Abstract
Improved diagnostics for acute infections could decrease morbidity and mortality by increasing early antibiotics for patients with bacterial infections and reducing unnecessary antibiotics for patients without bacterial infections. Several groups have used gene expression microarrays to build classifiers for acute infections, but these have been hampered by the size of the gene sets, use of overfit models, or lack of independent validation. We used multicohort analysis to derive a set of seven genes for robust discrimination of bacterial and viral infections, which we then validated in 30 independent cohorts. We next used our previously published 11-gene Sepsis MetaScore together with the new bacterial/viral classifier to build an integrated antibiotics decision model. In a pooled analysis of 1057 samples from 20 cohorts (excluding infants), the integrated antibiotics decision model had a sensitivity and specificity for bacterial infections of 94.0 and 59.8%, respectively (negative likelihood ratio, 0.10). Prospective clinical validation will be needed before these findings are implemented for patient care.
INTRODUCTION
Early and accurate diagnosis of infection is key to improving patient outcomes and reducing antibiotic resistance. The mortality rate of bacterial sepsis increases by 8% for each hour by which antibiotics are delayed (1); however, indiscriminate prescription of antibiotics to patients without bacterial infections increases rates of morbidity and antimicrobial resistance. The rate of inappropriate antibiotic prescriptions in the hospital setting is estimated at 30 to 50% and would be decreased by improved diagnostics (2, 3). In broader use, up to 95% of outpatients given antibiotics for suspected enteric fever have negative cultures (4). There is currently no gold standard point-of-care diagnostic that can broadly determine the presence and type of infection. Thus, the White House has established the National Action Plan for Combating Antibiotic-Resistant Bacteria, which called for “point-of-need diagnostic tests to distinguish rapidly between bacterial and viral infections” (5).
Although new polymerase chain reaction (PCR)–based molecular diagnostics can profile pathogens directly from a blood culture (6), such methods rely on the presence of adequate numbers of pathogens in the blood. Moreover, they are limited to detecting a discrete range of pathogens. As a result, there is a growing need for molecular diagnostics that profile the host gene response. These include diagnostics that can distinguish the presence of infection as compared to inflammation in the absence of infection, such as our 11-gene “Sepsis MetaScore” (SMS) (7), which has been validated across multiple cohorts (8), among others (9, 10). Other groups have focused on gene sets that can distinguish between types of infections, such as bacterial versus viral infections (11–13); however, these gene sets often contain too many genes to translate into a useful clinical tool. Tsalik et al. described a model that distinguishes among all three groups—noninfected patients and those with bacterial or viral illness—but this model required the measurement of 122 probes, presenting an implementation challenge (14). Similarly, we have described a “meta-virus signature” that describes a common response to viral infection but contained too many genes (396) for clinical application (15). Overall, although great promise has been shown in this field, no pragmatic infection diagnostic based on host gene expression has yet made it into clinical practice.
The data from these biomarker studies and dozens of other genome-wide expression studies in sepsis and acute infections have been published and deposited for further study in public databases such as the National Institutes of Health (NIH) Gene Expression Omnibus (GEO) and the European Bioinformatics Institute (EBI) ArrayExpress. These data are a largely untapped resource that can be used for both biomarker discovery and validation. We have previously shown that our integrated multicohort analysis of gene expression produces robust diagnostic tools for organ transplant (16), sepsis (7), specific types of viral infections (15), and active tuberculosis (17). Furthermore, these data are also useful as a benchmarking and validation tool for new host gene expression diagnostics. However, such validation using public data has previously been limited to only those cohorts that contain at least two classes of interest (those in which a direct comparison between classes is possible), because interstudy technical differences preclude direct comparison of diagnostic scores between cohorts.
Here, we sought to improve the diagnostic power of the SMS by adding the ability to discriminate bacterial from viral infections. Thus, to derive an improved biomarker for discriminating infection types, we applied our multicohort analysis framework to clinical microarray cohorts that compared the host response to bacterial and viral infections. We further developed a method to conormalize gene expression data among multiple cohorts, allowing direct comparison of a diagnostic score among multiple cohorts. Finally, we combined the previous SMS and the bacterial/viral diagnostic described here into an integrated antibiotics decision model (IADM) that can determine whether a patient with acute inflammation has an underlying bacterial infection.
RESULTS
Derivation of the seven-gene bacterial/viral metascore
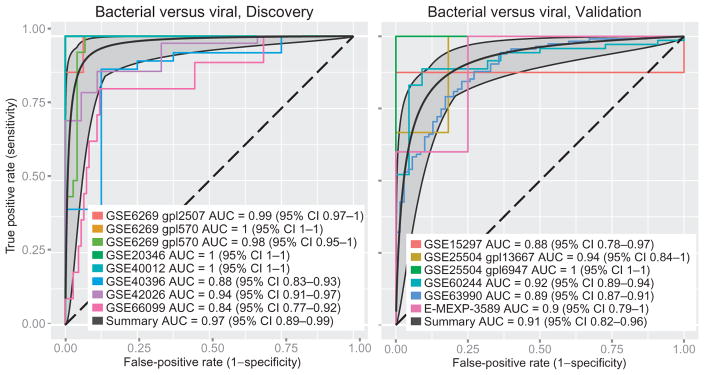
Our previously published 11-gene SMS cannot reliably distinguish between bacterial and viral infections, showing mostly nonsignificant differences in score distribution between patients with bacterial and viral infections (fig. S1). Having previously shown that there is a conserved host gene response to viral infections (15), we hypothesized that a classifier for bacterial versus viral infections would allow for an improved diagnostic model. We thus performed a systematic search for gene expression microarray cohorts that studied patients with viral and/or bacterial infections. We identified eight cohorts (11, 18–26) [both whole blood and peripheral blood mononuclear cells (PBMCs)] that included n > 5 patients with both viral and bacterial infections (Table 1A). The eight cohorts were composed of 426 patient samples (142 viral and 284 bacterial infections), including children and adults, medical and surgical patients, and those with multiple sites of infection. We performed multicohort analysis on the eight cohorts as described previously (fig. S2) (7, 15–17). We set significance thresholds at an effect size >2-fold and a false discovery rate (FDR) <1% in leave–one–data set–out round-robin analysis. However, to make sure that neither tissue type (whole blood or PBMCs) biased our results, we further selected only those genes that also had an effect size >1.5-fold in separate analyses of both PBMC and whole-blood cohorts. This process resulted in 72 differentially expressed genes significant at the above thresholds (table S1). We used a greedy forward search (7) to find a gene set optimized for diagnosis, resulting in seven genes [higher in viral infections (IFI27, JUP, and LAX1) and higher in bacterial infections (HK3, TNIP1, GPAA1, and CTSB); fig. S3)]. As expected, a “bacterial/viral metascore” based on these seven genes robustly distinguished viral from bacterial infections in all eight of the discovery cohorts [summary receiver operating characteristic (ROC) area under the curve (AUC), 0.97; 95% confidence interval (CI), 0.89 to 0.99; Fig. 1 and fig. S4].
Table 1.
Data sets used in the discovery and direct validation of the bacterial/viral metascore.
| Accession | Author | Tissue | Platform | Demographic | Bacteria | Viruses | Number healthy | Number bacterial | Number viral |
|---|---|---|---|---|---|---|---|---|---|
| A. Discovery data sets | |||||||||
| GSE6269 | Ramilo | PBMC | GPL2507 | Children admitted with infection | Escherichia coli, Staphylococcus aureus, Streptococcus pneumoniae | Influenza | 0 | 16 | 8 |
| GPL570 | S. aureus, S. pneumoniae | Influenza | 0 | 12 | 10 | ||||
| GPL96 | S. aureus, S. pneumoniae | Influenza | 6 | 73 | 18 | ||||
| GSE20346 | Parnell | Whole blood | GPL6947 | Adults with CAP | Unknown bacterial pneumonia | Influenza | 36 | 12 | 8 |
| GSE40012 | Parnell | Whole blood | GPL6947 | Adults with CAP | Unknown bacterial pneumonia | Influenza | 18 | 36 | 11 |
| GSE40396 | Hu | Whole blood | GPL10558 | Febrile children in emergency department | Multiple | Adenovirus, enterovirus, rhinovirus, HHV6 | 22 | 8 | 35 |
| GSE42026 | Herbeg | Whole blood | GPL6947 | Children admitted with infection | Streptococcus and Staphylococcus spp. | Influenza, RSV | 33 | 18 | 41 |
| GSE66099 | Wong | Whole blood | GPL570 | Septic children in PICU | Multiple | Influenza, HSV, CMV, BK, adenovirus | 47 | 109 | 11 |
| B. Validation data sets | |||||||||
| GSE15297 | Popper | Whole blood | GPL8328 | Febrile children | Scarlet fever (Streptococcus) | Adenovirus | 0 | 5 | 8 |
| GSE25504 | Smith | Whole blood | GPL13667 | Septic neonates | Multiple | Rhinovirus, CMV | 6 | 11 | 3 |
| GPL6947 | Multiple | CMV | 35 | 26 | 1 | ||||
| GSE60244 | Suarez | Whole blood | GPL10558 | Adults hospitalized with LRTI | Gram-positive and atypical | Influenza, RSV, MPV | 40 | 22 | 71 |
| GSE63990 | Tsalik | Whole blood | GPL571 | Adults with ARI | Multiple | Multiple | 0 | 70 | 115 |
| E-MEXP-3589 | Almansa | Whole blood | GPL10332 | Adults with COPD with infection | Gram-positive, Gram-negative, atypical | Influenza, RSV, MPV | 4 | 4 | 5 |
CAP, community-acquired pneumonia; HHV6, human herpesvirus 6; RSV, respiratory syncytial virus; HSV, herpes simplex virus; CMV, cytomegalovirus; MPV, metapneumovirus; PICU, pediatric intensive care unit; LRTI, lower respiratory tract infection; ARI, acute respiratory infection; COPD, chronic obstructive pulmonary disease.
Fig. 1. Summary ROC curves for discovery and direct validation data sets for the bacterial/viral meta-score.
Summary ROC curve is shown in black, with 95% CIs in dark gray.
We next tested the seven-gene set in the six remaining independent clinical cohorts (13, 14, 27–29) that directly compared bacterial and viral infections (138 bacterial and 203 viral infections, totaling 341 samples) and found a summary ROC AUC of 0.91 (95% CI, 0.82 to 0.96) (Fig. 1, Table 1B, and fig. S5; individual test characteristics in table S2). To measure the generalizability of our signature, we also tested whether cells stimulated in vitro with lipopolysaccharide (LPS) or influenza virus could be separated with the bacterial/viral metascore [GSE53166 (30), n = 75; AUC, 0.99; fig. S6].
Global validation via COCONUT conormalization
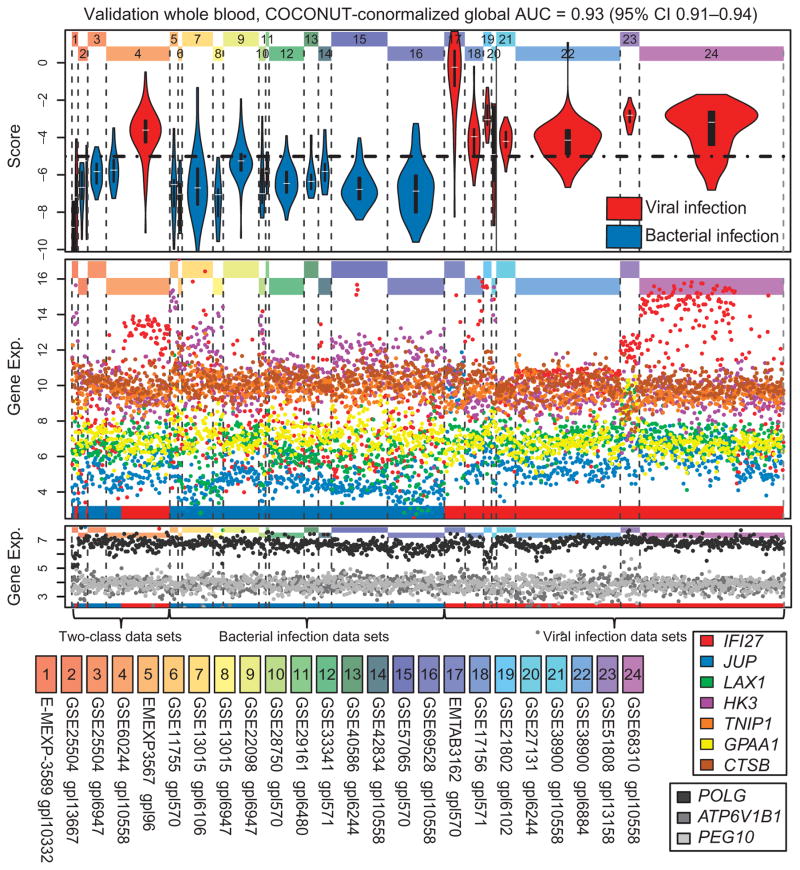
There are dozens of microarray cohorts in the public domain that studied either bacterial or viral infections, but not both, thus precluding a direct (within data set) estimate of diagnostic power for separating bacterial and viral illness. To apply and compare a gene score across these cohorts, we needed a method that could remove inter–data set batch effects while remaining unbiased to the diagnosis of the diseased patients. We designed and implemented a modified type of array normalization that uses the ComBat (31) empirical Bayes normalization methods on healthy controls to obtain bias-free corrections of disease samples (a method we call COmbat CO-Normalization Using conTrols or “COCONUT”; fig. S7). Housekeeping genes remained invariant across both diseases and cohorts after COCONUT conormalization, and each gene still retained the same distribution between diseases and controls within each data set (fig. S8). Because our method assumes that all healthy samples are derived from the same distribution, we separately normalized the whole-blood and PBMC samples because different immune cell types have different baseline gene expression distributions. Using COCONUT conormalization, the bacterial/viral metascore has a global AUC of 0.92 (95% CI, 0.89 to 0.96) in the whole-blood discovery cohorts (figs. S9 to S10). We then applied this method to test the bacterial/viral metascore in all public domain microarray cohorts that matched inclusion criteria and used whole blood. These data sets included the four direct validation cohorts that included control patients and an additional 20 cohorts that measured either bacterial or viral infections but not both (n = 143 + 897 = 1040) (32–49). These data sets represent a wide variety of clinical conditions, including a range of infection types (Gram-positive, Gram-negative, atypical bacterial, common respiratory viruses, and dengue) and severities (mild infections to septic shock). The bacterial/viral metascore showed an overall ROC AUC of 0.93 (95% CI, 0.91 to 0.94) across these data, which allowed us to set a single global score cutoff (Fig. 2, Table 1B, and fig. S11). Finally, we performed the same procedure on PBMC validation cohorts [six cohorts (50–54), n = 259; global AUC, 0.92 (95% CI, 0.87 to 0.97; figs. S12 to S13)]. All three global ROC AUCs using COCONUT conormalization (discovery whole blood, 0.92; validation whole blood, 0.93; validation PBMCs, 0.92) approximately matched the summary AUC of the direct validation cohorts (0.91), giving high confidence in the diagnostic power of this method.
Fig. 2. Bacterial/viral score in COCONUT-conormalized whole-blood validation data sets.
The global AUC across all whole-blood discovery data sets is 0.93. Top: Score distribution by data set (blue, bacterial; red, viral). Middle: Individual gene expression (exp.). Bottom: Housekeeping genes (grayscale). The dotted line at the top shows a possible global threshold for discriminating infection type.
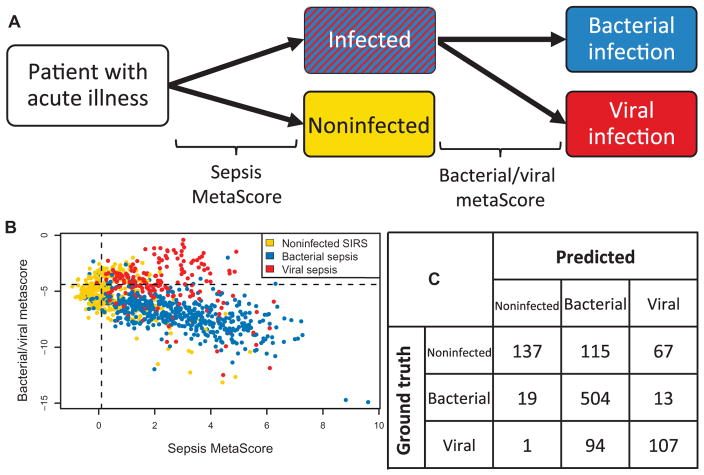
Integrated antibiotics decision model
A key clinical need is diagnosing whether a patient with signs and symptoms of inflammation has an underlying bacterial infection, because rapid and judicious administration of antibiotics is key to improving patient outcomes. Neither the SMS nor the bacterial/viral metascore alone can robustly distinguish between all three classes of (i) noninfected inflammation, (ii) bacterial illness, and (iii) viral illness. Thus, to increase clinical relevance, we developed an “integrated antibiotics decision model” (IADM), whereby we first apply our previously described SMS (7) to test for the presence of an infection and then apply the bacterial/viral metascore to the samples that test positive for infection (Fig. 3A). As described previously, the only way to establish test characteristics for the IADM simultaneously across cohorts is to use COCONUT conormalization. We found that the SMS in COCONUT-conormalized data is strongly influenced by age, which could be caused by age-dependent differences in healthy subjects, infected patients, or both (fig. S14). We thus excluded cohorts focused on infants (children <1 year old) from the IADM. We also removed the low-severity outpatient viral illness cohorts (GSE17156 and GSE68310) because in outpatient settings, the history and physical exam findings make noninfectious causes of acute inflammation less likely. This resulted in a total of 20 cohorts for testing the IADM (n = 1057). The resulting global AUC for the SMS across the available data was 0.86 (95% CI, 0.84 to 0.89) (fig. S15 and table S3). We set global thresholds for an SMS sensitivity for infection of 95% and a bacterial/viral metascore sensitivity for bacterial infection of 95%. Considering all three classes of noninfectious inflammation, bacterial infection, and viral infection, this yielded an overall sensitivity and specificity of 94.0 and 59.8% for bacterial infections and 53.0 and 90.6% for viral infections, respectively (Fig. 3). These performance characteristics were largely unchanged if healthy patients were included in the noninfected class (fig. S16). The overall positive and negative likelihood ratios for bacterial infection in the IADM were thus 2.34 (LR+) and 0.10 (LR−), respectively, compared to a recent meta-analysis of procalcitonin that showed a negative likelihood ratio of 0.29 (95% CI, 0.22 to 0.38) (55). We plotted negative predictive value (NPV) and positive predictive value (PPV) versus prevalence for these test characteristics; the NPV and PPV for bacterial infection at a prevalence of 15% were 98.3 and 29.2%, respectively (fig. S17).
Fig. 3. IADM across COCONUT-conormalized public gene expression data that matched inclusion criteria.
(A) IADM schematic. (B) Distribution of scores and cutoffs for IADM in COCONUT-conormalized data. SIRS, systemic inflammatory response syndrome. (C) Confusion matrix for diagnosis. Bacterial infection sensitivity, 94.0%; bacterial infection specificity, 59.8%; viral infection sensitivity, 53.0%; viral infection specificity, 90.6%.
There was only one data set [GSE63990 (14)] that included non-infectious inflammation patients and patients with both bacterial and viral illness but did not include healthy controls, precluding its addition to the global calculations. We thus tested the IADM on this cohort with locally derived test thresholds. We found an overall bacterial infection sensitivity and specificity of 94.3 and 52.2%, respectively (fig. S18).
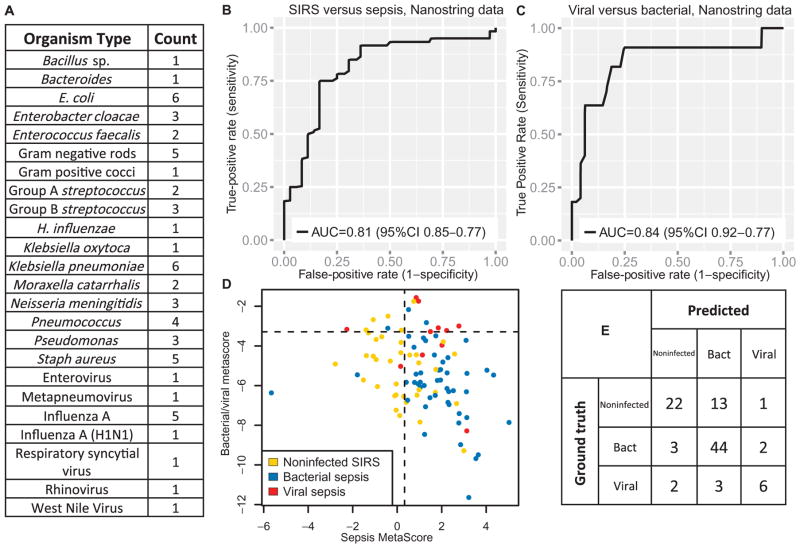
Validation in independent samples using NanoString nCounter
Finally, we used targeted quantitative NanoString nCounter (56) gene expression assays to prospectively validate these results in independent whole-blood samples from children with sepsis from the Genomics of Pediatric SIRS and Septic Shock Investigators (GPSSSI) cohort (total n = 96, with 36 SIRS, 49 bacterial sepsis, and 11 viral sepsis patients; Fig. 4 and table S4). The GPSSSI cohort was also used by data set GSE66099, but the children profiled here were never profiled via micro-array and are thus not part of the discovery data sets. In the NanoString validation cohort, the SMS AUC was 0.81 (AUC of 0.80 in GSE66099). Similarly, the bacterial/viral metascore AUC was 0.84 (AUC of 0.83 in GSE66099). The microarray AUCs were thus preserved when tested with a targeted, quantitative gene expression assay in new patients. Applying the same IADM, the sensitivity and specificity were 89.7 and 70.0% for bacterial infections (LR−, 0.15; LR+, 3.0) and 54.5 and 96.5% for viral infections (LR−, 0.47; LR+, 15.6), respectively.
Fig. 4. Targeted NanoString gene expression data for children with SIRS/sepsis from the GPSSSI cohort never tested with microarrays.
Total n = 96, of which SIRS = 36, bacterial sepsis = 49, and viral sepsis = 11. (A) Breakdown of infected patients by organism type. (B and C) ROC curves for the SMS and the bacterial/viral metascore. (D) Distribution of scores and cutoffs for IADM. (E) Confusion matrix for IADM. Bacterial infection sensitivity, 89.7%; bacterial infection specificity, 70.0%; viral infection sensitivity, 54.5%; viral infection specificity, 96.5%.
DISCUSSION
Better diagnostics for acute infections are needed in both the inpatient and outpatient settings. In low-acuity outpatient settings, a simple diagnostic that can discriminate bacterial from viral infections may be enough to assist in appropriate antibiotic usage. In higher-acuity settings, causes of noninfectious inflammation become more important to rule out, and so a decision model for antibiotic prescriptions must include a noninfected, nonhealthy case. Thus, a reliable diagnostic needs to distinguish all three cases (noninfected inflammation, bacterial infection, and viral infection). Here, using 426 samples from 8 cohorts, we derived a parsimonious set of only seven genes that can accurately discriminate bacterial from viral infections across a very broad range of clinical conditions in independent cohorts (total of 30 cohorts composed of 1299 patients). We further demonstrated that by integrating our published SMS (7) (to distinguish the presence or absence of infection) with the bacterial/viral metascore (to determine infection type) into a single IADM, we can determine with high accuracy which patients do not require antibiotics. Finally, we confirmed the diagnostic power of both the seven-gene set and the IADM in independent samples using a targeted NanoString assay, showing that the signatures retain diagnostic power when not relying on microarrays.
The IADM has a low negative likelihood ratio (0.10) and high estimated NPV, meaning it would be potentially effective as a rule-out test. A meta-analysis of procalcitonin that included 3244 patients from 30 studies resulted in an overall estimated negative likelihood ratio of 0.29 (95% CI, 0.22 to 0.38) (55). Thus, the IADM negative likelihood ratio is much lower than the estimate for procalcitonin (on the basis of nonoverlapping 95% CIs), indicating clinical utility. Moreover, these test characteristics assume no knowledge of the patient and so are only estimates of the real-world clinical utility of such a test, because patient history, physical examination, vital signs, and laboratory values would all assist in a diagnosis as well. Even given these caveats, a recent economic decision model of screening ICU patients for hospital-acquired infections suggested that a test such as the IADM that can accurately diagnose bacterial and viral infections could be cost-effective (57). Ultimately, only interventional trials will be able to establish cost-effectiveness and clinical utility of a diagnostic test.
We validated our diagnostic in pediatric sepsis patients from the GPSSSI cohort using a NanoString assay. NanoString is highly accurate and is a useful tool for measuring the expression of multiple genes at once; however, it is also likely too slow for clinical application (4 to 6 hours per assay). Thus, although the assay confirms that our gene set is robust in targeted measurements, further work will be needed to improve the turnaround time. There are multiple possibilities for developing a commercial assay based on rapid multiplexed quantitative PCR that meets the time-sensitive demands of an infection diagnostic test. However, this technical hurdle is something that all gene expression–based infection diagnostics must overcome to gain clinical relevance.
A simple linear score generally cannot adequately separate the three classes (noninfected inflammation, bacterial infections, and viral infections); other machine-learning techniques, such as multiple regression or tree-based methods, are typically used. However, because the location and scale of different genes vary greatly between microarray types, such techniques cannot usually be truly validated across microarray platforms. Although COCONUT partially circumvents this issue by allowing for discovery of a model across several cohorts with application of the same model in COCONUT-conormalized validation cohorts, such method would be forced to leave out data sets that did not include healthy controls. We thus opted to use our simple, scale-free, difference-of-geometric-means scores. This allowed us to both discover and validate our diagnostic models across multiple cohorts. It may be possible to discover a gene set using multiple classification techniques, instead of our current method of starting with the SMS and then adding the bacterial/viral metascore. However, to discover and then validate in multiple cohorts requires that all cohorts be appropriate for COCONUT normalization, and we have instead erred on the side of maximizing the utility of multiple clinically heterogeneous cohorts.
Several groups have published models for diagnosing infections on the basis of host gene expression; none have yet made it into clinical practice. Most of these classifiers were not tested in multiple independent cohorts, had too many genes to allow rapid profiling necessary for useful diagnosis, or both. For instance, Suarez et al. created a 10-gene k-nearest-neighbor classifier but did not test it outside their published data set (GSE60244) (13). Tsalik et al. created a 122-probe (120-gene) classifier on the basis of multiple regression models, but in testing it in external GEO cohorts, they retrained their regression coefficients in each new data set (14). Such model retraining results in a strong upward bias to these validation numbers (assuming that a final model would not be locally retrained). Other groups have made gene expression classifiers for infection but did not include models for discriminating viral infections (7, 9, 10). Our IADM is robust across a wide range of disease types and severities but has a relatively lower sensitivity for viral infections. Non–gene expression biomarkers have also been used for infection diagnosis. Procalcitonin has been studied extensively in the setting of sepsis diagnosis but cannot distinguish between noninfected individuals and those with viral infections (58). Protein-panel assays have been shown to discriminate bacterial from viral infections but cannot discriminate patients with noninfectious inflammation (59, 60). Thus, all of these classifiers have certain strengths and weaknesses that will become more apparent with further prospective testing and direct comparison.
Although our goal in this study was to identify biomarkers and not necessarily mechanistic biology, it is still important for a biomarker set to have biological plausibility. Of the seven genes in the bacterial/viral meta-score, six have previously been linked to infections or leukocyte activation. Both IFI27 and JUP were shown in single-cohort genome-wide expression studies to be induced in response to viral infection (52, 61), whereas TNIP1 and CTSB are important in modulating the nuclear factor kB and necrotic responses to bacterial infection (62, 63). Finally, LAX1 (up-regulated in viral infections) is involved in activation of T and B cells (64), and HK3 is instrumental in the neutrophil differentiation pathway (65). Thus, the role of these transcripts as biomarkers for infection type is not coincidental.
Here, we developed a method, COCONUT, to directly compare our model across a large pool of one-class cohorts that would otherwise be unusable for benchmarking a diagnostic gene set. COCONUT assumes that all controls come from the same distribution; that is, the genes in each group of controls are reset to have the same mean and variance, with batch parameters learned empirically from gene groups. This method corrects for microarray and batch processing differences between cohorts and thus allows for the creation of a global ROC curve with a single threshold. This is a more “real-world” measure of diagnostic power than reporting multiple validation ROC curves, because no single cutoff could attain the same test characteristics in the different cohorts (17). However, the COCONUT-conormalized data showed differences in infants as compared to older children and adults. Therefore, infants were excluded from the validation, meaning that the efficacy of the IADM in infants remains unknown. These data also had few children with viral infections between the toddler and teenage years; this is an age group that will require further study. The most important takeaway from the COCONUT-conormalized data is that both the bacterial/viral metascore and the IADM retain diagnostic power across a very broad range of infection types and severities, with overall AUCs that are similar to the summary AUCs from head-to-head comparisons within cohorts.
Overall, we have leveraged our proven multicohort analysis pipeline to derive a highly robust model for improving infection diagnosis. Using our method, we were able to validate this in dozens of independent microarray cohorts. We have also validated using a targeted NanoString assay in pediatric sepsis patients. Although the IADM still needs to undergo optimization for rapid turnaround as well as a prospective interventional trial, it seems clear that molecular profiling of the host genome will become part of the clinical toolkit in the future.
MATERIALS AND METHODS
Study design
The purpose of this study was to use an integrated multicohort analysis framework to analyze multiple gene expression data sets to identify a biomarker that can classify patients with bacterial or viral infections. This framework has been described previously (7, 16, 17).
Systematic search and multicohort analysis
We performed a systematic search in NIH GEO and EBI ArrayExpress for public human microarray genome-wide expression studies using the following search terms: bact[wildcard], vir[wildcard], infection, sepsis, SIRS, ICU, nosocomial, fever, and pneumonia. Abstracts were screened to remove all studies that were (i) nonclinical, (ii) performed using tissues other than whole blood or PBMCs, or (iii) comparing patients who were not matched for clinical time.
In all data sets that included sample-level microbiology data, we retained only those samples for which pathogens of a single type (either bacterial or viral) had been identified. Data sets for which sample-level microbiology data were not available were still retained if the corresponding paper described the cohort as being infected with only bacteria or only viruses. In three cohorts, the diagnosis was not necessarily micro-biologically confirmed: GSE11755 (one of six patients described as culture-negative meningitis), GSE42834 (described as a cohort of bacterial pneumonia meeting clinical criteria), and GSE57065 (described as a cohort of bacterial sepsis for which 86% of patients had microbiological confirmation). All other samples used in our analysis had confirmed microbiological diagnoses at the sample or cohort level.
All microarray data were renormalized from raw data (when available) using standardized methods. Affymetrix arrays were renormalized using GC robust multiarray average (gcRMA) (on arrays with mismatch probes) or RMA. Illumina, Agilent, GE, and other commercial arrays were renormalized via normal-exponential background correction followed by quantile normalization. Custom arrays were not renormalized. Data were log2-transformed, and a fixed-effect model was used to summarize probes to genes within each study. Within each study, cohorts assayed with different microarray types were treated as independent.
We performed multicohort meta-analysis as described previously (7, 15–17). Briefly, genes were summarized using Hedges’ g, and the DerSimonian-Laird random-effects model was used for meta-analysis, followed by Benjamini-Hochberg multiple hypothesis correction (66). Patients with bacterial infections were compared to patients with viral infections within studies, such that a positive effect size indicates that a gene was more highly expressed in virus-infected patients and a negative effect size indicates that a gene was more highly expressed in bacteria-infected patients. All data sets that matched criteria with n > 5 in both bacterial and viral cohorts that were published by the time of the initial search (1 April 2015) were used for discovery; data sets published after this time were used in validation.
To find a set of genes highly conserved in differential expression between bacterial and viral infections, we selected all cohorts that directly compared patients with bacterial and viral infections. Patients with documented co-infections (both bacterial and viral) were removed. Cohorts were required to have >5 patients in each group to be included in the meta-analysis. Both PBMC and whole-blood cohorts were included. We only included genes present in 7 (k − 1) data sets; thus, there were 14,729 genes included in the analysis. Significant genes were those that had an effect size >2-fold and an FDR <1% in a leave–one–data set–out round-robin analysis. However, to ensure that genes from both whole blood and PBMCs were represented in the final gene set, we also performed separate meta-analyses of the PBMC and whole-blood cohorts and removed all genes that had an effect size <1.5-fold in either whole blood or PBMCs separately. The remaining genes were considered significant.
Derivation of the seven-gene set
To find a set of highly diagnostic genes, the significant genes from the meta-analysis were run through a greedy forward search as described previously (7). Briefly, this algorithm starts with zero gene and adds one gene in each cycle that best improves the AUC for diagnosis in the discovery cohorts, until a new gene cannot improve the discovery AUCs more than some threshold (an increase in the weighted discovery AUC of ≥1). The resulting genes are used to calculate a single bacterial/viral metascore, calculated as the geometric mean of the “viral” response genes minus the geometric mean of the “bacterial” response genes, times the ratio of the number of genes in each set. The resulting continuous score can then be tested for diagnostic power using ROC curves.
Direct validation of the seven-gene set
The resulting gene set was first validated in the remaining public gene expression cohorts that directly compared bacterial to viral infections but were too small to be used for meta-analysis. Two cohorts [GSE60244 (13) and GSE63990 (14)] were made public after our meta-analysis was completed and so were used for validation. To show generalizability, we also examined one large in vitro data set comparing LPS to influenza exposure in monocyte-derived dendritic cells, but this was not included in the summary AUC because it is not expected to come from the same distribution as the clinical studies.
Summary ROC curves
For both discovery and validation cohorts, summary ROC curves were constructed according to the method of Kester and Buntinx (67) and as previously described (17). Briefly, linear-exponential models were made for each ROC curve, and the parameters of these individual curves were summarized using a random-effects model to estimate the overall summary ROC curve parameters. The α-parameter controls AUC (in particular, distance of the line from the line of identity), and the β-parameter controls skewness of the ROC curve. Summary AUC CIs were estimated from the SE of the α and β in meta-analysis.
COCONUT conormalization
There are dozens of public microarray cohorts that profiled patients with either bacterial or viral infections, but not both. It would be advantageous to be able to compare a gene score across these cohorts, but it has not previously been possible because each different microarray has widely different background measurements for each gene, and among studies using the same types of microarrays, there are large batch effects. To make use of these data, we needed to conormalize these cohorts in such a way that (i) no bias is introduced that could influence final classification (the normalization protocol should be blind to diagnosis), (ii) there should be no change to the distribution of a gene within a study, and (iii) a gene should show the same distributions between studies after normalization. A method with these characteristics would allow our gene score to be calculated and compared across multiple studies and thus allow us to broadly test its generalizability.
The ComBat empirical Bayes normalization method (31) is popular for cross-platform normalization but crucially falls short of our desired criteria because it assumes an equal distribution across disease states. We thus developed a modified version of the ComBat method, which conormalizes control samples from different cohorts to allow for direct comparison of diseased samples from those same cohorts. We call this method COmbat CO-Normalization Using conTrols or COCONUT. COCONUT makes one strong assumption that it forces control/healthy patients from different cohorts to represent the same distribution. Briefly, all cohorts are split into healthy and diseased components. The healthy components undergo ComBat conormalization without covariates. The ComBat estimated parameters α̂, β̂, σ̂, δ*, and γ* are obtained for each data set for the healthy component and then applied to the diseased component (fig. S7). This forces the diseased components of all cohorts to be from the same background distribution but retains their relative distance from the healthy component (t statistics within data sets are only different post-COCONUT because of floating-point math). It also does not require any a priori knowledge of disease classification (such as bacterial or viral infection), thus meeting our prespecified criteria. This method does have the notable requirement that healthy/control patients are required to be present in a data set for it to be pooled with other available data. Also, because healthy/control patients are set to be in the same distribution, it should only be used where such an assumption is reasonable (such as within the same tissue type, among the same species, etc.).
The ComBat model and the COCONUT method
As described by Johnson et al., the ComBat model corrects for location and scale of each gene by first solving an ordinary least-squares model for gene expression and then shrinking the resulting parameters using an empirical Bayes estimator, solved iteratively (31). Formally, each gene expression level Yijg (for gene g for sample j in batch i) is assumed to be composed of overall gene expression αg, design matrix of sample conditions X with regression coefficients βg, additive and multiplicative batch effects γig and δig, and an error term εijg
Estimating parameters using ordinary least-squares regression standardizes Yijg to a new term Zijg (where σ̂g is the SD of εijg)
The standardized data are now distributed according to , where and .
The inverse γ is assumed as a standard uninformative Bayesian prior. The remaining hyperparameters are estimated empirically, with the derivation and solution found in the original reference (31). The estimated batch effects and can then be used to adjust the standardized data to an empirical Bayes batch-adjusted final output
In our modified version of this method (COCONUT), all of the above are performed according to the original method without modification. However, it is applied to only the healthy/control patients in each data set (Y is a matrix of only healthy patient samples). The estimated parameters α̂, β̂, σ̂, δ*, and γ* are all taken and applied directly to a matrix D that consists only of diseased patient sample (which must be ordered in the same manner as Y)
We can thus obtain a batch-corrected version of diseased samples D*, which corrects for the differences between healthy controls but does not change each submatrix Di with respect to each Yi.
Global ROCs
We used COCONUT conormalization to test (i) all discovery cohorts and (ii) all validation cohorts, even those containing only bacterial or only viral illness. We did this separately for the PBMC and whole-blood data, for reasons described previously. After conormalization, the distributions for the individual cohorts were plotted together to allow for direct comparison. For each plot, we show (i) the distribution of scores for each data set, (ii) the normalized gene expression for each gene within the diagnostic test, and (iii) the housekeeping genes that are expected to show no difference between classes based on meta-analysis. The healthy patients have been removed from these plots. However, to show that the distributions of genes between healthy and diseased patients within cohorts do not change after COCONUT conormalization, we have also shown plots with both patient types with both target genes and housekeeping genes (fig. S8). Genes with minimal effect size and minimal variance in meta-analysis were selected as housekeeping genes.
For each comparison, a single global ROC AUC was calculated, and a single threshold was set to allow for an estimate of the real-world diagnostic performance of the tests. Thresholds for the cutoffs for bacterial versus viral infection were set to approximate a sensitivity of 90% for bacterial infection, because a bacterial infection false-negative (the recommendation not to give antibiotics when antibiotics are needed) can be devastating.
Integrated antibiotics decision model
The SMS can discriminate between patients with severe acute infections and those with inflammation from other sources, but it cannot distinguish between types of infection (fig. S1). We thus tested an IADM, in which the 11-gene SMS is applied, followed by the 7-gene bacterial/viral metascore. This model thus identifies (i) whether a patient has an infection and, if so, (ii) what type of infection is present (bacterial or viral). We were unable to identify enough validation cohorts with patients with noninfected inflammation that also included healthy controls; thus, in constructing the global ROCs, we used both discovery and validation cohorts. Using COCONUT conormalization, we set global thresholds across all included cohorts, and these were applied to each individual data set to test the ability of the IADM to correctly distinguish patients with noninfectious inflammation, bacterial infection, and viral infection. Healthy patients were not included as a diagnostic class because they were used in the conormalization procedure. The IADM was also applied separately to all cohorts that had no healthy controls but that included (i) noninfected SIRS patients and (ii) patients with both bacterial and viral infections.
Because the PPV and NPV are dependent on prevalence and the prevalence of infections in the data used here does not match the prevalence of infections in a hospital setting, we calculated PPV and NPV curves on the basis of the sensitivity and specificity for bacterial infections attained with the IADM. Formally, NPV = specificity × (1 − prevalence)/((1 − sensitivity) × prevalence + specificity × (1 − prevalence)); PPV = sensitivity × prevalence/(sensitivity × prevalence + (1 − specificity) × (1 − prevalence)).
NanoString validation
We tested 96 samples from independent patients (those never profiled via microarray) from the GPSSSI trials (18–22) using a targeted NanoString (56) digital multiplex gene quantitation assay. The 18 genes were re-normalized to housekeeping genes (FRAS1 and LRRC17). The SMS and bacterial/viral metascore genes were both assayed, and the diagnostic performance of the IADM was calculated.
Data and source code availability
All analyses were conducted in the R statistical computing language (version 3.1.1). Code to recreate the multicohort meta-analysis, the COCONUT R package source code, and the COCONUT-normalized data used here have been deposited and are available at http://khatrilab.stanford.edu/sepsis.
Supplementary Material
Table S4. NanoString data.
Fig. S1. The SMS and pathogen type.
Fig. S2. Study schematic.
Fig. S3. Forest plots of the seven-gene set.
Fig. S4. Summary ROC forest plots for discovery data.
Fig. S5. Summary ROC forest plots for direct validation data.
Fig. S6. Bacterial/viral metascore ROC in GSE53166.
Fig. S7. Schematic of COCONUT conormalization.
Fig. S8. COCONUT conormalization of whole-blood discovery data sets.
Fig. S9. Bacterial/viral score in global ROC of non-conormalized whole-blood discovery data sets.
Fig. S10. Bacterial/viral score in global ROC of COCONUT-conormalized whole-blood discovery data sets.
Fig. S11. Bacterial/viral score in global ROC of non-conormalized whole-blood validation data sets.
Fig. S12. Bacterial/viral score in global ROC of non-conormalized PBMC validation data sets.
Fig. S13. Bacterial/viral score in global ROC of COCONUT-conormalized PBMC validation data sets.
Fig. S14. The effects of age on SMS in COCONUT-conormalized data.
Fig. S15. SMS across all COCONUT-conormalized whole-blood data (both discovery and validation).
Fig. S16. IADM across COCONUT-conormalized public gene expression data including healthy controls.
Fig. S17. NPV and PPV for the IADM.
Fig. S18. GSE63990, adults with ARIs.
Table S1. Significant gene list.
Table S2. Test characteristics of the bacterial/viral metascore in direct validation data sets.
Table S3. Data sets with noninfected inflammatory conditions used to test the IADM.
Table 2.
Validation data sets that matched inclusion criteria and have a single known pathogen type (viral or bacterial).
| Accession | Author | Tissue | Platform | Demographic | Specific pathogens | Number healthy | Number bacterial | Number viral |
|---|---|---|---|---|---|---|---|---|
| E-MEXP-3567 | Irwin | Whole blood | GPL96 | Malawian children with bacterial meningitis or pneumonia | S. pneumoniae, Neisseria meningitidis, or Haemophilus influenzae | 3 | 12 | 0 |
| GSE11755 | Emonts | Whole blood | GPL570 | Children in PICU with meningococcal sepsis | N. meningitidis | 3 | 6 | 0 |
| GSE13015 | Pankla | Whole blood | GPL6106 | Adults with bacterial sepsis | Burkholderia pseudomallei and others | 10 | 45 | 0 |
| GPL6947 | 10 | 15 | 0 | |||||
| GSE22098 | Berry | Whole blood | GPL6947 | Children with Gram-positive infections | Staphylococcus and Streptococcus | 81 | 52 | 0 |
| GSE28750 | Sutherland | Whole blood | GPL570 | Adults with community-acquired bacterial sepsis | Multiple bacteria | 20 | 10 | 0 |
| GSE29161 | Thuny | Whole blood | GPL6480 | Adults with native valve-infected endocarditis | Staphylococcus and Streptococcus | 5 | 5 | 0 |
| GSE33341 | Ahn | Whole blood | GPl571 | Adults with septic bloodstream infections | S. aureus or E. coli | 43 | 51 | 0 |
| GSE40586 | Lill | Whole blood | GPL6244 | Bacterial meningitis | Multiple bacteria | 18 | 21 | 0 |
| GSE42834 | Bloom | Whole blood | GPL10558 | Bacterial pneumonia | Unknown | 118 | 19 | 0 |
| GSE57065 | Cazalis | Whole blood | GPL570 | Adults with bacterial septic shock | Multiple bacteria | 25 | 82 | 0 |
| GSE69528 | Conejero | Whole blood | GPL10558 | Adults with bacterial sepsis | B. pseudomallei and others | 55 | 83 | 0 |
| E-MTAB-3162 | van de Weg | Whole blood | GPL570 | Indonesian patients >14 years old with uncomplicated and severe dengue | Dengue | 15 | 0 | 30 |
| GSE17156 | Zaas | Whole blood | GPL571 | Volunteers with viral challenge peak symptoms | Influenza, RSV, rhinovirus | 56 | 0 | 27 |
| GSE21802 | Bermejo-Martin | Whole blood | GPL6102 | Adults with septic influenza | Influenza (H1N1) | 4 | 0 | 12 |
| GSE27131 | Berdal | Whole blood | GPL6244 | Adults with septic influenza with mechanical ventilation | Influenza (H1N1) | 7 | 0 | 7 |
| GSE38900 | Mejias | Whole blood | GPL10558 | Children with acute LRTI | RSV | 8 | 0 | 28 |
| GPL6884 | Influenza, RSV, rhinovirus | 31 | 0 | 153 | ||||
| GSE51808 | Kwissa | Whole blood | GPL13158 | Children and adults with uncomplicated dengue and DHF | Dengue | 9 | 0 | 28 |
| GSE68310 | Zhai | Whole blood | GPL10558 | Adults with ARIs | Mostly influenza and rhinovirus | 243 | 0 | 211 |
| GSE16129 | Ardura | PBMC | GPL6106 | Children with invasive staph infections | S. aureus | 9 | 9 | 0 |
| GPL96 | 10 | 46 | 0 | |||||
| GSE23140 | Liu | PBMC | GPL6254 | Children with acute otitis media | S. pneumoniae | 4 | 4 | 0 |
| GSE34205 | Ioannidis | PBMC | GPL570 | Infants and children with ARIs | Influenza, RSV | 22 | 0 | 79 |
| GSE38246 | Popper | PBMC | GPL15615 | Nicaraguan children with un-complicated dengue, DHF, and DSS | Dengue | 8 | 0 | 95 |
| GSE69606 | Brand | PBMC | GPL570 | Children with mild-to-severe RSV | RSV | 17 | 0 | 26 |
DHF, dengue hemorrhagic fever; DSS, dengue shock syndrome.
Acknowledgments
We thank the patients who contributed clinical samples to the studies herein and the researchers who gathered, analyzed, and shared their data (see Supplementary Acknowledgments). We are sure that the push for open data will substantially improve translational medicine.
Funding: T.E.S. was funded by a Stanford Child Health Research Institute Young Investigator award (through the Institute for Immunity, Transplantation and Infection) and the Society of University Surgeons. P.K. was funded by the Bill & Melinda Gates Foundation and National Institute of Allergy and Infectious Diseases grants 1U19AI109662, U19AI057229, U54I117925, and U01AI089859.
Footnotes
Author contributions: T.E.S. and P.K. were in charge of the study conception and design; H.R.W. contributed to the materials; T.E.S. performed the experiments; T.E.S. and P.K. drafted the manuscript; and T.E.S., H.R.W., and P.K. critically revised the manuscript.
Competing interests: The seven-gene set and the IADM have been disclosed for possible patent protection to the Stanford Office of Technology and Licensing by T.E.S. and P.K. T.E.S. also serves as a scientific advisor to Multerra Bio, which had no role in this manuscript. H.R.W. declares that he has no competing interests.
Data and materials availability: The post–COCONUT-normalized data for this study have been deposited at khatrilab.stanford.edu/sepsis. Original MIAME (minimum information about a microarray experiment)–compliant microarray data are all available under their respective accession numbers in National Center for Biotechnology Information GEO or EBI ArrayExpress. COCONUT is available as an R package on CRAN (Comprehensive R Archive Network).
REFERENCES AND NOTES
- 1.Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP, Artigas A, Schorr C, Levy MM. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: Results from a guideline-based performance improvement program. Crit Care Med. 2014;42:1749–1755. doi: 10.1097/CCM.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 2.Fridkin S, Baggs J, Fagan R, Magill S, Pollack LA, Malpiedi P, Slayton R, Khader K, Rubin MA, Jones M, Samore MH, Dumyati G, Dodds-Ashley E, Meek J, Yousey-Hindes K, Jernigan J, Shehab N, Herrera R, McDonald CL, Schneider A, Srinivasan A Centers for Disease Control and Prevention (CDC) Vital signs: Improving antibiotic use among hospitalized patients. Morb Mortal Wkly Rep. 2014;63:194–200. [PMC free article] [PubMed] [Google Scholar]
- 3.Grijalva CG, Nuorti JP, Griffin MR. Antibiotic prescription rates for acute respiratory tract infections in US ambulatory settings. JAMA. 2009;302:758–766. doi: 10.1001/jama.2009.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews J. 9th International Conference on Typhoid and Invasive NTS Disease; Bali, Indonesia. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Strategy and Action Plan for Combating Antibiotic Resistant Bacteria. Nova Publishers; New York: 2015. [Google Scholar]
- 6.Liesenfeld O, Lehman L, Hunfeld K-P, Kost G. Molecular diagnosis of sepsis: New aspects and recent developments. Eur J Microbiol Immunol. 2014;4:1–25. doi: 10.1556/EuJMI.4.2014.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course–based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015;7:287ra271. doi: 10.1126/scitranslmed.aaa5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweeney TE, Khatri P. Comprehensive validation of the FAIM3:PLAC8 ratio in time-matched public gene expression aata. Am J Respir Crit Care Med. 2015;192:1260–1261. doi: 10.1164/rccm.201507-1321LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McHugh L, Seldon TA, Brandon RA, Kirk JT, Rapisarda A, Sutherland AJ, Presneill JJ, Venter DJ, Lipman J, Thomas MR, Klein Klouwenberg PMC, van Vught L, Scicluna B, Bonten M, Cremer OL, Schultz MJ, van der Poll T, Yager TD, Brandon RB. A molecular host response assay to discriminate between sepsis and infection-negative systemic inflammation in critically ill patients: Discovery and validation in independent cohorts. PLOS Med. 2015;12:e1001916. doi: 10.1371/journal.pmed.1001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scicluna BP, Klein Klouwenberg PMC, van Vught LA, Wiewel MA, Ong DSY, Zwinderman AH, Franitza M, Toliat MR, Nürnberg P, Hoogendijk AJ, Horn J, Cremer OL, Schultz MJ, Bonten MJ, van der Poll T. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am J Respir Crit Care Med. 2015;192:826–835. doi: 10.1164/rccm.201502-0355OC. [DOI] [PubMed] [Google Scholar]
- 11.Hu X, Yu J, Crosby SD, Storch GA. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc Natl Acad Sci USA. 2013;110:12792–12797. doi: 10.1073/pnas.1302968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaas AK, Burke T, Chen M, McClain M, Nicholson B, Veldman T, Tsalik EL, Fowler V, Rivers EP, Otero R, Kingsmore SF, Voora D, Lucas J, Hero AO, Carin L, Woods CW, Ginsburg GS. A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci Transl Med. 2013;5:203ra126. doi: 10.1126/scitranslmed.3006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suarez NM, Bunsow E, Falsey AR, Walsh EE, Mejias A, Ramilo O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J Infect Dis. 2015;212:213–222. doi: 10.1093/infdis/jiv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsalik EL, Henao R, Nichols M, Burke T, Ko ER, McClain MT, Hudson LL, Mazur A, Freeman DH, Veldman T, Langley RJ, Quackenbush EB, Glickman SW, Cairns CB, Jaehne AK, Rivers EP, Otero RM, Zaas AK, Kingsmore SF, Lucas J, Fowler VG, Jr, Carin L, Ginsburg GS, Woods CW. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci Transl Med. 2016;8:322ra311. doi: 10.1126/scitranslmed.aad6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andres-Terre M, McGuire HM, Pouliot Y, Bongen E, Sweeney TE, Tato CM, Khatri P. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity. 2015;43:1199–1211. doi: 10.1016/j.immuni.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khatri P, Roedder S, Kimura N, De Vusser K, Morgan AA, Gong Y, Fischbein MP, Robbins RC, Naesens M, Butte AJ, Sarwal MM. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210:2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweeney TE, Braviak L, Tato CM, Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: A multicohort analysis. Lancet Respir Med. 2016;4:213–224. doi: 10.1016/S2213-2600(16)00048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanley TP, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Odoms K, Barnes M, Sakthivel B, Aronow BJ, Wong HR. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med. 2007;13:495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Tagavilla MA, Odoms K, Dunsmore K, Barnes M, Aronow BJ Genomics of Pediatric SIRS. Septic Shock Investigators. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cvijanovich N, Shanley TP, Lin R, Allen GL, Thomas NJ, Checchia P, Anas N, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Wong HR Genomics Pediatric SIRS/Septic Shock Investigators. Validating the genomic signature of pediatric septic shock. Physiol Genomics. 2008;34:127–134. doi: 10.1152/physiolgenomics.00025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong HR, Cvijanovich N, Allen GL, Lin R, Anas N, Meyer K, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Shanley TP Genomics of Pediatic SIRS/Septc Shock Investigators. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med. 2009;37:1558–1566. doi: 10.1097/CCM.0b013e31819fcc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong HR, Cvijanovich NZ, Hall M, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Bigham MT, Sen A, Nowak J, Quasney M, Henricksen JW, Chopra A, Banschbach S, Beckman E, Harmon K, Lahni P, Shanley TP. Interleukin-27 is a novel candidate diagnostic biomarker for bacterial infection in critically ill children. Crit Care. 2012;16:R213. doi: 10.1186/cc11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, Chaussabel D. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parnell G, McLean A, Booth D, Huang S, Nalos M, Tang B. Aberrant cell cycle and apoptotic changes characterise severe influenza A infection – a meta-analysis of genomic signatures in circulating leukocytes. PLOS One. 2011;6:e17186. doi: 10.1371/journal.pone.0017186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parnell GP, McLean AS, Booth DR, Armstrong NJ, Nalos M, Huang SJ, Manak J, Tang W, Tam O-Y, Chan S, Tang BM. A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit Care. 2012;16:R157. doi: 10.1186/cc11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herberg JA, Kaforou M, Gormley S, Sumner ER, Patel S, Jones KDJ, Paulus S, Fink C, Martinon-Torres F, Montana G, Wright VJ, Levin M. Transcriptomic profiling in childhood H1N1/09 influenza reveals reduced expression of protein synthesis genes. J Infect Dis. 2013;208:1664–1668. doi: 10.1093/infdis/jit348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popper SJ, Watson VE, Shimizu C, Kanegaye JT, Burns JC, Relman DA. Gene transcript abundance profiles distinguish Kawasaki disease from adenovirus infection. J Infect Dis. 2009;200:657–666. doi: 10.1086/603538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CL, Dickinson P, Forster T, Craigon M, Ross A, Khondoker MR, France R, Ivens A, Lynn DJ, Orme J, Jackson A, Lacaze P, Flanagan KL, Stenson BJ, Ghazal P. Identification of a human neonatal immune-metabolic network associated with bacterial infection. Nat Commun. 2014;5:4649. doi: 10.1038/ncomms5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almansa R, Socias L, Sanchez-Garcia M, Martín-Loeches I, del Olmo M, Andaluz-Ojeda D, Bobillo F, Rico L, Herrero A, Roig V, San-Jose CA, Rosich S, Barbado J, Disdier C, de Lejarazu RO, Gallegos MC, Fernandez V, Bermejo-Martin JF. Critical COPD respiratory illness is linked to increased transcriptomic activity of neutrophil proteases genes. BMC Res Notes. 2012;5:401. doi: 10.1186/1756-0500-5-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MN, Ye C, Villani A-C, Raj T, Li W, Eisenhaure TM, Imboywa SH, Chipendo PI, Ran FA, Slowikowski K, Ward LD, Raddassi K, McCabe C, Lee MH, Frohlich IY, Hafler DA, Kellis M, Raychaudhuri S, Zhang F, Stranger BE, Benoist CO, De Jager PL, Regev A, Hacohen N. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 32.Zhai Y, Franco LM, Atmar RL, Quarles JM, Arden N, Bucasas KL, Wells JM, Niño D, Wang X, Zapata GE, Shaw CA, Belmont JW, Couch RB. Host transcriptional response to influenza and other acute respiratory viral infections – a prospective cohort study. PLOS Pathog. 2015;11:e1004869. doi: 10.1371/journal.ppat.1004869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwissa M, Nakaya HI, Onlamoon N, Wrammert J, Villinger F, Perng GC, Yoksan S, Pattanapanyasat K, Chokephaibulkit K, Ahmed R, Pulendran B. Dengue virus infection induces expansion of a CD14+CD16+ monocyte population that stimulates plasmablast differentiation. Cell Host Microbe. 2014;16:115–127. doi: 10.1016/j.chom.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mejias A, Dimo B, Suarez NM, Garcia C, Suarez-Arrabal MC, Jartti T, Blankenship D, Jordan-Villegas A, Ardura MI, Xu Z, Banchereau J, Chaussabel D, Ramilo O. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLOS Med. 2013;10:e1001549. doi: 10.1371/journal.pmed.1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berdal J-E, Mollnes TE, Wæhre T, Olstad OK, Halvorsen B, Ueland T, Laake JH, Furuseth MT, Maagaard A, Kjekshus H, Aukrust P, Jonassen CM. Excessive innate immune response and mutant D222G/N in severe A (H1N1) pandemic influenza. J Infect. 2011;63:308–316. doi: 10.1016/j.jinf.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Bermejo-Martin JF, Martin-Loeches I, Rello J, Antón A, Almansa R, Xu L, Lopez-Campos G, Pumarola T, Ran L, Ramirez P, Banner D, Ng DC, Socias L, Loza A, Andaluz D, Maravi E, Gómez-Sánchez MJ, Gordón M, Gallegos MC, Fernandez V, Aldunate S, León C, Merino P, Blanco J, Martin-Sanchez F, Rico L, Varillas D, Iglesias V, Marcos MÁ, Gandía F, Bobillo F, Nogueira B, Rojo S, Resino S, Castro C, Ortiz de Lejarazu R, Kelvin D. Host adaptive immunity deficiency in severe pandemic influenza. Crit Care. 2010;14:R167. doi: 10.1186/cc9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaas AK, Chen M, Varkey J, Veldman T, Hero AO, III, Lucas J, Huang Y, Turner R, Gilbert A, Lambkin-Williams R, Øien NC, Nicholson B, Kingsmore S, Carin L, Woods CW, Ginsburg GS. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Weg CAM, van den Ham H-J, Bijl MA, Anfasa F, Zaaraoui-Boutahar F, Dewi BE, Nainggolan L, van IJcken WFJ, Osterhaus ADME, Martina BE, van Gorp ECM, Andeweg AC. Time since onset of disease and individual clinical markers associate with transcriptional changes in uncomplicated dengue. PLOS Negl Trop Dis. 2015;9:e0003522. doi: 10.1371/journal.pntd.0003522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conejero L, Potempa K, Graham CM, Spink N, Blankley S, Salguero FJ, Pankla-Sranujit R, Khaenam P, Banchereau JF, Pascual V, Chaussabel D, Lertmemongkolchai G, O’Garra A, Bancroft GJ. The blood transcriptome of experimental melioidosis reflects disease severity and shows considerable similarity with the human disease. J Immunol. 2015;195:3248–3261. doi: 10.4049/jimmunol.1500641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cazalis M-A, Lepape A, Venet F, Frager F, Mougin B, Vallin H, Paye M, Pachot A, Monneret G. Early and dynamic changes in gene expression in septic shock patients: A genome-wide approach. Intensive Care Med Exp. 2014;2:20. doi: 10.1186/s40635-014-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lill M, Kõks S, Soomets U, Schalkwyk LC, Fernandes C, Lutsar I, Taba P. Peripheral blood RNA gene expression profiling in patients with bacterial meningitis. Front Neurosci. 2013;7:33. doi: 10.3389/fnins.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn SH, Tsalik EL, Cyr DD, Zhang Y, van Velkinburgh JC, Langley RJ, Glickman SW, Cairns CB, Zaas AK, Rivers EP, Otero RM, Veldman T, Kingsmore SF, Lucas J, Woods CW, Ginsburg GS, Fowler VG., Jr Gene expression-based classifiers identify Staphylococcus aureus infection in mice and humans. PLOS One. 2013;8:e48979. doi: 10.1371/journal.pone.0048979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thuny F, Textoris J, Amara AB, Filali AE, Capo C, Habib G, Raoult D, Mege J-L. The gene expression analysis of blood reveals S100A11 and AQP9 as potential biomarkers of infective endocarditis. PLOS One. 2012;7:e31490. doi: 10.1371/journal.pone.0031490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutherland A, Thomas M, Brandon RA, Brandon RB, Lipman J, Tang B, McLean A, Pascoe R, Price G, Nguyen T, Stone G, Venter D. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit Care. 2011;15:R149. doi: 10.1186/cc10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berry MPR, Graham CM, McNab FW, Xu Z, Bloch SAA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O’Garra A. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pankla R, Buddhisa S, Berry M, Blankenship DM, Bancroft GJ, Banchereau J, Lertmemongkolchai G, Chaussabel D. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol. 2009;10:R127. doi: 10.1186/gb-2009-10-11-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irwin AD, Marriage F, Mankhambo LA, Jeffers G, Kolamunnage-Dona R, Guiver M, Denis B, Molyneux EM, Molyneux ME, Day PJ, Carrol ED. Novel biomarker combination improves the diagnosis of serious bacterial infections in Malawian children. BMC Med Genomics. 2012;5:13. doi: 10.1186/1755-8794-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloom CI, Graham CM, Berry MPR, Rozakeas F, Redford PS, Wang Y, Xu Z, Wilkinson KA, Wilkinson RJ, Kendrick Y, Devouassoux G, Ferry T, Miyara M, Bouvry D, Valeyre D, Dominique V, Gorochov G, Blankenship D, Saadatian M, Vanhems P, Beynon H, Vancheeswaran R, Wickremasinghe M, Chaussabel D, Banchereau J, Pascual V, Ho L-p, Lipman M, O’Garra A. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLOS One. 2013;8:e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emonts M. thesis. Erasmus University; Rotterdam, Rotterdam, Netherlands: 2008. Polymorphisms in Immune Response Genes in Infectious Diseases and Autoimmune Diseases. [Google Scholar]
- 50.Ardura MI, Banchereau R, Mejias A, Di Pucchio T, Glaser C, Allantaz F, Pascual V, Banchereau J, Chaussabel D, Ramilo O. Enhanced monocyte response and decreased central memory T cells in children with invasive Staphylococcus aureus infections. PLOS One. 2009;4:e5446. doi: 10.1371/journal.pone.0005446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu K, Chen L, Kaur R, Pichichero M. Transcriptome signature in young children with acute otitis media due toStreptococcus pneumoniae. Microbes Infect. 2012;14:600–609. doi: 10.1016/j.micinf.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ioannidis I, McNally B, Willette M, Peeples ME, Chaussabel D, Durbin JE, Ramilo O, Mejias A, Flaño E. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J Virol. 2012;86:5422–5436. doi: 10.1128/JVI.06757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Popper SJ, Gordon A, Liu M, Balmaseda A, Harris E, Relman DA. Temporal dynamics of the transcriptional response to dengue virus infection in Nicaraguan children. PLOS Negl Trop Dis. 2012;6:e1966. doi: 10.1371/journal.pntd.0001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brand HK, Ahout IM, de Ridder D, van Diepen A, Li Y, Zaalberg M, Andeweg A, Roeleveld N, de Groot R, Warris A, Hermans PWM, Ferwerda G, Staal FJ. Olfactomedin 4 serves as a marker for disease severity in pediatric Respiratory Syncytial Virus (RSV) infection. PLOS One. 2015;10:e0131927. doi: 10.1371/journal.pone.0131927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wacker C, Prkno A, Brunkhorst FM, Schlattmann P. Procalcitonin as a diagnostic marker for sepsis: A systematic review and meta-analysis. Lancet Infect Dis. 2013;13:426–435. doi: 10.1016/S1473-3099(12)70323-7. [DOI] [PubMed] [Google Scholar]
- 56.Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol. 2011;Chapter 25(Unit25B.10) doi: 10.1002/0471142727.mb25b10s94. [DOI] [PubMed] [Google Scholar]
- 57.Tsalik EL, Li Y, Hudson LL, Chu VH, Himmel T, Limkakeng AT, Katz JN, Glickman SW, McClain MT, Welty-Wolf KE, Fowler VG, Ginsburg GS, Woods CW, Reed SD. Potential cost-effectiveness of early identification of hospital-acquired infection in critically Ill patients. Ann Am Thorac Soc. 2016;13:401–413. doi: 10.1513/AnnalsATS.201504-205OC. [DOI] [PubMed] [Google Scholar]
- 58.Gilbert DN. Procalcitonin as a biomarker in respiratory tract infection. Clin Infect Dis. 2011;52(suppl 4):S346–S350. doi: 10.1093/cid/cir050. [DOI] [PubMed] [Google Scholar]
- 59.Oved K, Cohen A, Boico O, Navon R, Friedman T, Etshtein L, Kriger O, Bamberger E, Fonar Y, Yacobov R, Wolchinsky R, Denkberg G, Dotan Y, Hochberg A, Reiter Y, Grupper M, Srugo I, Feigin P, Gorfine M, Chistyakov I, Dagan R, Klein A, Potasman I, Eden E. A novel host-proteome signature for distinguishing between acute bacterial and viral infections. PLOS One. 2015;10:e0120012. doi: 10.1371/journal.pone.0120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valim C, Ahmad R, Lanaspa M, Tan Y, Acácio S, Gillette MA, Almendinger KD, Milner DA, Jr, Madrid L, Pellé K, Harezlak J, Silterra J, Alonso PL, Carr SA, Mesirov JP, Wirth DF, Wiegand RC, Bassat Q. Responses to bacteria, virus, and malaria distinguish the etiology of pediatric clinical pneumonia. Am J Respir Crit Care Med. 2016;193:448–459. doi: 10.1164/rccm.201506-1100OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tolfvenstam T, Lindblom A, Schreiber MJ, Ling L, Chow A, Ooi EE, Hibberd ML. Characterization of early host responses in adults with dengue disease. BMC Infect Dis. 2011;11:209. doi: 10.1186/1471-2334-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: The expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 63.Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kB-mediated inflammatory response. Nat Cell Biol. 2010;12:66–73. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu M, Granillo O, Wen R, Yang K, Dai X, Wang D, Zhang W. Negative regulation of lymphocyte activation by the adaptor protein LAX. J Immunol. 2005;174:5612–5619. doi: 10.4049/jimmunol.174.9.5612. [DOI] [PubMed] [Google Scholar]
- 65.Federzoni EA, Valk PJM, Torbett BE, Haferlach T, Löwenberg B, Fey MF, Tschan MP. PU.1 is linking the glycolytic enzyme HK3 in neutrophil differentiation and survival of APL cells. Blood. 2012;119:4963–4970. doi: 10.1182/blood-2011-09-378117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 67.Kester ADM, Buntinx F. Meta-analysis of ROC curves. Med Decis Making. 2000;20:430–439. doi: 10.1177/0272989X0002000407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. NanoString data.
Fig. S1. The SMS and pathogen type.
Fig. S2. Study schematic.
Fig. S3. Forest plots of the seven-gene set.
Fig. S4. Summary ROC forest plots for discovery data.
Fig. S5. Summary ROC forest plots for direct validation data.
Fig. S6. Bacterial/viral metascore ROC in GSE53166.
Fig. S7. Schematic of COCONUT conormalization.
Fig. S8. COCONUT conormalization of whole-blood discovery data sets.
Fig. S9. Bacterial/viral score in global ROC of non-conormalized whole-blood discovery data sets.
Fig. S10. Bacterial/viral score in global ROC of COCONUT-conormalized whole-blood discovery data sets.
Fig. S11. Bacterial/viral score in global ROC of non-conormalized whole-blood validation data sets.
Fig. S12. Bacterial/viral score in global ROC of non-conormalized PBMC validation data sets.
Fig. S13. Bacterial/viral score in global ROC of COCONUT-conormalized PBMC validation data sets.
Fig. S14. The effects of age on SMS in COCONUT-conormalized data.
Fig. S15. SMS across all COCONUT-conormalized whole-blood data (both discovery and validation).
Fig. S16. IADM across COCONUT-conormalized public gene expression data including healthy controls.
Fig. S17. NPV and PPV for the IADM.
Fig. S18. GSE63990, adults with ARIs.
Table S1. Significant gene list.
Table S2. Test characteristics of the bacterial/viral metascore in direct validation data sets.
Table S3. Data sets with noninfected inflammatory conditions used to test the IADM.