‘IRF7-driven inflammatory network’) as a major contributor to T1DM risk111. MDA5 initiates virus-induced chemokine production via activation of the transcription factors IRF3, IRF7 and NF-κB112, whereas PTPN2 and USP18 (a member of the IDIN network) provide negative feedback by inhibiting activation of the STAT signal transduction pathway and preventing β-cell apoptosis induced by interferons84–86,113.
The hygiene hypothesis
In addition to genetic background, early education of the immune system by microbial exposure during childhood can influence an individual's response to viral infection.
Accumulating data indicate that the progressive decrease in infections among high-income countries, which has occurred secondary to improved sanitation, socioeconomic status, vaccination and the use of antibiotics, might be an important contributory factor to the increased incidence of allergic and autoimmune diseases (for example, T1DM, coeliac disease and multiple sclerosis) reported in the past 50 years. This observation led to the hygiene hypothesis, which suggests that decreased microbial exposure in early childhood increases the risk of allergic and autoimmune diseases114–116.
Definitive proof for the hygiene hypothesis based on intervention trials in humans is still lacking; however, epidemiological and experimental data from rodents support this theory116–118. For instance, infections with Schistosoma mansoni, Trichinella spiralis or Mycobacterium bovis prevent spontaneous diabetes mellitus in NOD mice117. Possible reasons why early decreases in microbial exposure increases the risk of autoimmune diseases include defective development of the regulatory T cells that produce TGF-β and IL-10 and have the capacity to prevent excessive innate and adaptive immune responses116,118,119; lack of microorganism-induced maturation of dendritic cells that favours the development of regulatory T cells120; and modifications in the gut microbiota121. Immunoregulatory mechanisms triggered by viral infections in NOD mice protect against diabetes mellitus through both increasing the number of regulatory T cells and preventing expansion of diabetogenic CD8+ T cells43,44.
The first indication of autoimmunity against β cells (the appearance of islet autoantibodies) occurs before 12 months of age among children who will eventually develop T1DM122–124, which suggests that a putative protective effect of nonspecific infections and consequent immune system ‘education’ should take place before the first 6 months of life or even in utero125.
We propose that the putative role for viral infections in triggering insulitis and T1DM must be understood in the context of genetic predisposition and/or defective immune system education secondary to decreased infections in early life. Indeed, if we take into account the variables involved — namely, the host–microorganism interactions determined by the nature of the virus invading the β cells; the genetically determined and immune-education-determined inflammatory responses by islet and immune cells; and a putative interaction between these factors (FIG. 2) — we can envisage different scenarios for the role of environmental pressure on the increasing incidence of T1DM. Thus, in a population with high genetic risk of T1DM, removal of a protective factor, such as nonspecific infections in early life, might be sufficient to increase disease risk. Conversely, in populations with low genetic risk of T1DM, a combination of protection removal and increased infections by coxsackieviruses might be required for increased prevalence of T1DM. If this hypothesis is correct, one should not expect that similar environmental factors are driving the observed increase of T1DM incidence in different parts of the world.
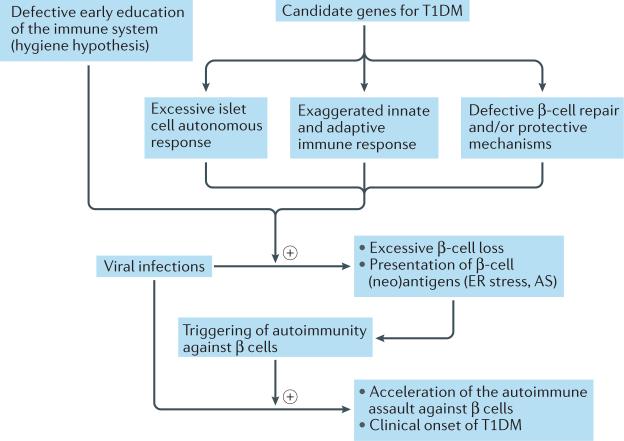
Figure 2. Crosstalk between viral infection, genetic background and early education of the immune system.
Genetic background and early immune education, either alone or in combination, define the individual's capacity to modulate the cell autonomous response upon viral infection. These diverse responses to viral infection can lead to different outcomes, including excessive β-cell loss (with or without viral persistence) and triggering of an autoimmune response. Repeated viral infection might accelerate the ongoing autoimmune assault against β cells, culminating in clinical disease. AS, alternative splicing; ER, endoplasmic reticulum; T1DM, type 1 diabetes mellitus.
Viral infection — why the β cells?
As discussed above, putative viral infection of the islet cells might induce local inflammation and potential presentation of β-cell antigens (native or modified) to the immune system. Such viral infection and antigen presentation could be the starting point of autoimmunity, eventually leading to T1DM126. A crucial unanswered question in this context is why only β cells become infected and then targeted by the immune system, whereas pancreatic α cells remain mostly unaffected by both viruses and autoimmunity30–32.
The host defence against microorganisms in vertebrates is usually considered to be the task of specialized immune cells, a view that underestimates the capacity of nonimmune cells to activate self-defence or cell autonomous immune responses against infection. The required mechanisms are present at a basal level in many cell types but are upregulated upon viral infection127,128. These responses rely on detection of microbial signatures by pattern recognition receptors127. By comparing microarray and RNA sequencing data obtained in human islets81,129 with known pattern recognition receptors and other antiviral or antibacterial factors127,130, we found that human islets exposed to virus or cytokines express several antiviral proteins (TLR3, MDA5, RIG-1, APOBEC36, SAMHD1, TRIM22, CNP, TETHERIN and VIPERIN), whereas antibacterial factors (TLR4, NLRP1, CLEC6A and CLEC7A) are poorly expressed, which suggests that islet cells are under evolutionary pressure to counteract viral but not bacterial infections, probably because they are seldom confronted with bacteria68.
Rat α cells and β cells purified using fluorescence- activated cell sorting131 are equally sensitive to apoptosis induced by cytokines or the viral mimic dsRNA; however, these cells display differing permissiveness to replication of the coxsackievirus CVB4 and CVB5 serotypes68, despite similar expression of the viral receptors Car (coxsackie–adenovirus receptor) and Daf. Accordingly, adenovirus tagged with green fluorescent protein (a tool that enables researchers to count the number of infected cells and measure expression levels based on the intensity of fluorescence) infects α cells and β cells via CAR with similar efficacy but leads to a reduced expression of green fluorescent protein in α cells68. This finding suggests that adenoviruses can enter both cell types but express viral genes and/or replicates less efficiently in α cells than in β cells owing to efficient blocking of translation in the former cell type. Consistent with productive entry of coxsackievirus in α cells, a clear increase in expression of the viral protein VP1 occurs 8 h after infection in both α cells and β cells from dispersed human islets (FIG. 3a,b) and in the pancreata of two of three children who died during the course of an acute and severe coxsackievirus infection68. Noncytopathogenic infection by three different coxsackievirus strains has been reported in both primary human α cells and β cells50. These infections were long-lasting in both cell types but production of INF-α was only detected in the β cells50. A potential concern is that islet dispersion could modify the infection and its outcome. However, coxsackievirus infection of α cells was detected in rat primary α cells purified using fluorescence-activated cell sorting68, dispersed human islets50,68, whole human islets50 and in pancreatic islets obtained from acutely infected patients68.
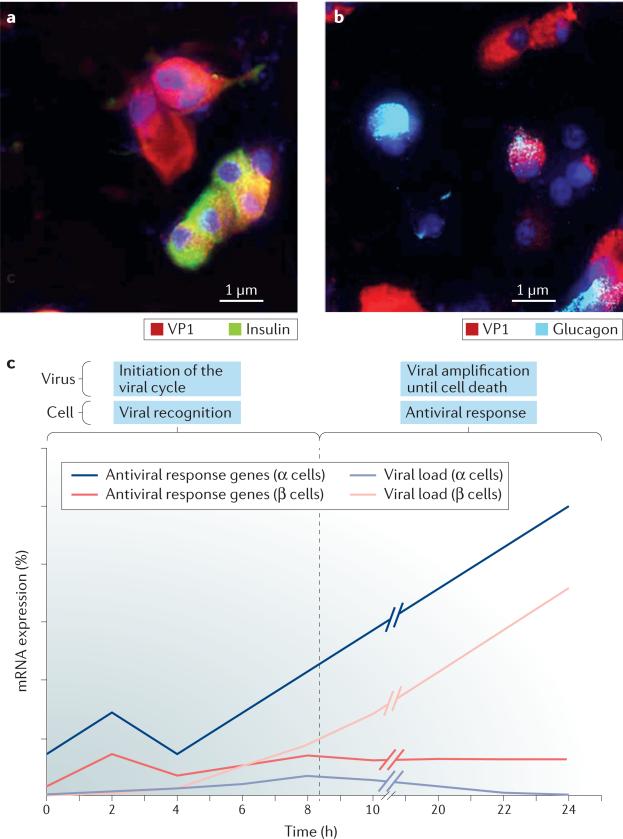
Figure 3. Differential autonomous antiviral response determines the outcome of infection in pancreatic α cells and β cells.
a | and b | Viral protein expression in human islet cells infected by coxsackievirus B5 serotype (CVB5) for 8 h. Immunocytochemistry labelling of VP1 (red), insulin (green), glucagon (light blue) and nuclei (dark blue) indicates expression of the viral protein at an early time point of infection in both insulin-producing-β cells (part a) and glucagon-producing-α cells (part b). Scale bar, 1 μm. c | Time course of CVB5 proliferation versus expression of antiviral response genes in rat α cells and β cells68. Purified rat α cells and β cells were infected with CVB5 and examined at 1, 2, 4, 6, 8 and 24 h after infection. The mRNA expression levels of antiviral genes (Stat1 and Mx1) and viral genes (VP1) are shown. Basal expression of antiviral genes is higher in α cells than in β cells and rapidly increases, which enables this cell type to eradicate the viral infection and survive. By contrast, β cells exhibit lower and less effective antiviral responses than α cells, which enables the viral load to increase and eventually kill the infected cells.
These observations suggest that the viral cycle is initiated in both α cells and β cells but that α cells control viral amplification, which leads to an abortive infection. This difference might explain why VP1 labelling is not detected in the α cells of infected human islets after long infection times132,133 or among patients with T1DM30,32. The resistance of α cells to viruses could be the consequence of both lowered competence of these cells to replicate coxsackievirus, compared with β cells, owing to shortage in crucial cellular factors used by the virus and/or of a more efficient antiviral response in α cells versus β cells. Despite low expression of VP1, α cells induce a more rapid and marked antiviral response than do β cells and, in α cells, viral expression declines after 8 h of infection, which suggests that α cells take the control of the viral amplification whereas β cells do not68. Furthermore, an unrelated DNA virus (adenovirus) is also translated less efficiently in α cells that in β cells, which argues for an efficient antiviral response in α cells. Supporting the latter hypothesis, antiviral factors (Rig-1, Mda5, Pkr, Mx1 and Viperin), chemokines (Cxcl-10 and Ccl2), cytokines (Ifn-α) and downstream transcription factors (Stat1) that are required to control viral infections are more highly expressed in rat α cells than in β cells68 (FIG. 3c), similar to the findings observed when comparing virus-resistant granule neurons with virus-sensitive cortical neurons (cortical neurons are preferentially destroyed during West Nile virus encephalitis, compared with granule cell neurons)134. This potent antiviral response remains to be demonstrated in human α cells. Considering that viral markers are present in the α cells of acutely infected patients68, but not during chronic infection30–33, it might be suggested that human α cells are also more efficient at clearing infection than human β cells.
In agreement with these observations, human β cells are also more sensitive than α cells to coxsackievirus-induced infection and functional impairment30,32,132. Taken together, these converging data suggest that pancreatic α cells and β cells have different cell autonomous signatures. This divergence could explain their differential ability to clear viral infections and potentially explain why chronically infected pancreatic β cells, but not α cells, are targeted by an autoimmune response and killed during T1DM68,135.
Prevention of T1DM
The identification of key viruses involved in the development of T1DM might enable a preventive approach by vaccination18,136. Similarly, evidence for the persistence of a particular virus in β cells acting as a crucial trigger for T1DM might offer the possibility of pharmacological approaches to clear viral infection and prevent disease onset.
Vaccination
As discussed above, an excessive inflammatory response seems deleterious to virally infected pancreatic β cells. Thus, a vaccine developed to boost the immune response could aggravate the problem. Ideally, a vaccine against coxsackievirus should generate a neutralizing immune response that aims to avoid the primary infection of target cells. That is, the vaccine should contain non-infectious (inactivated) viruses and be administered early in life. In line with this approach, vaccination of NOD mice with formalin-fixed CVB1 leads to an efficient production of neutralizing antibodies, which prevent both viral replication and the acceleration of diabetes mellitus onset in these animals70.
Antiviral drugs
At present, no licensed drugs for the treatment of enter-oviral infection exist. Nevertheless, antiviral drugs that act at different levels of the viral cycle (reviewed elsewhere137) can successfully reduce enterovirus amplification. Pleconaril and its analogues bind to the viral capsid, thereby inhibiting entry of the virus in the target cells. Antiviral drugs such as guanidine hydrochloride efficiently interfere with viral replication; peptide and non-peptide inhibitors block the viral proteases 3C and 2A, whereas hydantoin prevents viral assembly. The availability of drugs targeting different steps of the viral cycle enables combined treatment to avoid viral resistance to the antiviral therapy. Some compounds have shown efficacy to reduce coxsackievirus amplification and to protect pancreatic islets from coxsackievirus-induced cytopathogenic effects138–141; to eradicate the virus in infected pancreatic β cell lines142; and to prevent and/or reduce the incidence of virally-induced diabetes mellitus in mice140,141. Adapted antiviral treatment at the pre-diabetic stage could eliminate persistent infection, thus reducing inflammation and the risk of autoimmunity and T1DM.
The problem remains to identify the correct target(s). A growing body of evidence suggests that infection by coxsackievirus B serotypes is an early event in the development of T1DM among genetically susceptible individuals17,18. Both insulin-positive and MDA5-positive islet cells are detected in patients with T1DM. This observation, coupled with local expression of VP1 (REF. 32), supports the idea of chronic and persistent low-level viral infection in the β cells35,68.
Conclusions
Several viral infectious events probably contribute to auto-immunity and the progressive β-cell death that leads to T1DM. This contribution could occur at various stages during disease evolution and the eventual disappearance of the pathogenic virus after the onset of autoimmunity might complicate the goal of defining a unique causal virus. Identifying the viruses associated with the development of T1DM and determining their contribution to pathogenesis must be done in the context of genetic background. Furthermore, cell autonomous responses and putative differences in the early education of the immune system should also be considered as factors that might affect the degree of β-cell damage and the transition (or not) from innate immunity to adaptive immunity (and autoimmunity). Future research in the field should take these points into careful account. Indeed, only by tackling the pathogenesis of T1DM at its true level of complexity will it eventually be possible to develop preventive and curative therapies for this disease.
Key points.
Viral infections — particularly by enteroviruses (for example, coxsackievirus) — have been implicated in the development of type 1 diabetes mellitus (T1DM)
Many candidate genes for T1DM regulate antiviral responses in pancreatic β cells
Pancreatic islet α cells trigger a more efficient antiviral response than β cells following infection with diabetogenic viruses, thus enabling α cells to eradicate viral infections without undergoing apoptosis
An inability to clear viral infections could explain why chronically infected β cells, but not α cells, are targeted by an autoimmune response and killed during development of T1DM
The identification of key diabetogenic viruses and the downstream mechanisms leading to insulitis might enable a preventive approach to T1DM by vaccination
Coxsackieviruses.
Coxsackieviruses belong to the genus enterovirus in the Picornaviridae family. These nonenveloped viruses have a linear positive-sense single-stranded RNA genome. They are present worldwide and cause infections that are predominantly asymptomatic or associated with mild respiratory or gastrointestinal symptoms. In some cases, however, infection can lead to severe myocarditis or, as presently discussed, contribute to triggering autoimmunity against pancreatic β cells143,144. Among the two subtypes (A and B), only coxsackieviruses of the B subtype have been associated with the development of diabetes mellitus143. Six different serotypes (CVB1 to CVB6) are able to replicate in pancreatic β cells. The serotypes enter the β cells through CAR29, a component of the tight junction. At present, neither specific treatments nor vaccination are available to prevent or cure coxsackievirus infections.
Acknowledgements
The research by A.O.d.B. and D.L.E. that is discussed in this Review was supported by the Belgian Fonds National de la Recherche Scientifique (FNRS; grants T.0036.13 and FRFS-Welbio CR-2015A-06); the European Union (projects Naimit and BetaBat in the Seventh Framework Programme of the European Commission); the Juvenile Diabetes Foundation; the Helmsley Type 1 Diabetes Program; and the NIH–NIDDK–HIRN Consortium. A.O.d.B. and D.L.E. also receive support from the Network for Pancreatic Organ Donors with Diabetes (nPOD), a collaborative type 1 diabetes research project sponsored by the Juvenile Diabetes Research Foundation International (JDRF). Organ procurement organisations partnering with nPOD to provide research resources are listed at http://www.jdrfnpod.org/our-partners.php. F. Grieco (Center for Diabetes Research, Universite Libre de Bruxelles, Belgium) provided the micrographs shown in FIG. 3.
Footnotes
Competing interests statement
The authors declare no competing interests.
Review criteria
Relevant publications were identified by searching the PubMed database using combinations for the following search terms: “diabetes”, “type 1 diabetes”, “pancreatic β cells”, “pancreatic islets”, “virus”, ”coxsackievirus”, “diabetes candidate genes”, “MDA5”, “RIG-I”, “TLR3”, “PTPN2”, “TYK2”, “GLIS3”, and “STAT1”. A manual search of some references cited in these papers, or in relevant articles related to the pathogenesis of diabetes mellitus, was also performed. All selected papers were English-language, full-text articles. Many of the references identified could not be included owing to space restrictions.
References
- 1.Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383:69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and β-cell loss in type 1 diabetes. Nat. Rev. Endocrinol. 2009;5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- 3.Butler AE, et al. Modestly increased β-cell apoptosis but no increased β-cell replication in recent onset type 1 diabetic patients who died of diabetic ketoacidosis. Diabetologia. 2007;50:2323–2331. doi: 10.1007/s00125-007-0794-x. [DOI] [PubMed] [Google Scholar]
- 4.Meier JJ, Bhushan A, Butler AE, Rizza RA, Butler PC. Sustained β-cell apoptosis in patients with long standing type 1 diabetes: indirect evidence for islet regeneration? Diabetologia. 2005;48:2221–2228. doi: 10.1007/s00125-005-1949-2. [DOI] [PubMed] [Google Scholar]
- 5.Gepts W. Pathologic anatomy of the pancreas in juvenile diabetes mellitus. Diabetes. 1965;14:619–633. doi: 10.2337/diab.14.10.619. [DOI] [PubMed] [Google Scholar]
- 6.Kloppel G, Drenck CR, Oberholzer M, Heitz PU. Morphometric evidence for a striking β-cell reduction at the clinical onset of type 1 diabetes. Virchows Arch. A Pathol. Anat. Histopathol. 1984;403:441–452. doi: 10.1007/BF00737292. [DOI] [PubMed] [Google Scholar]
- 7.Foulis AK, Stewart JA. The pancreas in recent onset type 1 (insulin dependent) diabetes mellitus: insulin content of islets, insulitis and associated changes in the exocrine acinar tissue. Diabetologia. 1984;26:456–461. doi: 10.1007/BF00262221. [DOI] [PubMed] [Google Scholar]
- 8.Krogvold L, et al. Function of isolated pancreatic islets fom patients at onset of type 1 diabetes: insulin secretion can be restored after some days in a nondiabetogenic environment in vitro: results from the DiViD study. Diabetes. 2015;64:2506–2512. doi: 10.2337/db14-1911. [DOI] [PubMed] [Google Scholar]
- 9.Strandell E, Eizirik DL, Sandler S. Reversal of β-cell suppression in vitro in pancreatic islets isolated from nonobese diabetic mice during the phase preceding insulin dependent diabetes mellitus. J. Clin. Invest. 1990;85:1944–1950. doi: 10.1172/JCI114657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell Thompson M, et al. Insulitis and β-cell mass in the natural history of type 1 diabetes. Diabetes. 2016;65:719–731. doi: 10.2337/db15-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Santin I, Eizirik DL. Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and β-cell apoptosis. Diabetes Obes. Metab. 2013;15(Suppl. 3):71–81. doi: 10.1111/dom.12162. [DOI] [PubMed] [Google Scholar]
- 13.Patterson CC, Dahlquist GG, Gyurus E, Green A, Soltesz G. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–20: a multicentre prospective registration study. Lancet. 2009;373:2027–2033. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- 14.Bodansky HJ, Staines A, Stephenson C, Haigh D, Cartwright R. Evidence for an environmental effect in the aetiology of insulin dependent diabetes in a transmigratory population. BMJ. 1992;304:1020–1022. doi: 10.1136/bmj.304.6833.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redondo MJ, Jeffrey J, Fain PR, Eisenbarth GS, Orban T. Concordance for islet autoimmunity among monozygotic twins. N. Engl. J. Med. 2008;359:2849–2850. doi: 10.1056/NEJMc0805398. [DOI] [PubMed] [Google Scholar]
- 16.Richardson SJ, Morgan NG, Foulis AK. Pancreatic pathology in type 1 diabetes mellitus. Endocr. Pathol. 2014;25:80–92. doi: 10.1007/s12022-014-9297-8. [DOI] [PubMed] [Google Scholar]
- 17.Kondrashova A, Hyoty H. Role of viruses and other microbes in the pathogenesis of type 1 diabetes. Int. Rev. Immunol. 2014;33:284–295. doi: 10.3109/08830185.2014.889130. [DOI] [PubMed] [Google Scholar]
- 18.Drescher KM, von Herrath M, Tracy S. Enteroviruses, hygiene and type 1 diabetes: toward a preventive vaccine. Rev. Med. Virol. 2015;25:19–32. doi: 10.1002/rmv.1815. [DOI] [PubMed] [Google Scholar]
- 19.Hober D, Sauter P. Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nat. Rev. Endocrinol. 2010;6:279–289. doi: 10.1038/nrendo.2010.27. [DOI] [PubMed] [Google Scholar]
- 20.Ghazarian L, Diana J, Simoni Y, Beaudoin L, Lehuen A. Prevention or acceleration of type 1 diabetes by viruses. Cell. Mol. Life Sci. 2013;70:239–255. doi: 10.1007/s00018-012-1042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imagawa A, Hanafusa T. Fulminant type 1 diabetes — an important subtype in East Asia. Diabetes Metab. Res. Rev. 2011;27:959–964. doi: 10.1002/dmrr.1236. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka S, Aida K, Nishida Y, Kobayashi T. Pathophysiological mechanisms involving aggressive islet cell destruction in fulminant type 1 diabetes. Endocr. J. 2013;60:837–845. doi: 10.1507/endocrj.ej13-0222. [DOI] [PubMed] [Google Scholar]
- 23.Gamble DR, Taylor KW. Seasonal incidence of diabetes mellitus. Br. Med. J. 1969;3:631–633. doi: 10.1136/bmj.3.5671.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulte BM, et al. Detection of enterovirus RNA in peripheral blood mononuclear cells of type 1 diabetic patients beyond the stage of acute infection. Viral Immunol. 2010;23:99–104. doi: 10.1089/vim.2009.0072. [DOI] [PubMed] [Google Scholar]
- 25.Gamble DR, Kinsley ML, FitzGerald MG, Bolton R, Taylor KW. Viral antibodies in diabetes mellitus. Br. Med. J. 1969;3:627–630. doi: 10.1136/bmj.3.5671.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oikarinen S, et al. Virus antibody survey in different European populations indicates risk association between coxsackievirus B1 and type 1 diabetes. Diabetes. 2014;63:655–662. doi: 10.2337/db13-0620. [DOI] [PubMed] [Google Scholar]
- 27.Laitinen OH, et al. Coxsackievirus B1 is associated with induction of β-cell autoimmunity that portends type 1 diabetes. Diabetes. 2014;63:446–455. doi: 10.2337/db13-0619. [DOI] [PubMed] [Google Scholar]
- 28.Yeung WC, Rawlinson WD, Craig ME. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta analysis of observational molecular studies. BMJ. 2011;342:d35. doi: 10.1136/bmj.d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ylipaasto P, et al. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet β-cells. Diabetologia. 2004;47:225–239. doi: 10.1007/s00125-003-1297-z. [DOI] [PubMed] [Google Scholar]
- 30.Dotta F, et al. Coxsackie B4 virus infection of β-cells and natural killer cell insulitis in recent onset type 1 diabetic patients. Proc. Natl Acad. Sci. USA. 2007;104:5115–5120. doi: 10.1073/pnas.0700442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson SJ, Leete P, Bone AJ, Foulis AK, Morgan NG. Expression of the enteroviral capsid protein VP1 in the islet cells of patients with type 1 diabetes is associated with induction of protein kinase R and downregulation of Mcl 1. Diabetologia. 2013;56:185–193. doi: 10.1007/s00125-012-2745-4. [DOI] [PubMed] [Google Scholar]
- 32.Richardson SJ, Willcox A, Bone AJ, Foulis AK, Morgan NG. The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia. 2009;52:1143–1151. doi: 10.1007/s00125-009-1276-0. [DOI] [PubMed] [Google Scholar]
- 33.Krogvold L, et al. Detection of a low grade enteroviral infection in the islets of langerhans of living patients newly diagnosed with type 1 diabetes. Diabetes. 2015;64:1682–1687. doi: 10.2337/db14-1370. [DOI] [PubMed] [Google Scholar]
- 34.Chapman NM, Kim KS. Persistent coxsackievirus infection: enterovirus persistence in chronic myocarditis and dilated cardiomyopathy. Curr. Top. Microbiol. Immunol. 2008;323:275–292. doi: 10.1007/978-3-540-75546-3_13. [DOI] [PubMed] [Google Scholar]
- 35.Tracy S, Smithee S, Alhazmi A, Chapman N. Coxsackievirus can persist in murine pancreas by deletion of 5′ terminal genomic sequences. J. Med. Virol. 2015;87:240–247. doi: 10.1002/jmv.24039. [DOI] [PubMed] [Google Scholar]
- 36.Willcox A, Richardson SJ, Bone AJ, Foulis AK, Morgan NG. Immunohistochemical analysis of the relationship between islet cell proliferation and the production of the enteroviral capsid protein, VP1, in the islets of patients with recent onset type 1 diabetes. Diabetologia. 2011;54:2417–2420. doi: 10.1007/s00125-011-2192-7. [DOI] [PubMed] [Google Scholar]
- 37.Oikarinen M, et al. Analysis of pancreas tissue in a child positive for islet cell antibodies. Diabetologia. 2008;51:1796–1802. doi: 10.1007/s00125-008-1107-8. [DOI] [PubMed] [Google Scholar]
- 38.Cnop M, et al. The long lifespan and low turnover of human islet β-cells estimated by mathematical modelling of lipofuscin accumulation. Diabetologia. 2010;53:321–330. doi: 10.1007/s00125-009-1562-x. [DOI] [PubMed] [Google Scholar]
- 39.Bonifacio E, Lampasona V, Genovese S, Ferrari M, Bosi E. Identification of protein tyrosine phosphatase like IA2 (islet cell antigen 512) as the insulin dependent diabetes related 37/40K autoantigen and a target of islet cell antibodies. J. Immunol. 1995;155:5419–5426. [PubMed] [Google Scholar]
- 40.Coppieters KT, et al. Demonstration of islet autoreactive CD8 T cells in insulitic lesions from recent onset and long term type 1 diabetes patients. J. Exp. Med. 2012;209:51–60. doi: 10.1084/jem.20111187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savinov AY, Wong FS, Stonebraker AC, Chervonsky AV. Presentation of antigen by endothelial cells and chemoattraction are required for homing of insulin specific CD8+ T cells. J. Exp. Med. 2003;197:643–656. doi: 10.1084/jem.20021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chabot S, et al. Mouse liver specific CD8+ T cells encounter their cognate antigen and acquire capacity to destroy target hepatocytes. J. Autoimmun. 2013;42:19–28. doi: 10.1016/j.jaut.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christen U, et al. Cure of prediabetic mice by viral infections involves lymphocyte recruitment along an IP 10 gradient. J. Clin. Invest. 2004;113:74–84. doi: 10.1172/JCI200417005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filippi CM, Estes EA, Oldham JE, von Herrath MG. Immunoregulatory mechanisms triggered by viral infections protect from type 1 diabetes in mice. J. Clin. Invest. 2009;119:1515–1523. doi: 10.1172/JCI38503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drescher KM, Kono K, Bopegamage S, Carson SD, Tracy S. Coxsackievirus B3 infection and type 1 diabetes development in NOD mice: insulitis determines susceptibility of pancreatic islets to virus infection. Virology. 2004;329:381–394. doi: 10.1016/j.virol.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 46.Flodstrom M, et al. Target cell defense prevents the development of diabetes after viral infection. Nat. Immunol. 2002;3:373–382. doi: 10.1038/ni771. [DOI] [PubMed] [Google Scholar]
- 47.Thorel F, et al. Conversion of adult pancreatic α-cells to β-cells after extreme β-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horwitz MS, et al. Diabetes induced by Coxsackie virus: initiation by bystander damage and not molecular mimicry. Nat. Med. 1998;4:781–785. doi: 10.1038/nm0798-781. [DOI] [PubMed] [Google Scholar]
- 49.Sarmiento L, Cubas Duenas I, Cabrera Rode E. Evidence of association between type 1 diabetes and exposure to enterovirus in Cuban children and adolescents. MEDICC Rev. 2013;15:29–32. doi: 10.37757/MR2013V15.N1.7. [DOI] [PubMed] [Google Scholar]
- 50.Chehadeh W, et al. Persistent infection of human pancreatic islets by coxsackievirus B is associated with α-interferon synthesis in β-cells. J. Virol. 2000;74:10153–10164. doi: 10.1128/jvi.74.21.10153-10164.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marroqui L, et al. TYK2, a candidate gene for type 1 diabetes, modulates apoptosis and the innate immune response in human pancreatic β-cells. Diabetes. 2015;64:3808–3811. doi: 10.2337/db15-0362. [DOI] [PubMed] [Google Scholar]
- 52.Slifka MK, Rodriguez F, Whitton JL. Rapid on/off cycling of cytokine production by virus specific CD8+ T cells. Nature. 1999;401:76–79. doi: 10.1038/43454. [DOI] [PubMed] [Google Scholar]
- 53.Campbell IL, Kay TW, Oxbrow L, Harrison LC. Essential role for interferon γ and interleukin 6 in autoimmune insulin dependent diabetes in NOD/Wehi mice. J. Clin. Invest. 1991;87:739–742. doi: 10.1172/JCI115055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kay TW, Campbell IL, Oxbrow L, Harrison LC. Overexpression of class I major histocompatibility complex accompanies insulitis in the non obese diabetic mouse and is prevented by anti interferon γ antibody. Diabetologia. 1991;34:779–785. doi: 10.1007/BF00408350. [DOI] [PubMed] [Google Scholar]
- 55.von Herrath MG, Oldstone MB. Interferon γ is essential for destruction of β-cells and development of insulin dependent diabetes mellitus. J. Exp. Med. 1997;185:531–539. doi: 10.1084/jem.185.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eizirik DL, Mandrup Poulsen T. A choice of death — the signal transduction of immune mediated β-cell apoptosis. Diabetologia. 2001;44:2115–2133. doi: 10.1007/s001250100021. [DOI] [PubMed] [Google Scholar]
- 57.Sandberg JO, Eizirik DL, Sandler S. IL 1 receptor antagonist inhibits recurrence of disease after syngeneic pancreatic islet transplantation to spontaneously diabetic non obese diabetic (NOD) mice. Clin. Exp. Immunol. 1997;108:314–317. doi: 10.1046/j.1365-2249.1997.3771275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gurzov EN, Eizirik DL. Bcl 2 proteins in diabetes: mitochondrial pathways of β-cell death and dysfunction. Trends Cell Biol. 2011;21:424–431. doi: 10.1016/j.tcb.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Atkinson MA, et al. Cellular immunity to a determinant common to glutamate decarboxylase and coxsackie virus in insulin dependent diabetes. J. Clin. Invest. 1994;94:2125–2129. doi: 10.1172/JCI117567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Honeyman MC, Stone NL, Harrison LC. T cell epitopes in type 1 diabetes autoantigen tyrosine phosphatase IA 2: potential for mimicry with rotavirus and other environmental agents. Mol. Med. 1998;4:231–239. [PMC free article] [PubMed] [Google Scholar]
- 61.Hiemstra HS, et al. Cytomegalovirus in autoimmunity: T cell crossreactivity to viral antigen and autoantigen glutamic acid decarboxylase. Proc. Natl Acad. Sci. USA. 2001;98:3988–3991. doi: 10.1073/pnas.071050898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richter W, et al. Sequence homology of the diabetes associated autoantigen glutamate decarboxylase with coxsackie B4 2C protein and heat shock protein 60 mediates no molecular mimicry of autoantibodies. J. Exp. Med. 1994;180:721–726. doi: 10.1084/jem.180.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schloot NC, et al. Molecular mimicry in type 1 diabetes mellitus revisited: T cell clones to GAD65 peptides with sequence homology to coxsackie or proinsulin peptides do not crossreact with homologous counterpart. Hum. Immunol. 2001;62:299–309. doi: 10.1016/s0198-8859(01)00223-3. [DOI] [PubMed] [Google Scholar]
- 64.Christen U, et al. A viral epitope that mimics a self antigen can accelerate but not initiate autoimmune diabetes. J. Clin. Invest. 2004;114:1290–1298. doi: 10.1172/JCI22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ehl S, Hombach J, Aichele P, Hengartner H, Zinkernagel RM. Bystander activation of cytotoxic T cells: studies on the mechanism and evaluation of in vivo significance in a transgenic mouse model. J. Exp. Med. 1997;185:1241–1251. doi: 10.1084/jem.185.7.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zarozinski CC, Welsh RM. Minimal bystander activation of CD8 T cells during the virus induced polyclonal T cell response. J. Exp. Med. 1997;185:1629–1639. doi: 10.1084/jem.185.9.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pane JA, Coulson BS. Lessons from the mouse: potential contribution of bystander lymphocyte activation by viruses to human type 1 diabetes. Diabetologia. 2015;58:1149–1159. doi: 10.1007/s00125-015-3562-3. [DOI] [PubMed] [Google Scholar]
- 68.Marroqui L, et al. Differential cell autonomous responses determine the outcome of coxsackievirus infections in murine pancreatic α and β-cells. eLIFE. 2015;4:e06990. doi: 10.7554/eLife.06990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Serreze DV, et al. Diabetes acceleration or prevention by a coxsackievirus B4 infection: critical requirements for both interleukin 4 and γ interferon. J. Virol. 2005;79:1045–1052. doi: 10.1128/JVI.79.2.1045-1052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larsson PG, et al. A preclinical study on the efficacy and safety of a new vaccine against Coxsackievirus B1 reveals no risk for accelerated diabetes development in mouse models. Diabetologia. 2015;58:346–354. doi: 10.1007/s00125-014-3436-0. [DOI] [PubMed] [Google Scholar]
- 71.Serreze DV, Ottendorfer EW, Ellis TM, Gauntt CJ, Atkinson MA. Acceleration of type 1 diabetes by a coxsackievirus infection requires a preexisting critical mass of autoreactive T cells in pancreatic islets. Diabetes. 2000;49:708–711. doi: 10.2337/diabetes.49.5.708. [DOI] [PubMed] [Google Scholar]
- 72.Fattovich G, Giustina G, Favarato S, Ruol A. A survey of adverse events in 11,241 patients with chronic viral hepatitis treated with α-interferon. J. Hepatol. 1996;24:38–47. doi: 10.1016/s0168-8278(96)80184-x. [DOI] [PubMed] [Google Scholar]
- 73.Nakamura K, et al. Type 1 diabetes and interferon therapy: a nationwide survey in Japan. Diabetes Care. 2011;34:2084–2089. doi: 10.2337/dc10-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zornitzki T, Malnick S, Lysyy L, Knobler H. Interferon therapy in hepatitis C leading to chronic type 1 diabetes. World J. Gastroenterol. 2015;21:233–239. doi: 10.3748/wjg.v21.i1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casadevall A, Pirofski LA. Microbiology: ditch the term pathogen. Nature. 2014;516:165–166. doi: 10.1038/516165a. [DOI] [PubMed] [Google Scholar]
- 76.Pociot F, et al. Genetics of type 1 diabetes: what's next? Diabetes. 2010;59:1561–1571. doi: 10.2337/db10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bonifacio E, Krumsiek J, Winkler C, Theis FJ, Ziegler AG. A strategy to find gene combinations that identify children who progress rapidly to type 1 diabetes after islet autoantibody seroconversion. Acta Diabetol. 2014;51:403–411. doi: 10.1007/s00592-013-0526-2. [DOI] [PubMed] [Google Scholar]
- 78.Chen YG, et al. Molecular signatures differentiate immune states in type 1 diabetic families. Diabetes. 2014;63:3960–3973. doi: 10.2337/db14-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Onengut Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N. Engl. J. Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 81.Eizirik DL, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro inflammatory cytokines. PLoS Genet. 2012;8:e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bergholdt R, et al. Identification of novel type 1 diabetes candidate genes by integrating genome wide association data, protein protein interactions, and human pancreatic islet gene expression. Diabetes. 2012;61:954–962. doi: 10.2337/db11-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moore F, et al. PTPN2, a candidate gene for type 1 diabetes, modulates interferon γ induced pancreatic β-cell apoptosis. Diabetes. 2009;58:1283–1291. doi: 10.2337/db08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Colli ML, Moore F, Gurzov EN, Ortis F, Eizirik DL. MDA5 and PTPN2, two candidate genes for type 1 diabetes, modify pancreatic β-cell responses to the viral by product double stranded RNA. Hum. Mol. Genet. 2010;19:135–146. doi: 10.1093/hmg/ddp474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Santin I, et al. PTPN2, a candidate gene for type 1 diabetes, modulates pancreatic β-cell apoptosis via regulation of the BH3 only protein Bim. Diabetes. 2011;60:3279–3288. doi: 10.2337/db11-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nogueira TC, et al. GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic β-cell apoptosis via regulation of a splice variant of the BH3 only protein Bim. PLoS Genet. 2013;9:e1003532. doi: 10.1371/journal.pgen.1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Floyel T, et al. CTSH regulates β-cell function and disease progression in newly diagnosed type 1 diabetes patients. Proc. Natl Acad. Sci. USA. 2014;111:10305–10310. doi: 10.1073/pnas.1402571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marroqui L, et al. BACH2, a candidate risk gene for type 1 diabetes, regulates apoptosis in pancreatic β-cells via JNK1 modulation and crosstalk with the candidate gene PTPN2. Diabetes. 2014;63:2516–2527. doi: 10.2337/db13-1443. [DOI] [PubMed] [Google Scholar]
- 90.Coccia EM, Battistini A. Early IFN type I response: learning from microbial evasion strategies. Semin. Immunol. 2015;27:85–101. doi: 10.1016/j.smim.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang X, et al. Interferon expression in the pancreases of patients with type I diabetes. Diabetes. 1995;44:658–664. doi: 10.2337/diab.44.6.658. [DOI] [PubMed] [Google Scholar]
- 92.Ferreira RC, et al. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes. 2014;63:2538–2550. doi: 10.2337/db13-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reynier F, et al. Specific gene expression signature associated with development of autoimmune type I diabetes using whole blood microarray analysis. Genes Immun. 2010;11:269–278. doi: 10.1038/gene.2009.112. [DOI] [PubMed] [Google Scholar]
- 94.Li Q, et al. Interferon α-initiates type 1 diabetes in nonobese diabetic mice. Proc. Natl Acad. Sci. USA. 2008;105:12439–12444. doi: 10.1073/pnas.0806439105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Diana J, et al. Crosstalk between neutrophils, B 1a cells and plasmacytoid dendritic cells initiates autoimmune diabetes. Nat. Med. 2013;19:65–73. doi: 10.1038/nm.3042. [DOI] [PubMed] [Google Scholar]
- 96.Quah HS, et al. Deficiency in type I interferon signaling prevents the early interferon induced gene signature in pancreatic islets but not type 1 diabetes in NOD mice. Diabetes. 2014;63:1032–1040. doi: 10.2337/db13-1210. [DOI] [PubMed] [Google Scholar]
- 97.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Downes K, et al. Reduced expression of IFIH1 is protective for type 1 diabetes. PLoS ONE. 2010;5:e12646. doi: 10.1371/journal.pone.0012646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Winkler C, et al. An interferon induced helicase (IFIH1) gene polymorphism associates with different rates of progression from autoimmunity to type 1 diabetes. Diabetes. 2011;60:685–690. doi: 10.2337/db10-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lempainen J, et al. Non HLA gene effects on the disease process of type 1 diabetes: from HLA susceptibility to overt disease. J. Autoimmun. 2015;61:45–53. doi: 10.1016/j.jaut.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 101.Lincez PJ, Shanina I, Horwitz MS. Reduced expression of the MDA5 gene IFIH1 prevents autoimmune diabetes. Diabetes. 2015;64:2184–2193. doi: 10.2337/db14-1223. [DOI] [PubMed] [Google Scholar]
- 102.Wallace C, et al. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tao JH, et al. Meta analysis of TYK2 gene polymorphisms association with susceptibility to autoimmune and inflammatory diseases. Mol. Biol. Rep. 2011;38:4663–4672. doi: 10.1007/s11033-010-0601-5. [DOI] [PubMed] [Google Scholar]
- 104.Richter MF, Dumenil G, Uze G, Fellous M, Pellegrini S. Specific contribution of Tyk2 JH regions to the binding and the expression of the interferon α/β-receptor component IFNAR1. J. Biol. Chem. 1998;273:24723–24729. doi: 10.1074/jbc.273.38.24723. [DOI] [PubMed] [Google Scholar]
- 105.Izumi K, et al. Reduced Tyk2 gene expression in β-cells due to natural mutation determines susceptibility to virus induced diabetes. Nat. Commun. 2015;6:6748. doi: 10.1038/ncomms7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nagafuchi S, et al. TYK2 promoter variant and diabetes mellitus in the Japanese. EBioMedicine. 2015;2:744–749. doi: 10.1016/j.ebiom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smyth DJ, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Espino Paisan L, et al. A polymorphism in PTPN2 gene is associated with an earlier onset of type 1 diabetes. Immunogenetics. 2011;63:255–258. doi: 10.1007/s00251-010-0500-x. [DOI] [PubMed] [Google Scholar]
- 109.Long SA, et al. An autoimmune associated variant in PTPN2 reveals an impairment of IL 2R signaling in CD4+ T cells. Genes Immun. 2011;12:116–125. doi: 10.1038/gene.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhernakova A, et al. Meta analysis of genome wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Heinig M, et al. A trans acting locus regulates an anti viral expression network and type 1 diabetes risk. Nature. 2010;467:460–464. doi: 10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takeuchi O, Akira S. MDA5/RIG I and virus recognition. Curr. Opin. Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 113.Santin I, et al. USP18 is a key regulator of the interferon driven gene network modulating pancreatic β-cell inflammation and apoptosis. Cell Death Dis. 2012;3:e419. doi: 10.1038/cddis.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kamradt T, Goggel R, Erb KJ. Induction, exacerbation and inhibition of allergic and autoimmune diseases by infection. Trends Immunol. 2005;26:260–267. doi: 10.1016/j.it.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 116.Kondrashova A, Seiskari T, Ilonen J, Knip M, Hyoty H. The ‘hygiene hypothesis’ and the sharp gradient in the incidence of autoimmune and allergic diseases between Russian Karelia and Finland. Apmis. 2013;121:478–493. doi: 10.1111/apm.12023. [DOI] [PubMed] [Google Scholar]
- 117.Cooke A. Review series on helminths, immune modulation and the hygiene hypothesis: how might infection modulate the onset of type 1 diabetes? Immunology. 2009;126:12–17. doi: 10.1111/j.1365-2567.2008.03009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bach JF, Chatenoud L. The hygiene hypothesis: an explanation for the increased frequency of insulin dependent diabetes. Cold Spring Harb. Perspect. Med. 2012;2:a007799. doi: 10.1101/cshperspect.a007799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lehuen A, Diana J, Zaccone P, Cooke A. Immune cell crosstalk in type 1 diabetes. Nat. Rev. Immunol. 2010;10:501–513. doi: 10.1038/nri2787. [DOI] [PubMed] [Google Scholar]
- 120.Rook GA, Brunet LR. Old friends for breakfast. Clin. Exp. Allergy. 2005;35:841–842. doi: 10.1111/j.1365-2222.2005.02112.x. [DOI] [PubMed] [Google Scholar]
- 121.Gulden E, Wong FS, Wen L. The gut microbiota and type 1 diabetes. Clin. Immunol. 2015;159:143–153. doi: 10.1016/j.clim.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ziegler AG, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA. 2013;309:2473–2479. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Krischer JP, et al. The 6 year incidence of diabetes associated autoantibodies in genetically at risk children: the TEDDY study. Diabetologia. 2015;58:980–987. doi: 10.1007/s00125-015-3514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ilonen J, et al. Patterns of β-cell autoantibody appearance and genetic associations during the first years of life. Diabetes. 2013;62:3636–3640. doi: 10.2337/db13-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vercelli D. Mechanisms of the hygiene hypothesis — molecular and otherwise. Curr. Opin. Immunol. 2006;18:733–737. doi: 10.1016/j.coi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 126.Morgan NG, Richardson SJ. Enteroviruses as causative agents in type 1 diabetes: loose ends or lost cause? Trends Endocrinol. Metab. 2014;25:611–619. doi: 10.1016/j.tem.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 127.Randow F, MacMicking JD, James LC. Cellular self defense: how cell autonomous immunity protects against pathogens. Science. 2013;340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat. Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cnop M, et al. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63:1978–1993. doi: 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- 130.Carty M, Reinert L, Paludan SR, Bowie AG. Innate antiviral signalling in the central nervous system. Trends Immunol. 2014;35:79–87. doi: 10.1016/j.it.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 131.Marroqui L, et al. Pancreatic α-cells are resistant to metabolic stress induced apoptosis in type 2 diabetes. EBioMedicine. 2015;2:378–385. doi: 10.1016/j.ebiom.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Anagandula M, et al. Infection of human islets of Langerhans with two strains of coxsackie B virus serotype 1: assessment of virus replication, degree of cell death and induction of genes involved in the innate immunity pathway. J. Med. Virol. 2014;86:1402–1411. doi: 10.1002/jmv.23835. [DOI] [PubMed] [Google Scholar]
- 133.Gallagher GR, et al. Viral infection of engrafted human islets leads to diabetes. Diabetes. 2015;64:1358–1369. doi: 10.2337/db14-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cho H, et al. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive stranded RNA viruses. Nat. Med. 2013;19:458–464. doi: 10.1038/nm.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Atkinson MA, von Herrath M, Powers AC, Clare Salzler M. Current concepts on the pathogenesis of type 1 diabetes — considerations for attempts to prevent and reverse the disease. Diabetes Care. 2015;38:979–988. doi: 10.2337/dc15-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hyoty H, Knip M. Developing a vaccine for type 1 diabetes through targeting enteroviral infections. Expert Rev. Vaccines. 2014;13:989–999. doi: 10.1586/14760584.2014.933078. [DOI] [PubMed] [Google Scholar]
- 137.De Palma AM, Vliegen I, De Clercq E, Neyts J. Selective inhibitors of picornavirus replication. Med. Res. Rev. 2008;28:823–884. doi: 10.1002/med.20125. [DOI] [PubMed] [Google Scholar]
- 138.Moell A, Skog O, Ahlin E, Korsgren O, Frisk G. Antiviral effect of nicotinamide on enterovirus infected human islets in vitro: effect on virus replication and chemokine secretion. J. Med. Virol. 2009;81:1082–1087. doi: 10.1002/jmv.21476. [DOI] [PubMed] [Google Scholar]
- 139.Berg AK, Olsson A, Korsgren O, Frisk G. Antiviral treatment of coxsackie B virus infection in human pancreatic islets. Antiviral Res. 2007;74:65–71. doi: 10.1016/j.antiviral.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 140.See DM, Tilles JG. WIN 54954 treatment of mice infected with a diabetogenic strain of group B coxsackievirus. Antimicrob. Agents Chemother. 1993;37:1593–1598. doi: 10.1128/aac.37.8.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Powers RD, Dotson WM, Jr, Hayden FG. Modification of encephalomyocarditis virus induced diabetes in mice by antiviral agents. Antiviral Res. 1983;3:151–159. doi: 10.1016/0166-3542(83)90022-0. [DOI] [PubMed] [Google Scholar]
- 142.Alidjinou EK, Sane F, Bertin A, Caloone D, Hober D. Persistent infection of human pancreatic cells with coxsackievirus B4 is cured by fluoxetine. Antiviral Res. 2015;116:51–54. doi: 10.1016/j.antiviral.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 143.Schneider DA, von Herrath MG. Potential viral pathogenic mechanism in human type 1 diabetes. Diabetologia. 2014;57:2009–2018. doi: 10.1007/s00125-014-3340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sin J, Mangale V, Thienphrapa W, Gottlieb RA, Feuer R. Recent progress in understanding coxsackievirus replication, dissemination, and pathogenesis. Virology. 2015;484:288–304. doi: 10.1016/j.virol.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]