Abstract
Background
Hydrogen sulfide (H2S) is a toxic foul-smelling gas produced by subgingival biofilms in patients with periodontal disease and is suggested to be part of the pathogenesis of the disease. We studied the H2S-producing protein expression of bacterial strains associated with periodontal disease. Further, we examined the effect of a cysteine-rich growth environment on the synthesis of intracellular enzymes in F. nucleatum polymorphum ATCC 10953. The proteins were subjected to one-dimensional (1DE) and two-dimensional (2DE) gel electrophoresis An in-gel activity assay was used to detect the H2S-producing enzymes; Sulfide from H2S, produced by the enzymes in the gel, reacted with bismuth forming bismuth sulfide, illustrated as brown bands (1D) or spots (2D) in the gel. The discovered proteins were identified with liquid chromatography – tandem mass spectrometry (LC-MS/MS).
Results
Cysteine synthase and proteins involved in the production of the coenzyme pyridoxal 5′phosphate (that catalyzes the production of H2S) were frequently found among the discovered enzymes. Interestingly, a higher expression of H2S-producing enzymes was detected from bacteria incubated without cysteine prior to the experiment.
Conclusions
Numerous enzymes, identified as cysteine synthase, were involved in the production of H2S from cysteine and the expression varied among Fusobacterium spp. and strains. No enzymes were detected with the in-gel activity assay among the other periodontitis-associated bacteria tested. The expression of the H2S-producing enzymes was dependent on environmental conditions such as cysteine concentration and pH but less dependent on the presence of serum and hemin.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-017-0967-9) contains supplementary material, which is available to authorized users.
Keywords: Periodontitis, Hydrogen sulfide, Fusobacterium spp., Enzymes, Bismuth sulfide, Proteomics, 2D gel electrophoresis, LC-MS/MS
Background
Oral biofilms differ in composition depending on their niche within the mouth. The biofilms occupying the periodontal pocket, the area between the tooth and the surrounding connective tissue, are usually dominated by Gram-positive, facultative anaerobic bacteria but can undergo a compositional change towards Gram-negative, anaerobic and motile bacteria when oral hygiene is insufficient [1]. The latter biofilms utilize the gingival crevicular fluid as a nutrient source and metabolize proteins, peptides and amino acids to various carboxylic acids and volatile sulfur compounds (VSC). This shift in bacterial ecology along with a host inflammatory response is believed to explain the etiology of periodontal disease where the supportive tissue of teeth is affected by a host immune reaction leading to destruction of alveolar bone (periodontitis).
Hydrogen sulfide (H2S) is the most common VSC formed by bacterial degradation of mainly the sulfur-containing amino acid cysteine in the oral cavity. It is a low-molecular weight and volatile gas compound detected in halitosis (bad breath) patients and in periodontal pockets in patients with periodontitis [2–4]. H2S is regarded as one of the most toxic metabolites produced in the periodontal pocket. In vitro laboratory studies have shown that H2S can damage epithelial cells [5], enhance permeability of the oral mucosa [6] and cause apoptosis of gingival fibroblasts [7]. However, the exact mechanism by which H2S exerts its effect on cells is not known. Likewise, the pathogenesis of periodontal disease is poorly understood but it is usually accepted that bacterial metabolites in general, and H2S in particular, are of importance in the development and activity of the disease.
Various oral bacterial species are known to be producers of H2S. Previous studies by Persson et al. [8] showed that Porphyromonas endodontalis, Porphyromonas gingivalis, Prevotella intermedia and Treponema denticola were the strongest H2S producers when incubated in serum, which contain many of the plasma proteins found in gingival crevicular fluid. In that study, all 163 strains tested were able to produce H2S when L-cysteine was used as substrate. Moreover, Fusobacterium nucleatum and Parvimonas micra were able to generate H2S not only from amino acids but also peptides such as glutathione [9, 10]. In our previous in vitro study Fusobacterium spp. were the strongest and most rapid producers of H2S from L-cysteine, and used the coenzyme pyridoxal 5′phosphate (PLP) [11].
The activity of L-cysteine desulfhydrase, the intracellular enzyme that catalyzes the degradation of cysteine into H2S, pyruvate and ammonia, has been shown to vary among different strains of Fusobacterium [12]. F. nucleatum ATCC 25586 possesses L-cysteine desulfhydrases [13], but these are not the most abundant enzymes involved in the production of H2S. The production of a greater amount of H2S compared to ammonia and pyruvate suggests that other enzymatic pathways for generation of H2S exist. So far, four genes encoding different enzymes involved in H2S production have been identified [14–18]. The highest molecular weight enzymes Fn0625 and Fn1419 (47 and 43 kDa respectively) generate H2S with pyruvate and ammonia. Fn1220 (the cdl gene homologue) is the smallest (33 kDa) but most frequently used enzyme in the formation of H2S. It is a L-cysteine desulfhydrase, also known as “L-cysteine lyase”, that catalyzes the β-replacement of L-cysteine giving rise to H2S and L-lanthionine [12]. Fn1055 is a 37 kDa protein that catalyzes a reaction that yields H2S and L-serine.
In this study we investigated the expression of H2S-producing enzymes in 14 bacterial strains associated with periodontal diseases. In addition, we undertook to examine whether a cysteine-rich growth environment induced synthesis of H2S producing enzymes and other intracellular enzymes in F. nucleatum polymorphum ATCC 10953.
Methods
Bacterial strains and culture conditions
Bacterial strains used in this study included type collection strains of Fusobacterium spp., Parvimonas sp., Porphyromonas spp., Prevotella spp. and Treponema sp. (Table 1), but also fresh clinical isolates from subgingival plaque samples taken from two young adolescents suffering from periodontitis in Ghana [19]. The clinical isolates were typed at Culture Collection University of Gothenburg (CCUG). All strains were recovered on Brucella agar (BBL Microbiology Systems Cockeysville, MD, USA) with 50 ml/l defibrinated horse blood, 20 ml/l hemolyzed human blood and 0.5 mg/l menadione after 5 days incubation under anaerobic conditions (5% CO2, 10% H2 in N2) at 37 °C.
Table 1.
Bacteria examined for hydrogen sulfide (H2S)-producing enzymes, identified with in-gel cysteine digestion and bismuth staininga
| Species | Subspecies | Strain | Broth |
|---|---|---|---|
| Fusobacterium canifelinum | CCUGb 66382 | Todd Hewitt | |
| Fusobacterium necrophorum | funduliforme | ATCCc 51357 | Todd Hewitt |
| Fusobacterium necrophorum | CCUG 48192 | Todd Hewitt | |
| Fusobacterium nucleatum | polymorphum | ATCC 10953 | Todd Hewitt |
| Fusobacterium nucleatum | OMGSd 3938e | Todd Hewitt | |
| Fusobacterium periodonticum | ATCC 33693 | Todd Hewitt | |
| Fusobacterium periodonticum | CCUG 66383 | Todd Hewitt | |
| Parvimonas micra | ATCC 33270 | BHIf + 10% serum | |
| Porphyromonas endodontalis | OMGS 1205 | BHI + 10% serum | |
| Porphyromonas gingivalis (W83) | OMGS 197 | BHI | |
| Porphyromonas gingivalis (381 F) | CCUG 14449 | BHI | |
| Prevotella intermedia | ATCC 25611 | BHI | |
| Prevotella tannerae | ATCC 51259 | BHI | |
| Treponema denticola | OMGS 3271g | Spirochete brothh |
aBacterial species were grown in broth until OD600 of approximately 0.8. After washing and centrifugation, the cells were lysed and the proteins were separated in gel by molecular weight, before staining in bismuth(III)chloride solution containing cysteine. The cysteine-degrading proteins that produced H2S were identified in the assay by color change; Sulfide from H2S reacted with bismuth and formed bismuth sulfide, a black precipitate. Another set of gels were also stained with conventional Coomassie staining. All experiments were repeated at least once
bCulture Collection University of Gothenburg
cAmerican Type Culture Collection
dOral Microbiology Gothenburg Sweden
eOriginally received from Malmö (Badersten 5U)
fBrain Heart Infusion broth with 2 mL/L menadione and 10 mL/L hemin
gOriginally received from Dr R. Ellen, University of Toronto, Toronto, Canada
hDawson JR, Ellen RP. Tip-oriented adherence to Treponema denticola to fibronectin. Infect Immun. 1990//;58(12):3924–8
Fusobacterium spp. were cultured in Todd Hewitt (TH) broth (Becton Dickinson, Sparks, MD, USA) while P. micra, Porphyromonas spp. and Prevotella spp. were grown in Brain Heart Infusion broth supplemented with menadione (2 ml/l) and hemin (10 ml/l). For growth of P. micra and Prevotella tannerae the medium was also containing 10% serum.
To investigate the significance of cysteine for the expression of H2S-producing enzymes during growth, strains were incubated in the presence of L-cysteine (1 mg/ml) in the appropriate media stated above. For F. nucleatum ATCC 10953, the influence of other environmental conditions were tested in TH broth buffered to pH 6, pH 7, pH 8 or TH broth, pH 7.8 supplemented with glutathione (2.5 mg/ml), sodium sulfide (0.46 mg/ml), 5% serum, 50% serum or 50% serum with hemin (10 ml/l). In all cases, the cultures were grown under anaerobic conditions at 37 °C and made in duplicate.
Preparation of cell extracts (crude enzyme extracts)
Each strain was grown anaerobically in 50 ml culture medium until mid-exponential phase (OD600 approximately 0.8) was reached. Cells were harvested by centrifugation (3000 g for 15 min, 4 °C), washed twice in 40 mM Tris pH 9,5 (Sigma-Aldrich Sweden AB, Stockholm Sweden) and resuspended in 1 ml of lysis buffer (5 M urea (Merck KGaA, Darmstadt, Germany), 2 M thiourea (MP Biomedicals, LLC, Illkirch, France), 2% CHAPS (GE Healthcare Bio-Sciences AB, Uppsala, Sweden), 2% sulfobetaine (G-Biosciences, St. Louis, MO, USA), 2 mM tributyl phosphine (Sigma-Aldrich Sweden AB), 40 mM Tris-base pH 9,5 and 2% IPG (GE Healthcare Bio-Sciences AB)). The cell suspensions were shaken gently in room temperature for 1 h with vortexing every 10 min. The extracts were centrifuged (6000 g for 10 min, 4 °C) to remove intact cells and the supernatants were stored separately at −20 °C. The concentration of proteins in the crude enzyme extract was determined with 2-D Quant Kit (GE Healthcare Bio-Sciences AB) following the manufacturer’s instructions.
One-dimensional gel electrophoresis (1DE)
A 7.5 μl aliquot of the crude enzyme extract (5 – 20 μg protein/sample) was mixed with 2.5 μl of sample buffer NuPAGE LDS (Novex, Carlsbad, CA, USA). Proteins were separated by SDS-PAGE in 4–12% gradient Bis-Tris gels (NuPAGE, Novex) at constant voltage of 200 V for 60 min using NuPAGE SDS MES (Novex) as running buffer. Amersham High-Range Rainbow Molecular Weight Markers (GE Healthcare Bio-Sciences AB) was used as standard.
Two-dimensional gel electrophoresis (2DE)
Samples of crude enzyme extracts (300 μg protein in 200 μl) were diluted with 130 μl buffer containing 8 M urea, 2% CHAPS, 10 mM dithiothreitol (GE Healthcare Bio-Sciences AB), 2% IPG and 0.01% bromophenol blue and placed in re-swelling cassettes under Immobiline dry gel (IPG) Strips (pH 4–7, 18 cm; GE Healthcare Bio-Sciences AB). The loading and rehydration of IPG strips took place at room temperature for 24 h under silicone oil. Isoelectric focusing was conducted using Multiphor II (GE Healthcare Bio-Sciences AB) with supply of cooling water at 15 °C. Isoelectric focusing was initiated at 150 V for 1 h, the voltage increased gradually during 18 h to 1200 V and maintained at 3500 V for 20 h. After focusing, the strips were stored at −80 °C. Before separation of proteins in the second dimension, the IPG strips were equilibrated first in 50 mM Tris–HCl (pH 6.8), 2% SDS, 26% glycerol and 16 mM dithiothreitol for 15 min and then for another 15 min in the same buffer but containing 250 mM iodoacetamide (GE Healthcare Bio-Sciences AB) and 0.005% bromophenol blue instead of dithiothreitol. The IPG strips were embedded, using 0.5% (w/v) molten agarose, on top of 14% polyacrylamide gels (0.38 M Tris buffer pH 8.8, 14% Bis-acrylamide (Bio-Rad Laboratories, Sundbyberg, Sweden), 0.1% SDS, 4.6% glycerol, 0.05% TEMED (Bio-Rad Laboratories) and 0.05% ammonium persulfate (Bio-Rad Laboratories)). SDS-PAGE was run in PROTEAN II xi Cell (Bio-Rad Laboratories) at constant current (19 mA) overnight with running buffer containing 50 mM Tris (pH 8.3), 0.1% SDS and 0.384 M glycine.
Detection of H2S-producing enzymes
The enzymes degrading L-cysteine and forming H2S were detected through precipitation of bismuth sulfide using an in-gel activity assay, essentially as described previously [12, 16]. H2S-producing enzymes appeared as brown to black bands in the 1DE gels and as spots in the 2DE gels. Before bismuth staining, the gels were subjected to a renaturation process where SDS was removed and replaced with nonionic detergents. The renaturation took place during gentle shaking at 4 °C with the following solutions: (i) 25 mM triethanolamine-HCl pH 8.0, 0.05% SDS and 0.5% Triton-X-100 for 1 h; (ii) 25 mM triethanolamine-HCl pH 8.0, 0.5% Triton-X-100 and 0.5% Lubrol PX for 2 × 1 h; (iii) 25 mM triethanolamine-HCl pH 7.0 and 0.5% Lubrol PX for 2 × 0.5 h. For activity staining, the gels were incubated in 100 mM triethanolamine-HCl pH 7.6, 10 μM pyridoxal 5-phosphate monohydrate (VWR, Stockholm, Sweden), 0.5 or 1.0 mM bismuth trichloride (Fisher Scientific GTF AB, Gothenburg, Sweden), 10 mM EDTA (Sigma-Aldrich Sweden AB) and 5 or 20 mM L-cysteine (Sigma-Aldrich Sweden AB) at 37 °C for 2 h. All the activity assays, including both 1DE and 2DE gels, were performed at least twice, with double sets of gels for staining with bismuth and Coomassie staining.
Coomassie and silver staining
Before staining, 1DE gels were fixed in 40% ethanol and 2% acetic acid for 1 h, and 2DE gels in 40% ethanol and 5% acetic acid for 0.5 h. Gels were stained with 16% Coomassie brilliant blue G colloidal concentrate (Sigma-Aldrich Sweden AB) in 20% ethanol overnight at room temperature. After rinsing in 5% acetic acid and 25% ethanol for 1 min, gels were destained in 25% ethanol for 1–3 h and washed with ultra-high quality water. The 2DE gels were also stained with silver according to the protocol of the manufacturer (GE Healthcare Bio-Sciences AB).
Identification of proteins by mass spectrometry
Protein spots of interest were excised manually from Coomassie brilliant blue stained 2DE gels of crude cell extract and subjected to LC-MS/MS as described previously [20]. Briefly, proteins in gels were reduced with dithiothreitol, alkylated with iodoacetamide and then digested with trypsin. Tryptic peptides were separated and analyzed by mass spectrometry. The peaks were later identified by creating Mascot Generic Files and by database searching using Matrix science web server (www.matrixscience.com).
Results
H2S-producing enzymes among bacterial strains
Cell extracts of 14 strains of bacteria associated with periodontitis (Table 1) were screened for H2S-producing enzymes with in-gel activity assay after renaturation. All Fusobacterium spp. except F. necrophorum ATCC 51357 had enzymes producing H2S, detected as brownish bands on the gels (Fig. 1). F. nucleatum OMGS 3938 displayed one band around 37 kDa while F. nucleatum ATCC 10953 showed 37 kDa and 47 kDa enzymes illustrating differences within the same subspecies. The F. necrophorum strain that showed activity had three bands with sizes around 47, 43 and 33 kDa. F. periodonticum ATCC 33693 displayed bands at 47, 43, 37 and 33 kDa and the remaining two clinical isolates of Fusobacterium spp. had low activity at 37 and 33 kDa (Fig. 1). Other bacterial species associated with periodontitis such as P. gingivalis, P. intermedia, P. micra, P. tannerae and T. denticola were also examined with this method and no H2S-producing enzymes could be detected.
Fig. 1.
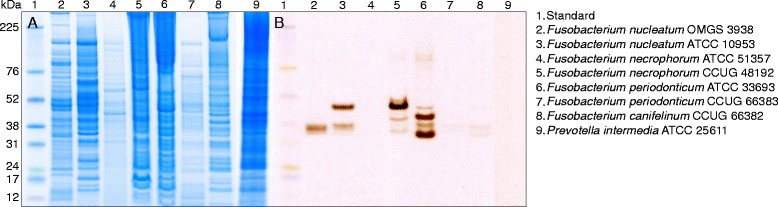
The protein expression of different bacterial strains grown in broth without cysteine was examined with gel electrophoresis. With Coomassie staining (a) all proteins were stained. However, the in-gel activity assay with bismuth staining (b) only detected the proteins that produced hydrogen sulfide (H2S) from cysteine. Sulfide from H2S reacted with bismuth and formed bismuth sulfide, a brown to black precipitate
Cell extracts of selected Fusobacterium spp., were also separated by isoelectric focusing and molecular weight in two-dimensional gel electrophoresis. The gels were, after protein separation, stained with silver staining for better resolution or Coomassie staining for protein extraction and identification. The total protein expression for different subspecies and strains of Fusobacterium differed in both pattern and intensity (Fig. 2). The H2S-producing enzymes were colored in the bismuth solution and extracted from the Coomassie stained gels. The most frequently detected enzymes were identified as cysteine synthase, involved in cysteine metabolism. Also a protein involved in the biosynthesis of the coenzyme pyridoxal phosphate was identified (Additional file 1: Table S1).
Fig. 2.
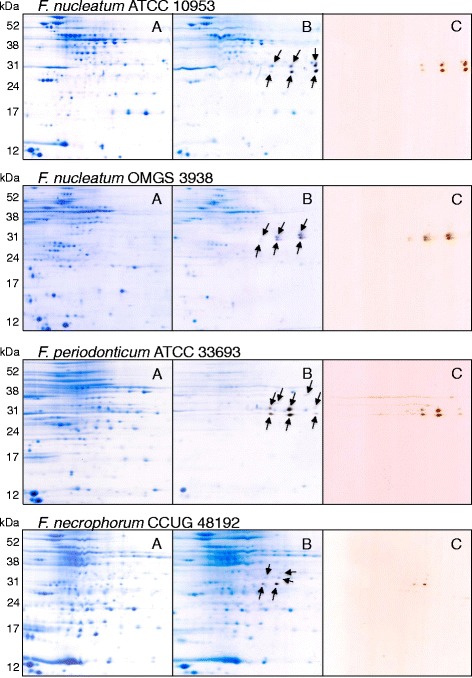
Two-dimensional gel-electrophoresis of proteins extracted from different Fusobacterium spp. The bacteria were grown in Todd Hewitt broth without cysteine prior to protein extraction and separation. One gel was stained with Coomassie blue (a) for protein detection and extraction and another with in-gel activity bismuth staining (c) for detection of proteins producing H2S. Bismuth reacts with sulfide and produces a precipitate, bismuth sulfide, shown as brown spots in the gel. After bismuth staining the same gel was stained with Coomassie blue (b). As illustrated in the figure the protein expression of the subspecies differed
The effect of environmental conditions on enzyme expression in Fusobacterium spp.
To investigate the significance of cysteine for the expression of H2S-producing enzymes, all strains were grown in the presence of L-cysteine (1 mg/ml) in appropriate growth media. A difference in protein expression, both with regard to the number of bands and the intensity of the color, between bacteria grown in broth with and without cysteine prior to the experiment was seen on the bismuth-stained gels (Figs. 1 and 3). When bacteria were grown in broth without cysteine, additional and stronger bands appeared on the gels. For F. nucleatum ATCC 10953 the 47 kDa band was shown in both environments while the 37 kDa band was only clearly seen after growth in broth without cysteine. Similarly, the band with the enzyme of the smallest size, around 33 kDa was seen for F. necrophorum CCUG 48192 without cysteine. In addition, the largest enzyme (47 kDa) was enhanced without cysteine. For F. periodonticum ATCC 33693 a fourth band was seen (37 kDa) without cysteine and the smallest enzyme (around 33 kDa) was enhanced. The clinical isolate F. canifelinum CCUG 66382 showed lowest activity and had bands of the size 47 kDa and 33 kDa when grown with cysteine and 37 kDa and 33 kDa without cysteine.
Fig. 3.
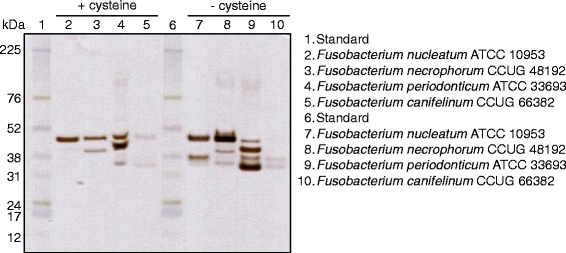
A protein separation by molecular weight of enzymes from Fusobacterium spp. Bacteria were grown in broth with and without cysteine prior to protein extraction and in-gel activity assay that stained the proteins that produced H2S
F. nucleatum polymorphum ATCC 10953 was selected for further studies on the influence of environmental conditions on expression of H2S-producing enzymes and therefore incubated in TH broth buffered to pH 6, pH 7, pH 8 or TH broth supplemented with glutathione, NaHS, 5% serum, 50% serum or 50% serum with hemin (Fig. 4). All tested modifications of the broth resulted in at least two clear bands on the bismuth stained gels. When the bacteria were incubated in broth containing sodium hydrosulfide (NaHS) the strongest bismuth sulfide precipitation was detected and 2–3 clear bands were shown. Also bacteria incubated in TH broth without any additives produced strong bands compared with all but NaHS. When L-cysteine was added the bands with lower molecular weight values produced less H2S, compared with the broth without any additives. The readers should note that a higher concentration of bismuth trichloride and L-cysteine was used here (Fig. 4) compared to the initial experiments (Fig. 3), which may explain the band with lower molecular weight seen in Fig. 4 but not initially in Fig. 3. The different pH values tested, all displaying the same bands, showed minor trends toward a more pronounced staining intensity when incubated at a higher pH. A higher enzymatic activity was not seen when serum, glutathione or hemin were added to the broth.
Fig. 4.
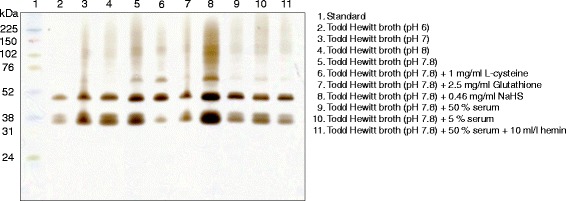
The effect of the environmental conditions on the activity of H2S-producing enzymes was tested for Fusobacterium nucleatum polymorphum ATCC 10953. The bacteria were grown in different broths before protein extraction and 1D gel electrophoresis followed by in-gel activity assay
The cellular response to cysteine-rich environment
Differences in protein expression between bacteria grown in cysteine-rich and poor broth were also examined by protein extraction of spots enhanced in the gels where the bacteria were grown in cysteine-rich broth (Fig. 5). The extracted proteins, identified with LS-MS/MS (Table 2), were glycolytic proteins, proteins involved in butyrate metabolism and oxidoreductase. Also a protein involved in pyridoxal 5′phosphate biosynthesis was identified (a coenzyme for the degradation of cysteine and production of H2S).
Fig. 5.
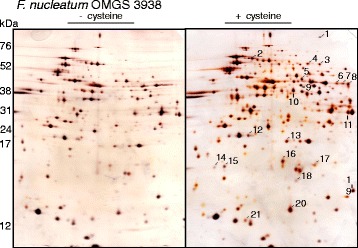
Two-dimensional gel-electrophoresis of Fusobacterium OMGS 3938 grown in Todd Hewitt broth with and without cysteine prior to protein extraction and separation. Silver staining was used to detect the proteins. The proteins enhanced when the bacteria was grown in cysteine, compared to the protein expression when they were grown without cysteine, were extracted (line) and identified (Table 2)
Table 2.
Proteins of Fusobacterium nucleatum enhanced when incubated in cysteine-rich broth prior to protein extraction*
| Spot no. | Protein | Protein function |
|---|---|---|
| 1 | Glyceraldehyde-3-phosphate dehydrogenasea, b | Glycolytic protein |
| 2 | Bifunctional penicillin tolerance protein LytB/ribosomal protein S1 RpsAa | Translation |
| 3, 4 | Pyruvate kinaseb, c | Glycolytic protein |
| 5 | Recombination protein Ad /Histidyl-tRNA synthetasee, f | DNA repair/histidyl-tRNA aminoacylation |
| 6, 8 | Acetate kinaseb, g | Acetyl-CoA biosynthetic process |
| 7 | Electron transfer flavoprotein subunit alphab | Electron carrier activity |
| 9 | Phosphoglycerate kinaseb, h | Glycolytic protein |
| 10 | Zn-dependent alcohol dehydrogenase and related dehydrogenaseb | Oxidoreductase, zinc ion binding |
| 11 | Pyridoxal biosynthesis lyase PdxSb | Pyridoxal 5′-phosphate |
| 12 | Butyrate-acetoacetate CoA-transferase subunit Bb | Butyrate metabolism |
| 13 | Acetoacetate: butyrate/acetate coenzyme A transferaseb | Butyrate metabolism |
| 14 | Iron-sulfur cluster-binding proteinb | Iron and sulphur binding |
| 15 | (S)-2-hydroxy-acid oxidase chain Di /glycolate oxidase, subunit GlcDg | Oxidoreductase |
| 16, 17 | Rubrerythrina | Oxidoreductase, iron ion binding |
| 18 | PTS-system, N-acetylglucosamine-specific IIA componentb | Phosphotransferase system |
| 19 | Mannose-1-phosphate guanylyl transferase (GDP)b | GDP-mannose biosynthetic process, lipopolysaccharide biosynthetic process |
| 20 | Translation initiation inhibitorb | Deaminase activity |
| 21 | Anti-sigma F factor antagonistb | Regulation of transcription |
*Fusobacterium nucleatum OMGS 3938 was incubated in Todd Hewitt broth with and without cysteine. The spots that were enhanced when incubated in cysteine-rich broth were extracted for identification with LC- MS/MS
a Fusobacterium nucleatum subsp. polymorphum ATCC 10953
b Fusobacterium nucleatum subsp. nucleatum ATCC 25586
c Fusobacterium sp. 7_1
d Fusobacterium nucleatum subsp. nucleatum ATCC 23726
e Fusobacterium sp. 4_1_13
f Fusobacterium nucleatum ChDC F128
g Fusobacterium periodonticum ATCC 33693
h Desulfosporosinus sp. OT
i Fusobacterium nucelatum subsp. vicentii ATCC 49256
Discussion
The production of H2S is complex and involves different enzymatic pathways for different bacterial species and strains. The literature on this subject is rather sparse as opposed to the production of eukaryotic cells, where H2S is produced by three PLP dependent enzymes; cystathionine β-synthase, cystathionine γ-lyase and 3-mercaptopyruvate sulfurtransferase, that use L-cysteine as their principle substrate [21]. The bacterial production of H2S is mainly due to the degradation of the sulfur-containing amino acid cysteine and results in different metabolic end products depending on the enzymes participating. One common cysteine degradation pathway involves the PLP dependent L-cysteine desulfhydrases, including α, β-elimination activity, that results in the production of H2S, pyruvate and ammonia [22, 23]. L-cysteine desulfhydrases have been identified in many oral bacterial species and are known to be encoded by the cdl gene in F. nucleatum [14], the hly gene in T. denticola [24] and the lcs gene in P. intermedia [25]. Moreover, Streptococcus anginosus and S. intermedius are capable to produce H2S from L-cysteine using a cystathionase, encoded by the lcd gene, that uses L-cystathionine as well as cysteine as substrate [26–28]. In the current study, the in-gel activity assay for detection of H2S-producing enzymes revealed a variety of enzymes with molecular weights between 30 and 50 kDa in F. nucleatum, F. necrophorum and F. periodonticum. The sizes of these enzymes are in line with the desulfhydrases previously reported for F. nucleatum ATCC 25586; 33 kDa (Fn1220, cdl), 37 kDa (Fn1055), 43 kDa (Fn1419) and 47 kDa (Fn0625) [18]. It is therefore tempting to suggest that similar desulfhydrases are also involved in H2S-production in F. necrophorum and F. periodonticum.
When in-gel activity assays were used to investigate the H2S-producing enzyme profile in cell extracts of P. gingivalis, P. intermedia, P. micra, P. tannerae and T. denticola no H2S-producing protein bands could be detected despite previous reports of the ability to produce H2S for these bacterial species [8, 11]. The lack of activity may be due to several factors such as strain differences, suboptimal conditions for enzyme reactivation after SDS-PAGE or a lower affinity of the enzyme to bind cysteine. The reported Km-values of enzymes extracted from T. denticola are high compared to Fusobacterium spp. [14, 17], which suggests that the method used in this study is not sensitive enough to detect the enzymes with lower affinity to L-cysteine.
The most prominent H2S-producing enzymes in F. nucleatum, F. necrophorum and F. periodonticum were found around 30 kDa on 2DE gels (Fig. 2). The majority of protein spots exhibiting precipitates of bismuth sulfide were excised from 2DE-gels and subjected to mass spectrometric analysis. The results revealed that all proteins could be allocated to cysteine synthases. Further analysis of the amino acid sequences of cysteine synthase from the three species showed almost complete homologies with the sequence reported for cdl (Fn1220) in F. nucleatum. Yoshida and coworkers reported approximately 40% identity of the H2S producing gene Fn1220 from F. nucleatum to cysteine synthases A and B in E. coli and suggested that both these enzymes may catalyze both of the reactions that result in the production of H2S and L-lantionine and of L-cysteine and acetate respectively [15]. One can therefore assume that H2S production in different species of Fusobacterium is the result of the condensation of cysteine molecules with lanthionine as a byproduct.
In this study, enzymatic H2S-producing activity was detected for F. necrophorum CCUG 48192 but not for strain ATCC 51357 (Fig. 1). This confirms results from previous reports of the differences in H2S producing capacity among different strains of Fusobacterium [12, 16]. However, the variance seen does not seem to be something unique for this genus. Similar variations in H2S production have been reported for different subspecies of Streptococcus [27, 28]. L-cysteine desulfhydrase activity for some Fusobacterium spp. and L-cysteine lyase activity for other strains adds on the complexity by the diverse enzymes being active under aerobic and anaerobic conditions [12].
The expression of H2S-producing enzymes was not significantly affected by the presence of serum proteins or the pH of growth medium (Fig. 4). However, as illustrated in Figs. 3 and 4, lower expression of H2S-producing enzymes was demonstrated for F. nucleatum, F. necrophorum and F. periodonticum when cells were grown in broth supplemented with cysteine compared to without cysteine. These results indicate cysteine-mediated down-regulation of these enzymes in the genus Fusobacterium. In all species, the enzyme expression mostly affected was that with the lowest molecular weight, which probably correspond to Fn1220. Of interest is that the Fn1220 enzyme is known to exhibit the highest H2S-producing activity and is responsible for more than 85% of the H2S production in F. nucleatum [18]. In addition, the ability of the enzyme to degrade cysteine is inversely related to cysteine concentration. When comparing different concentrations of cysteine as substrate, a higher sulfide production was observed at 0.5 mM L-cysteine-HCl than at 2 mM and 6 mM, which suggests that desulfuration is inhibited by the excess of substrate also on the enzyme activity level [13]. This might be indicative of a mechanism that supports bacterial survival and limits production of toxic H2S in cysteine-rich environments.
Proteomics has recently been reviewed [29]. Despite some drawbacks with the method, such as that some proteins are excluded because of very high and low isoelectric point and molecular weight, a majority of the proteins expressed by bacteria that have been exposed to changed environmental factors can be studied [30]. When F. nucleatum OMGS 3938 was grown in the presence of cysteine more than one hundred proteins were differently expressed compared to cells grown without cysteine. The observed down-regulation of H2S-producing enzymes in cells grown in cysteine-rich environment, as previously demonstrated by SDS-PAGE followed by in-gel activity staining (Fig. 3), was supported by the observation of a higher expression on 2DE gels from cells grown in the absence of cysteine (data not shown). Twenty-one abundant protein spots exhibited more than a two-fold increase in optical intensity and these were subjected to identification with LC-MS/MS. Many of these proteins were identified as glycolytic enzymes, oxidoreductases or proteins involved in the butyrate metabolism (Table 2). These results suggest that the primary metabolic pathway for carbohydrate metabolism is activated during growth in a cysteine-rich environment. Of interest is that nine of the up-regulated proteins identified in this study (1, 6, 7, 8, 10, 11, 12, 13, 14 in Table 2) were down-regulated when anaerobically grown cells of F. nucleatum were exposed to oxygen [31]. This confirms the reducing potential of cysteine and thus the avoiding of oxidative stress. Cysteine has many functions besides being a substrate in the formation of H2S; it contributes to a more anaerobic environment by reduction.
Conclusions
Periodontal disease is defined as an infectious disease but the role of the biofilm and the host-parasite interaction is still unknown. The bacterial metabolism and the net effect of a biofilm is of importance in the understanding of the mechanisms involved where biofilms are contributing to disease development. In this study we focused on bacterial production of H2S from cysteine. Numerous enzymes, identified as cysteine synthase, were involved in the production of H2S from cysteine and the expression varied among Fusobacterium spp. and strains. No enzymes were detected with the in-gel activity assay among the other periodontitis-associated bacteria tested. The expression of the H2S-producing enzymes was dependent on environmental conditions such as cysteine concentration and pH but less dependent on the presence of serum and hemin. Knowledge of H2S-production and the possible affect it may have on host cells is needed to elucidate its potential role in the pathogenesis of periodontal disease.
Acknowledgements
Special thanks to Agnethe Henriksson for technical assistance.
Funding
TUA-Grant (TUAGBG-89621)
Swedish Dental Society (STS)
Gothenburg Dental Society (GTS)
None of the funding bodies were involved in the design of the study nor the collection, analysis and interpretations of data or in writing the manuscript.
Availability of data and materials
All data is presented in the Tables and Additional file 1: Table S1.
Authors’ contribution
AB contributed to design, interpretation, drafted the manuscript. MB refined the method, interpretation, critically revised the manuscript. GD contributed to conception, design and interpretation and critically revised the manuscript. GS contributed to conception, design and interpretation and critically revised the manuscript. All authors have read and approved of the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable. This is an in vitro study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- 1DE
One dimensional gel electrophoresis
- 2DE
Two dimensional gel electrophoresis
- H2S
Hydrogen sulfide
- LC-MS/MS
Liquid chromatography – tandem mass spectrometry
- PLP
Pyridoxal 5′phosphate
- VSC
Volatile sulfur compounds
Additional file
Identification of proteins of Fusobacterium spp. involved in hydrogen sulfide (H2S) production, detected with in-gel cysteine digestion and bismuth staining*. (DOC 47 kb)
Contributor Information
Amina Basic, Phone: +46 31 7863266, Phone: +46 735075959, Email: amina.basic@gu.se.
Madeleine Blomqvist, Email: Madeleine.Blomqvist@mah.se.
Gunnar Dahlén, Email: Gunnar.Dahlen@odontologi.gu.se.
Gunnel Svensäter, Email: Gunnel.Svensater@mah.se.
References
- 1.Marsh PD, Devine DA. How is the development of dental biofilms influenced by the host? J Clin Periodontol. 2011;38(Suppl 11):28–35. doi: 10.1111/j.1600-051X.2010.01673.x. [DOI] [PubMed] [Google Scholar]
- 2.Rizzo AA. The possible role of hydrogen sulfide in human periodontal disease. I. Hydrogen sulfide production in periodontal pockets. Periodontics. 1967;5(5):233–236. [PubMed] [Google Scholar]
- 3.Horowitz A, Folke LEA. Hydrogen sulfide production in the periodontal environment. J Periodontol. 1973;44(7):390–395. doi: 10.1902/jop.1973.44.7.390. [DOI] [PubMed] [Google Scholar]
- 4.Persson S. Hydrogen sulfide and methyl mercaptan in periodontal pockets. Oral Microbiol Immunol. 1992;7(6):378–379. doi: 10.1111/j.1399-302X.1992.tb00641.x. [DOI] [PubMed] [Google Scholar]
- 5.Morhart RE, Mata LJ, Sinskey AJ, Harris RS. A microbiological and biochemical study of gingival crevice debris obtained from Guatemalan Mayan Indians. J Periodontol. 1970;41(11):644–649. doi: 10.1902/jop.1970.41.11.644. [DOI] [PubMed] [Google Scholar]
- 6.Ng W, Tonzetich J. Effect of hydrogen sulfide and methyl mercaptan on the permeability of oral mucosa. J Dent Res. 1984;63(7):994–997. doi: 10.1177/00220345840630071701. [DOI] [PubMed] [Google Scholar]
- 7.Yaegaki K, Qian W, Murata T, Imai T, Sato T, Tanaka T, Kamoda T. Oral malodorous compound causes apoptosis and genomic DNA damage in human gingival fibroblasts. J Periodontal Res. 2008;43(4):391–399. doi: 10.1111/j.1600-0765.2007.01052.x. [DOI] [PubMed] [Google Scholar]
- 8.Persson S, Edlund MB, Claesson R, Carlsson J. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol Immunol. 1990;5(4):195–201. doi: 10.1111/j.1399-302X.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 9.Carlsson J, Larsen JT, Edlund MB. Peptostreptococcus micros has a uniquely high capacity to form hydrogen sulfide from glutathione. Oral Microbiol Immunol. 1993;8(1):42–45. doi: 10.1111/j.1399-302X.1993.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 10.Carlsson J, Larsen JT, Edlund MB. Utilization of glutathione (L-gamma-glutamyl-L-cysteinylglycine) by Fusobacterium nucleatum subspecies nucleatum. Oral Microbiol Immunol. 1994;9(5):297–300. doi: 10.1111/j.1399-302X.1994.tb00074.x. [DOI] [PubMed] [Google Scholar]
- 11.Basic A, Blomqvist S, Carlén A, Dahlén G. Estimation of bacterial hydrogen sulfide production in vitro. J Oral Microbiol. 2015;7:28166. doi:10.3402/jom.v7.28166. [DOI] [PMC free article] [PubMed]
- 12.Claesson R, Edlund MB, Persson S, Carlsson J. Production of volatile sulfur compounds by various Fusobacterium species. Oral Microbiol Immunol. 1990;5(3):137–142. doi: 10.1111/j.1399-302X.1990.tb00411.x. [DOI] [PubMed] [Google Scholar]
- 13.Pianotti R, Lachette S, Dills S. Desulfuration of cysteine and methionine by Fusobacterium nucleatum. J Dent Res. 1986;65(6):913–917. doi: 10.1177/00220345860650061101. [DOI] [PubMed] [Google Scholar]
- 14.Fukamachi H, Nakano Y, Yoshimura M, Koga T. Cloning and characterization of the L-cysteine desulfhydrase gene of Fusobacterium nucleatum. FEMS Microbiol Lett. 2002;215(1):75–80. doi: 10.1111/j.1574-6968.2002.tb11373.x. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida Y, Ito S, Kamo M, Kezuka Y, Tamura H, Kunimatsu K, Kato H. Production of hydrogen sulfide by two enzymes associated with biosynthesis of homocysteine and lanthionine in Fusobacterium nucleatum subsp. nucleatum ATCC 25586. Microbiology. 2010;156(7):2260–2269. doi: 10.1099/mic.0.039180-0. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida Y, Ito S, Tamura H, Kunimatsu K. Use of a novel assay to evaluate enzymes that produce hydrogen sulfide in Fusobacterium nucleatum. J Microbiol Methods. 2010;80(3):313–315. doi: 10.1016/j.mimet.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida Y, Suwabe K, Nagano K, Kezuka Y, Kato H, Yoshimura F. Identification and enzymic analysis of a novel protein associated with production of hydrogen sulfide and l-serine from l-cysteine in fusobacterium nucleatum subsp. Nucleatum ATCC 25586. Microbiology. 2011;157(7):2164–2171. doi: 10.1099/mic.0.048934-0. [DOI] [PubMed] [Google Scholar]
- 18.Suwabe K, Yoshida Y, Nagano K, Yoshimura F. Identification of an L-methionine γ-lyase involved in the production of hydrogen sulfide from L-cysteine in Fusobacterium nucleatum subsp. nucleatum ATCC 25586. Microbiology. 2011;157(10):2992–3000. doi: 10.1099/mic.0.051813-0. [DOI] [PubMed] [Google Scholar]
- 19.Dahlén G, Claesson R, Åberg CH, Haubek D, Johansson A, Kwamin F: Subgingival bacteria in Ghanaian adolescents with or without progression of attachment loss. J Oral Microbiol. 2014;6(1). http://www.tandfonline.com/doi/full/10.3402/jom.v6.23977. [DOI] [PMC free article] [PubMed]
- 20.Davies JR, Svensäter G, Herzberg MC. Identification of novel LPXTG-linked surface proteins from Streptococcus gordonii. Microbiology. 2009;155(6):1977–1988. doi: 10.1099/mic.0.027854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura H. Hydrogen sulfide: its production, release and functions. Amino Acids. 2011;41(1):113–121. doi: 10.1007/s00726-010-0510-x. [DOI] [PubMed] [Google Scholar]
- 22.Ohkishi H, Nishikawa D, Kumagai H, Yamada H. Distribution of cysteine desulfhydrase in microorganisms. Agric Biol Chem. 1981;45(1):253–257. [Google Scholar]
- 23.Awano N, Wada M, Mori H, Nakamori S, Takagi H. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl Environ Microbiol. 2005;71(7):4149–4152. doi: 10.1128/AEM.71.7.4149-4152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu L, Ebersole JL, Kurzban GP, Holt SC. Cystalysin, a 46-kilodalton cysteine desulfhydrase from Treponema denticola, with hemolytic and hemoxidative activities. Infect Immun. 1997;65(8):3231–3238. doi: 10.1128/iai.65.8.3231-3238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yano T, Fukamachi H, Yamamoto M, Igarashi T. Characterization of L-cysteine desulfhydrase from Prevotella intermedia. Oral Microbiol Immunol. 2009;24(6):485–492. doi: 10.1111/j.1399-302X.2009.00546.x. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida Y, Nakano Y, Amano A, Yoshimura M, Fukamachi H, Oho T, Koga T. Icd from Streptococcus encodes a C-S lyase with α, β-elimination activity that degrades L-cysteine. Microbiology. 2002;148(12):3961–3970. doi: 10.1099/00221287-148-12-3961. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida Y, Negishi M, Amano A, Oho T, Nakano Y. Differences in the βC-S lyase activities of viridans group streptococci. Biochem Biophys Res Commun. 2003;300(1):55–60. doi: 10.1016/S0006-291X(02)02803-6. [DOI] [PubMed] [Google Scholar]
- 28.Ito S, Nagamune H, Tamura H, Yoshida Y. Identification and molecular analysis of βC-S lyase producing hydrogen sulfide in Streptococcus intermedius. J Med Microbiol. 2008;57(11):1411–1419. doi: 10.1099/jmm.0.2008/001677-0. [DOI] [PubMed] [Google Scholar]
- 29.Guzman YA, Sakellari D, Arsenakis M, Floudas CA. Proteomics for the discovery of biomarkers and diagnosis of periodontitis: a critical review. Expert Rev Proteomics. 2014;11(1):31–41. doi: 10.1586/14789450.2014.864953. [DOI] [PubMed] [Google Scholar]
- 30.Macarthur DJ, Jacques NA. Proteome analysis of oral pathogens. J Dent Res. 2003;82(11):870–876. doi: 10.1177/154405910308201105. [DOI] [PubMed] [Google Scholar]
- 31.Steeves CH, Potrykus J, Barnett DA, Bearne SL. Oxidative stress response in the opportunistic oral pathogen Fusobacterium nucleatum. Proteomics. 2011;11(10):2027–2037. doi: 10.1002/pmic.201000631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data is presented in the Tables and Additional file 1: Table S1.


