Abstract
Background
The Rhodococcus ruber strain Chol-4 genome contains at least three putative 3-ketosteroid Δ1-dehydrogenase ORFs (kstD1, kstD2 and kstD3) that code for flavoenzymes involved in the steroid ring degradation. The aim of this work is the functional characterization of these enzymes prior to the developing of different biotechnological applications.
Results
The three R. ruber KstD enzymes have different substrate profiles. KstD1 shows preference for 9OHAD and testosterone, followed by progesterone, deoxy corticosterone AD and, finally, 4-BNC, corticosterone and 19OHAD. KstD2 shows maximum preference for progesterone followed by 5α-Tes, DOC, AD testosterone, 4-BNC and lastly 19OHAD, corticosterone and 9OHAD. KstD3 preference is for saturated steroid substrates (5α-Tes) followed by progesterone and DOC. A preliminary attempt to model the catalytic pocket of the KstD proteins revealed some structural differences probably related to their catalytic differences. The expression of kstD genes has been studied by RT-PCR and RT-qPCR. All the kstD genes are transcribed under all the conditions assayed, although an additional induction in cholesterol and AD could be observed for kstD1 and in cholesterol for kstD3. Co-transcription of some correlative genes could be stated. The transcription initiation signals have been searched, both in silico and in vivo. Putative promoters in the intergenic regions upstream the kstD1, kstD2 and kstD3 genes were identified and probed in an apramycin-promoter-test vector, leading to the functional evidence of those R. ruber kstD promoters.
Conclusions
At least three putative 3-ketosteroid Δ1-dehydrogenase ORFs (kstD1, kstD2 and kstD3) have been identified and functionally confirmed in R. ruber strain Chol-4. KstD1 and KstD2 display a wide range of substrate preferences regarding to well-known intermediaries of the cholesterol degradation pathway (9OHAD and AD) and other steroid compounds. KstD3 shows a narrower substrate range with a preference for saturated substrates. KstDs differences in their catalytic properties was somehow related to structural differences revealed by a preliminary structural modelling. Transcription of R. ruber kstD genes is driven from specific promoters. The three genes are constitutively transcribed, although an additional induction is observed in kstD1 and kstD3. These enzymes have a wide versatility and allow a fine tuning-up of the KstD cellular activity.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-017-0657-1) contains supplementary material, which is available to authorized users.
Keywords: Rhodococcus ruber, 3-Ketosteroid-∆1-dehydrogenase, Promoters, Expression, Steroids
Background
Rhodococci are aerobic Gram-positive soil bacteria belonging to the Actinomycetes group. They show a broad catabolic diversity over different substrates, from pollutants to many aromatic compounds, including steroids and sterols [1–3]. Steroids are a source of contamination of soil and waters and their presence has been detected even in drinking water, threatening many ways of life and public health [4–6]. Rhodococci can be useful in this biodegradation field due to their metabolic versatility and steroids degradation capability. On the other hand Rhodococcus spp. are potential biotechnological tools [3, 7] as they can provide with key enzymes essential for certain reactions that yield industrial needed intermediaries such as 4-androstene-3,17-dione (AD) and 1,4-androstadiene-3,17-dione (ADD) [8].
But before exploiting all the advantages the different rhodococci offer, it is essential to know how these bacteria degrade steroids and which enzymes are involved in this process.
Steroids are molecules with a carbon skeleton of 4 fused rings (A to D) and a side chain up to 10 carbons. During the last years, the increasing number of studies concerning steroid degradation, and more concretely the degradation of cholesterol in bacteria, have clarified some of the catabolic steps (e.g. initiation of the ring degradation by either a NAD+-dependent 3β-hydroxysteroid dehydrogenase or a cholesterol oxidase) although other steps still remain unclear (e.g. the processing of the C and D rings of the steroid structure or the relative order in which the different steps of the degradation of ring and chain occurs) [3, 9–12].
In the general scheme of steroid degradation, there are two key enzymes that initiate the opening of the steroid ring: the 3-ketosteroid-∆1-dehydrogenase [4-ene-3-oxosteroid: (acceptor)-1-ene-oxoreductase; EC 1.3.99.4)], also known as KstD and the 3-ketosteroid 9α-hydroxylase [Androsta-1,4-diene-3,17-dione; EC 1.14.13.142], also known as Ksh [13]. KstD is a flavoenzyme involved in the ∆1-dehydrogenation of the steroid molecule leading to the initiation of the breakdown of the steroid nucleus by introducing a double bond into the A-ring of 3-ketosteroids [14, 15]. This flavoprotein converts 4-ene-3-oxosteroids (e.g. AD) to 1,4-diene-3-oxosteroids (e.g. ADD) by trans-axial elimination of the C-1(α) and C-2(β) hydrogen atoms [16]. KstD homologs have been identified in 100 different bacterial species (78 actinobacteria, 20 proteobacteria and 2 firmicutes) and at least in one fungus, Aspergillus fumigatus CICC 40167 [17, 18]. Most of these KstD-containing bacteria occur in soil, marine or river sediments and are also able to degrade polycyclic aromatic hydrocarbons [19]. Phylogenetic analysis leads to classify the KstD-like enzymes in at least 4 different groups, in which KstD1, KstD2, KstD3 of Rhodococcus erythropolis SQ1 are representatives of three of them [20]. The crystal structure of the enzyme KstD1 of R. erythropolis SQ1 has been elucidated [21] confirming the presence of the two domains previously described, namely a N-terminal flavin adenine dinucleotide (FAD) binding motif and a substrate-binding domain [14, 20, 22, 23].
The substrate range of different KstD proteins has been studied in R. erythropolis SQ1, being 3-ketosteroids with a saturated A-ring (e.g. 5α-androstane-3,17-dione and 5α-testosterone) the preferred substrates for KstD3 and (9α-hydroxy-)4-androstene-3,17-dione the favourite one for both KstD1 and KstD2 [20]. It should be mentioned that, apart from their role in steroids degradation, KstD proteins could have specific roles depending of their origin; for instance, the KstD of A. fumigatus CICC 40167 is involved in fusidane antibiotic biosynthesis [17].
We have previously reported the occurrence of three KstD enzymes in R. ruber (NCBI::AFH57399 for KstD1; NCBI::AFH57395 for KstD2 and NCBI::ACS73883 for KstD3) [24]. Growth experiments with single, double or triple kstD mutants proved that KstD2 is a key enzyme in the transformation of both AD to ADD and 9α-hydroxy-4-androstene-3,17-dione (9OHAD) to 9α-hydroxy-1,4-androstadiene-3,17-dione (9OHADD) while both KstD2 and KstD3 are involved in the cholesterol catabolism in R. ruber. On the other hand, the role of KstD1 on the steroids catabolism remains unclear as kstD1 mutation did not affect growing of this strain in steroids [24]. In this study, we cloned the three kstD ORFs and heterologously expressed them in R. erythropolis CECT3014, in order to initiate the biochemical characterization of the encoded enzymes, as the basis for further studies on their applications. The results revealed that KstD3 uses more actively substrates with a saturated ring in contrast to KstD1 and KstD2. Additionally, we located and functionally defined the promoters of the three kstD ORFS in order to provide a basis for future research on the regulation of these genes.
Methods
Bacterial strains, plasmids and growth conditions
Rhodococcus ruber strain Chol-4 (CECT 7469; DSM 45280) was isolated from a sewage sludge sample [25]. This strain was routinely grown in Luria-Bertani (LB) or minimal medium (M457 of the DSMZ, Braunschweig, Germany) containing the desired carbon and energy source under aerobic conditions at 30 °C in a rotary shaker (250 rpm) for 1–3 days. For the steroids growth experiments, a LB pre-grown culture was washed two times with minimal medium prior to inoculation. Cholesterol or AD (Sigma), were added directly to the minimal medium culture for growing and/or induction at 0.6 and 0.44 g/L, respectively. Escherichia coli strains were grown in LB broth at 37 °C, 250 rpm. For the promoter growth experiments, cells were plated in minimal medium M457 plates containing the desired carbon source and incubated at 30 °C for 3 days. Cholesterol and AD were previously dissolved in methyl-β-cyclodextrin (CD) [26] and prepared as described [27]. Plasmids and bacterial strains used are listed in Additional file 1. Competent cells of E. coli DH5αF’ and BL21 (DE3) were prepared and transformed by standard protocols [28].
Cloning of kstD1, kstD2 and kstD3 of R. ruber strain Chol-4 and heterologous expression in Rhodococcus erythropolis CECT3014 cells
Chromosomal DNA extraction of R. ruber grown in a LB agar plate was performed using the hexadecyltrimethylammonium bromide (CTAB) procedure [29] with the following modifications. Bacterial cells were collected, suspended in 400 µL Tris–EDTA buffer (10 mM Tris/HCl, pH 8.1 mM EDTA) and incubated at 80 °C for 20 min. Afterwards, a lysozyme treatment (50 µL of 10 mg/mL stock) was carried out at 37 °C for 1–12 h, and then 75 µL of SDS containing proteinase K (70 µL SDS 10% wt/vol plus 5 µL proteinase K 10 mg/mL) was added and incubated for 10 min at 65 °C. Proteins were precipitated with 100 µL of 5 M NaOH and 100 µL CTAB (0.1 g/mL suspended in 0.7 M NaOH) for 10 min at 65 °C. DNA was purified by extraction with chloroform-isoamyl alcohol (24:1) and phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with 0.6 vol of isopropanol at room temperature for 30 min. After centrifugation, DNA was washed with 70% ethanol and suspended in distilled water.
The kstD ORFs were previously identified using the Bioedit program [25] and they were PCR amplified, from start to stop codon, using primers from Additional file 2. PCR was performed under standard conditions using High Fidelity PCR Enzyme Mix (Fermentas) with a specific high GC buffer (Roche) at 30 cycles of 1 min at 95 °C, 1 min at the desired Tm and 0.5–3 min at 72 °C (unless stated otherwise).
NdeI-BamHI (kstD3), NdeI-BglII (kstD2 and kstD1) restricted PCR products were first cloned into pGEM-T Easy Vector (Promega) and then moved to the shuttle vector pTip-QC1 [30]. Expression plasmids either with or without a kstD ORF were used to electroporate Rhodococcus erythropolis CECT3014. Selection was made in LB with 34 µg/mL Cm. Cultures of R. erythropolis harbouring expression plasmids (pTip-KsTD1, pTip-KstD2, pTip-KstD3 or pTip-QC1) were grown (30 °C, 200 rpm) in 50 mL LB broth supplemented with Cm until an OD600nm of 0.6–0.8. After that, expression was induced by adding 1 µg/mL thiostrepton (Sigma) to the culture. Cells were kept growing for 24 h more and they were collected at 5000 rpm for 10 min, washed twice in 50 mM phosphate buffer pH 7.0, concentrated to 5 mL and sonicated. The resulting cell extracts were used for analysis of KstD activity with a range of steroid substrates. Total protein content was measured by Bradford assay [31]. Samples were analysed by PAGE-SDS in 12.5% wt/vol gels and using 10 µg of total protein per lane.
KstD enzymatic assay
The kinetic of the enzymatic extracts was determined as previously stated [20, 32]. The cell-free extracts were incubated with AD (Sigma), 9OHAD (Organon Biosciences), 17β-hydroxy-5α-androstan-3-one or 5α-Tes, DOC (deoxycorticosterone), testosterone, corticosterone, 19OHAD, progesterone (Sigma), 4-BNC (4-pregnen-3-one-20β-carboxylic acid) (Steraloids), 5β-androstane-3,17-dione (Steraloids), 4-cholestene-3-one (Sigma) or 5α-cholestan-3-one (Acros Organics). Structures of steroids used in this study are shown in Additional file 3. Enzyme activities were measured spectrophotometrically at 30 °C using 2,6-dichlorophenol-indophenol (DCPIP, Sigma) as an artificial electron acceptor. The reaction mixture (1 mL) consisted of 50 mM Tris (pH 7), 80 μM DCPIP, cell-free extract and 200 μM steroid in ethanol (methanol in the case of 5α-cholestane-3-one). Four replicates were analysed. Activities are expressed as mean values ± SD in units per milligram of protein; one unit is defined as the amount of enzyme which causes the reduction of 1 µmol of DCPIP/min (ε600 = 21 mM−1 cm−1) after taking into account the value of the activity of control (cells harbouring the empty pTip-QC1 vector) for a certain steroid. Total protein concentration (mg/mL) was measured by Bradford assay [31]. The kinetics of the KstD enzymes were determined by incubating the cell-free extracts with varying concentrations of steroid substrates. The kinetics parameters were analysed by nonlinear regression curve fitting of the data to the Michaelis- Menten equation using Hyper32 1.0 software (Informer Technologies, http://hyper32.software.informer.com/).
Expression analysis by RT-PCR and RT-qPCR
RNA samples for RT-PCR experiments were obtained from mid-log exponential phase cultures (OD600nm 0.7–0.8). Total RNA was prepared with the RNeasy Mini Kit (Qiagen) following the manufacturer’s indications with the following modification: 50 mg of acid-washed glass beads (150 μm diameter) were added in the first step and each sample was shaken at maximum speed in a Bullet Blender for 5 min. The cell debris was removed by centrifugation. The supernatant was subjected to the RNeasy Mini Kit (Qiagen) protocol. The total RNA obtained (0.5–1 μg) was treated once with 5 U of Turbo DNase RNase-Free (Ambion) in a 700 μL volume for 2 h at 37 °C. RNA samples were extracted with 1 volume of acid phenol (Sigma), vigorously shaken and incubated at room temperature for 15 min. After 15 min centrifugation, the upper phase was precipitated by addition of 0.12 volumes of 5 M NH4Ac, 0.02 volumes of glycogen (5 mg/mL) and 1 volume of isopropanol, washed twice in 70% ethanol and dissolved in water. Samples were treated with DNase until no DNA was detected by PCR to avoid DNA contamination. The RNA concentration was then evaluated using a NanoDrop Spectrophotometer ND-1000.
For the RT-PCR the cDNA was synthesized using SuperScrip III Reverse Transcriptase (Invitrogen) following the manufacturer’s indications. cDNA was used as template (25 ng) for PCR reactions (20 μL final volume). Controls without reverse transcriptase (RT-) were used to detect any contamination of undigested DNA in the RNA preparations. PCR products were analysed in 0.8% agarose gels.
To quantify the expression of the three KstD genes, a RT-qPCR analysis was performed using RNA from wild-type strains cultured in M457 minimal medium containing the desired carbon and energy source (2 g/L sodium acetate, 0.44 g/L AD or 0.6 g/L cholesterol). The RNA quality was assessed by using Bioanalyzer 2100 (Agilent). cDNA was synthetized using 1 μg of RNA with the high capacity RNA to cDNA Kit (Applied Biosystems). The RT-qPCR analysis of cDNA was performed on Applied Biosystems QuantStudio 12K Flex Real-Time PCR Systems. The reaction conditions were 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C for extension. The temperature of the melting curve was from 60 to 95 °C. The FAD-binding dehydrogenase D092_14375 gene was used as an internal control to normalize messenger RNA levels. All reactions were performed in triplicate. The RT-qPCR experiment and the analysis of the relative fold difference of each gene using the 2−∆∆Ct algorithm was performed in the Genomic unit of Universidad Complutense de Madrid.
The sodium acetate grown culture was used as the reference medium. Therefore, the relative expression indicates how many times the expression level of a certain gene is detected respect to the levels detected when growing on sodium acetate.
In silico analyses
DNASTAR (Lasergene) programs were used to analyse sequences and to design primers. The R. ruber strain Chol-4 genomic DNA has been previously sequenced [33]. BioEdit program was used to perform local-blast alignments within the genome data (NCIB::ANGC01000000). Putative signal peptides were predicted by SignalP 4.1 server using a model trained on Gram-positive bacteria [34]. Sigma 70 putative promoters predictions were performed using the BPROM server, a bacterial sigma 70 promoter recognition program with about 80% accuracy and specificity [35], the Neural Network Promoter Prediction (NNPP) based on prokaryotes [36] in all cases with a score value of ≥80%, or the webserver PePPER for prediction of prokaryote promoter elements and regulons [37].
For the protein modelling we employed different software. I-Tasser (http://zhanglab.ccmb.med.umich.edu/I-TASSER) was used for an approach to protein structure and function prediction [38] and PredictProtein (www.predictprotein.org/home) was used for the secondary structure, solvent accessibility and transmembrane helix prediction [39]. COBALT was used as a multiple sequence alignment tool to find similarities among the catalytic residues (www.ncbi.nlm.nih.gov/tools/cobalt/re_cobalt.cgi) [40] and PyMOL (www.pymol.org/) as a molecular visualization system (PyMOL Molecular Graphics System, Version 0.99, Schrödinger).
Promoter cloning and characterization
The NheI-PciI 0.4 Kb multiple cloning site (mcs) from pSEVA351 [41, 42] was cloned into pNV119 vector [43], from now on named as pNVS (Additional file 4).
The putative kstD promoter sequences were amplified by PCR, from the end of the upstream flanking gene to the end of the first six amino acid codifying sequence. The XbaI, PstI-flanked kstD promoter regions (kstD1p, kstD2p and kstD3p) and the KpnI, PstI-flanked kstD3 b p minimal promoter were cloned into pNVS. The resulting vectors were designated pNVSP1, pNVSP2, pNVSP3 and pNVSP3b, respectively.
Apramycin resistance gene (Amr) was amplified by PCR from plasmid pIJ773 [44], from start to stop codon. The NruI/HindII digested Amr fragment was cloned in each one of the previous kstD constructions. The resulting plasmids were checked by sequencing (Secugen) and named pNVSP1-A, pNVSP2-A, pNVSP3-A and pNVSP3b-A, respectively.
All the primers used are listed in Additional file 2. PCR was performed under standard conditions using High Fidelity PCR (Roche) with glycerol 5% and a basic program unless stated otherwise.
As a control, a plasmid without any promoter but carrying the Amr gene (pNVSA) was made by digesting pNVSP1-A with NruI-XbaI; the resulting 4.2 Kb fragment was cut from a 1% agarose gel and purified with GENECLEAN Turbo Kit. Blunt ends suitable for ligation with T4 DNA ligase (Takara) were generated using the End repair kit (DNA terminator, Lucigen). The final ligated product was used to transform E. coli DH5αF′. Deletion was checked by sequencing (Secugen).
Every one of the plasmid set was introduced in R. ruber strain Chol-4 by electroporation (200 or 400 µL cells with 1 µg DNA at 400 Ω, 25 mA, 2.5 µF; 10–11 ms), the resulting cells were suspended in 800 µL of LB and kept for 6 min at 46 °C, and then for 5 h at 30 °C without shaking. Finally, they were plated on LB Agar with 200 μg/mL kanamycin and kept at 30 °C. To verify the presence of plasmids, two colonies of each plate were picked and grown in 3 mL of LB-200 μg/mL kanamycin. Plasmid were extracted using the method described in Hopwood et al. [45] and used to transform E. coli strain DH5αF’ [28]. Plasmids obtained from E. coli colonies grown at 37 °C in 50 μg/mL kanamycin were verified by sequencing (Secugen). Finally, those colonies of R. ruber strain Chol-4 harbouring the right recombinant plasmids were picked and grown in agar minimal media at 30 °C with different carbon source with or without apramycin (300 μg/mL) or kanamycin (200 μg/mL).
To define the transcriptional start sites (TSS) of kstD promoters, a transcription start point protocol (ARF-TSS) [46] was used on R. ruber cells with the following modifications.
RNA from R. ruber cells growing in different carbons sources cultures was isolated. The culture media were: LB for kstD1 TSS, minimal medium supplemented with AD for kstD2 TSS and minimal medium supplemented with cholesterol for kstD3 TSS. Total RNA was isolated as described previously [47]. It was qualified by electrophoresis and quantified by Nanodrop 1000 (NanoDrop Technologies). 20 µg of RNA were reverse transcribed with a gene specific phosphorylated 5′-end primer (R1 for each kstD) using SuperScrip III Reverse Transcriptase from Invitrogen. As a result, cDNA fragments with the TSS as 3′-end were generated and purified. After removing of RNA with 0.5 µg/µL RNase A (37 °C for 30 min), cDNA was purified using the GENECLEAN turbo Kit (MPI) and final products were treated with T4 RNA ligase from Fermentas (10U, 37 °C for 30 min). This T4 RNA ligase catalyses the ATP-dependent intra- and intermolecular formation of phosphodiester bonds between 5′-phosphate and 3′-hydroxyl termini of oligonucleotides, single-stranded RNA and DNA. It was used in this study to circularize the cDNA. The circularized cDNAs were then used as template for PCR amplification with Expand High Fidelity (Roche) using R2 and F3 primers specific for each kstD ORF. PCR products were purified using the GENECLEAN turbo Kit and sequenced (Secugen). The nucleotide upstream the 5′-end of the R1 primer is the transcription initiation site.
Results and discussion
As we have published earlier, the R. ruber strain Chol-4 genome contains three putative 3-ketosteroid Δ1-dehydrogenase ORFs (kstD1, kstD2 and kstD3) that code for flavoenzymes involved in the steroid ring degradation [24]. Growth experiments with kstD mutants proved that KstD2 is the main enzyme involved in the transformation of AD to ADD. R. ruber kstD2 mutants accumulate 9OHAD from AD due to the action of a 3-ketosteroid-9α-hydroxylase (KshAB). On the other hand, only the strains lacking both KstD2 and KstD3 were unable to grow in minimal medium with cholesterol as the only carbon source. In order to know more about these R. ruber enzymes, we have performed transcriptional studies and followed their heterologous expression in R. erythropolis to detect their activities on a set of different substrates.
In silico analyses of kstD promoter regions of R. ruber strain Chol-4
A scheme of the three R. ruber kstD ORFs and their genomic surroundings is depicted in Fig. 1 showing also the in silico predicted pP1, pP2, pP3, pP4 and pP5 promoters. The available programs (see “Methods”) yielded no putative promoter region just upstream either kstD1 or kstD2 ORFs, although it should be noted here that promoter prediction programs are not specific for Gram-positive species.
Fig. 1.
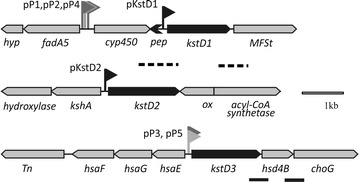
Schematic representation of R. ruber strain Chol-4 DNA kstD regions. Putative promoters (pP) predicted using the BPROM (pP1: TTCCTT−35..TGCTTGAAT−10), PePPER (pP2: TTGAATGCTTTTAGAACGTGTTCCACATCgcgaC+1 and pP3: TGGACTCACCGCGCCATCATTCTATAACgtgtT+1) or NNPP programs (pP4: GGTTGTCGTGGCGGACAAGGTGTGGTCCGAATGATCGGGAC+1TTGGCGATT and pP5: GAAGGGATGGACTCACCGCGCCATCATTC+1TATAACGTG) are shown in grey flags. TSS derived promoters (pKstD1, pKstD2) appear in black flags. Positive (solid line) and negative (dotted line) results of the amplification of co-transcribed products are depicted. hyp hypothetical protein, fadA5 acetyl-CoA acyltransferase, cyp450 cytochrome P450, pep hypothetical peptide, MFSt major facilitator superfamily transporter, kshA 3-ketosteroid 9α-hydroxylase, ox oxidoreductase, Tn transposase, hsaF 4-hydroxy-2-oxovalerate aldolase, hsaG acetaldehyde dehydrogenase, hsaE 2-hydroxypenta-2,4-dienoate hydratase, hsd4B 2-enoyl acyl-CoA hydratase, choG cholesterol oxidase
However, these programs detected putative promoters (pP1, pP2 and pP4) upstream the cyp450 gene, lying in the intergenic region of the fadA5-hyp and cyp450-kstD1-MFSt opposite clusters. Flanking the pP1 putative promoter there are two palindromic sequences (TagAACagGTTgtc and TagAACgtGTTccA) (Fig. 2), one of them rather similar and the second one identical to the consensus binding region reported for the KstR regulatory protein of Mycobacterium (TnnAACnnGTTnnA) [48]. KstR and KstR2, two TetR family repressor regulators, have been found to control most of the steroid pathways in actinobacteria [48–50]. KstR is a highly conserved TetR family repressor that regulates the transcription of genes related to the upper and central pathway of cholesterol catabolism, namely the membrane transport of cholesterol, the degradation of the steroid side chain and the opening of the A and B rings [12, 48]. Upon binding to a 3-oxo-4-cholestenoic acid or to a CoA thioester cholesterol metabolite, KstR releases the DNA and allows transcription to begin [51, 52]. The KstR binding motif has been proved to be conserved within actinobacteria [48, 53].
Fig. 2.
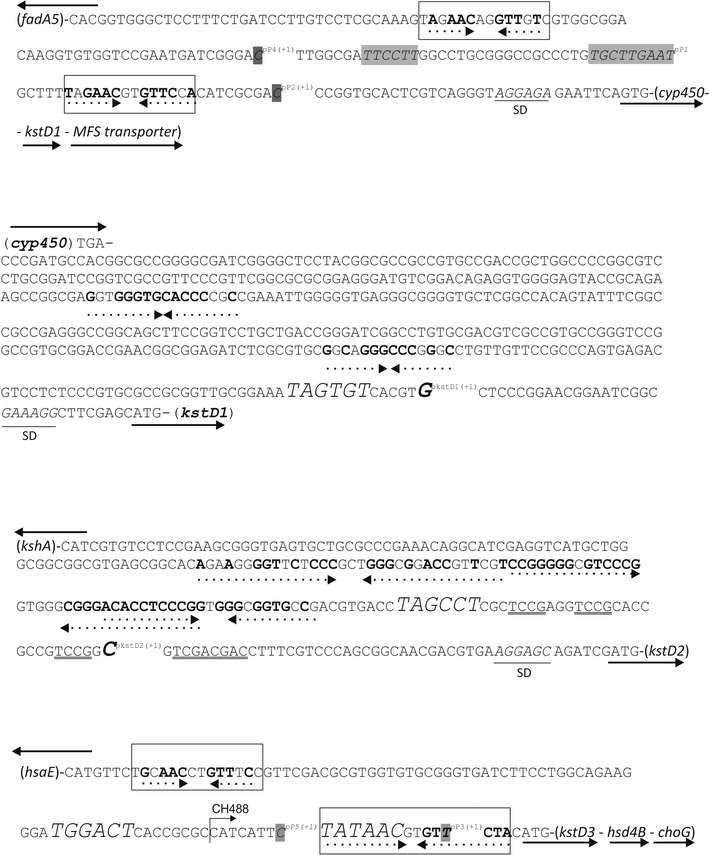
Sequence of regions upstream kstD ORFs. Solid arrows represent the orientation of the different ORFs from the initiation to the final codon. Sequences similar to Mycobacterium KstR binding sites (TnnAACnnGTTnnA) are within a square. Palindromic sequences appear in bold characters and dotted underlined. Shine Dalgarno (SD) sequences are in italics and underlined. The −10, −35 boxes are marked in grey. The in silico transcription initiation point of the putative pP1, pP2, pP3 and pP4 promoters are in italics and marked as pP(+1). The transcription start sites of kstD1 and kstD2 ORF obtained by the ARF-TSS method are shown in bold italics and marked as pKstD(+1). The TCCG repeats and the Sal box upstream kstD2 are double underlined. Promoter signals similar to others described (e.g. M. tuberculosis) [55] appear in bigger size. Primer CH488 indicating the beginning of the minimum kstD3 promoter is also shown
The possible co-transcription of the cyp450-kstD1-MSFt ORF cluster to a polycistronic mRNA would come into contradiction with the transcription of a non-yet described short putative ORF (pep, Fig. 1) located in opposite sense in the 425 bp cyp450-kstD1 intergenic region. This short ORF might code for a 35 amino acid peptide which shows a 98% amino acid identity with part of the hypothetical 78 aa protein RHRU231_750039 (R. ruber, 78 aa). Therefore, kstD1 might be independently transcribed while pP1/pP2/pP4 putative promoters might be only involved in cyp450 transcription. Moreover, a putative ribosome binding site (GAAAGG) was found 9 bp upstream the kstD1 initiation codon (Fig. 2) that is identical to the proposed one for sigA of R. ruber TH [54].
In the case of the kstD2 ORF, none of the online programs recognized a promoter consensus, although this region contains some quasi-palindromic sequences and a Shine-Dalgarno-like motif (AGGAGC) (Fig. 2).
There are two putative promoters for the kstD3 ORF (pP3 and pP5, see Figs. 1 and 2) that lie in the intergenic region between hsaE and kstD3 ORFs. The putative promoter pP3 contains the sequence TATAAC similar to the −10 consensus motif described for M. smegmatis promoters (T100%A93%T50%A57%A43%T71%) [55], and a −35 region (TGGACT) that resembles the E. coli promoter consensus motif TTGACA. In this region, there are also a tandem of two putative KstR binding sequences around this promoter (TgcAACctGTTtcc and TatAACgtGTTctA), one quite similar and the other identical to the KstR binding consensus, in a similar way to the cyp450 pP1/pP2 putative promoter (Fig. 2). The arrangement of these promoter and regulatory sequences, lying between opposite cluster genes, also occurs in the R. ruber kstD1 region and it is similar to that found in Mycobacterium and Rhodococcus jostii RHA1 genomes [48, 56]. On the other hand, Shell et al. have recently described that the abundance of leaderless transcripts (that lack a 5′ UTR and a Shine-Dalgarno sequence and that begin with ATG or GTG) is a major feature of mycobacterial that accounts for around one-quarter of the transcripts [57]. kstD3 could be a leaderless ORF: there is no evidence of a Shine-Dalgarno sequence in its 5′ region and the putative promoter is quite near to the ATG initiation codon. Moreover, as it will be stated later, the promoter of this intergenic region is functional in Rhodococcus but not in E. coli, a fact that has also been confirmed in the mycobacterial leaderless messenger translation [57].
Promoter cloning and characterization
To go further in the characterization of the promoter regions of the R. ruber kstD ORFs, a promoter-test vector suitable for R. ruber strain Chol-4 was constructed. R. ruber is sensitive to apramycin so we chose the expression of a gene encoding this resistance as a proof of the promoter activity.
pNV119 [43], a Nocardian shuttle vector shown to replicate in R. ruber [24], was modified by adding the mcs of pSEVA351 that contains a transcriptional terminator in each extreme [41, 42]. The resulting pNVS plasmid (see Additional file 4) was used to study the activity of the Chol-4 putative promoter regions. The three kstD intergenic regions (Fig. 2) plus the first 21 bases of each kstD ORF were PCR amplified, transcriptionally fused to the apramycin resistance gene obtained from pIJ773 and cloned into the mcs of pNVS. The recombinant plasmids were introduced into R. ruber by electroporation and kanamycin resistant clones were selected. R. ruber clones, harbouring the plasmids pNVSP1-A, pNVSP2-A or pNVSP3-A, were then plated in minimal medium supplemented with either 1.5 mM cholesterol, 1.5 mM AD or 10 mM sodium acetate, and in the presence of either 200 µg/mL kanamycin or 300 µg/mL apramycin. The apramycin resistance gene (Amr) without any upstream promoter region was cloned in pNVS mcs generating the vector pNVSA that was used as a negative control. As a second control, a set of pNVSP vectors that contain every promoter region but do not carry the apramycin resistance gene was used. Figure 3 shows that only the cells harbouring the double system formed by a putative promoter and the apramycin resistance gene were able to grow on apramycin and kanamycin while cells harbouring the pNVSA or the pNVSPs vector were only able to grow in kanamycin plates. These results unambiguously confirm that all the three checked DNA regions contain R. ruber promoter sequences functionally active in the conditions used. On the other hand, E. coli harbouring the plasmids pNVSP1-A or pNVSP2-A were also able to grow in apramycin, in contrast with those harbouring pNVSP3-A (data not shown). Other actinobacteria promoters (e.g. some of Mycobacterium and Streptomyces spp.; [55, 58] are also not functional in E. coli strains; this fact could related to the occurrence of leaderless genes [57].
Fig. 3.
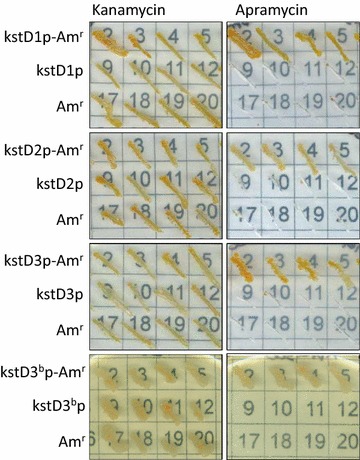
Comparative assessment of R. ruber kstD promoters. Cells of R. ruber strain Chol-4 harbouring different recombinant plasmids were grown in minimal medium supplemented with either 1.5 mM cholesterol (kstD2, kstD3 or kstD3 b promoters) or 2 mM AD (kstD1 promoter) in the presence of either 200 µg/mL kanamycin or 300 µg/mL apramycin. kstDp: R. ruber cells harbouring pNVSP plasmids (kstD promoters cloned in pNVS vector). kstDp- Amr: R. ruber cells harbouring pNVSP-A plasmids (apramycin resistance gene fused to kstD promoters in pNVSP plasmids). Amr: pNVS vector containing the promoter less apramycin resistance gene
Although none of the online programs recognized a promoter consensus for the kstD2 ORF, the promoter-less vector pNVS has enabled us to check the promoter activity of the intergenic regions. The construction of an improved version of this plasmid that allows a quantitative analysis of promoter strength is under work.
To define the transcription start sites (TSS) of the kstD genes, the transcription start point protocol (ARF-TSS) indicated in “Methods” section was followed in R. ruber cells. We could conclude that the 5′ terminal base in kstD1 messenger RNA is a G resulting from the transcription that starts 34 bp upstream the kstD1 initiation codon. Similarly, the transcription start site of the kstD2 gene is a C 48 bp upstream the kstD2 initiation codon (Figs. 2, 4). However, this approach did not yield any result in the case of the kstD3 gene. In order to better define the limits of the kstD3 promoter, progressively shorter sections of the intergenic region, keeping the first 21 bases of kstD3 ORF, were PCR amplified and transcriptionally fused to the apramycin resistance gene. As it can be seen in Figs. 2 and 3, just a minimum region of only 23 pb upstream the ATG of kstD3 ORF (pNVSP3b-A vector) was enough to act as a promoter as the Rhodococcus cells harbouring this vector were able to grow on medium with either apramycin or kanamycin. This promoter region is partially similar to the putative pP3 and pP5 promoters mentioned before, containing the TATAAC sequence similar to the −10 consensus motif described for Mycobacterium smegmatis promoters (Fig. 1).
Fig. 4.
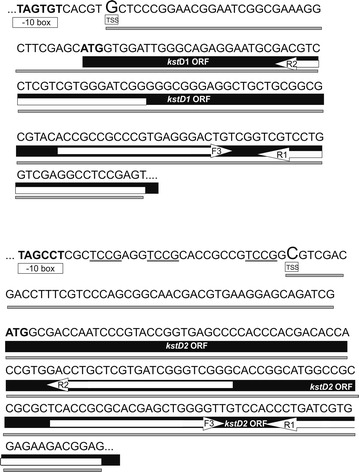
ARF-TSS analysis of kstD transcripts. The translation initiation codon ATG, the TSS identified by ARF-TSS and the proposed −10 box appear in black. R1, R2 and F3 primers are indicated. A grey bar shows the sequence obtained by ARF-TSS from the circularized cDNA and the subsequent PCR amplification with R2 and F3
Putative −35 and −10 hexamers identification was based on the sequence of other actinobacteria promoters. The −10 region motif (TAGTGT) found 40 bases upstream the kstD1 initiation codon is similar to the −10 consensus region described for Mycobacterium tuberculosis promoters (T80%A90%Y60%G40%A60%T100%) and identical to the T101 promoter described by Bashyam et al. in the same bacteria [55]. No sequence similar to any −35 motif was found in the kstD1 region. The absence of −35 motifs seems to be a characteristic of actinobacterial genes [55, 58].
There is a putative −10 sequence (TAGCCT) in the intergenic region between kshA and kstD2 that resembles the T6 promoter of M. tuberculosis (TAGGCT) [55]. However, it is a bit far away from the kstD2 TSS. There is a repetitive motif TCCG in this region (Fig. 2) that also appears in the human Sal box element present in the 3′ terminal spacer of rDNA and that constitutes a termination signal for RNA polymerase I (TCCGCACGGGTCGACCAG) [59]. In this kstD2 upstream region we found something similar: TCCGAGGTCCGCACCGCCGTCCGGCGTCGACGAC, where black-underlined letters marked the resemblance between them. Such promoter proximal terminators can appear also upstream of the transcription start site and in this case, they are described to positively affect transcription initiation and to prevent transcriptional interference by reading through of polymerases from the spacer that separates each rDNA repetition [60–62]. The biological significance of this motif in this intergenic kstD2 region should be further determined.
Transcriptional analysis of kstD genes in R. ruber strain Chol-4
The transcription of the three kstD genes found in R. ruber strain Chol-4 was analysed by RT-PCR of RNA samples prepared from cultures grown in either M457 mineral medium or LB medium supplemented with either AD or cholesterol as possible inductors.
Control cultures were grown in either M457 supplemented with sodium acetate (2 g/L NaAc) or LB, both in the absence of any steroid. Using the specific primer pairs designed to search for the transcript of each different ORF (Additional file 2), we could show that all the kstD genes are transcribed in all the conditions used in our assays (Fig. 5) and that transcription of some of them is induced by cholesterol or AD. There are also other Rhodococcus metabolic genes reported to show a low-level constitutive transcription that can be strongly induced under the presence of a determinate substrate. For instance, phenol degradation genes in R. erythropolis are constitutively transcribed and also highly induced by phenol [63].
Fig. 5.
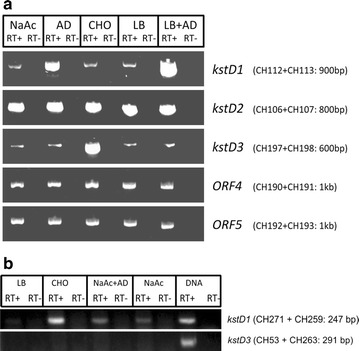
a Analysis of transcription products of kstD genes in R. ruber strain Chol-4 by specific RT-PCR amplification and agarose gel electrophoresis. RNA samples were isolated from cultures grown in: M457 minimal medium supplemented with sodium acetate (lane NaAc), AD (lane AD) or cholesterol (lane CHO) and LB medium supplemented with (lane LB + AD) or without AD (lane LB). RT—refers to negative controls (not incubated with retrotranscriptase) to exclude DNA contamination in RNA samples. Primers used in each PCR reaction and amplified fragment size are shown in brackets. As non-induced expression controls, the transcription of two R. ruber Chol-4 enzyme genes was followed in the same conditions: ORF4, that codes for a FAD-binding dehydrogenase D092_14375 (99% identity with the protein NCIB::WP_010594120.1 of Rhodococcus sp. P14) and ORF5 (D092_13875) that codes for a fumarate reductase (99% identity with NCIB::WP_010594021.1 of Rhodococcus sp. P14; see sequence in Additional file 6). the FAD-binding dehydrogenase D092_14375 and the fumarate reductase D092_13875. b RT-PCR assays of R. ruber Chol-4 kstD1 and kstD3 transcription carried out in kstD2 R. ruber mutant growing in: LB (lane LB) or M457 plus CHO (lane CHO), NaAc and AD (lane NaAc + AD) or NaAc (lane NaAc). Lane DNA shows the result of the control experiment: PCR made on R. ruber DNA
cDNA templates concentrations were adjusted to the same value in RT-PCR experiments, so we can consider the differences observed in the thickness of some amplification bands to be meaningful (Fig. 5). Specific amplification of kstD1 cDNA was higher in AD induced than in non-induced cultures, while amplification of kstD3 cDNA was higher in cultures induced with cholesterol as compared to the other conditions assayed. So we can conclude that these two genes, although constitutively transcribed, they can also be additionally induced by the presence of AD (kstD1) and cholesterol (kstD3). The induction of kstD genes by cholesterol or AD has also been reported in other microorganisms: a 3.3 expression ratio (cholesterol/pyruvate) was reported for one kstD in R. jostii RHA1 (ro04532) [64, 65], while other putative kstD genes of the same strain (e.g. ro02483, ro05798 and ro05813) were up-regulated in 7-ketocholesterol but not in cholesterol [64]; the main kstD in M. smegmatis (MSMEG_5941) was 13-fold up-regulated in cholesterol respect to glycerol [12]; a 1.8, 4.1 or 1.2 -fold up-regulation (AD/glycerol) was found for kstD1, kstD3 and kstD2 genes respectively in Mycobacterium neoaurum [66]. These differences in regulation among the different kstD genes within the same strain highlight the view that KstD proteins may be acting in different metabolic steps and/or pathways, each one having a particular catalytic role.
The amplification of R. ruber strain Chol-4 kstD2 cDNA yielded thick amplification bands in all cases (Fig. 5), which clearly leads to propose a constitutive expression of kstD2. In a similar way, two putative kstD genes in R. jostii RHA1 genome (ro90203 and ro09040, belonging to the KstD2-branch of the KstD phylogenetic tree) [20] were expressed but not up-regulated neither in cholesterol nor in 7-ketocholesterol, when compared to pyruvate [64].
The expression profile of R. ruber kstD1, kstD2 and kstD3 genes was determined by real-time PCR. Taking as 1 the expression levels on sodium acetate (unexposed steroid culture), the values obtained for the expression of the three genes were: 7.6, 2.0 and 240.5-fold for kstD1, kstD2 and kstD3, respectively, in cultures grown in cholesterol; and 13.6, 0.7 and 0.6-fold for kstD1, kstD2 and kstD3, respectively, in cultures grown in AD.
The particular organization of R. ruber Chol-4 kstD1 and kstD3 genes (Fig. 1) opens the possibility that polycistronic kstD mRNAs could be synthesized by the co-transcription of the cyp450-kstD1-MFS transporter and kstD3-hsd4B-choG gene clusters. The results of the RT-PCR experiments did not show the occurrence in AD culture medium of either cyp450-kstD1 or kstD1-MFS transporter RNA sequences, indicating that kstD1 gene is independently transcribed.
In contrast, co-transcripts from both kstD3-hsd4B ORFs and hsd4B-choG ORFs could be amplified from cultures grown in the presence of cholesterol, strongly suggesting that kstD3-hsd4B-choG ORFs are co-transcribed into a polycistronic mRNA (Fig. 1). This group of three genes are also described to be co-transcribed in R. erythropolis [27].
The adjacent location of kstD3-hsd4B ORFs is highly conserved among rhodococci. The hsd4B ORF encodes a 2-enoyl acyl-CoA hydratase involved in the β-oxidative cycle of the C-17 cholesterol side chain [65]. The choG ORF encodes an extracellular cholesterol oxidase that it is involved in the first step of cholesterol catabolism that implies its conversion to 4-cholesten-3-one [27, 47, 67–69]. All this suggests that kstD3, hsd4B and choG genes are mainly involved in the steroid catabolism.
In this work we showed that kstD2 is constitutively transcribed, while kstD1 and kstD3 are also constitutively but faintly transcribed, although they can be highly induced by AD or CHO. In a previous work [24], we reported the construction of a kstD2 deletion mutant of R. ruber that is unable to grow in minimal medium supplemented with AD. The question is why KstD1 and/or KstD3 cannot substitute KstD2 allowing the kstD2 deletion mutant to grow on AD. To partially advance in this subject, RT-PCR experiments were also performed on RNA from the kstD2 R. ruber mutant to check the expression of the other kstD genes (Fig. 5b).
The results showed that the transcription pattern of the kstD genes of the mutant is far different than that of the same genes in the wild type. Namely, kstD1 gene in the mutant was constitutively and slightly transcribed and its expression is induced in CHO. A more noticeable change affects to the kstD3 gene, which is not transcribed at all in any of the conditions used. These data reveal a complex relationship among the KstD enzymes and their expression control mechanisms. Modification of the kstD transcription levels of genes remaining in the cell have also been described in a M. neoaurum kstD mutant: the transcription ratio of the kstD1 ORF (similar to the kstD3 ORF from R. ruber) in AD induced cultures respect to glycerol cultures increases from 1.8 (in the wild type strain) to 2.7-fold (in the kstD3 M. neoaurum mutant strain) [66].
An appealing conclusion of that complex situation is that the three KstD proteins of R. ruber strain Chol-4 may be differentially involved in distinct pathways of steroid degradation, and that their expression could also be differentially and specifically controlled.
Heterologous expression of KstD1, KstD2 and KstD3 of R. ruber strain Chol-4
The three R. ruber kstD genes (kstD1, kstD2 and kstD3) were cloned into the pTip-QC1 expression vector. R. erythropolis CECT3014 cells were electroporated with these constructions and clones harbouring each of those recombinant plasmids were isolated. Expression of the KstD proteins from these vectors in the CECT3014 transformed cells was followed by SDS-PAGE analysis. Molecular weights were 54.8, 60.8 and 61.8 kDa for KstD1, KstD2 and KstD3 respectively (Additional file 5). Cell-free extracts of cultures grown from these clones were used for the analysis of KstDs activities, and the kinetic parameters of the heterologously expressed KstDs from R. ruber were followed for different substrates (Table 1; see also Additional file 3 for substrates structure). Control cell extract from the R. erythropolis culture harbouring an empty pTip-QC1 vector yielded none or very low basal levels when acting on all the substrates used in the assay and were taken into account for final activities.
Table 1.
Substrate profiles of R. ruber KstDs expressed an analyzed in cell-free extracts of R. erythropolis
| Substrate | KstD1 | KstD2 | KstD3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rel. act % | Km (µM) | RCE | RCE/RCEAD | Rel. act % | Km (µM) | RCE | RCE/RCEAD | Rel. act % | Km (µM) | RCE | RCE/RCEAD | |
| AD | 100.0 ± 11.6 | 34.2 ± 3.8 | 2.92 | 1.00 | 100.0 ± 11.8 | 40.1 ± 8.7 | 2.60 | 1.00 | nd | nd | ||
| 9OHAD | 107.4 ± 13.4 | 22.1 ± 7.0 | 4.84 | 1.66 | 29.8 ± 6.5 | 543.2 ± 77.8 | 0.05 | 0.02 | nd | nd | ||
| 4BNC | 52.3 ± 7.4 | 76.1 ± 15.7 | 0.69 | 0.24 | 30.8 ± 7.2 | 38.2 ± 6.6 | 0.81 | 0.31 | nd | nd | ||
| Prog | 89.7 ± 7.9 | 27.9 ± 9.5 | 3.22 | 1.10 | 182.0 ± 7.0 | 33.8 ± 4.9 | 5.38 | 2.07 | 18.6 ± 3.7 | 43.6 ± 2.9 | 0.42 | 0.78 |
| Cort | 20.3 ± 5.8 | 161.6 ± 7.6 | 0.13 | 0.04 | 19.0 ± 3.0 | 374.3 ± 74.5 | 0.05 | 0.02 | nd | nd | ||
| Tes | 134.6 ± 22.6 | 28.8 ± 7.4 | 4.67 | 1.60 | 233.3 ± 23.3 | 107.9 ± 18.5 | 2.16 | 0.83 | nd | nd | ||
| 19OHAD | 39.0 ± 10.6 | 368.8 ± 102.4 | 0.11 | 0.04 | 24.6 ± 7.8 | 347.4 ± 41.7 | 0.07 | 0.03 | nd | nd | ||
| DOC | 67.0 ± 7.4 | 21.6 ± 4.5 | 3.10 | 1.06 | 124.2 ± 17.2 | 42.5 ± 5.8 | 2.92 | 1.12 | 21.6 ± 8.8 | 111.3 ± 4.0 | 0.19 | 0.36 |
| 5α-Tes | nd | nd | 75.2 ± 4.8 | 24.57 ± 7.8 | 3.06 | 1.18 | 100.0 ± 21.9 | 181.1 ± 42.6 | 0.55 | 1.00 | ||
Rel. act: relative activity values. Enzyme activities are expressed as percentage of activity of AD (for KstD1 and KstD2 with 3.2 U/mg and 1.4 U/mg respectively) or 5α-tes (for KstD3 with 0.3 U/mg) that were set as 100%
nd enzyme activity was not detected for this substrate, RCE relative catalytic efficiency given by the ratio Rel. act/km, Prog progesterone, Cort corticosterone, Tes testosterone, 5α-tes 5-alpha-testosterone, 19OHAD 19-hydroxy-4-androstene-3,17-dione, DOC deoxycorticosterone, 4-BNC 4-pregnen-3-one-20β-carboxylic acid
The substrate profile of KstD1 showed a clear preference to 9OHAD and testosterone, followed by progesterone, Deoxy corticosterone (DOC) and AD (Table 1). All these compounds display a keto group at C3, a C4-C5 double bond and an electronegative side-chain at C17 (Additional file 3). When comparing to the substrate preference order of R. erythropolis SQ1 KstD1 (Prog > 9OHAD, AD > 5α-Tes > BNC > 11βCort), some details highlights: (i) KstD1SQ1 has a relative catalytic efficiency (RCE) on progesterone 3.4 times higher than on AD, a very big difference to the ratio 9OHAD/AD (1.6) showed by R. ruber KstD1 (Table 1); (ii) R. ruber KstD1 is not active on 5α-Tes in contrast to KstD1SQ1.
The order of substrate preference of R. ruber KstD2 placed progesterone in the first position, followed by 5α-Tes, DOC, AD and testosterone (Table 1), displaying a substrate profile similar to KstD2SQ1 enzyme (Prog > AD > 5α-Tes > 9OHAD, BNC > 11βCort). It is noteworthy the similarity of both KstD profiles, having in mind that they have been expressed in different cellular context (a Rhodococcus strain, and E. coli). R. ruber KstD2 has a broader range of substrates than R. ruber KstD1 as it can act on all the KstD1 substrates and also on 5α-Tes that contains a saturated A ring (Table 1).
Rhodococcus ruber KstD3 did not show any activity at all when acting on AD or 9OHAD, which are considered the natural substrates for KstD enzymes, but it showed the highest activity when using 5α-Tes as substrate followed by progesterone and lastly DOC (Table 1). R. ruber KstD3 has a very narrow substrate range similarly to the R. erythropolis SQ1 and M. tuberculosis H37Rv isoforms, being the A-ring saturated 5α-Tes the preferred substrate for all KstD3 enzymes (Table 1, [20]). However, KstD3 affinity for 5α-Tes differs from 33–36 µM in the cases of KstD3H37RV and KstD3SQ1 respectively, to 181 µM in R. ruber KstD3 (Table 1). Despite this low affinity for 5α-Tes, this is the best substrate for the R. ruber enzyme among those that were assayed (Table 1).
On the other hand, although it has been proposed that only steroids carrying a small or no aliphatic side chain at C-17 are suitable substrates for KstD3 [20], a minor activity, not very different to that obtained with progesterone, has been observed in the assays of R. ruber KstD3 enzyme on DOC (Additional file 3; Table 1). R. ruber KstD3 seems to be more related to the cholesterol metabolism than to the AD metabolism [24] and then it could be acting on some not yet neatly defined intermediaries of the bacterial cholesterol metabolism.
None of the three R. ruber KstD proteins displayed detectable activity on 4-cholestene-3-one, 5α-cholestane-3-one or 5β-androstane-3,17-dione (5β-AD), ADD, cholesterol, cholestenone, cholic acid, DHEA, ergosterol, stigmasterol, β-estradiol, sodium deoxycholate or 5-pregen-3-β-nolone. Therefore, these KstDs catalyse preferentially 4-ene-3-oxosteroids.
Molecular modelling of KstD1 of R. ruber strain Chol-4
The sequence of the three KstDs of R. ruber were described previously [24]. In an attempt to go deeper inside the reasons of the catalytic and kinetic differences among some of these enzymes, we performed protein sequence analysis and modelling studies on the R. ruber KstDs using different approaches. The published three-dimensional structure and the catalytic mechanism proposed for KstD1 from R. erythropolis SQ1 has been used as a suitable model [21, 70]. The I-Tasser and PredictProtein programmes predicted that none of the three R. ruber KstDs contain sulphur bridges or transmembrane segments.
The catalytic mechanism of R. erythropolis SQ1 KstD1 is based in the keto-enol tautomerization of the substrate caused by Tyr487 and Gly491 residues that increases the acidity of the C2 hydrogen atoms of the substrate. Then Tyr119 and Tyr318 capture the axial β-hydrogen from C2 as a proton whereas the FAD molecule accepts the axial α-hydrogen from the C1 atom of the substrate as a hydride ion [21, 70]. Tables 2 and 3 collects the residues involved in the active site of KstD1SQ1 described for Rohman et al. [21] and the homologue residues of the three KstDs from R. ruber found by COBALT programme. The four key residues: Tyr119, Tyr318, Tyr487 and Gly491 of the KstD1SQ1 active site have a counterpart residue in the R. ruber KstDs. However, the I-TASSER model prediction of these R. ruber enzymes shows that the orientations of the side chain of the tyrosine residues differ within the catalytic pocket site being specific of each enzyme. The variation in orientation of the key residues inside the catalytic poked is shown in Fig. 6. Particularly, the orientation of KstD3 Y354 and Y530 residues is almost opposite to that of its homologous KstD1 and KstD2 residues. Moreover, amino acid Y128 from KstD2 has a position highly separated compared to its homologous from KstD1 (Y121) and KstD3 (Y119). These differences could justify the different affinity and catalytic properties among the three KstDs. The current shortage of crystalline structures of these proteins greatly limit more detailed conclusions.
Table 2.
Homologous residues of KstDs implicated in the catalytic pocket attending to Rohman et al. [21]
| KstD1-SQ1 | KstD1-ruber | KstD2-ruber | KstD3-ruber |
|---|---|---|---|
| S52 | G53 | A59 | G50 |
| F116 | W118 | Y125 | Y116 |
| Y119 | Y121 | Y128 | Y119 |
| F294 | F295 | P337 | F327 |
| V296 | L297 | V339 | S330 |
| Y318 | Y319 | Y366 | Y354 |
| I352 | T356 | I395 | |
| I354 | V358 | F405 | P397 |
| L447 | L447 | L499 | L490 |
| Y487 | Y487 | Y539 | Y530 |
| P490 | G490 | A542 | P533 |
| G491 | G491 | G543 | G534 |
| V492 | N492 | A544 | A535 |
| P493 | P493 | T545 | T537 |
Italic files contain the four key residues of the KstD1 active site reported for R. erythropolis SQ1: Tyr119, Tyr318, Tyr487 and Gly491
Table 3.
PredictProtein prediction of the secondary structure and solvent accessibility in % of the three KstDs from R. ruber
| Structure | KstD1 | KstD2 | KstD3 |
|---|---|---|---|
| Strand | 12.72 | 11.88 | 64.74 |
| Loop | 63.99 | 64.01 | 14.38 |
| Helix | 23.29 | 24.11 | 20.88 |
| Accessibility | KstD1 | KstD2 | KstD3 |
|---|---|---|---|
| Exposed | 24.1 | 21.81 | 22.11 |
| Buried | 67.7 | 71.63 | 70.5 |
| Intermediate | 8.22 | 6.56 | 7.54 |
Fig. 6.
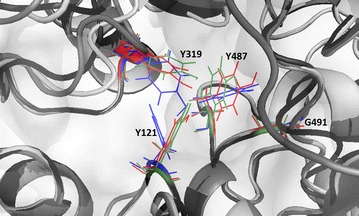
Modelling of the active site of KstDs. The orientation of the four key residues in the KstD binding pocket is shown. They are superposed and depicted in different colours: green for KstD1, blue for KstD2 and red for KstD3. Only KsD1 residues are named, their counterpart homologues residues of KstD2 and KstD3 being listed in Tables 2 and 3
A redundancy of KstD and Ksh enzymes have been described in the actinobacteria genomes [20, 24, 64, 70]. This redundancy could provide the cell with a bigger metabolic versatility and a fine-tuned response to the challenging environment. In the case of KstD enzymes, three homologues have been found in R. erythropolis strain SQ1 and in M. neoaurum that displayed different substrate preferences and that could be involved in different metabolic steps in a strain-dependent way [20, 32, 66].
Even more, it has been shown recently that mutations in the KstDs provokes a different ADD/AD molar ratio [71] and that environmental factors such as an increase of temperature can inhibit the KstD/Ksh action [72] on phytosterol in Mycobacterium sp. These multiplicity and versatility give to these enzymes a substantial role in the catalytic dehydrogenation of several related steroid molecules and pose many difficulties to clarify the particular role and way of acting of every single KstD.
Our results suggest that both KstD1 and KstD2 of R. ruber could act in the conversion of AD to ADD being KstD1 mainly involved in the 9OHAD to 9OHADD conversion, in a similar way to what has been described in R. erythropolis SQ1 [32]. However, there are differences between these two strains, as the necessity of a double kstD1 and kstD2 mutation to prevent the growth in AD in the case of R. erythropolis SQ1 [32] or R. rhodochrous DSM43269 [73], while the same effect is obtained by the single kstD2 deletion in R. ruber strain Chol-4 [24], suggesting that this last mutation affects in some way the activity of the KstD1 protein.
kstD3 ORF occurs in the R. ruber genome in a quite conserved location within Rhodococcus species, clustered with hsd4B (which encodes a 2-enoyl acyl-CoA hydratase proposed to be involved in cholesterol side-chain shortening) [65] and choG (coding for a cholesterol oxidase that converts cholesterol into cholestenone) [27, 47, 68]. Recently it has been proposed that sterols can be catabolised in R. equi USA-18 by two partially different pathways, namely via AD or via Δ1,4-BNC, that converge in the intermediary 9OHADD [74]. Given its substrate preference, the fact that the growth in AD is independent of the KstD3 activity while the growth in cholesterol needs the presence of either KstD2 or KstD3 in R. ruber [24], lead us to suggest that KstD3 may be involved in an alternative AD-independent cholesterol catabolic pathway.
Conclusions
To sum up, this study provides biochemical and genetic insights into the three KstD proteins found in R. ruber. The kinetic differences between the three KstDs suggest that each enzyme could act on different steps of the steroid catabolic routes. Both KstD1 and KstD2 could be involved in the AD catabolism while KstD1 would have a preference for 9OHAD and KstD2 for progesterone. KstD2 seems to be a more versatile enzyme than KstD1 in R. ruber as it can act also on saturated steroid substrates such as 5α-Tes. On the other hand, the narrower range of substrates for KstD3 and its preference for saturated steroid made this enzyme different to KstD1 and KstD2 and suggest that it may be involved in AD-independent steroid catabolism. The differences found in the orientation of catalytic residues of each KstD within the binding pocket site could explain the substrates preferences of each enzyme.
The promoter regions that support transcription of kstD genes have been cloned and functionally identified. The three promoter boxes contain different expression patterns, from the TCCG motif found in the kshA-kstD2 ORF intergenic region to the KstR boxes in the hsaE-kstD3 intergenic region. Moreover, kstD3 ORF was transcribed as a polycistronic kstD3-hsd4B-choG mRNA and was induced in cholesterol growing media, reinforcing the role of KstD3 in the cholesterol metabolism. Potential functions of R. ruber strain Chol-4 KstDs in other steroid pathways remain to be elucidated.
Authors’ contributions
GG made the heterologous expression in R. erythropolis, the coexpression study, the RT-qPCR and the promoter characterization. JMNLL and GG performed the bioinformatic analyses. LFH carried out the RT-PCR experiment and collaborated with GG in the heterologous expression experiments. JP and JMNLL drafted the manuscript. JP, JMNLL and GG participated in the design and coordination of the study. All authors read and approved the final manuscript.
Acknowledgements
We are in debt to R. van der Geize (Netherland) and to the group of J. L. García (CIB, CSIC) for their scientific support and invaluable help. We thank Rodrigo Velasco for his help on molecular modelling and Sara Montero for her help on kinetics.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its supplementary materials.
Funding
This work was funded by projects BFU2009-11545-C03-02, BIO2012-39695-CO2-01 and RTC-2014-2249-1 (GG is under contract of this last project) from the Spanish Ministry of Education and Science.
Abbreviations
- AD
4-androstene-3,17-dione
- ADD
1,4-androstadiene-3,17-dione
- 5β-AD
5β-androstane-3,17-dione
- 9OHAD
9α-hydroxy-4-androstene-3,17-dione
- 9OHADD
9α-hydroxy-1,4-androstadiene-3,17-dione
- 4-BNC
4-pregnen-3-one-20β-carboxylic acid
- 5α-Tes
17β-hydroxy-5α-androstan-3-one
- DOC
deoxy corticosterone
- ORF
open reading frame
- DCPIP
2,6-dichlorophenol-indophenol
- KstD
3-ketosteroid-Δ1-dehydrogenase
- LB
Luria-Bertani
- mcs
multiple cloning site
- TSS
transcriptional start site
Additional files
Additional file 1. Bacterial strains and plasmids used in this work.
Additional file 2. Primers used in this work. Restriction sites are marked in bold.
Additional file 3. Structure of the steroids used in this work.
Additional file 4. Scheme of the pNVSP vectors construction. The shuttle vector pNV119 was modified to include a mcs from pSEVA351 (pNVS) and after that coupled with the apramycin resistance ORF. Intergenic regions containing the putative kstD promoters were cloned in the mcs to obtain pNVSP vectors.
Additional file 5. Expression of KstDs from ptip-QC1 vectors in induced R. erythropolis CECT3014 cells. SDS-PAGE analysis on a 12.5% gel was performed using 10 µg of the cell-free extracts. The band corresponding to KstD overexpression is marked with a rectangle. Precision plus protein standard from Bio-Rad was used as size marker.
Additional file 6. R. ruber strain Chol-4 ORF4 and ORF5 protein sequences.
Contributor Information
Govinda Guevara, Email: fguevara@ucm.es.
Laura Fernández de las Heras, Phone: +31 50 363 2150, Email: lauritafh@gmail.com, Email: l.fernandez.de.las.heras@rug.nl.
Julián Perera, Email: jpererag@bio.ucm.es.
Juana María Navarro Llorens, Phone: (34) 913944145, Email: joana@bio.ucm.es.
References
- 1.Iwabuchi N, Sunairi M, Urai M, Itoh C, Anzai H, Nakajima M, Harayama S. Extracellular polysaccharides of Rhodococcus rhodochrous S-2 stimulate the degradation of aromatic components in crude oil by indigenous marine bacteria. Appl Environ Microbiol. 2002;68:2337–2343. doi: 10.1128/AEM.68.5.2337-2343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLeod MP, Warren RL, Hsiao WW, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci USA. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yam KC, Okamoto S. Adventures in Rhodococcus—from steroids to explosives. Can J Microbiol. 2011;57:155–168. doi: 10.1139/W10-115. [DOI] [PubMed] [Google Scholar]
- 4.Barel-Cohen K, Shore LS, Shemesh M, Wenzel A, Mueller J, Kronfeld-Schor N. Monitoring of natural and synthetic hormones in a polluted river. J Environ Manag. 2006;78:16–23. doi: 10.1016/j.jenvman.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Gracia T, Jones PD, Higley EB, Hilscherova K, Newsted JL, Murphy MB, Chan AK, Zhang X, Hecker M, Lam PK, et al. Modulation of steroidogenesis by coastal waters and sewage effluents of Hong Kong, China, using the H295R assay. Environ Sci Pollut Res Int. 2008;15:332–343. doi: 10.1007/s11356-008-0011-6. [DOI] [PubMed] [Google Scholar]
- 6.Soto AM, Calabro JM, Prechtl NV, Yau AY, Orlando EF, Daxenberger A, Kolok AS, Guillette LJ, Jr, le Bizec B, Lange IG, et al. Androgenic and estrogenic activity in water bodies receiving cattle feedlot effluent in Eastern Nebraska, USA. Environ Health Perspect. 2004;112:346–352. doi: 10.1289/ehp.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Geize R, Dijkhuizen L. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr Opin Microbiol. 2004;7:255–261. doi: 10.1016/j.mib.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Malaviya A, Gomes J. Androstenedione production by biotransformation of phytosterols. Bioresour Technol. 2008;99:6725–6737. doi: 10.1016/j.biortech.2008.01.039. [DOI] [PubMed] [Google Scholar]
- 9.García JL, Uhía I, Galan B. Catabolism and biotechnological applications of cholesterol degrading bacteria. Microb Biotechnol. 2012;5:679–699. doi: 10.1111/j.1751-7915.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horinouchi M, Hayashi T, Kudo T. Steroid degradation in Comamonas testosteroni. J Steroid Biochem Mol Biol. 2012;129:4–14. doi: 10.1016/j.jsbmb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Philipp B. Bacterial degradation of bile salts. Appl Microbiol Biotechnol. 2011;89:903–915. doi: 10.1007/s00253-010-2998-0. [DOI] [PubMed] [Google Scholar]
- 12.Uhía I, Galán B, Kendall SL, Stoker NG, García JL. Cholesterol metabolism in Mycobacterium smegmatis. Environ Microbiol Rep. 2012;4:168–182. doi: 10.1111/j.1758-2229.2011.00314.x. [DOI] [PubMed] [Google Scholar]
- 13.Petrusma M, van der Geize R, Dijkhuizen L. 3-Ketosteroid 9alpha-hydroxylase enzymes: rieske non-heme monooxygenases essential for bacterial steroid degradation. Antonie Van Leeuwenhoek. 2014;106:157–172. doi: 10.1007/s10482-014-0188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florin C, Kohler T, Grandguillot M, Plesiat P. Comamonas testosteroni 3-ketosteroid-delta 4(5 alpha)-dehydrogenase: gene and protein characterization. J Bacteriol. 1996;178:3322–3330. doi: 10.1128/jb.178.11.3322-3330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itagaki E, Hatta T, Wakabayashi T, Suzuki K. Spectral properties of 3-ketosteroid-delta 1-dehydrogenase from Nocardia corallina. Biochim Biophys Acta. 1990;1040:281–286. doi: 10.1016/0167-4838(90)90088-W. [DOI] [PubMed] [Google Scholar]
- 16.Itagaki E, Wakabayashi T, Hatta T. Purification and characterization of 3-ketosteroid-delta 1-dehydrogenase from Nocardia corallina. Biochim Biophys Acta. 1990;1038:60–67. doi: 10.1016/0167-4838(90)90010-D. [DOI] [PubMed] [Google Scholar]
- 17.Chen MM, Wang FQ, Lin LC, Yao K, Wei DZ. Characterization and application of fusidane antibiotic biosynthesis enzyme 3-ketosteroid-1-dehydrogenase in steroid transformation. Appl Microbiol Biotechnol. 2012;96:133–142. doi: 10.1007/s00253-011-3855-5. [DOI] [PubMed] [Google Scholar]
- 18.Kisiela M, Skarka A, Ebert B, Maser E. Hydroxysteroid dehydrogenases (HSDs) in bacteria—a bioinformatic perspective. J Steroid Biochem Mol Biol. 2012;129:31–46. doi: 10.1016/j.jsbmb.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Hilyard EJ, Jones-Meehan JM, Spargo BJ, Hill RT. Enrichment, isolation, and phylogenetic identification of polycyclic aromatic hydrocarbon-degrading bacteria from Elizabeth River sediments. Appl Environ Microbiol. 2008;74:1176–1182. doi: 10.1128/AEM.01518-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knol J, Bodewits K, Hessels GI, Dijkhuizen L, van der Geize R. 3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem J. 2008;410:339–346. doi: 10.1042/BJ20071130. [DOI] [PubMed] [Google Scholar]
- 21.Rohman A, van Oosterwijk N, Thunnissen AM, Dijkstra BW. Crystal structure and site-directed mutagenesis of 3-ketosteroid Delta1-dehydrogenase from Rhodococcus erythropolis SQ1 explain its catalytic mechanism. J Biol Chem. 2013;288:35559–35568. doi: 10.1074/jbc.M113.522771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molnár I, Choi KP, Yamashita M, Murooka Y. Molecular cloning, expression in Streptomyces lividans, and analysis of a gene cluster from Arthrobacter simplex encoding 3-ketosteroid-delta 1-dehydrogenase, 3-ketosteroid-delta 5-isomerase and a hypothetical regulatory protein. Mol Microbiol. 1995;15:895–905. doi: 10.1111/j.1365-2958.1995.tb02359.x. [DOI] [PubMed] [Google Scholar]
- 23.Wierenga RK, Terpstra P, Hol WG. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 24.de las Heras LF, van der Geize R, Drzyzga O, Perera J, Navarro Llorens JM. Molecular characterization of three 3-ketosteroid-∆1-dehydrogenase isoenzymes of Rhodococcus ruber strain Chol-4. J Steroid Biochem Mol Biol. 2012;132:271–281. doi: 10.1016/j.jsbmb.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Fernández de las Heras L, García Fernández E, Navarro Llorens JM, Perera J, Drzyzga O. Morphological, physiological, and molecular characterization of a newly isolated steroid-degrading actinomycete, identified as Rhodococcus ruber strain Chol-4. Curr Microbiol. 2009;59:548–553. doi: 10.1007/s00284-009-9474-z. [DOI] [PubMed] [Google Scholar]
- 26.Klein U, Gimpl G, Fahrenholz F. Alteration of the myometrial plasma membrane cholesterol content with b-cyclodextrin modulates the binding affinity of the oxytocin receptor. Biochemistry. 1995;34:13784–13793. doi: 10.1021/bi00042a009. [DOI] [PubMed] [Google Scholar]
- 27.Fernández de las Heras L, Mascaraque V, García Fernández E, Navarro-Llorens JM, Perera J, Drzyzga O. ChoG is the main inducible extracellular cholesterol oxidase of Rhodococcus sp. strain CECT3014. Microbiol Res. 2011;166:403–418. doi: 10.1016/j.micres.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Hosek J, Svastova P, Moravkova M, Pavlik I, Bartos M. Methods of mycobacterial DNA isolation from different biological material: a review. Vet Res. 2006;524:180–192. [Google Scholar]
- 30.Nakashima N, Tamura T. Isolation and characterization of a rolling-circle-type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. Appl Environ Microbiol. 2004;70:5557–5568. doi: 10.1128/AEM.70.9.5557-5568.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.van der Geize R, Hessels GI, Dijkhuizen L. Molecular and functional characterization of the kstD2 gene of Rhodococcus erythropolis SQ1 encoding a second 3-ketosteroid ∆1-dehydrogenase isoenzyme. Microbiology. 2002;148:3285–3292. doi: 10.1099/00221287-148-10-3285. [DOI] [PubMed] [Google Scholar]
- 33.de las Heras LF, Alonso S, de Leon AD, Xavier D, Perera J, Navarro Llorens JM. Draft genome sequence of the steroid degrader Rhodococcus ruber strain Chol-4. Genome Announc. 2013 doi: 10.1128/genomeA.00215-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen TN, Brunak S, von Heijne G, von Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 35.Solovyev V, Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. In: Li RW, editor. Metagenomics and its applications in agriculture, biomedicine and environmental studies. New York: Nova Science Publishers; 2011. pp. 61–78. [Google Scholar]
- 36.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26:51–56. doi: 10.1016/S0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 37.de Jong A, Pietersma H, Cordes M, Kuipers OP, Kok J. PePPER: a webserver for prediction of prokaryote promoter elements and regulons. BMC Genom. 2012;13:299. doi: 10.1186/1471-2164-13-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Honigschmid P, Schafferhans A, Roos M, Bernhofer M, et al. PredictProtein-an open resource for online prediction of protein structural and functional features. Nucl Acids Res. 2014;42:W337–W343. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 41.Durante-Rodriguez G, de Lorenzo V, Martinez-Garcia E. The Standard European Vector Architecture (SEVA) plasmid toolkit. Methods Mol Biol. 2014;1149:469–478. doi: 10.1007/978-1-4939-0473-0_36. [DOI] [PubMed] [Google Scholar]
- 42.Silva-Rocha R, Martínez-García E, Calles B, Chavarría M, Arce-Rodríguez A, de Las Heras A, Paez-Espino AD, Durante-Rodríguez G, Kim J, Nikel PI, et al. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucl Acids Res. 2013;41:D666–D675. doi: 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiba K, Hoshino Y, Ishino K, Kogure T, Mikami Y, Uehara Y, Ishikawa J. Construction of a pair of practical Nocardia-Escherichia coli shuttle vectors. Jpn J Infect Dis. 2007;60:45–47. [PubMed] [Google Scholar]
- 44.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H. Genetic manipulation of Streptomyces. Norwich: The John Innes Foundation, Cold Spring Harbour Laboratory; 1985. [Google Scholar]
- 46.Wang C, Lee J, Deng Y, Tao F, Zhang LH. ARF-TSS: an alternative method for identification of transcription start site in bacteria. Biotechniques. 2012 doi: 10.2144/000113858. [DOI] [PubMed] [Google Scholar]
- 47.de las Heras LF, Perera J, Navarro Llorens JM. Cholesterol to cholestenone oxidation by ChoG, the main extracellular cholesterol oxidase of Rhodococcus ruber strain Chol-4. J Steroid Biochem Mol Biol. 2014;139:33–44. doi: 10.1016/j.jsbmb.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Kendall SL, Withers M, Soffair CN, Moreland NJ, Gurcha S, Sidders B, Frita R, Ten Bokum A, Besra GS, Lott JS, et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol Microbiol. 2007;65:684–699. doi: 10.1111/j.1365-2958.2007.05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crowe AM, Stogios PJ, Casabon I, Evdokimova E, Savchenko A, Eltis LD. Structural and functional characterization of a ketosteroid transcriptional regulator of Mycobacterium tuberculosis. J Biol Chem. 2015;290:872–882. doi: 10.1074/jbc.M114.607481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhía I, Galán B, Medrano FJ, García JL. Characterization of the KstR-dependent promoter of the first step of cholesterol degradative pathway in Mycobacterium smegmatis. Microbiology. 2011;157:2670–2680. doi: 10.1099/mic.0.049213-0. [DOI] [PubMed] [Google Scholar]
- 51.García-Fernández E, Medrano FJ, Galán B, García JL. Deciphering the transcriptional regulation of cholesterol catabolic pathway in mycobacteria: identification of the inducer of KstR repressor. J Biol Chem. 2014;289:17576–17588. doi: 10.1074/jbc.M113.545715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho NA, Dawes SS, Crowe AM, Casabon I, Gao C, Kendall SL, Baker EN, Eltis LD, Lott JS. The structure of the transcriptional repressor KstR in complex with CoA thioester cholesterol metabolites sheds light on the regulation of cholesterol catabolism in Mycobacterium tuberculosis. J Biol Chem. 2016;291:7256–7266. doi: 10.1074/jbc.M115.707760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shtratnikova VY, Schelkunov MI, Fokina VV, Pekov YA, Ivashina T, Donova MV. Genome-wide bioinformatics analysis of steroid metabolism-associated genes in Nocardioides simplex VKM Ac-2033D. Curr Genet. 2016;62:643–656. doi: 10.1007/s00294-016-0568-4. [DOI] [PubMed] [Google Scholar]
- 54.Ma Y, Yu H. Engineering of Rhodococcus cell catalysts for tolerance improvement by sigma factor mutation and active plasmid partition. J Ind Microbiol Biotechnol. 2012;39:1421–1430. doi: 10.1007/s10295-012-1146-5. [DOI] [PubMed] [Google Scholar]
- 55.Bashyam MD, Kaushal D, Dasgupta SK, Tyagi AK. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J Bacteriol. 1996;178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kendall SL, Burgess P, Balhana R, Withers M, Ten Bokum A, Lott JS, Gao C, Uhia-Castro I, Stoker NG. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2. Microbiology. 2010;156:1362–1371. doi: 10.1099/mic.0.034538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, et al. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015;11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strohl WR. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfleiderer C, Smid A, Bartsch I, Grummt I. An undecamer DNA sequence directs termination of human ribosomal gene transcription. Nucleic Acids Res. 1990;18:4727–4736. doi: 10.1093/nar/18.16.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grummt I, Kuhn A, Bartsch I, Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986;47:901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- 61.Henderson S, Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986;47:891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- 62.Henderson SL, Ryan K, Sollner-Webb B. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable transcription complex caused by polymerase read-in. Genes Dev. 1989;3:212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- 63.Szoköl J, Rucká L, Simciková M, Halada P, Nesvera J, Pátek M. Induction and carbon catabolite repression of phenol degradation genes in Rhodococcus erythropolis and Rhodococcus jostii. Appl Microbiol Biotechnol. 2014;98:8267–8279. doi: 10.1007/s00253-014-5881-6. [DOI] [PubMed] [Google Scholar]
- 64.Mathieu JM, Mohn WW, Eltis LD, LeBlanc JC, Stewart GR, Dresen C, Okamoto K, Alvarez PJ. 7-ketocholesterol catabolism by Rhodococcus jostii RHA1. Appl Environ Microbiol. 2010;76:352–355. doi: 10.1128/AEM.02538-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci USA. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yao K, Xu LQ, Wang FQ, Wei DZ. Characterization and engineering of 3-ketosteroid- big up tri, open1-dehydrogenase and 3-ketosteroid-9alpha-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9alpha-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab Eng. 2014;24:181–191. doi: 10.1016/j.ymben.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 67.Kreit J, Sampson NS. Cholesterol oxidase: physiological functions. FEBS J. 2009;276:6844–6856. doi: 10.1111/j.1742-4658.2009.07378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pollegioni L, Piubelli L, Molla G. Cholesterol oxidase: biotechnological applications. FEBS J. 2009;276:6857–6870. doi: 10.1111/j.1742-4658.2009.07379.x. [DOI] [PubMed] [Google Scholar]
- 69.Vrielink A, Ghisla S. Cholesterol oxidase: biochemistry and structural features. FEBS J. 2009;276:6826–6843. doi: 10.1111/j.1742-4658.2009.07377.x. [DOI] [PubMed] [Google Scholar]
- 70.Petrusma M, Hessels G, Dijkhuizen L, van der Geize R. Multiplicity of 3-ketosteroid-9α-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. J Bacteriol. 2011;193:3931–3940. doi: 10.1128/JB.00274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shao M, Zhang X, Rao Z, Xu M, Yang T, Li H, Xu Z, Yang S. A mutant form of 3-ketosteroid-∆1-dehydrogenase gives altered androst-1,4-diene-3, 17-dione/androst-4-ene-3,17-dione molar ratios in steroid biotransformations by Mycobacterium neoaurum ST-095. J Ind Microbiol Biotechnol. 2016;43:691–701. doi: 10.1007/s10295-016-1743-9. [DOI] [PubMed] [Google Scholar]
- 72.Xu XW, Gao XQ, Feng JX, Wang XD, Wei DZ. Influence of temperature on nucleus degradation of 4-androstene-3, 17-dione in phytosterol biotransformation by Mycobacterium sp. Lett Appl Microbiol. 2015;61:63–68. doi: 10.1111/lam.12428. [DOI] [PubMed] [Google Scholar]
- 73.Liu Y, Shen Y, Qiao Y, Su L, Li C, Wang M. The effect of 3-ketosteroid-∆1-dehydrogenase isoenzymes on the transformation of AD to 9alpha-OH-AD by Rhodococcus rhodochrous DSM43269. J Ind Microbiol Biotechnol. 2016;43:1303–1311. doi: 10.1007/s10295-016-1804-0. [DOI] [PubMed] [Google Scholar]
- 74.Yeh CH, Kuo YS, Chang CM, Liu WH, Sheu ML, Meng M. Deletion of the gene encoding the reductase component of 3-ketosteroid 9α-hydroxylase in Rhodococcus equi USA-18 disrupts sterol catabolism, leading to the accumulation of 3-oxo-23,24-bisnorchola-1,4-dien-22-oic acid and 1,4-androstadiene-3,17-dione. Microb Cell Fact. 2014;13:130. doi: 10.1186/s12934-014-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its supplementary materials.


