Abstract
The baker’s yeast Saccharomyces cerevisiae has been extensively explored for our understanding of fundamental cell biology processes highly conserved in the eukaryotic kingdom. In this context, they have proven invaluable in the study of complex mechanisms such as those involved in a variety of human disorders. Here, we first provide a brief historical perspective on the emergence of yeast as an experimental model and on how the field evolved to exploit the potential of the model for tackling the intricacies of various human diseases. In particular, we focus on existing yeast models of the molecular underpinnings of Parkinson’s disease (PD), focusing primarily on the central role of protein quality control systems. Finally, we compile and discuss the major discoveries derived from these studies, highlighting their far-reaching impact on the elucidation of PD-associated mechanisms as well as in the identification of candidate therapeutic targets and compounds with therapeutic potential.
Keywords: protein misfolding, neurodegeneration, alpha-synuclein, Parkinson’s disease, synucleinopathies
INTRODUCTION
The unicellular eukaryote Saccharomyces cerevisiae has been extensively used as an industrial microorganism. The first records indicating its use in fermentation processes to produce alcoholic beverages and to leaven bread date back to ancient Egypt, over 5,000 years ago 1,2. Ever since, S. cerevisiae has been used for making bread, being therefore also frequently referred as baker’s yeast.
The first uses of S. cerevisiae as an experimental model organism date back to the mid-thirties of the 20th century 3, and its consequent establishment as a robust model system in diverse areas of biology was largely fueled by its unique features. They include short generation time, easy handling which is further simplified by its nonpathogenic nature, inexpensive culture conditions, and, most importantly, its amenability for genetic manipulation. Being very versatile for biological and genetic studies, these attributes placed yeast at the forefront for the development of countless genetic tools to address major biological issues. Hence, for almost a century, S. cerevisiae has served as a remarkable experimental model for several seminal discoveries in biology, revealing important aspects of microbiology and biochemistry. For example, studies using yeast have contributed to the elucidation of fundamental cellular mechanisms involved in DNA replication, recombination and repair 4, in RNA metabolism 5, and in cell division and cell cycle progression 6.
The discovery of the high-degree of evolutionary conservation of disease genes and of fundamental biological processes among eukaryotes, combined with the power of yeast genetics, has brought S. cerevisiae from the baker to the bedside, as a model organism with an unprecedented potential to decipher the intricacies of devastating human pathologies such as Parkinson’s disease (PD), as well as to help in the identification of molecular targets and lead molecules with therapeutic potential.
In this review, we summarize the impact of yeast models on the current knowledge and understanding of the molecular underpinnings of PD. We first discuss the most relevant findings in the yeast field and the extent to which they have paved the way for the use of S. cerevisiae in biomedical research. We then briefly review the main aspects of PD, emphasizing the molecular players and pathways governing disease pathology. Finally, we cover the most important yeast models generated thus far, and discuss their contribution to the elucidation of PD-related mechanisms, as well as to the identification of molecular targets and compounds with therapeutic potential.
THE POWER OF YEAST GENETICS
The peculiar life cycle of S. cerevisiae constitutes, in itself, an invitation for performing genetic studies. In the wild, it can be found in both haploid and diploid forms that reproduce vegetatively, by budding. In nutrient-poor environments, a condition easily mimicked experimentally, diploid cells undergo meiosis and sporulate, yielding a progeny of four haploid cells. Hence, under controlled laboratory conditions, sporulation of a particular diploid cell allows the generation of different combinations of genotypes with desired genetic traits. Additionally, the life cycle of budding yeast also greatly simplifies the study of lethal mutations in heterozygous diploids as well as recessive mutations in haploid cells 7,8.
Yeast research has definitely won a place in history after the demonstration that yeast strains with a mutation in LEU2 locus, therefore unable to grow in media depleted of the amino acid leucine, can be transformed with a chimeric ColE1 plasmid encoding the wild type (WT) yeast LEU2 gene, and that this sequence can integrate into the yeast chromosome restoring leucine prototrophy 9. The discovery of the amenability of yeast cells for transformation opened new avenues for manipulation of yeast genome, allowing insertion or deletion of genes to generate recombinant strains. The high efficiency of the transformation process, aided by a very effective homologous recombination system, has provided yeast geneticists a tremendous flexibility in experimental design, which is currently incremented by the availability of a large collection of recombination-based Gateway vectors 7,10,11,12.
S. cerevisiae was also the host organism for the development of pioneering approaches to investigate the interaction between biomolecules. Taking advantage of the bi-modular nature of the yeast transcription factor Gal4, researchers generated a novel genetic system to study protein-protein interactions in which two known proteins are separately fused to the DNA-binding and transcriptional activation domains of Gal4 13. The principle of the methodology relies on the premise that the interaction between proteins reconstitutes a functional Gal4, which in turn activates expression of reporter genes. After its original description, a number of “variations on the theme” has been described to allow the study of DNA-protein (one-hybrid), RNA-protein (RNA-based three-hybrid) and small molecule-protein interactions (ligand-based three-hybrid), as well as to identify mutations, peptides or small molecules that dissociate macromolecular interactions - the reverse n-hybrid systems 14. Additionally, a split-ubiquitin membrane-based two-hybrid assay (also referred as membrane yeast two-hybrid - MYTH) was designed to overcome the limitation of the original system on the assessment of protein interactions forced to occur within the nucleus, for membrane-embedded proteins 15,16. The fact that novel hits for a target protein, from both yeast and other organisms, can be identified in the screening of libraries, without any prior bias or knowledge of their identity, is the most powerful application of these techniques.
Another tool allowing the study of protein interactions in vivo is the bimolecular fluorescence complementation (BiFC). This method, originally developed in mammalian cells 17,18, has been efficiently used in yeast to visualize protein interactions with minimal perturbation of the normal cellular environment 19,20. It is based on the principle that two fragments of a fluorescent protein are each fused to target proteins. The reassembly of these non-fluorescent fragments into a fluorescent complex is mediated by the interaction between the target proteins, thereby constituting a powerful tool to resolve spatial and temporal aspects of many molecular interactions 21.
The genetic features highlighted have distinguished yeast as a versatile model organism. As such, S. cerevisiae was the first eukaryotic organism to have its genome fully sequenced 22. Thus, the completion of the yeast genome in 1996 represented a landmark achievement in the history of eukaryotic biology. This has been providing, in the course of the last two decades, a wealth of information allowing the development of several biological resources such as the yeast gene deletion strains 23, the tetracycline (tet)-repressible 24 and heat-inducible shutoff set of strains 25 to generate conditional mutants of essential genes, the GFP- 26 and TAP-tagged 27 collection of strains, collections of other protein tags, and collections engineered for protein overexpression 28,29,30. A compendium of existing tools and resources available for the yeast research community is provided by Tenreiro and Outeiro 31 and by Duina and coauthors 7.
As the most facile eukaryotic model organism for experimental biology, yeasts were placed at the forefront for the establishment of new research fields: (1) functional genomics, in particular transcriptomics (DNA microarrays and RNA-sequencing), proteomics, metabolomics, interactomics and locasomics (protein subcellular localization) 11, employing the resources detailed above in a high-throughput automated setup; and (2) systems biology, to scrutinize the huge amount of data generated towards building a comprehensive model of eukaryotic cell functioning 7,8,32. Compilations of genetic and biological data from these analyses, including information regarding predicted orthologues in humans, are easily accessible at the comprehensively annotated Saccharomyces Genome Database online resource (http://www.yeastgenome.org) 33 and other public databases 7,31,34.
The power of yeast genetics has fueled all the achievements discussed, rendering S. cerevisiae as the best-understood eukaryotic organism. A surprising, but delightful finding, emerging from the cumulative knowledge on yeast genomics and biology, was the unpredictable gene homology and functional conservation of key fundamental cellular processes between yeasts and higher eukaryotes 4,5,8.
Importantly, the S. cerevisiae genome encodes nearly 1000 genes which are members of orthologous gene families related to human disease, representing about 20% of the total yeast genes 35. The mammalian orthologues of most of these genes are functional in yeast and complement the respective yeast deletion mutant. In line with the evolutionary conservation of disease genes in eukaryotes, it has been extensively shown that several disease-associated cellular pathways are also highly conserved from yeast to humans 11,34,36, enabling the modeling of specific disease aspects in this model organism. Protein quality control systems 37, vesicular trafficking and secretion 38, autophagic pathways 39, the unfolded protein response 40, and mitochondrial biogenesis and metabolism 41 are among the conserved cellular mechanisms, allowing the study of fundamental mechanisms associated with neurodegenerative diseases, such as PD, in yeast cell models 11.
Modeling particular molecular aspects of human diseases in yeast models can be achieved using distinct strategies, depending on the presence or absence, of disease-genes orthologues in the yeast genome 42. If the genes of interest have yeast counterparts, a unique opportunity to directly study their function is offered, either through their deletion or overexpression. In a more physiological context, human wild type (WT) and mutant alleles of favorite genes can be heterologously expressed in the respective yeast mutant backgrounds, since they are capable of replacing the function of the endogenous yeast gene product. Otherwise, if the disease-associated genes do not have a yeast orthologue, a functional analysis can still be conveniently performed via heterologous expression in WT strains 31,43 to provide paradigms of their function on cellular physiology and metabolism. Insights into the function of most PD-associated genes have been obtained using the latter strategy. However, the study of endogenous yeast proteins has been also providing clues on the role of key PD players, as discussed in detail bellow.
Once comprehensively validated as reliable experimental model systems to recapitulate specific aspects of human diseases, “humanized” yeasts constitute powerful toolboxes for high-throughput screenings of genes that may constitute therapeutic targets, and as robust primary drug-screening platforms to filter for cytoprotective compounds 31,44.
Despite several singularities and idiosyncrasies of yeast cells, their simplicity can be turned from a limitation into an asset, by virtue of all the attributes mentioned above. Thus, S. cerevisiae is uniquely suited for the task of assisting our understating of the cellular mechanisms underlying human diseases as well as in the search for novel molecular targets and compounds prone for therapeutic intervention.
PARKINSON'S DISEASE AND THE KEY GENETIC PLAYERS
PD, described by James Parkinson in 1817 45 and then further refined by Jean-Martin Charcot 46, is one of the most common neurodegenerative disorders. Currently, it is estimated that there are 4 million diagnosed PD patients worldwide. However, it is estimated that 7 to 10 million people live with this devastating chronic disease (data from the Parkinson’s Disease Foundation). The typical motor symptoms of PD include tremor at rest, bradykinesia, stiffness, and postural instability 47. These symptoms are caused by the progressive degeneration of nigrostriatal dopaminergic neurons from the substantia nigra pars compacta of the brain. Nevertheless, PD is currently considered as a whole-brain disorder, affecting multiple brain areas and presenting a broad variety of symptoms 48. The histopathological hallmark of PD, and other synucleinopathies, is the appearance of proteinaceous intraneuronal cytoplasmic inclusions termed as Lewy bodies (LB) and Lewy neurites 49. These insoluble aggregates are predominantly composed of alpha-synuclein (aSyn) 49, a protein encoded by the SNCA gene, and are decorated with components of protein quality control systems such as molecular chaperones, and proteasomal and lysosomal subunits 50.
The SNCA gene was the first genetic locus to be associated with familial forms of PD, and was later also implicated in sporadic cases of the disease 51. It encodes aSyn, a protein whose function remains unclear, but that has been proposed to be linked to diverse functions ranging from transcriptional regulation 52,53,54,55,56,57, mitochondrial homeostasis 58 and vesicle trafficking 59,60,61, possibly regulating dopamine neurotransmission, synaptic function and synaptic plasticity 62,63,64,65.
Genetic alterations in the SNCA gene linked to PD include duplication or triplication of the SNCA locus 66,67,68, as well as missense mutations A30P 69, E46K 70, H50Q 71,72, G51D 73, A53T 51,74,75, and A53E 76, causing autosomal dominant forms of the disease. The precise effect of each of these mutations is unclear, but they seem to affect the interaction of aSyn with membranes 77,78,79,80,81,82, and to alter the propensity of the protein to aggregate, at least in vitro 81,83,84,85,86,87,88,89.
As a common genetic determinant of both idiopathic and inherited forms of PD, and the major component of LB, much of the work in the PD field converges on aSyn (also called PARK1). However, several genes linked to heritable, monogenic PD, have been described. These include the leucine-rich repeat kinase 2 LRRK2 (PARK8), the E3 ubiquitin-ligase Parkin (PARK2), the mitochondrial PTEN-induced putative kinase 1 Pink1 (PARK6), the oxidation-sensitive chaperone DJ-1 (PARK7), and the lysosomal ATPase ATP13A2 (PARK9) 90. Additionally, several genes are known to be associated with an increased risk of developing PD, such as the vacuolar protein sorting 35 homolog VPS35 (PARK17), the ubiquitin carboxyl-terminal esterase L1 UCH-L1 (PARK5), the translation initiation factor 4-gamma 1 EIF4G1 (PARK18), and beta-glucocerebrosidase (GBA) 90. A comprehensive list of PD genetic risk factors, including the six risk loci associated with proximal gene expression or DNA methylation recently identified in large-scale meta-analysis of genome-wide association studies, are available online at ‘PDGene’ (http://www.pdgene.org) 91,92.
Mutations in LRRK2, encoding a 2527-amino acid cytosolic kinase, are the most frequent genetic cause of PD 93,94. The role of this kinase has been associated with biological processes such as endocytosis of synaptic vesicles 95,96, autophagy 97, and neurite outgrowth 98. The recent discovery of LRRK2 interactions with members of the dynamin superfamily of large GTPases, by yeast two-hybrid analyses, implicates its function in the regulation of membrane dynamics relevant for endocytosis and mitochondrial morphology 99. In line with these findings, LRRK2 appears to modulate the cellular protection against oxidative insults in a mechanism that is dependent on endocytosis and mitochondrial function 100. LRRK2 was also shown to interact with aSyn 101,102, possibly mediating its phosphorylation on S129 103. The pathogenic mechanisms triggered by mutant versions of LRRK2 are still unclear 94. However, it is speculated that mutations may affect its interactions with other proteins 90, possibly also with aSyn.
Parkin homozygous mutations are the most frequent cause of juvenile PD. Parkin has been associated with various cellular pathways but special importance has been given to its role in mitochondrial quality control, where it participates in common pathways with Pink1 to regulate the formation of mitochondrial-derived vesicles and mitophagy 104,105,106,107,108,109,110. Indeed, the MVD pathway has been referred as the primary defense mechanism against mitochondrial damage. Mitophagy only plays a role once MVDs are overwhelmed. Parkin-assisted Pink1 translocation into mitochondria is associated with autophagy of damaged mitochondria 107,109, and was further supported by yeast studies on the modulation of mitochondrial degradation upon oxidative injury and chronological aging 111.
The DJ-1 protein, the most extensively studied member of the DJ-1 superfamily, is a multifunctional protein associated with numerous cellular functions including oxidative stress responses 112. The cellular mechanisms by which mutations in DJ-1 cause PD are still unclear, but DJ-1 may act as a redox-dependent chaperone preventing aSyn aggregation 113. DJ-1 overexpression confers protection against neurodegeneration in model organisms, in a mechanism that is dependent on protein-protein interactions between DJ-1 and disease proteins 114,115. This suggests that the direct interaction between DJ-1 and its targets constitutes the basis for the neuroprotective effect of DJ-1. Further insights into putative roles of members of the DJ-1 superfamily were obtained in yeast, where Hsp31-like chaperones may serve as regulators of autophagy 116.
Finally, it is important to stress that most of the genes associated with PD encode proteins whose functions appear to be related to mitochondrial function, membrane trafficking and protein quality control systems, underscoring the importance of these mechanisms in the pathophysiology of PD.
MODELLING PD IN YEAST
The identification of genes associated with Mendelian forms of PD enabled major leaps forward in our understanding of the molecular mechanisms involved in the disease. This also led to the development of various cell and animal models that are widely used. Among these, yeast models have proven extremely useful to dissect the basic molecular mechanisms associated with PD and other synucleinopathies (Table 1). These models are based either on the heterologous expression of the human genes, or by studying the function and pathological role of the yeast counterparts, when these are represented in the yeast genome (Fig. 1).
Table 1.
Yeast models of PD.
| Human gene | Human protein | Yeast homolog | Yeast model | References |
| SNCA | alpha-synuclein | __ | heterologous expression of human aSyn | 117,121 |
| PARK8 | LRRK2 | __ | heterologous expression of human LRRK2 | 135 |
| PARK7 | DJ-1 | HSP31, HSP32, HSP33 and HSP34 | functional analysis of the yeast homologs; co-expression of DJ-1 with aSyn; co-expression of DJ-1 with PD associated mutations | 114,116 |
| PARK2 | Parkin | __ | heterologous expression of human Parkin | 111,139 |
| PINK1 | Pink1 | __ | heterologous expression of human Pink1 | 111,139 |
| VPS35 | VPS35, or PARK17 | VPS35 | functional analysis of the yeast homolog | 144 |
| EIF4G1 | EIF4G1 | TIF4631 and TIF4632 | functional analysis of the yeast homologs | 144 |
| ATP13A2 | ATP13A2 | YPK9 | co-expression of Ypk9 with human aSyn; co-expression of Ypk9 with PD associated mutations | 127,146,147,191 |
Figure 1. FIGURE 1: Schematic representation of yeast PD models.
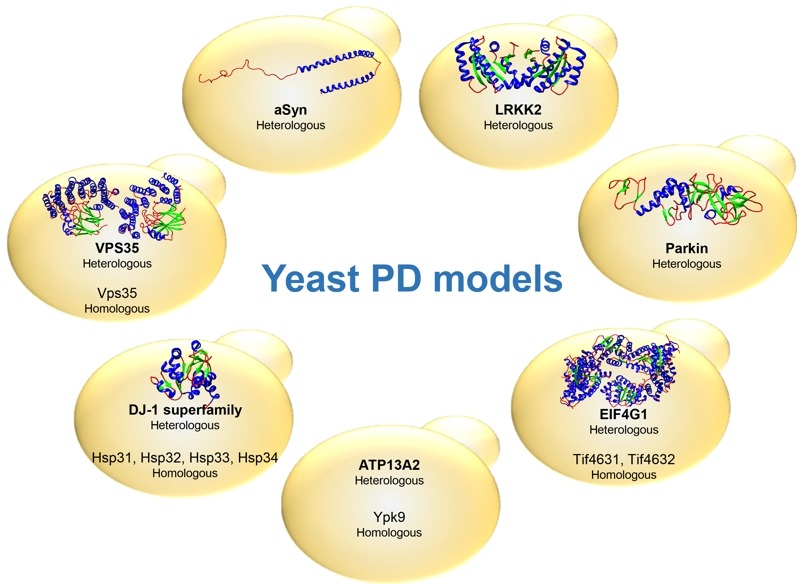
The PD-associated proteins are indicated as well as the type of expression (heterologous or homologous). The structure of the proteins is represented for aSyn, LRRK2, Parkin, EIF4G1, DJ-1 and VPS35. Protein Data Bank ID: 1XQ8, 2ZEJ, 4I1H, 2VSX, 4OQ4 and 2R17 respectively. The structure of ATP13A2 is still undetermined.
aSyn
The first yeast model of PD was based on the heterologous expression of human aSyn 117. Since then, many other studies used this approach to investigate the molecular mechanisms underlying aSyn toxicity and to identify novel compounds of therapeutic interest, as described below.
The expression of aSyn in yeast results in dose-dependent cytotoxicity 117, as observed in other model systems. This is also in line with the identification of familial forms of PD associated with duplications 67 and triplications 66 of the aSyn locus. In addition, expression of aSyn in yeast also resulted in the formation of intracellular inclusions. The nature of these inclusions was matter of debate, as it was observed that aSyn leads to the accumulation of vesicles 118,119,120, raising doubts on whether the inclusions observed were indeed aggregated aSyn. However, amyloid-like aggregates of aSyn seem to be also formed in yeast cells, as some inclusions are positively stained by thioflavin S 121 or thioflavin T 122. More recently, the formation of large oligomeric species of aSyn in yeast cells was also demonstrated using both sucrose gradients and size exclusion chromatography 123.
The various yeast models based on the heterologous expression of aSyn present different phenotypes, depending on the expression system, given that this affects the level of aSyn expression 117,124,125. This feature has been also explored according to the objectives of the studies.
The use of multicopy plasmids revealed that yeast cells reduce the average plasmid copy number in order to reduce aSyn expression and toxicity 117. To avoid this, insertions of the aSyn coding sequence in the yeast genome enabled more stable expression, and the levels of toxicity could be manipulated by varying the number of copies of the aSyn cDNA inserted in the genome 125,126. The use of a galactose-inducible promoter provided additional control for the synchronous induction of expression of aSyn, avoiding the negative pressure during routine cell manipulations.
Using these various expression systems, several genetic modifiers (enhancers and suppressors) of aSyn toxicity were identified in genetic screenings in yeast 124,125.
In other studies, yeast cells expressing different levels of aSyn, hence displaying different levels of cytotoxicity, revealed the involvement of multiple cellular pathways in the toxicity 117,119,125,126,127,128,129. In turn, this facilitated the identification of synergistic effects between different genetic suppressors of toxicity 126,127. Overall, these studies greatly expanded our understanding of the cellular pathways affected by aSyn. Among these, ER-to-Golgi trafficking appeared to be significantly disrupted 125. Also, mitochondrial stress was identified as an early signature of aSyn toxicity 126. Indeed, the aSyn expression system controlled by the yeast MET25 promoter 121 enabled the verification that aSyn cytotoxicity requires functional mitochondria 130. Moreover, mitochondria has been pointed out as the site of enhanced ROS production in response to Pmr1-dependent Ca2+ overload leading to cellular death in yeast, flies and worms aSyn models 131.
Other effects of aSyn overexpression in yeast are impairment of proteasome activity 117, accumulation of cytoplasmic lipid droplets 117, ER stress 125, activation of the heat-shock response 128, mitochondrial dysfunction 126, shorter chronological life span and induction of autophagy and mitophagy (mediated by Sir2) 132, impairment of endocytosis 117, ROS production and induction of apoptosis 126,133. Recently, it was shown that apoptosis is dependent on the translocation of Endonuclease G (EndoG) from the mitochondria to the nucleus, where it mediates DNA degradation 134.
LRRK2
The role of other genes associated with monogenic forms of PD has also been studied in yeast models. Namely, important insights into the function and the pathogenic mechanisms of LRRK2 mutations were obtained in yeast cells heterologously expressing human LRRK2 100,135. Higher levels of expression of the full-length LRRK2, based on a multicopy expression vector and on a galactose inducible promoter, resulted in the accumulation of insoluble protein but no alterations in the phenotype 100,135. However, with lower levels of LRRK2 expression, a phenotype of protection against oxidative stress is conferred by LRRK2 100. Interestingly, the protective effect of LRRK2 is lost when PD-associated mutants are used instead of the WT protein 100.
The effects of the overexpression of various functional domains of human LRRK2 were also analyzed in yeast 135. The GTPase activity of LRRK2 was found to be inversely correlated with cytotoxicity 135. This toxicity was correlated with defects in endocytic trafficking and autophagy 135. These results were further validated in mouse primary cortical neurons were the overexpression of full-length LRRK2 causes defects in both synaptic vesicle endocytosis and exocytosis 135. This study also revealed that aSyn and LRRK2 cause vesicular trafficking-associated toxicity through distinct pathways that, nevertheless, culminate in trafficking defects to the vacuole, the yeast counterpart of the lysosome 135.
DJ-1
In yeast, there are four homologous and highly conserved genes that belong to the DJ-1 superfamily: Hsp31, Hsp32, Hsp33 and Hsp34. Recently, it was found that these proteins are required for metabolic reprogramming triggered by glucose limitation, known in yeast as diauxic-shift 116. It was also found that these DJ-1 homologs contribute to target of rapamycin complex 1 (TORC1) regulation 116, a central player in diauxic-shift reprogramming and, importantly, in autophagy 136. Both TORC1 and autophagy are dysfunctional in several pathologies including PD 137,138. Thus, this study constitutes an example of how the functional analysis of yeast homologs provides important insight into the putative functions of human genes associated with disease.
In a separate study, the human DJ-1 gene was expressed in yeast cells using a multicopy vector 114. It was found that DJ-1 interacts with aSyn and that PD-associated mutations impair this interaction 114. Interestingly, it was found that DJ-1 and the yeast homologs attenuate aSyn toxicity in yeast 114. Thus, these findings suggest that the physical interaction between DJ-1 and aSyn might represent a neuroprotective mechanism that is disrupted by familial mutations in DJ-1, thereby contributing to PD 114.
Parkin and Pink1
Recently, human Parkin was expressed in yeast and, although the protein was found to be cytosolic, it was translocated to mitochondria under oxidative stress or aging, accelerating mitochondrial degradation 111. Moreover, Parkin promotes chronological longevity and oxidative stress resistance through a mitochondria-dependent pathway 111. In the same study, Pink1 was also expressed in yeast and was found to promote resistance to oxidative stress 111. However, co-expression of both proteins does not show a synergistic effect, suggesting the two proteins affect the same pathway independently 111.
Recently, the mechanism underlying Parkin activation by Pink1 was dissected in an elegant study where a yeast model was used 139. In particular, full activation of the E3 activity of Parkin E3, in response to mitochondrial damage, was found to occur in a two-step mechanism, involving the phosphorylation of both Parkin and ubiquitin, by Pink1 139.
VPS35 and EIF4G1
The first indications highlighting the importance of vesicle-trafficking genes as modulators of aSyn toxicity resulted from genetic screens performed in yeast. Deletion of VPS24, VPS28, VPS60 or SAC2 was found to increase aSyn toxicity 124. These genes are involved in protein sorting in the late Golgi, sorting to the prevacuolar endosomes, for protein sorting, and trafficking to the vacuole, respectively. In another screen, ENT3, involved in protein transport between the trans-Golgi network and the vacuole, was also identified as a suppressor of aSyn toxicity using a yeast model 140. Only more recently, next-generation sequencing and genome-wide association studies implicated mutations in VPS35 (PARK17) in familial cases of PD 141,142. Genome-wide association studies also allowed the identification of EIF4G1 (PARK18) as a novel autosomal dominant PD gene 143. These genes are known to function in regulating vacuolar transport (VPS35) and transcription/translation (EIF4G1), and are highly conserved from yeast to humans. Yeast has a VPS35 homolog, also called VPS35, and two EIF4G1 homologs, TIF4631 and TIF4632. These two genes were found to interact functionally and genetically, and to converge on aSyn, in a study that combined yeast, worms and transgenic mouse models 144. This study started with two independent genetic screens in strains deleted for VPS35 or TIF4631 and enabled the identification of synthetic sick or lethal genes. The results pointed to a common pathway associated with both genes: the retromer complex function, important in the regulation of recycling, sorting, and trafficking between the endosomal and Golgi network. The impairment of a functional retromer complex results in the accumulation of protein misfolding, thereby exacerbating the accumulation and toxicity of aSyn 144. In particular, it was found that overexpression of EIF4G1 (or the yeast homolog TIF4631) in cells lacking VPS35 was highly toxic. This toxicity was found to be due to the loss of retromer function, which could be restored by WT VPS35 but not by the PD-associated mutant D620N 144.
ATP13A2
Mutations in the gene encoding the lysosomal P-type ATPase ATP13A2 cause Kufor-Rakeb syndrome and early-onset PD 145. Yeast has an orthologue of ATP13A2, the YPK9 gene, which encodes a vacuolar transporter with a possible role in sequestering heavy metals 146. The understanding of role of ATP13A2 in PD and how missense mutations could lead to a loss-of-function of the protein was facilitated by studying the yeast homolog, followed by validation in other model systems. Namely, YPK9 was found to be a suppressor of aSyn toxicity in yeast 127. This protective function depends on the vacuolar localization and ATPase activity of Vps35 127, and is probably related to its role in homeostasis of manganese and other divalent heavy metal ions 146,147, which are recognized environmental risk factors for PD 148. Among the most common PD genetic players, ATP13A2 is the unique whose structure has not yet been unveiled.
aSYN AND PROTEIN QUALITY CONTROL SYSTEMS IN YEAST
Alterations in proteostasis occur in several types of disorders 149. When the accumulation of misfolded proteins surpasses the capacity of the cell to cope with the protein load, diverse quality control mechanisms are called to action, to actively sequester, refold, and/or degrade these proteins 150,151,152,153 (Fig. 2). These cellular protein quality control mechanisms are conserved from yeast to mammalian cells 37.
Figure 2. FIGURE 2: Quality control systems and aSyn aggregation.
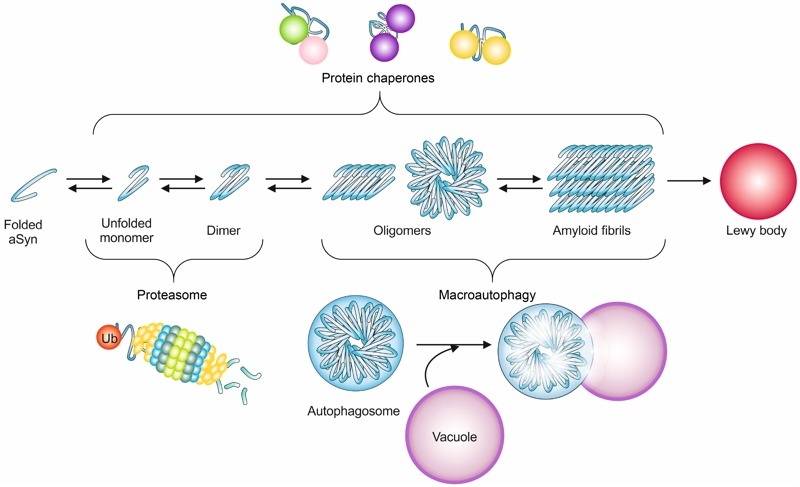
aSyn can misfold and form oligomeric species that fibrillate and deposit into larger aggregates, ultimately forming Lewy bodies. In healthy cells, the cellular quality control systems are able to maintain proteostasis, avoiding this cascade of events. The first steps of aggregation can be prevented or reversed by promoting the degradation of misfolded proteins by the ubiquitin-proteasome system, while the later ones are counteracted by degradation mediated by macroautophagy. Chaperones are thought to assist in the proper folding of aSyn at different stages of the aggregation process.
Strong evidence on the involvement of the quality control systems in neurodegeneration came from studies of familial forms of PD, as mutations in several genes playing a role in these pathways are intimately associated with the disease, as mention above.
Besides those players, several molecular chaperones were also found to have an important role in PD. Namely, the involvement of molecular chaperones in aSyn yeast toxicity was evidenced by the enhanced aSyn inclusion formation observed upon deletion of individual chaperones 154. Concomitantly, pharmacological activation of the heat shock response upon treatment with geldanamycin or overexpression of the chaperones Ssa3 155, Jem1 or Hsp90 140 protected yeast cells against aSyn-induced ROS and subsequent toxicity. These data elegantly recapitulated results obtained with neuronal cell lines 156, transgenic flies 157 and mice 158,159.
The protein Gip2, an activator of the heat-shock transcription factor Hsf1, was also identified as a multicopy suppressor of aSyn toxicity, by triggering a heat-shock response in yeast 128. A screening of a mouse brain-specific cDNA library identified the mouse chaperone RPS3A as a suppressor of aSyn WT and A53T toxicity in yeast 160. Moreover, co-expression of RPS3A delayed the formation of aSyn inclusions 160. Overexpression of DJ-1, which has protein chaperone-like activity, was also described as a negative regulator of aSyn dimerization 114.
Finally, the heat shock-induced protective mechanism may involve Hsp104 and its co-chaperones, which were described to relieve cells from ER-stress 161. It was shown that Hsp104 degraded aSyn in a concentration-dependent manner and decreased aSyn fibrillation in vitro 162,163,164. In agreement, Hsp104 antagonized aSyn aggregation and reduced dopaminergic degeneration in a rat model of PD 165. However, the studies in yeast are still scarce. It was shown that at endogenous levels the presence of Hsp104 had a deleterious effect on aSyn aggregation, and deletion of Hsp104 in yeast expressing WT or mutant aSyn resulted in lower oxidative stress, cytotoxicity, increased cell viability and rescue of endocytic defects 166. Nevertheless, another study reported that it is possible to reprogram Hsp104 to rescue aSyn proteotoxicity in yeast by mutating single residues 167. These potentiated Hsp104 variants enhanced aggregate dissolution, restored protein localization, suppressed proteotoxicity in yeast 167, and attenuate dopaminergic neurodegeneration in a C. elegans PD model 167.
Additional strong evidences of the relevance of the clearance pathways in PD arrive from yeast models. Namely, the expression of aSyn in yeast promotes proteasome impairment 117. This reduced proteasome activity was found to be the result of a deficient proteasome composition 168. Furthermore, the failure of the UPS (ubiquitin-proteasome system) enhances aSyn toxicity 169 170 and leads to the accumulation of inclusions 121.
Despite the clear involvement of proteasome dysfunction in PD, the degradation of aSyn inclusions is more dependent on autophagy than on proteasome function, at least in yeast 171. This is consistent with the proteasome being responsible for the degradation of soluble forms of aSyn, and suggests a complex cross-talk between the different proteolytic pathways involved in the degradation of aSyn 172 (Fig. 2).
Autophagy involves the formation of an autophagosomal vesicle that transports the misfolded and aggregated proteins to degradation, in the lysosome in higher eukaryotes, or in the vacuole in yeast. The first molecular insights into autophagy were learned from yeast 173, and as in mammalians, the process is regulated by the kinase “target of rapamycin” (TOR) pathway 174. Concomitantly Lst8, a TOR-interactor was identified as modulator of aSyn toxicity in yeast 128. Moreover, Ypk9, a vacuolar P-type ATPase that is the orthologue of ATP13A2, was identified as a suppressor of aSyn toxicity 128 and aggregation 127.
More recently, it was reported that deletion of the autophagy related genes ATG1 or ATG7 lead to impaired degradation of aSyn and increased toxicity 123. In agreement with the beneficial role of autophagy on aSyn toxicity, rapamycin treatment, which induces autophagy by inhibiting TOR, was reported to reduce aSyn aggregation 121. It also appears that toxic forms of aSyn lead to the impairment of autophagy and that inducing autophagy or increasing the autophagic flux is protective against aSyn toxicity in yeast 123,175.
Nevertheless, there is still controversy regarding the role of autophagy on aSyn toxicity. A study reported that rapamycin treatment increases aSyn toxicity in yeast 128. It was also shown that WT or A53T mutant aSyn were not able to enter the vacuole and promoted vacuolar fusion defects in yeast 154. Additionally, aSyn-mediated mitophagy, a specific degradation of mitochondria through autophagy, was reported to be deleterious in aged yeast cells 132. To intensify the discussion, aSyn Lewy body-like aggregates resisted degradation and impaired autophagy in mammalian cell models 132.
It is clear that the interplay between autophagy, aSyn toxicity and aggregation is still elusive. A reasonable explanation for the toxicity of aSyn mediated by autophagy induction is that the excessive activation of a dysfunctional autophagy will lead to a loss of selectivity, resulting in the trapping of functional competent proteins and organelles in autophagosomes. Ultimately, this could lead to a loss of function and cell toxicity. Thus, the beneficial or detrimental role of autophagy should be studied having in consideration its functionality and selectivity, as well as the size and nature of the aSyn aggregates.
YEAST AS A DRUG DISCOVERY PLATFORM FOR PD
The identification of therapeutic compounds for neurodegenerative disorders is of utmost importance. These devastating illnesses only have, in some cases, symptomatic therapies being therefore disruptive and costly for society. Thus, intense efforts are being made to understand the molecular underpinnings of neurodegenerative diseases and to identify novel therapeutic strategies. Nevertheless, given the complexity of the mechanisms leading to neurodegeneration, and the limitations in the models available, drug discovery is often slow, challenging and with limited success.
In the context of PD, drug discovery efforts focused on aSyn are complicated by the fact that it is a ”natively unstructured protein”, lacking defined secondary structure under physiological conditions 176. Thus, cell-based high-throughput phenotypic assays afford important possibilities, as they are based on relevant disease-associated phenotypes induced by aSyn expression. The readouts may include viability/toxicity, aggregation, mitochondrial function, proteasome activity, among others. Once relevant molecules and targets are identified, then it is fundamental to scrutinize the mechanism of action of the potential small molecules and candidate compounds. This is where yeast cells offer a remarkable advantage, as they enable the identification of target genes and mechanisms through diverse and complementary genetic approaches, accelerating the selection of pre-clinical candidates.
In mammalian cell systems, aSyn-associated phenotypes are often mild or inconsistent, complicating the development of reliable screening platforms 11,177. Primary rat neurons, infected with lentiviruses encoding for aSyn-expressing, have been used, but they also present technical limitations. Many of these systems are more suited for secondary validation steps, focusing on candidate genes or molecules identified in yeast, for example.
Yeast affords numerous advantages at the early stages of the drug development process, in comparison to mammalian cells and animal models. Several major drugs hit the same targets and elicit the same responses in yeast as they do in humans, including statins, methotrexate, omeprazole, tacrolimus (FK506) and bortezomib (Velcade) 177,178. In spite of the obvious limitations, such as the absence of a nervous system, and the absence of numerous gene products that are only present in mammalian cells and neurons, yeast cells are ideally suited for investigating the primordial molecular events triggering cell dysfunction and pathology. In this context, yeast cells can be regarded as living test tubes, where genetic manipulations are faster and more straightforward, with rapid growth and reduced cost, and functional similarity to higher eukaryotes.
The presence of a cell wall in yeast cells poses challenges that need to be considered in the context of drug screening efforts. However, this can be minimized by genetic manipulation of the efflux pump system or the ergosterol biosynthesis, reducing the capability of yeast cells to export drugs or by increasing the permeability of the cells, respectively.
Yeast is considered a robust primary drug-screening platform to filter for compounds with cytoprotective activity, for further complementation with assays in more physiologically relevant models. Approaches involving the sequential use of different model systems, starting with simpler cellular models and ending with more complex animal models, as schematized in Fig. 3, already resulted in the discovery of promising small molecules with therapeutic potential (described below). Recently, a yeast-to-human discovery platform for synucleinopathies was established, where genes and small molecules identified in yeast were validated in PD-patient derived neurons. Subsequently, yeast cells were again used for clarification of the mechanism of action, due to unmatched genetic tools available in S. cerevisae 179 (Fig. 3).
Figure 3. FIGURE 3: Yeast as a discovery platform for PD.
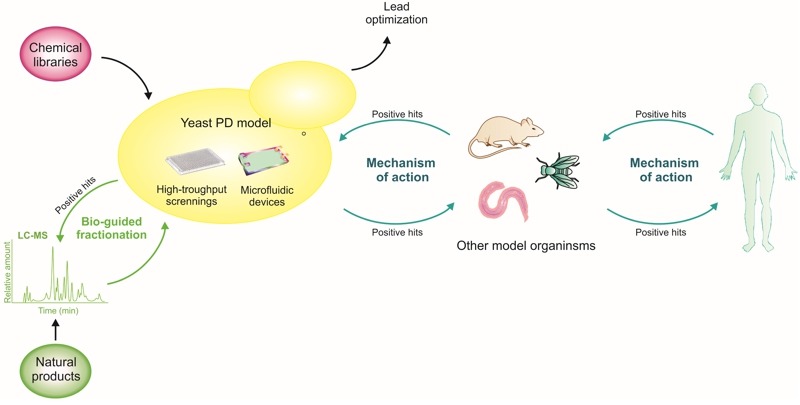
The discovery of small molecules, or natural products, which are able to rescue aSyn toxicity and aggregation benefits from the combination of approaches in yeast with those in other model systems. In particular, libraries of small molecules or natural products can first be screened in yeast models of PD, and combined with LC-MS approaches in order to enable an interactive bio-guided fractionation of complex mixtures to identify candidate compounds. The development of microfluidic devices, together with yeast genetics and high-throughput approaches, are also important to elucidate the mechanisms of action of the candidate compounds. Ultimately, the mechanisms and molecules identified can be further scrutinized in an iterative process between yeast, other model systems and, ultimately, in humans.
By iteratively moving between simple cellular models and patient derived cells, we will be able to elucidate mechanisms and evaluate patient-specific drug targets. Ultimately, this will enable scientists to conduct more significant animal and clinical trials in various neurodegenerative diseases.
Genetic screens
Large-scale genetic and chemical genetic approaches in yeast have provided important insight into the molecular basis of various neurodegenerative disorders. The first large-scale genetic screen used 4850 deletion yeast strains and successfully identified 86 genes enhancers of aSyn toxicity, of which 29% were involved in vesicular transport and lipid metabolism 124. Later, the same yeast collection enabled the identification of 185 modifiers of aSyn aggregation, using fluorescence microscopy 154. This study revealed that proteins involved in vesicular transport altered aSyn subcellular localization.
Using an overexpression screen, aSyn was found to block ER-to-Golgi trafficking due to the identification of enhancers and suppressors of toxicity 125. Importantly, these observations led to the identification of Rab1, the mammalian Ypt1 homolog, as a neuroprotector against dopaminergic neuron loss in animal models of PD 125.
In another genome wide-screen, using a high-expression library, Ypp1 was found to mediate the trafficking of aSyn A30P to the vacuole via the endocytic pathway, thus suppressing the toxicity of this aSyn mutant 133.
A separate yeast overexpression screen identified 40 genes that suppressed the toxicity of human WT aSyn 140. These genes were involved in ubiquitin-dependent protein catabolism, protein biosynthesis, vesicle trafficking and in the response to stress 140.
Using an integrative approach, the results from genetic screens were analyzed according to gene functionality and pathways. About ~3500 overexpression yeast strains were used and a cellular map of the proteins and genes responding to aSyn expression was obtained. Ergosterol biosynthesis and the TOR pathway were identified as modulators of aSyn cytotoxicity in yeast 128.
Drug discovery efforts in yeast models
The events leading to protein oligomerization are likely amenable to modulation by small molecules. Thus, yeast has also been used to screen for small molecules that can reduce aSyn aggregation and toxicity.
Screening of large libraries of compounds lead to the identification of aSyn toxicity suppressors in yeast. For instance, in a large-scale screen of small molecule, ~115.000 compounds were tested for their ability to reduce aSyn toxicity, resulting in the identification of a class of structurally related 1,2,3,4-tetrahydroquinolinones 126. These compounds reduced the formation of aSyn inclusions, re-established ER-to-Golgi trafficking, and ameliorated mitochondria-associated defects induced by aSyn. The targets were further confirmed in nematode neurons and in primary rat neuronal midbrain cultures. Interestingly, these compounds also rescued rotenone toxicity in neuronal cultures, a toxin used to study mitochondrial deficits in PD 126.
The ease of manipulation makes yeast a suitable tool to explore unconventional compounds and their mechanisms of action. Mannosylglycerate, a compatible solute typical of marine microorganisms thriving in hot environments, was found to reduce aSyn aggregation in a yeast model of PD 180. Latrepirdine, a drug in phase II clinical trials, was identified as protector against aSyn by inducing its degradation through autophagy, representing a novel scaffold for discovery of robust pro-autophagic/anti-neurodegeneration compounds 181.
A novel class of molecules, cyclic peptides (CPs), was also screened in yeast 182. CPs are natural-product-like chemicals with potent bioactivity. Yeast was exploited to express a plasmid-derived self-splicing intein that liberates a CP. This approach enabled the scale-up of high-throughput screens to 10-100 times the size of a typical small molecule screen. A pool of 5 million yeast transformants were screened and two related CP constructs with the ability to reduce aSyn toxicity were identified. These cyclic peptide constructs also prevented dopaminergic neuron loss in a nematode model of PD 182.
Due to their well-defined chemical nature, small molecules are the preferred molecules used in high-throughput screenings. Nevertheless, natural compounds have emerged as attractive molecules in the context of neurodegeneration. It is largely accepted that products such as green tea, small fruits and even olive oil have, in its constitution, compounds promoting health benefits. However, the major advances regarding their targets and mechanisms of action were only achieved in the last decade. Yeast models, together with chemical and animal studies, have significantly contributed for these discoveries 175.180,183,184
The first small compound screen in yeast tested ~10.000 compounds and identified a group of flavonoids, quercetin and epigallocatechin gallate as protectants against aSyn toxicity in the presence of iron 185. The protection promoted by these compounds was further analyzed, and the positive effect was due to their anti-oxidant and metal-chelating activities. Importantly, (poly)phenols, and particularly quercetin and epigallocatechin gallate, have proven beneficial in cellular and animal models of PD 186,187.
The advances in biochemical tools and the assembly of multidisciplinary teams also gave a major push to the drug discovery process. In fact, the benefits of green tea have been deciphered by combining HPLC fractionation in a microplate format with screening in yeast and parallel electrospray mass spectrometry (LC-MS) 188 (Fig. 3). This integrated process enabled the rapid assessment of the efficacy of the fractions and to systematically identify their bioactive constituents. The green tea metabolites were individually examined for their pharmacological effects and, interestingly, the protective properties of Camelia sinensis lied on the combination of multiple catechin metabolites 188. This study emphasizes the prominence of yeast high-throughput screenings to dissect natural extracts and to explore the numerous synergistic effects of its metabolites.
The bioactivities of plant (poly)phenol extracts in the yeast aSyn model were recently investigated using viability, oxidative stress, metabolic capacity and aSyn inclusion formation as phenotypic assays 175. The most promising extract was the one from Corema album leaves. The dissection of the mechanism of action of this extract, focused primarily on pathways related to proteostasis, showed that it promotes autophagic flux both in yeast and in a mammalian cell model of PD 175.
Ascorbic acid, a natural antioxidant, was found to promote a significant reduction in the percentage of yeast cells bearing aSyn inclusions 189. Remarkably, this study was performed using a new microfluidic device designed to validate compounds in yeast. Additional advantages are achieved by using this device, since it offers a powerful way for studying aSyn biology with single-cell resolution and high-throughput, using genetically modified yeast cells 189.
Screenings in other disease models, as in the amyotrophic lateral sclerosis yeast model (induced by the expression of the protein TDP-43), may also provide insight into candidate compounds to be tested in aSyn yeast models. Using yeast genetics, multiple protective 8-hydroxyquinolines, natural plant alkaloids, were identified 184. Some of these compounds were also found to be protective in aSyn yeast and nematode models. The putative protective mechanisms were related to their ionophore and intracellular metal chelation activities 184. From this screening, N-aryl benzimidazole proved more potent and effective against aSyn toxicity than against TDP-43 toxicity. Thus, the yeast aSyn platform was explored to identify the mechanism of action of N-aryl benzimidazole and it was found that it reverses diverse phenotypes induced by aSyn, including the accumulation of aSyn inclusions, the generation of ROS, the block of ER-Golgi trafficking and the nitration of proteins 190. Moreover, this compound was used in an iterative yeast-to-human neuron platform 129 (Fig. 3).
Taken together, identifying novel effective disease therapies is an incredible challenge. Nevertheless, rapidly improving methodologies and iterative processes, allied with an evolving mechanistic understanding of disease, is nurturing more interdisciplinary approaches to research and fostering drug discovery, with the ultimate goal of discovering novel therapeutics for humans.
CONCLUSIONS AND FUTURE PERSPECTIVES
The development of effective treatments and preventive therapies for PD is still a great challenge, mostly due to the scarcity of knowledge of disease-associated mechanisms that ultimately lead to neuronal dysfunction and death. As discussed herein, the versatile eukaryotic model organism S. cerevisiae has largely contributed to bridge this gap in PD medical research. By providing important insights into the molecular foundations of the disease as well as novel molecular targets and lead compounds with therapeutic potential. Notwithstanding, several fundamental aspects of PD pathophysiology remain to be elucidated. For example, yeast models were very helpful to clarify some facets of the still controversial role of aSyn phosphorylation, and will certainly further contribute to our understanding of other mechanisms associated with aSyn and other PD-associated genes.
Undoubtedly, research using S. cerevisiae as a model system enabled significant advancements in our understanding of the molecular mechanisms underlying PD. However, it should be noted that, as a simplified model system, it also has natural limitations that need to be obviated by further validations in more complex models. Indeed, iterative processes using models with different degrees of complexity have proven to be a powerful strategy to investigate the fundamental aspects of neurodegenerative diseases, thereby accelerating drug discovery.
Funding Statement
This work was supported by Fundação para a Ciência e Tecnologia project PTDC/BIA-BCM/117975/2010, fellowships SFRH/BPD/101646/2014 (ST) and SFRH/BD/73429/2010 (DM), and IF/01097/2013 (CNS). This work was also supported by the BacHBerry project, cofunded by the European Commission in the 7th Framework Programme (Project No. FP7613793). RM is supported by a BacHBerry fellowship. TFO is supported by the DFG Center for Nanoscale Microscopy and Molecular Physiology of the Brain (CNMPB).
References
- 1.Samuel D. Investigation of ancient egyptian baking and brewing methods by correlative microscopy. Science. 1996;273(5274):488–490. doi: 10.1126/science.273.5274.488. [DOI] [PubMed] [Google Scholar]
- 2.Legras JL, Merdinoglu D, Cornuet JM, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Molecular ecology. 2007;16(10):2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 3.Mortimer RK. Evolution and variation of the yeast (Saccharomyces) genome. Genome research. 2000;10(4):403–409. doi: 10.1101/gr.10.4.403. [DOI] [PubMed] [Google Scholar]
- 4.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438(7066):379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coller J, Parker R. Eukaryotic mRNA decapping. Annual review of biochemistry. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 6.Fields S, Johnston M. Cell biology. Whither model organism research? Science. 2005;307(5717):1885–1886. doi: 10.1126/science.1108872. [DOI] [PubMed] [Google Scholar]
- 7.Duina AA, Miller ME, Keeney JB. Budding yeast for budding geneticists: a primer on the Saccharomyces cerevisiae model system. Genetics. 2014;197(1):33–48. doi: 10.1534/genetics.114.163188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botstein D, Fink GR. Yeast: an experimental organism for 21st Century biology. Genetics. 2011;189(3):695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinnen A, Hicks JB, Fink GR. Transformation of yeast. Proceedings of the National Academy of Sciences of the United States of America. 1978;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24(10):913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenreiro S, Munder MC, Alberti S, Outeiro TF. Harnessing the power of yeast to unravel the molecular basis of neurodegeneration. Journal of neurochemistry. 2013;127(4):438–452. doi: 10.1111/jnc.12271. [DOI] [PubMed] [Google Scholar]
- 12.Van Mullem V, Wery M, De Bolle X, Vandenhaute J. Construction of a set of Saccharomyces cerevisiae vectors designed for recombinational cloning. Yeast. 2003;20(8):739–746. doi: 10.1002/yea.999. [DOI] [PubMed] [Google Scholar]
- 13.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 14.Vidal M, Legrain P. Yeast forward and reverse 'n'-hybrid systems. Nucleic acids research. 1999;27(4):919–929. doi: 10.1093/nar/27.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stagljar I, Korostensky C, Johnsson N, te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(9):5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer K, Burkle L, Auerbach D, Thaminy S, Dinkel M, Engels K, Stagljar I. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Science's STKE : signal transduction knowledge environment. 2005;2005(275):pl3. doi: 10.1126/stke.2752005pl3. [DOI] [PubMed] [Google Scholar]
- 17.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Molecular cell. 2002;9(4):789–798. doi: 10.1016/S1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 18.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nature protocols. 2006;1(3):1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24(9):767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 20.He F, Nie WC, Tong Z, Yuan SM, Gong T, Liao Y, Bi E, Gao XD. The GTPase-activating protein Rga1 interacts with Rho3 GTPase and may regulate its function in polarized growth in budding yeast. PloS one. 2015;10(4):e0123326. doi: 10.1371/journal.pone.0123326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goncalves SA, Matos JE, Outeiro TF. Zooming into protein oligomerization in neurodegeneration using BiFC. Trends in biochemical sciences. 2010;35(11):643–651. doi: 10.1016/j.tibs.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274(5287):563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 23.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P. Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. . Science. 1999;285(5429):901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 24.Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, Trochesset M, Morse D, Krogan NJ, Hiley SL, Li Z, Morris Q, Grigull J, Mitsakakis N, Roberts CJ, Greenblatt JF, Boone C, Kaiser CA, Andrews BJ, Hughes TR. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118(1):31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Dohmen RJ, Varshavsky A. Heat-inducible degron and the making of conditional mutants. Methods in enzymology. 2005;399:799–822. doi: 10.1016/S0076-6879(05)99052-6. [DOI] [PubMed] [Google Scholar]
- 26.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 27.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 28.Jones GM, Stalker J, Humphray S, West A, Cox T, Rogers J, Dunham I, Prelich G. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nature methods. 2008;5(3):239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- 29.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, Gerstein M, Dumont ME, Phizicky EM, Snyder M, Grayhack EJ. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes & development. 2005;19(23):2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Rolfs A, Bhullar B, Murthy TV, Zhu C, Berger MF, Camargo AA, Kelley F, McCarron S, Jepson D, Richardson A, Raphael J, Moreira D, Taycher E, Zuo D, Mohr S, Kane MF, Williamson J, Simpson A, Bulyk ML, Harlow E, Marsischky G, Kolodner RD, LaBaer J. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome research. 2007;17(4):536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenreiro S, Outeiro TF. Simple is good: yeast models of neurodegeneration. FEMS yeast research. 2010;10(8):970–979. doi: 10.1111/j.1567-1364.2010.00649.x. [DOI] [PubMed] [Google Scholar]
- 32.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320(5881):1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic acids research. 2012;40(Database issue):D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller-Fleming L, Giorgini F, Outeiro TF. Yeast as a model for studying human neurodegenerative disorders. Biotechnology journal. 2008;3(3):325–338. doi: 10.1002/biot.200700217. [DOI] [PubMed] [Google Scholar]
- 35.Heinicke S, Livstone MS, Lu C, Oughtred R, Kang F, Angiuoli SV, White O, Botstein D, Dolinski K. The Princeton Protein Orthology Database (P-POD): a comparative genomics analysis tool for biologists. PloS one. 2007;2(8):e766. doi: 10.1371/journal.pone.0000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karathia H, Vilaprinyo E, Sorribas A, Alves R. Saccharomyces cerevisiae as a model organism: a comparative study. PloS one. 2011;6(2):e16015. doi: 10.1371/journal.pone.0016015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brodsky JL, Skach WR. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Current opinion in cell biology. 2011;23(4):464–475. doi: 10.1016/j.ceb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116(2):153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 39.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goeckeler JL, Brodsky JL. Molecular chaperones and substrate ubiquitination control the efficiency of endoplasmic reticulum-associated degradation. Diabetes, obesity & metabolism 1. 2010;2:32–38. doi: 10.1111/j.1463-1326.2010.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinaldi T, Dallabona C, Ferrero I, Frontali L, Bolotin-Fukuhara M. Mitochondrial diseases and the role of the yeast models. FEMS yeast research. 2010;10(8):1006–1022. doi: 10.1111/j.1567-1364.2010.00685.x. [DOI] [PubMed] [Google Scholar]
- 42.Dunham MJ, Fowler DM. Contemporary, yeast-based approaches to understanding human genetic variation. Current opinion in genetics & development. 2013;23(6):658–664. doi: 10.1016/j.gde.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker's yeast? Nature reviews Neuroscience. 2010;11(6):436–449. doi: 10.1038/nrn2809. [DOI] [PubMed] [Google Scholar]
- 44.Outeiro TF, Giorgini F. Yeast as a drug discovery platform in Huntington's and Parkinson's diseases. Biotechnology journal. 2006;1(3):258–269. doi: 10.1002/biot.200500043. [DOI] [PubMed] [Google Scholar]
- 45.Lees AJ. Unresolved issues relating to the shaking palsy on the celebration of James Parkinson's 250th birthday. Movement disorders. 2007;Suppl 17:S327–S334. doi: 10.1002/mds.21684. [DOI] [PubMed] [Google Scholar]
- 46.Goetz CG. The history of Parkinson's disease: early clinical descriptions and neurological therapies. Cold Spring Harbor perspectives in medicine. 2011;1(1):a008862. doi: 10.1101/cshperspect.a008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jankovic J. Parkinson's disease: clinical features and diagnosis. Journal of neurology, neurosurgery, and psychiatry. 2008;79(4):368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 48.Braak H, Ghebremedhin E, Rub U, Bratzke H, Del Tredici K. Stages in the development of Parkinson's disease-related pathology. Cell and tissue research. 2004;318(1):121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- 49.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 50.Shults CW. Lewy bodies. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(6):1661–1668. doi: 10.1073/pnas.0509567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 52.Siddiqui A, Chinta SJ, Mallajosyula JK, Rajagopolan S, Hanson I, Rane A, Melov S, Andersen JK. Selective binding of nuclear alpha-synuclein to the PGC1alpha promoter under conditions of oxidative stress may contribute to losses in mitochondrial function: implications for Parkinson's disease. Free radical biology & medicine. 2012;53(4):993–1003. doi: 10.1016/j.freeradbiomed.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasudevaraju P, Guerrero E, Hegde ML, Collen TB, Britton GB, Rao KS. New evidence on alpha-synuclein and Tau binding to conformation and sequence specific GC* rich DNA: Relevance to neurological disorders. Journal of pharmacy < bioallied sciences. 2012;4(2):112–117. doi: 10.4103/0975-7406.94811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Human molecular genetics. 2006;15(20):3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 55.Zhou M, Xu S, Mi J, Ueda K, Chan P. Nuclear translocation of alpha-synuclein increases susceptibility of MES23. cells to oxidative stress. . Brain research. 2013;1500:19–27. doi: 10.1016/j.brainres.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 56.Liu X, Lee YJ, Liou LC, Ren Q, Zhang Z, Wang S, Witt SN. Alpha-synuclein functions in the nucleus to protect against hydroxyurea-induced replication stress in yeast. Human molecular genetics. 2011;20(17):3401–3414. doi: 10.1093/hmg/ddr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goers J, Manning-Bog AB, McCormack AL, Millett IS, Doniach S, Di Monte DA, Uversky VN, Fink AL. Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry. 2003;42(28):8465–8471. doi: 10.1021/bi0341152. [DOI] [PubMed] [Google Scholar]
- 58.Nakamura K. alpha-Synuclein and mitochondria: partners in crime? Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics. 2013;10(3):391–399. doi: 10.1007/s13311-013-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisbach SE, Outeiro TF. Alpha-synuclein and intracellular trafficking: impact on the spreading of Parkinson's disease pathology. Journal of molecular medicine. 2013;91(6):693–703. doi: 10.1007/s00109-013-1038-9. [DOI] [PubMed] [Google Scholar]
- 60.Jensen PH, Nielsen MS, Jakes R, Dotti CG, Goedert M. Binding of alpha-synuclein to brain vesicles is abolished by familial Parkinson's disease mutation. The Journal of biological chemistry. 1998;273(41):26292–26294. doi: 10.1074/jbc.273.41.26292. [DOI] [PubMed] [Google Scholar]
- 61.Lashuel HA, Overk CR, Oueslati A, Masliah E. The many faces of alpha-synuclein: from structure and toxicity to therapeutic target. Nature reviews Neuroscience. 2013;14(1):38–48. doi: 10.1038/nrn3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott DA, Tabarean I, Tang Y, Cartier A, Masliah E, Roy S. A pathologic cascade leading to synaptic dysfunction in alpha-synuclein-induced neurodegeneration. The Journal of neuroscience. 2010;30(24):8083–8095. doi: 10.1523/JNEUROSCI.1091-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yavich L, Tanila H, Vepsalainen S, Jakala P. Role of alpha-synuclein in presynaptic dopamine recruitment. The Journal of neuroscience. 2004;24(49):11165–11170. doi: 10.1523/JNEUROSCI.2559-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaugler MN, Genc O, Bobela W, Mohanna S, Ardah MT, El-Agnaf OM, Cantoni M, Bensadoun JC, Schneggenburger R, Knott GW, Aebischer P, Schneider BL. Nigrostriatal overabundance of alpha-synuclein leads to decreased vesicle density and deficits in dopamine release that correlate with reduced motor activity. Acta neuropathologica. 2012;123(5):653–669. doi: 10.1007/s00401-012-0963-y. [DOI] [PubMed] [Google Scholar]
- 65.Lundblad M, Decressac M, Mattsson B, Bjorklund A. Impaired neurotransmission caused by overexpression of alpha-synuclein in nigral dopamine neurons. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(9):3213–3219. doi: 10.1073/pnas.1200575109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 67.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, Waucquier N, Defebvre L, Amouyel P, Farrer M, Destee A. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364(9440):1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 68.Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, Pollak P, Agid Y, Durr A, Brice A. Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet. 2004;364(9440):1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 69.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson's disease. Nature genetics. 1998;18(2):106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 70.Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, Llorens V, Gomez Tortosa E, del Ser T, Munoz DG, de Yebenes JG. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55(2):164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 71.Appel-Cresswell S, Vilarino-Guell C, Encarnacion M, Sherman H, Yu I, Shah B, Weir D, Thompson C, Szu-Tu C, Trinh J, Aasly JO, Rajput A, Rajput AH, Jon Stoessl A, Farrer MJ. Alpha-synuclein p.H50Q, a novel pathogenic mutation for Parkinson's disease. Movement disorders. 2013;28(6):811–813. doi: 10.1002/mds.25421. [DOI] [PubMed] [Google Scholar]
- 72.Porcari R, Proukakis C, Waudby CA, Bolognesi B, Mangione PP, Paton JF, Mullin S, Cabrita LD, Penco A, Relini A, Verona G, Vendruscolo M, Stoppini M, Tartaglia GG, Camilloni C, Christodoulou J, Schapira AH, Bellotti V. The H50Q Mutation Induces a 10-fold Decrease in the Solubility of alpha-Synuclein. The Journal of biological chemistry. 2015;290(4):2395–2404. doi: 10.1074/jbc.M114.610527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lesage S, Anheim M, Letournel F, Bousset L, Honore A, Rozas N, Pieri L, Madiona K, Durr A, Melki R, Verny C, Brice A. G51D alpha-synuclein mutation causes a novel parkinsonian-pyramidal syndrome. Annals of neurology. 2013;73(4):459–471. doi: 10.1002/ana.23894. [DOI] [PubMed] [Google Scholar]
- 74.Golbe LI, Di Iorio G, Bonavita V, Miller DC, Duvoisin RC. A large kindred with autosomal dominant Parkinson's disease. Annals of neurology. 1990;27(3):276–282. doi: 10.1002/ana.410270309. [DOI] [PubMed] [Google Scholar]
- 75.Puschmann A, Ross OA, Vilarino-Guell C, Lincoln SJ, Kachergus JM, Cobb SA, Lindquist SG, Nielsen JE, Wszolek ZK, Farrer M, Widner H, van Westen D, Hagerstrom D, Markopoulou K, Chase BA, Nilsson K, Reimer J, Nilsson C. A Swedish family with de novo alpha-synuclein A53T mutation: evidence for early cortical dysfunction. Parkinsonism & related disorders. 2009;15(9):627–632. doi: 10.1016/j.parkreldis.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pasanen P, Myllykangas L, Siitonen M, Raunio A, Kaakkola S, Lyytinen J, Tienari PJ, Poyhonen M, Paetau A. Novel alpha-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson's disease-type pathology. Neurobiology of aging. 2014;35(9):e2181–e2185. doi: 10.1016/j.neurobiolaging.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 77.Bodner CR, Maltsev AS, Dobson CM, Bax A. Differential phospholipid binding of alpha-synuclein variants implicated in Parkinson's disease revealed by solution NMR spectroscopy. Biochemistry. 2010;49(5):862–871. doi: 10.1021/bi901723p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim YS, Laurine E, Woods W, Lee SJ. A novel mechanism of interaction between alpha-synuclein and biological membranes. Journal of molecular biology. 2006;360(2):386–397. doi: 10.1016/j.jmb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 79.Bussell Jr R, Eliezer D. Effects of Parkinson's disease-linked mutations on the structure of lipid-associated alpha-synuclein. Biochemistry. 2004;43(16):4810–4818. doi: 10.1021/bi036135. [DOI] [PubMed] [Google Scholar]
- 80.Jo E, Fuller N, Rand RP, St George-Hyslop P, Fraser PE. Defective membrane interactions of familial Parkinson's disease mutant A30P alpha-synuclein. Journal of molecular biology. 2002;315(4):799–807. doi: 10.1006/jmbi.2001.5269. [DOI] [PubMed] [Google Scholar]
- 81.Fares MB, Ait-Bouziad N, Dikiy I, Mbefo MK, Jovicic A, Kiely A, Holton JL, Lee SJ, Gitler AD, Eliezer D, Lashuel HA. The novel Parkinson's disease linked mutation G51D attenuates in vitro aggregation and membrane binding of alpha-synuclein, and enhances its secretion and nuclear localization in cells. Human molecular genetics. 2014;23(17):4491–4509. doi: 10.1093/hmg/ddu165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi W, Zibaee S, Jakes R, Serpell LC, Davletov B, Crowther RA, Goedert M. Mutation E46K increases phospholipid binding and assembly into filaments of human alpha-synuclein. FEBS letters. 2004;576(3):363–368. doi: 10.1016/j.febslet.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 83.El-Agnaf OM, Jakes R, Curran MD, Wallace A. Effects of the mutations Ala30 to Pro and Ala53 to Thr on the physical and morphological properties of alpha-synuclein protein implicated in Parkinson's disease. FEBS letters. 1998;440(1-2):67–70. doi: 10.1016/S0014-5793(98)01419-7. [DOI] [PubMed] [Google Scholar]
- 84.Narhi L, Wood SJ, Steavenson S, Jiang Y, Wu GM, Anafi D, Kaufman SA, Martin F, Sitney K, Denis P, Louis JC, Wypych J, Biere AL, Citron M. Both familial Parkinson's disease mutations accelerate alpha-synuclein aggregation. The Journal of biological chemistry. 1999;274(14):9843–9846. doi: 10.1074/jbc.274.14.9843. [DOI] [PubMed] [Google Scholar]
- 85.Greenbaum EA, Graves CL, Mishizen-Eberz AJ, Lupoli MA, Lynch DR, Englander SW, Axelsen PH, Giasson BI. The E46K mutation in alpha-synuclein increases amyloid fibril formation. The Journal of biological chemistry. 2005;280(9):7800–7807. doi: 10.1074/jbc.M411638200. [DOI] [PubMed] [Google Scholar]
- 86.Yonetani M, Nonaka T, Masuda M, Inukai Y, Oikawa T, Hisanaga S, Hasegawa M. Conversion of wild-type alpha-synuclein into mutant-type fibrils and its propagation in the presence of A30P mutant. The Journal of biological chemistry. 2009;284(12):7940–7950. doi: 10.1074/jbc.M807482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghosh D, Mondal M, Mohite GM, Singh PK, Ranjan P, Anoop A, Ghosh S, Jha NN, Kumar A, Maji SK. The Parkinson's disease-associated H50Q mutation accelerates alpha-Synuclein aggregation in vitro. Biochemistry. 2013;52(40):6925–6927. doi: 10.1021/bi400999d. [DOI] [PubMed] [Google Scholar]
- 88.Li J, Uversky VN, Fink AL. Effect of familial Parkinson's disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human alpha-synuclein. Biochemistry. 2001;40(38):11604–11613. doi: 10.1021/bi010616g. [DOI] [PubMed] [Google Scholar]
- 89.Ghosh D, Sahay S, Ranjan P, Salot S, Mohite GM, Singh PK, Dwivedi S, Carvalho E, Banerjee R, Kumar A, Maji SK. The newly discovered Parkinson's disease associated Finnish mutation (A53E) attenuates alpha-synuclein aggregation and membrane binding. Biochemistry. 2014;53(41):6419–6421. doi: 10.1021/bi5010365. [DOI] [PubMed] [Google Scholar]
- 90.Klein C, Westenberger A. Genetics of Parkinson's disease. Cold Spring Harbor perspectives in medicine. 2012;2(1):a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide BM, Schjeide LM, Meissner E, Zauft U, Allen NC, Liu T, Schilling M, Anderson KJ, Beecham G, Berg D, Biernacka JM, Brice A, DeStefano AL, Do CB, Eriksson N, Factor SA, Farrer MJ, Foroud T, Gasser T, Hamza T, Hardy JA, Heutink P, Hill-Burns EM, Klein C, Latourelle JC. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: The PDGene database. PLoS genetics. 2012;8(3):e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nature genetics. 2014;46(9):989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martin I, Kim JW, Dawson VL, Dawson TM. LRRK2 pathobiology in Parkinson's disease. Journal of neurochemistry. 2014;131(5):554–565. doi: 10.1111/jnc.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Esteves AR, Swerdlow RH, Cardoso SM. LRRK2, a puzzling protein: insights into Parkinson's disease pathogenesis. Experimental neurology. 2014;261:206–216. doi: 10.1016/j.expneurol.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shin N, Jeong H, Kwon J, Heo HY, Kwon JJ, Yun HJ, Kim CH, Han BS, Tong Y, Shen J, Hatano T, Hattori N, Kim KS, Chang S, Seol W. LRRK2 regulates synaptic vesicle endocytosis. Experimental cell research. 2008;314(10):2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 96.Matta S, Van Kolen K, da Cunha R, van den Bogaart G, Mandemakers W, Miskiewicz K, De Bock PJ, Morais VA, Vilain S, Haddad D, Delbroek L, Swerts J, Chavez-Gutierrez L, Esposito G, Daneels G, Karran E, Holt M, Gevaert K, Moechars DW, De Strooper B, Verstreken P. LRRK2 controls an EndoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75(6):1008–1021. doi: 10.1016/j.neuron.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 97.Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Human molecular genetics. 2009;18(21):4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52(4):587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 99.Stafa K, Tsika E, Moser R, Musso A, Glauser L, Jones A, Biskup S, Xiong Y, Bandopadhyay R, Dawson VL, Dawson TM, Moore DJ. Functional interaction of Parkinson's disease-associated LRRK2 with members of the dynamin GTPase superfamily. Human molecular genetics. 2014;23(8):2055–2077. doi: 10.1093/hmg/ddt600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pereira C, Miguel Martins L, Saraiva L. LRRK2, but not pathogenic mutants, protects against H2O2 stress depending on mitochondrial function and endocytosis in a yeast model. Biochimica et biophysica acta. 2014;1840(6):2025–2031. doi: 10.1016/j.bbagen.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 101.Qing H, Zhang Y, Deng Y, McGeer EG, McGeer PL. Lrrk2 interaction with alpha-synuclein in diffuse Lewy body disease. Biochemical and biophysical research communications. 2009;390(4):1229–1234. doi: 10.1016/j.bbrc.2009.10.126. [DOI] [PubMed] [Google Scholar]
- 102.Guerreiro PS, Huang Y, Gysbers A, Cheng D, Gai WP, Outeiro TF, Halliday GM. LRRK2 interactions with alpha-synuclein in Parkinson's disease brains and in cell models. Journal of molecular medicine. 2013;91(4):513–522. doi: 10.1007/s00109-012-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qing H, Wong W, McGeer EG, McGeer PL. Lrrk2 phosphorylates alpha synuclein at serine 129: Parkinson disease implications. Biochemical and biophysical research communications. 2009;387(1):149–152. doi: 10.1016/j.bbrc.2009.06.142. [DOI] [PubMed] [Google Scholar]
- 104.Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Parkin and PINK1: much more than mitophagy. Trends in neurosciences. 2014;37(6):315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441(7097):1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 106.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441(7097):1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 107.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS biology. 2010;8(1):e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of cell biology. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(1):378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. The EMBO journal. 2014;33(19):2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pereira C, Costa V, Martins LM, Saraiva L. A yeast model of the Parkinsons disease-associated protein Parkin. Experimental cell research. 2015;333(1):73–79. doi: 10.1016/j.yexcr.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 112.Cookson MR. Parkinsonism due to mutations in PINK1, parkin, and DJ-1 and oxidative stress and mitochondrial pathways. Cold Spring Harbor perspectives in medicine. 2012;2(9):a009415. doi: 10.1101/cshperspect.a009415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wilson MA. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxidants & redox signaling. 2011;15(1):111–122. doi: 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zondler L, Miller-Fleming L, Repici M, Goncalves S, Tenreiro S, Rosado-Ramos R, Betzer C, Straatman KR, Jensen PH, Giorgini F, Outeiro TF. DJ-1 interactions with alpha-synuclein attenuate aggregation and cellular toxicity in models of Parkinson's disease. Cell death & disease. 2014;5:e1350. doi: 10.1038/cddis.2014.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sajjad MU, Green EW, Miller-Fleming L, Hands S, Herrera F, Campesan S, Khoshnan A, Outeiro TF, Giorgini F, Wyttenbach A. DJ-1 modulates aggregation and pathogenesis in models of Huntington's disease. Human molecular genetics. 2014;23(3):755–766. doi: 10.1093/hmg/ddt466. [DOI] [PubMed] [Google Scholar]
- 116.Miller-Fleming L, Antas P, Pais TF, Smalley JL, Giorgini F, Outeiro TF. Yeast DJ-1 superfamily members are required for diauxic-shift reprogramming and cell survival in stationary phase. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(19):7012–7017. doi: 10.1073/pnas.1319221111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302(5651):1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Soper JH, Roy S, Stieber A, Lee E, Wilson RB, Trojanowski JQ, Burd CG, Lee VM. Alpha-synuclein-induced aggregation of cytoplasmic vesicles in Saccharomyces cerevisiae. Molecular biology of the cell. 2008;19(3):1093–1103. doi: 10.1091/mbc.E07-08-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, Barlowe C, Lindquist S. The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(1):145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sancenon V, Lee SA, Patrick C, Griffith J, Paulino A, Outeiro TF, Reggiori F, Masliah E, Muchowski PJ. Suppression of alpha-synuclein toxicity and vesicle trafficking defects by phosphorylation at S129 in yeast depends on genetic context. Human molecular genetics. 2012;21(11):2432–2449. doi: 10.1093/hmg/dds058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zabrocki P, Pellens K, Vanhelmont T, Vandebroek T, Griffioen G, Wera S, Van Leuven F, Winderickx J. Characterization of alpha-synuclein aggregation and synergistic toxicity with protein tau in yeast. The FEBS journal. 2005;272(6):1386–1400. doi: 10.1111/j.1742-4658.2005.04571.x. [DOI] [PubMed] [Google Scholar]
- 122.Oien DB, Shinogle HE, Moore DS, Moskovitz J. Clearance and phosphorylation of alpha-synuclein are inhibited in methionine sulfoxide reductase a null yeast cells. Journal of molecular neuroscience. 2009;39(3):323–332. doi: 10.1007/s12031-009-9274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tenreiro S, Reimao-Pinto MM, Antas P, Rino J, Wawrzycka D, Macedo D, Rosado-Ramos R, Amen T, Waiss M, Magalhaes F, Gomes A, Santos CN, Kaganovich D, Outeiro TF. Phosphorylation modulates clearance of alpha-synuclein inclusions in a yeast model of Parkinson's disease. PLoS genetics. 2014;10(5):e1004302. doi: 10.1371/journal.pgen.1004302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science. 2003;302(5651):1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- 125.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313(5785):324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Su LJ, Auluck PK, Outeiro TF, Yeger-Lotem E, Kritzer JA, Tardiff DF, Strathearn KE, Liu F, Cao S, Hamamichi S, Hill KJ, Caldwell KA, Bell GW, Fraenkel E, Cooper AA, Caldwell GA, McCaffery JM, Rochet JC, Lindquist S. Compounds from an unbiased chemical screen reverse both ER-to-Golgi trafficking defects and mitochondrial dysfunction in Parkinson's disease models. Disease models and mechanisms. 2010;3(3-4):194–208. doi: 10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gitler AD, Chesi A, Geddie ML, Strathearn KE, Hamamichi S, Hill KJ, Caldwell KA, Caldwell GA, Cooper AA, Rochet JC, Lindquist S. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nature genetics. 2009;41(3):308–315. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, Lindquist S, Fraenkel E. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nature genetics. 2009;41(3):316–323. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, Mungenast AE, Muffat J, Mitalipova M, Pluth MD, Jui NT, Schule B, Lippard SJ, Tsai LH, Krainc D, Buchwald SL, Jaenisch R, Lindquist S. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342(6161):983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Buttner S, Bitto A, Ring J, Augsten M, Zabrocki P, Eisenberg T, Jungwirth H, Hutter S, Carmona-Gutierrez D, Kroemer G, Winderickx J, Madeo F. Functional mitochondria are required for alpha-synuclein toxicity in aging yeast. The Journal of biological chemistry. 2008;283(12):7554–7560. doi: 10.1074/jbc.M708477200. [DOI] [PubMed] [Google Scholar]
- 131.Buttner S, Faes L, Reichelt WN, Broeskamp F, Habernig L, Benke S, Kourtis N, Ruli D, Carmona-Gutierrez D, Eisenberg T, D'Hooge P, Ghillebert R, Franssens V, Harger A, Pieber TR, Freudenberger P, Kroemer G, Sigrist SJ, Winderickx J, Callewaert G, Tavernarakis N, Madeo F. The Ca2+/Mn2+ ion-pump PMR1 links elevation of cytosolic Ca(2+) levels to alpha-synuclein toxicity in Parkinson's disease models. Cell death and differentiation. 2013;20(3):465–477. doi: 10.1038/cdd.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sampaio-Marques B, Felgueiras C, Silva A, Rodrigues M, Tenreiro S, Franssens V, Reichert AS, Outeiro TF, Winderickx J, Ludovico P. SNCA (alpha-synuclein)-induced toxicity in yeast cells is dependent on sirtuin 2 (Sir2)-mediated mitophagy. Autophagy. 2012;8(10):1494–1509. doi: 10.4161/auto.21275. [DOI] [PubMed] [Google Scholar]
- 133.Flower TR, Clark-Dixon C, Metoyer C, Yang H, Shi R, Zhang Z, Witt SN. YGR198w (YPP1) targets A30P alpha-synuclein to the vacuole for degradation. The Journal of cell biology. 2007;177(6):1091–1104. doi: 10.1083/jcb.200610071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Buttner S, Habernig L, Broeskamp F, Ruli D, Vogtle FN, Vlachos M, Macchi F, Kuttner V, Carmona-Gutierrez D, Eisenberg T, Ring J, Markaki M, Taskin AA, Benke S, Ruckenstuhl C, Braun R, Van den Haute C, Bammens T, van der Perren A, Frohlich KU, Winderickx J, Kroemer G, Baekelandt V, Tavernarakis N, Kovacs GG, Dengjel J, Meisinger C, Sigrist SJ, Madeo F. Endonuclease G mediates alpha-synuclein cytotoxicity during Parkinson's disease. The EMBO journal. 2013;32(23):3041–3054. doi: 10.1038/emboj.2013.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xiong Y, Coombes CE, Kilaru A, Li X, Gitler AD, Bowers WJ, Dawson VL, Dawson TM, Moore DJ. GTPase activity plays a key role in the pathobiology of LRRK2. PLoS genetics. 2010;6(4):e1000902. doi: 10.1371/journal.pgen.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS letters. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jiang P, Mizushima N. Autophagy and human diseases. Cell research. 2014;24(1):69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 140.Liang J, Clark-Dixon C, Wang S, Flower TR, Williams-Hart T, Zweig R, Robinson LC, Tatchell K, Witt SN. Novel suppressors of alpha-synuclein toxicity identified using yeast. Human molecular genetics. 2008;17(23):3784–3795. doi: 10.1093/hmg/ddn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zimprich A, Benet-Pages A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, Rossle SC, Klopp N, Wolf E, Seppi K, Pirker W, Presslauer S, Mollenhauer B, Katzenschlager R, Foki T, Hotzy C, Reinthaler E, Harutyunyan A, Kralovics R, Peters A, Zimprich F, Brucke T, Poewe W, Auff E, Trenkwalder C, Rost B. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. American journal of human genetics. 2011;89(1):168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, Behrouz B, Melrose HL, Hentati E, Puschmann A, Evans DM, Conibear E, Wasserman WW, Aasly JO, Burkhard PR, Djaldetti R, Ghika J, Hentati F, Krygowska-Wajs A, Lynch T, Melamed E, Rajput A, Rajput AH, Solida A, Wu RM, Uitti RJ. VPS35 mutations in Parkinson disease. American journal of human genetics. 2011;89(1):162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chartier-Harlin MC, Dachsel JC, Vilarino-Guell C, Lincoln SJ, Lepretre F, Hulihan MM, Kachergus J, Milnerwood AJ, Tapia L, Song MS, Le Rhun E, Mutez E, Larvor L, Duflot A, Vanbesien-Mailliot C, Kreisler A, Ross OA, Nishioka K, Soto-Ortolaza AI, Cobb SA, Melrose HL, Behrouz B, Keeling BH, Bacon JA, Hentati E, Williams L, Yanagiya A, Sonenberg N, Lockhart PJ, Zubair AC. Translation initiator EIF4G1 mutations in familial Parkinson disease. American journal of human genetics. 2011;89(3):398–406. doi: 10.1016/j.ajhg.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dhungel N, Eleuteri S, Li LB, Kramer NJ, Chartron JW, Spencer B, Kosberg K, Fields JA, Stafa K, Adame A, Lashuel H, Frydman J, Shen K, Masliah E, Gitler AD. Parkinson's disease genes VPS35 and EIF4G1 interact genetically and converge on alpha-synuclein. Neuron. 2015;85(1):76–87. doi: 10.1016/j.neuron.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Di Fonzo A, Chien HF, Socal M, Giraudo S, Tassorelli C, Iliceto G, Fabbrini G, Marconi R, Fincati E, Abbruzzese G, Marini P, Squitieri F, Horstink MW, Montagna P, Libera AD, Stocchi F, Goldwurm S, Ferreira JJ, Meco G, Martignoni E, Lopiano L, Jardim LB, Oostra BA, Barbosa ER, Italian Parkinson Genetics N, Bonifati V. ATP13A2 missense mutations in juvenile parkinsonism and young onset Parkinson disease. Neurology. 2007;68(19):1557–1562. doi: 10.1212/01.wnl.0000260963.08711.08. [DOI] [PubMed] [Google Scholar]
- 146.Schmidt K, Wolfe DM, Stiller B, Pearce DA. Cd2+, Mn2+, Ni2+ and Se2+ toxicity to Saccharomyces cerevisiae lacking YPK9p the orthologue of human ATP13A2. Biochemical and biophysical research communications. 2009;383(2):198–202. doi: 10.1016/j.bbrc.2009.03.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chesi A, Kilaru A, Fang X, Cooper AA, Gitler AD. The role of the Parkinson's disease gene PARK9 in essential cellular pathways and the manganese homeostasis network in yeast. PloS one. 2012;7(3):e34178. doi: 10.1371/journal.pone.0034178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Farina M, Avila DS, da Rocha JB, Aschner M. Metals, oxidative stress and neurodegeneration: a focus on iron, manganese and mercury. Neurochemistry international. 2013;62(5):575–594. doi: 10.1016/j.neuint.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annual review of biochemistry. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 150.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nature reviews Drug discovery. 2013;12(9):703–719. doi: 10.1038/nrd3976. [DOI] [PubMed] [Google Scholar]
- 151.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nature reviews Molecular cell biology. 2010;11(11):777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 152.Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332(6163):462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 153.Braun RJ. Ubiquitin-dependent proteolysis in yeast cells expressing neurotoxic proteins. Frontiers in molecular neuroscience. 2015;8:8. doi: 10.3389/fnmol.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zabrocki P, Bastiaens I, Delay C, Bammens T, Ghillebert R, Pellens K, De Virgilio C, Van Leuven F, Winderickx J. Phosphorylation, lipid raft interaction and traffic of alpha-synuclein in a yeast model for Parkinson. Biochimica et biophysica acta. 2008;1783(10):1767–1780. doi: 10.1016/j.bbamcr.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 155.Flower TR, Chesnokova LS, Froelich CA, Dixon C, Witt SN. Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson's disease. Journal of molecular biology. 2005;351(5):1081–1100. doi: 10.1016/j.jmb.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 156.Outeiro TF, Klucken J, Strathearn KE, Liu F, Nguyen P, Rochet JC, Hyman BT, McLean PJ. Small heat shock proteins protect against alpha-synuclein-induced toxicity and aggregation. Biochemical and biophysical research communications. 2006;351(3):631–638. doi: 10.1016/j.bbrc.2006.10.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Auluck PK, Meulener MC, Bonini NM. Mechanisms of Suppression of {alpha}-Synuclein Neurotoxicity by Geldanamycin in Drosophila. The Journal of biological chemistry. 2005;280(4):2873–2878. doi: 10.1074/jbc.M412106200. [DOI] [PubMed] [Google Scholar]
- 158.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 Reduces alpha-Synuclein Aggregation and Toxicity. The Journal of biological chemistry. 2004;279(24):25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 159.Shen HY, He JC, Wang Y, Huang QY, Chen JF. Geldanamycin induces heat shock protein 70 and protects against MPTP-induced dopaminergic neurotoxicity in mice. The Journal of biological chemistry. 2005;280(48):39962–39969. doi: 10.1074/jbc.M505524200. [DOI] [PubMed] [Google Scholar]
- 160.De Graeve S, Marinelli S, Stolz F, Hendrix J, Vandamme J, Engelborghs Y, Van Dijck P, Thevelein JM. Mammalian ribosomal and chaperone protein RPS3A counteracts alpha-synuclein aggregation and toxicity in a yeast model system. The Biochemical journal. 2013;455(3):295–306. doi: 10.1042/BJ20130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hanninen AL, Simola M, Saris N, Makarow M. The cytoplasmic chaperone hsp104 is required for conformational repair of heat-denatured proteins in the yeast endoplasmic reticulum. Molecular biology of the cell. 1999;10(11):3623–3632. doi: 10.1091/mbc.10.11.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kong B, Chae Y, Lee K. Degradation of wild-type alpha-synuclein by a molecular chaperone leads to reduced aggregate formation. Cell biochemistry and function. 2005;23(2):125–132. doi: 10.1002/cbf.1219. [DOI] [PubMed] [Google Scholar]
- 163.Kong B, Chae YK, Lee K. Regulation of in vitro fibril formation of synuclein mutant proteins by Hsp104p. Protein and peptide letters. 2003;10(5):491–495. doi: 10.2174/0929866033478717. [DOI] [PubMed] [Google Scholar]
- 164.Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS biology. 2012;10(6):e1001346. doi: 10.1371/journal.pbio.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lo Bianco C, Shorter J, Regulier E, Lashuel H, Iwatsubo T, Lindquist S, Aebischer P. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. The Journal of clinical investigation. 2008;118(9):3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gade VR, Kardani J, Roy I. Effect of endogenous Hsp104 chaperone in yeast models of sporadic and familial Parkinson's disease. The international journal of biochemistry & cell biology. 2014;55:87–92. doi: 10.1016/j.biocel.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 167.Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, Caldwell GA, Shorter J. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156(1-2):170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen Q, Thorpe J, Keller JN. Alpha-synuclein alters proteasome function, protein synthesis, and stationary phase viability. The Journal of biological chemistry. 2005;280(34):30009–30017. doi: 10.1074/jbc.M501308200. [DOI] [PubMed] [Google Scholar]
- 169.Dixon C, Mathias N, Zweig RM, Davis DA, Gross DS. Alpha-synuclein targets the plasma membrane via the secretory pathway and induces toxicity in yeast. Genetics. 2005;170(1):47–59. doi: 10.1534/genetics.104.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sharma N, Brandis KA, Herrera SK, Johnson BE, Vaidya T, Shrestha R, Debburman SK. alpha-Synuclein budding yeast model: toxicity enhanced by impaired proteasome and oxidative stress. Journal of molecular neuroscience. 2006;28(2):161–178. doi: 10.1385/JMN:28:2:161. [DOI] [PubMed] [Google Scholar]
- 171.Petroi D, Popova B, Taheri-Talesh N, Irniger S, Shahpasandzadeh H, Zweckstetter M, Outeiro TF, Braus GH. Aggregate clearance of alpha-synuclein in Saccharomyces cerevisiae depends more on autophagosome and vacuole function than on the proteasome. The Journal of biological chemistry. 2012;287(33):27567–27579. doi: 10.1074/jbc.M112.361865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang F, Yang YP, Mao CJ, Liu L, Zheng HF, Hu LF, Liu CF. Crosstalk between the proteasome system and autophagy in the clearance of alpha-synuclein. Acta pharmacologica Sinica. 2013;34(5):674–680. doi: 10.1038/aps.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ohsumi Y. Historical landmarks of autophagy research. Cell research. 2014;24(1):9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194(2):341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Macedo D, Tavares L, McDougall GJ, Vicente Miranda H, Stewart D, Ferreira RB, Tenreiro S, Outeiro TF, Santos CN. (Poly)phenols protect from alpha-synuclein toxicity by reducing oxidative stress and promoting autophagy. Human molecular genetics. 2015;24(6):1717–1732. doi: 10.1093/hmg/ddu585. [DOI] [PubMed] [Google Scholar]
- 176.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury Jr PT. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 1996;35(43):13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 177.Tardiff DF, Lindquist S. Phenotypic screens for compounds that target the cellular pathologies underlying Parkinson's disease. Drug discovery today Technologies. 2013;10(1):e121–e128. doi: 10.1016/j.ddtec.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mager WH, Winderickx J. Yeast as a model for medical and medicinal research. Trends in pharmacological sciences. 2005;26(5):265–273. doi: 10.1016/j.tips.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 179.Tardiff DF, Khurana V, Chung CY, Lindquist S. From yeast to patient neurons and back again: powerful new discovery platform. Movement disorders. 2014;29(10):1231–1240. doi: 10.1002/mds.25989. [DOI] [PubMed] [Google Scholar]
- 180.Faria C, Jorge CD, Borges N, Tenreiro S, Outeiro TF, Santos H. Inhibition of formation of alpha-synuclein inclusions by mannosylglycerate in a yeast model of Parkinson's disease. Biochimica et biophysica acta. 2013;1830(8):4065–4072. doi: 10.1016/j.bbagen.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 181.Steele JW, Ju S, Lachenmayer ML, Liken J, Stock A, Kim SH, Delgado LM, Alfaro IE, Bernales S, Verdile G, Bharadwaj P, Gupta V, Barr R, Friss A, Dolios G, Wang R, Ringe D, Protter AA, Martins RN, Ehrlich ME, Yue Z, Petsko GA, Gandy S. Latrepirdine stimulates autophagy and reduces accumulation of alpha-synuclein in cells and in mouse brain. Molecular psychiatry. 2013;18(8):882–888. doi: 10.1038/mp.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kritzer JA, Hamamichi S, McCaffery JM, Santagata S, Naumann TA, Caldwell KA, Caldwell GA, Lindquist S. Rapid selection of cyclic peptides that reduce alpha-synuclein toxicity in yeast and animal models. Nature chemical biology. 2009;5(9):655–663. doi: 10.1038/nchembio.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(17):6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tardiff DF, Tucci ML, Caldwell KA, Caldwell GA, Lindquist S. Different 8-hydroxyquinolines protect models of TDP-43 protein, alpha-synuclein, and polyglutamine proteotoxicity through distinct mechanisms. The Journal of biological chemistry. 2012;287(6):4107–4120. doi: 10.1074/jbc.M111.308668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Griffioen G, Duhamel H, Van Damme N, Pellens K, Zabrocki P, Pannecouque C, van Leuven F, Winderickx J, Wera S. A yeast-based model of alpha-synucleinopathy identifies compounds with therapeutic potential. Biochimica et biophysica acta. 2006;1762(3):312–318. doi: 10.1016/j.bbadis.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 186.Dajas F, Rivera F, Blasina F, Arredondo F, Echeverry C, Lafon L, Morquio A, Heinzen H. Cell culture protection and in vivo neuroprotective capacity of flavonoids. Neurotoxicity research. 2003;5(6):425–432. doi: 10.1007/BF03033172. [DOI] [PubMed] [Google Scholar]
- 187.Choi YT, Jung CH, Lee SR, Bae JH, Baek WK, Suh MH, Park J, Park CW, Suh SI. The green tea polyphenol (-)-epigallocatechin gallate attenuates beta-amyloid-induced neurotoxicity in cultured hippocampal neurons. Life sciences. 2001;70(5):603–614. doi: 10.1016/s0024-3205(01)01438-2. [DOI] [PubMed] [Google Scholar]
- 188.Williams RB, Gutekunst WR, Joyner PM, Duan W, Li Q, Ross CA, Williams TD, Cichewicz RH. Bioactivity profiling with parallel mass spectrometry reveals an assemblage of green tea metabolites affording protection against human huntingtin and alpha-synuclein toxicity. Journal of agricultural and food chemistry. 2007;55(23):9450–9456. doi: 10.1021/jf072241x. [DOI] [PubMed] [Google Scholar]
- 189.Fernandes JT, Tenreiro S, Gameiro A, Chu V, Outeiro TF, Conde JP. Modulation of alpha-synuclein toxicity in yeast using a novel microfluidic-based gradient generator. Lab on a chip. 2014;14(20):3949–3957. doi: 10.1039/c4lc00756e. [DOI] [PubMed] [Google Scholar]
- 190.Tardiff DF, Jui NT, Khurana V, Tambe MA, Thompson ML, Chung CY, Kamadurai HB, Kim HT, Lancaster AK, Caldwell KA, Caldwell GA, Rochet JC, Buchwald SL, Lindquist S. Yeast reveal a "druggable" Rsp5/Nedd4 network that ameliorates alpha-synuclein toxicity in neurons. Science. 2013;342(6161):979–983. doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Usenovic M, Knight AL, Ray A, Wong V, Brown KR, Caldwell GA, Caldwell KA, Stagljar I, Krainc D. Identification of novel ATP13A2 interactors and their role in alpha-synuclein misfolding and toxicity. Science. 2012;21(17):3785–3794. doi: 10.1093/hmg/dds206. [DOI] [PMC free article] [PubMed] [Google Scholar]


