Abstract
Essential genes are defined as required for the survival of an organism or a cell. They are of particular interests, not only for their essential biological functions, but also in practical applications, such as identifying effective drug targets to pathogenic bacteria and fungi. The budding yeast Saccharomyces cerevisiae has approximately 6,000 open reading frames, 15 to 20% of which are deemed as essential. Some of the essential genes, however, appear to perform non-essential functions, such as aging and cell death, while many of the non-essential genes play critical roles in cell survival. In this paper, we reviewed and analyzed the levels of essentiality of the Saccharomyces cerevisiae genes and have grouped the genes into four categories: (1) Conditional essential: essential only under certain circumstances or growth conditions; (2) Essential: required for survival under optimal growth conditions; (3) Redundant essential: synthetic lethal due to redundant pathways or gene duplication; and (4) Absolute essential: the minimal genes required for maintaining a cellular life under a stress-free environment. The essential and non-essential functions of the essential genes were further analyzed.
Keywords: cell death, cell survival, essential genes, growth conditions, Saccharomyces cerevisiae
DIFFERENT LEVELS OF ESSENTIALITY
Generally speaking, essential genes are genes required for a cell or an organism to survive. Disruption or deletion of such genes causes cell death, indicating that these genes perform essential biological functions. Some of the essential genes, however, appear to have non-essential functions. FAS2, for example, encodes an α-subunit of fatty acid synthetase, which catalyzes the synthesis of long-chain saturated fatty acids 1. The null mutant could be rescued by fatty acid supplement. It is not clear why FAS2 functions as an essential gene. This also makes the term essential somewhat confusing, because essentiality depends on the environment in which cells live 2. For example, a yeast gene required for respiration is essential only when ethanol or glycerol is the sole carbon source. Here, we have grouped the essential genes into four categories, depending on the circumstances or conditions these genes are required (Fig. 1).
Figure 1. FIGURE 1: A symbolic illustration of different levels of essentiality of the Saccharomyces cerevisiae genes.
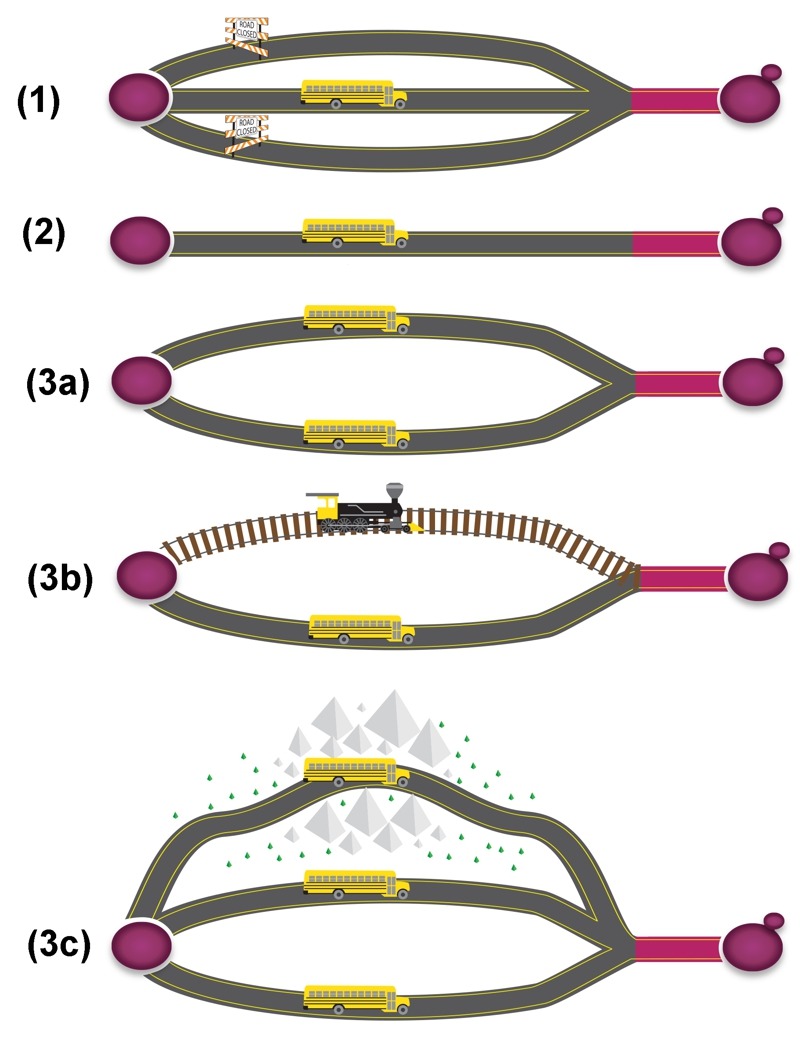
(1) Conditional essential: when all other roads are closed, this alternative road becomes essential.
(2) Essential: the only available road to your destination.
(3a) Redundant essential: two duplicated/similar roads to your destination; (3b) Redundant essential: two different roads (pathways), but to the same destination; (3c) Redundant essential (synthetic sick): Two similar/different roads, plus a less preferred road, when the main roads become unavailable. The last part of the road (marked as red) represents the absolute essential.
(1) Conditional essential: these genes are essential only under certain circumstances or growth conditions. Auxotrophic genes are among the best examples for conditional essential. In the absence of uracil in culture medium, the orotidine-5'-phosphate decarboxylase gene, URA3, which catalyzes the sixth enzymatic step in the de novo biosynthesis of pyrimidines, becomes essential 3. Many genes could be conditionally essential, but the “condition” is unknown (see below for discussion about PKC1). This could be one of the reasons why the molecular functions of more than 10% of the yeast protein-coding genes are still unknown. Many of these genes could be required or essential only under certain conditions, or stresses.
(2) Essential: refers to genes required for survival under the optimal growth condition. This is the most widely accepted definition of essential genes. The “optimal condition” though, is somewhat debatable. The yeast PKC1 gene, for example, encodes a protein serine/threonine kinase, which is essential for cell wall remodeling during growth 4. Deletion of PKC1 is lethal, but the lethality could be recovered by an osmotic stabilizer (such as sorbitol) 5, the “optimal growth condition” for this mutant. Another example is the Acetyl-CoA synthetase gene ACS2, which is required for fermentative growth on glucose. Null mutant is inviable in YPD medium, but is capable of respiratory growth on non-fermentable carbon sources, such as ethanol 6. Therefore, the “optimal growth condition” for acs2Δ, is non-fermentable carbon sources, rather than glucose.
The percentage of genes that are essential in the whole genome varies in different species, ranging from ~2% in Drosophila melanogaster 7 to about 80% in Mycoplasma genitalium 8. In the yeast Saccharomyces cerevisiae, about 20% of the whole genes are essential. The extremely low percentage of essential genes in Drosophila could be attributed to the methods (RNAi) used. RNAi is far less restrictive than the knock-outs used in S. cerevisiae and sometimes leads to hypomorphic results only 7. Because of the importance of essential genes, yeast may have even evolved a protection mechanism to reduce the mutation rate of essential genes by placing them in the cold spots for mutation 9.
Most of the gene products, essential or not, form protein complexes to carry out their molecular functions 10. A recent study showed that essential protein complexes tend to be formed by essential genes and nonessential complexes by nonessential genes, suggesting the existence of modular essentiality 11.
There are also cases where an essential gene becomes non-essential by deletion of another (non-essential) gene. FtsZ is a tubulin-like gene in bacteria that forms the cytoskeletal framework of the cytokinetic ring. It is essential for cell division in wild type bacteria. However, in the L-form of Bacillus subtilis, caused by a single amino acid mutation of yqiG gene, FtsZ becomes non-essential. The B. subtilis cells proliferate by extrusion and resolution, instead of binary fission 12.
(3) Redundant essential (synthetic lethal/sick): eukaryotic cells evolve multiple redundant pathways to resist environmental and genetic perturbations 13. One way of producing a redundant pathway is the so called whole genome duplication, a product of nondisjunction during meiosis, which results in additional copies of the entire genome 14. Gene duplication may also provide stability to an organism by buffering the effect of harmful mutations 15. The Saccharomyces genome contains ~1,060, or ~530 pairs of paralogs, homologous sequences evolved from gene duplication. Synthetic lethality occurs when mutations of both genes cause cell death, while single deletion of either gene is viable. The paralogous genes BDF1 and BDF2, for example, encode bromodomain-containing transcription factors 16. Double deletion is lethal, while single deletion of either BDF1 or BDF2 has no apparent phenotype 17. In addition, bdf1Δ is sensitive to salt stress and the sensitivity can be suppressed by over-expression of BDF2 18. Not all paralogous genes, however, are synthetic lethal. The paralogs CYC1 and CYC7 encode two isoforms of cytochrome c, iso-1-cytochrome c and iso-2-cytochrome c, respectively. Iso-1-cytochrome c and iso-2-cytochrome c constitute approximately 95% and 5% of the total amount of cytochrome c in aerobically grown cells 19. Double deletion of CYC1 and CYC7 completely blocks growth on non-fermentable carbon sources, but the growth is normal on rich glucose medium 20. As for essentiality, BDF1/BDF2 should be considered as redundant essential, while CYC1/CYC7 should be considered as conditional essential. Another example of conditional essential is PET9, which encodes the major ADP/ATP translocator of the mitochondrial inner membrane 21. PET9 has two paralogs, AAC1 and AAC3, both are expressed at a lower level compared to PET9. Similar to CYC1/CYC7 double deletion, the triple deletion of PET9/AAC1/AAC3 is inviable only under anaerobic conditions 22. A study in Caenorhabditis elegans showed that the non-essential (conditional essential) genes are actually duplicated more often, while essential genes (become redundant essential after duplication) are less often duplicated 15. It is unclear how many of the 530 pairs of paralogs in yeast were evolved from non-essential (conditional essential), or essential genes.
(4) Absolute essential (minimal genes): genes that are necessary and sufficient for supporting cellular life in an environment that provides all necessary nutrients and is free from any stress 23. These genes are often referred as minimal genes, and collectively as the minimal gene set. The “stress-free” environment, although easily defined, is difficult to implement in practice, since it is close to impossible to create such a “stress-free” environment.
The parasitic bacterium Mycoplasma genitalium has an extremely small genome of ~580 kb, containing only 482 protein-coding genes. It offers one of the best organisms to study minimal genes 24. Among the 482 genes, 382 were identified as essential and 256 were suggested as minimal genes 25. These minimal genes are mainly involved in DNA replication, protein translation and metabolism. A set of genes required for recombination and DNA repair was also identified in the minimal genes 25, suggesting that DNA repair is a fundamental process required for cell survival.
THE ESSENTIAL FUNCTIONS OF THE ESSENTIAL GENES
To survive, an organism must be able to perform at least two fundamental functions: obtain energy, and reproduce. Cells must convert nutrients from foods, such as proteins, fats and sugars, into adenosine triphosphate (ATP), the ultimate source of energy for living cells. This process of energy conversion is referred as metabolism. Gene ontology (GO) analysis 26 of the 1120 essential genes of Saccharomyces cerevisiae 27 indicates that about 74 % of the essential genes are involved in metabolic process and at least 14% in cell cycle regulation, the two essential functions for cell survival. To keep its simplicity, the discussion below only refers to these essential genes, unless noted otherwise. It should be noted that the genome-wide deletion set for Saccharomyces 27 also contains multiple mutations that can affect phenotypes.
Essential genes and metabolism
More than 700 essential genes are involved in this process that is essential for converting nutrients and providing energy for cells. It includes both catabolism (about 120 genes) and biosynthesis (about 450 genes) of nucleic acids, proteins, carbohydrate, lipids and other macromolecules. Among the ~120 catabolic related genes, 17 are involved in autophagy, a process for macromolecule recycling and intracellular structure remodeling. Recent studies have shown that autophagy plays a much greater physiological role, such as response to starvation and other stress conditions 28,29. In addition to the 17 essential genes, there are about 70 non-essential genes (including conditional essential and redundant essential, such as ATG33 and SCM4) involved in autophagy.
Essential genes and cell division/reproduction
Reproduction is one of the fundamental functions for cell survival. Among the 158 (14%) cell cycle related essential genes, the most important processes of these genes involved include: 1) Mitotic cell cycle, 2) Cytoskeleton organization, and 3) Response to stress and stimulus.
1) Mitotic cell cycle: Not surprisingly, more than half of the essential genes in mitosis are involved in nuclear division, including DNA replication, DNA repair, spindle assembly checkpoint, chromosome condensation and chromosome segregation. Yeast can reproduce either via asexual reproduction (mitosis) or sexual reproduction (meiosis). Meiosis, therefore, is not an essential function for cells to survive. It was puzzling to see 39 out of the 158 cell cycle-related essential genes regulate meiosis. Further analysis revealed that none of the 39 genes are meiotic specific. They are involved in both mitosis and meiosis. The cohesin maintenance factor gene PDS5, for example, is required for sister chromatid condensation and cohesion during mitosis 30,31. It is also required for sister chromatid cohesion in meiosis 32. Some genes, such as DSN1, which encodes an essential component of the MIND kinetochore complex 33, are essential for meiosis 34, but their essentiality for survival is more likely due to their functions in mitosis.
2) Cytoskeleton organization: Saccharomyces cerevisiae reproduces through budding, a stereotypical pattern of polarized growth. Cytoskeleton organization plays an essential role in all steps of cell growth and cell division. The microtubule cytoskeleton related proteins (such as IPL1, and SPC105) are mostly required for nuclear division, including spindle checkpoints. The actin cytoskeleton related proteins are involved in organelle localization, vesicle transport and cytokinesis, such as the septin ring organization. The actin cytoskeleton of budding yeast consists of both the actin cortical patch and the actin cables. The actin cortical patches show a polarized distribution while the actin cables are oriented along the mother-bud axis during cell division 35.
3) Response to stress or stimulus: Faithful DNA replication and transmission of hereditary information is critical to cell survival. Not surprisingly, the majority of these essential stress-response genes are involved in genome integrity checkpoints, DNA damage and DNA repair. Rad53, for example, is a DNA damage response protein. It is required for cell cycle checkpoint function in response to DNA damage 36.
THE “NON-ESSENTIAL” FUNCTIONS OF THE ESSENTIAL GENES
Essential genes and cell death
Cell death, as opposed to cell survival, is not an essential function. GO analysis, however, revealed that at least five essential genes, BIR1, MCD1, ESP1, PDS5 and CDC48 are involved in cell death in budding yeast. Cautions should be taken when using the GO analysis. While it works well on large scales, it may provide inaccurate information on specific genes. PET9 for example, is listed as one of the essential genes involved in cell death. PET9 is required for mitochondrial outer membrane permeabilization and cytochrome c release 37, it is a conditional essential, rather than an essential gene.
Interestingly, all five cell death-related genes play critical roles in mitotic nuclear division. Induction of apoptotic cell death is likely a result of their “secondary” functions or simply by loss of function. Bir1, for example, is a homolog of human survivin and a subunit of the chromosomal passenger complex, which regulates chromosome segregation 38,39. Under oxidative stress, however, the yeast Bir1 becomes a substrate for Nma111, the homologue of the human pro-apoptotic serine protease Omi/HtrA2BIR1, which mediates apoptotic cell death 40. Deletion of BIR1 also causes malfunction of the spindle assembly checkpoint and apoptosis 41, suggesting a possible connection between the functions in checkpoint regulation and anti-apoptosis.
The yeast Esp1 is a caspase-like cysteine protease 42. During metaphase-anaphase transition, Esp1 is released by its inhibitor Pds1, triggering sister chromatid separation by cleavage of the cohesin protein Mcd1. Upon oxidative stress, the Esp1 is prematurely released and cleaves Mcd1. The C-terminal fragment of Mcd1 is translocated into mitochondria and triggers the release of cytochrome c and consequently apoptotic cell death (Fig. 2) 43, suggesting that Mcd1 actively participates in cell death. It would be interesting to see if any other essential proteins perform similar functions, i.e., triggering cell death under stress conditions, to ensure their essentiality.
Figure 2. FIGURE 2: Involvement of essential genes (ESP1, MCD1 and BIR1) in apoptotic cell death. Modified according to 43.

Because of the nature of the essential genes - cells die without them, a still unanswered question is how cells die in the absence of an essential gene. Studies indicate that mutants of yeast essential genes could die apoptotically. In fact, the very first paper published on yeast apoptosis by Frank Madeo and his colleagues involved CDC48, an essential gene that regulates mitotic spindle disassembly. In this pioneer study, they reported that mutants in CDC48 show typical markers of apoptosis 44. Mutation of another essential gene, PDS5, was also reported to lead to features of apoptosis (Fig. 3) 45,46. What is unclear is whether the apoptotic cell death caused by mutation of CDC48 or PDS5 is due to the loss of functions of the essential gene, or like MCD1, the essential gene actively participates in the cell death. If it is due to the loss of function, what are the triggers or inducers of the cell death?
Figure 3. FIGURE 3: TEM image of pds5-1 mutant showing the nucleus unable to divide when switched to non-permissive temperature.
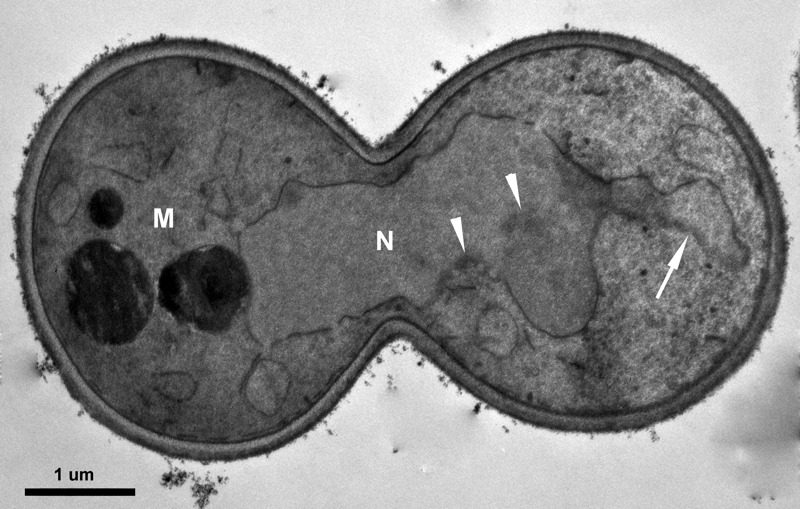
It also shows chromatin condensation (arrowhead) and nucleus with protruding tube (arrow), both typical markers of apoptotic cell death. N = nucleus. M = mother cell, as suggested by the vacuole.
In addition to cell death, GO analysis revealed that five essential genes are related to aging, another “non-essential” function for cell survival. Similar to genes in cell death, all these five essential genes (ACT1, DNA2, NMT1, CDC25 and PGA3) are involved in other essential functions and defects could lead to accumulation of damage that leads to aging. ACT1 for example, encodes the essential gene for actin, a cytoskeleton protein critical for many cellular processes, such as polarized cell growth 47. It has been shown that enhanced actin dynamics can increase the yeast lifespan, while decrease of actin dynamics causes depolarization of the mitochondrial membrane, production of reactive oxygen species (ROS) and cell death 48. Mutation of CDC25, on the other hand, enhances the lifespan in both yeast 49 and mice 50. CDC25 encodes a guanine nucleotide exchange factor, which is required for glucose metabolism by activing the Ras GTPases 51. Mutation of CDC25 reduces glucose/calorie consumption and possibly reduces the level of ROS 49.
Essential genes and mitochondrial organization
Mitochondria are organelles present in virtually all eukaryotic cells. They are the site for numerous important cellular metabolic processes. Although S. cerevisiae is able to carry out fermentative growth on carbon sources such as glucose in the absence of mitochondrial DNA, maintenance of the mitochondrial compartment is essential for survival. GO analysis revealed that about 50 essential genes are involved in mitochondrial organization. A systematic large screening of the 768 essential genes with a regulatable promoter identified about 120 genes that are essential for mitochondrial morphology 52. The majority of these genes are involved in protein targeting/import/transport to mitochondria. Most prominently are the TOM (translocase of outer membrane) proteins (TOM20, TOM22 and TOM40), and the TIM (translocase of inner membrane) proteins (TIM10, TIM12, TIM17, TIM22, TIM23, TIM44, TIM50 and TIM54). These are essential proteins of either the outer or inner mitochondrial membrane that are important for import/insertion of proteins into the mitochondrial inner membrane and to the mitochondrial matrix.
Besides producing ATP as the energy source, mitochondria are involved in a number of essential biosynthetic pathways, such as the ergosterol biosynthetic pathway (encoded by ERG29), which is essential to yeast viability 53. Mitochondria also play a central function in the biogenesis of cellular iron-sulphur proteins, such as Rli1, which is required for ribosome biogenesis and protein translation 54. Recent studies also demonstrated that mitochondria are key regulators of apoptotic cell death 55,56,57.
CONCLUSIONS AND PERSPECTIVES
The reason essential genes are essential is that these genes carry out biological functions that are necessary for cell survival. Many of the “non-essential genes” also perform essential functions. They are deemed non-essential, not because of their functions, but rather their functions are backed up with redundant genes or pathways. Therefore, the definition of essential genes should be expanded to include all genes that carry out essential functions, rather than simply required for survival. The difficulty of using this term is that a lot of essential biological functions are yet to be uncovered, a challenge and a future direction towards a better understanding of cell biology. Another challenge is to decipher the essential function(s) each gene carries. The problem for the “non-essential”, or conditionally essential genes, is to identify the unknown conditions/circumstances under which the genes are required, because mutants deleted for these genes lack any obvious phenotype under normal conditions. The challenge for essential genes is that death is the only “phenotype” in the absence of any essential gene. The generation of the tetracycline-regulatable alleles of essential genes 58 greatly facilitates large scale studies on essential genes, such as those involved in secretion 59, mitochondria 52 or G-protein signaling 60. These studies often identify the non-essential, or secondary functions of the essential genes, which may help further identify the essential functions. The essential gene YPP1/YGR198W was first discovered as a gene required for protection from α-synuclein, a protein that causes Parkinson’s disease in human 61. It is also shown that YPP1 interacts with genes involved in protein sorting and secretion, which further led the discovery of its primary essential functions 62,63. In addition to discovery of the essential functions, it is important to understand their involvement in cell death. Do all the essential genes actively participate in cell death? Is there a universal trigger of cell death in the absence of an essential gene?
Funding Statement
This study was supported in part by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P30 GM103398. The authors thank Ms. Cassidy Mittelstadt (University of Wyoming Graphic design services) for her help designing the illustration (Figure 1).
References
- 1.Fichtlscherer F, Wellein C, Mittag M, Schweizer E. A novel function of yeast fatty acid synthase. Subunit alpha is capable of self-pantetheinylation. Eur J Biochem. 2000;267:2666–2671. doi: 10.1046/j.1432-1327.2000.01282.x. [DOI] [PubMed] [Google Scholar]
- 2.D'Elia MA, Pereira MP, Brown ED. Are essential genes really essential? Trends Microbiol. 2009;17:433–438. doi: 10.1016/j.tim.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Umezu K, Amaya T, Yoshimoto A, Tomita K. Purification and properties of orotidine-5'-phosphate pyrophosphorylase and orotidine-5'-phosphate decarboxylase from baker's yeast. J Biochem. 1971;70:249–262. doi: 10.1093/oxfordjournals.jbchem.a129637. [DOI] [PubMed] [Google Scholar]
- 4.Lee KS, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin DE. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signaling by protein kinase C. Mol Cell Biol. 1993;13:3067–3075. doi: 10.1128/MCB.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sussman A, Huss K, Chio LC, Heidler S, Shaw M, Ma D, Zhu G, Campbell RM, Park TS, Kulanthaivel P, Scott JE, Carpenter JW, Strege MA, Belvo MD, Swartling JR, Fischl A, Yeh WK, Shih C, Ye XS. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Eukaryot Cell. 2004;3:932–943. doi: 10.1128/EC.3.4.932-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Berg MA, Steensma HY. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur J Biochem. 1995;231:704–713. doi: 10.1111/j.1432-1033.1995.0704d.x. [DOI] [PubMed] [Google Scholar]
- 7.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N; Heidelberg Fly Array Consortium. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 8. Chen WH, Minguez P, Lercher MJ, Bork P. OGEE: an online gene essentiality database. Nucleic Acids Res. 2012;40:D901–D906. doi: 10.1093/nar/gkr986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rattray A, Santoyo G, Shafer B, Strathern JN. Elevated mutation rate during meiosis in Saccharomyces cerevisiae. PLoS Genet. 2015;11:e1004910. doi: 10.1371/journal.pgen.1004910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 11.Ryan CJ, Krogan NJ, Cunningham P, Cagney G. All or nothing: protein complexes flip essentiality between distantly related eukaryotes. Genome Biol Evol. 2013;5:1049–1059. doi: 10.1093/gbe/evt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leaver M, Domínguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. Life without a wall or division machine in Bacillus subtilis. Nature. 2009;457:849–853. doi: 10.1038/nature07742. [DOI] [PubMed] [Google Scholar]
- 13.Hartwell L. Genetics: Robust interactions. Science. 2004;303:774–775. doi: 10.1126/science.1094731. [DOI] [PubMed] [Google Scholar]
- 14.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 15.Woods S, Coghlan A, Rivers D, Warnecke T, Jeffries SJ, Kwon T, Rogers A, Hurst LD, Ahringer J. Duplication and retention biases of essential and non-essential genes revealed by systematic knockdown analyses. PLoS Genet. 2013;(5):e1003330. doi: 10.1371/journal.pgen.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lygerou Z, Conesa C, Lesage P, Swanson RN, Ruet A, Carlson M, Sentenac A, Séraphin B. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 1994;22:5332–5340. doi: 10.1093/nar/22.24.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawa C, Nedea E, Krogan N, Wada T, Handa H, Greenblatt J, Buratowski S. Bromodomain factor 1 (Bdf1) is phosphorylated by protein kinase CK2. Mol Cell Biol. 2004;24:4734–4742. doi: 10.1128/MCB.24.11.4734-4742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu J, Hou J, Liu L, Chen L, Wang M, Shen Y, Zhang Z, Bao X. Interplay between BDF1 and BDF2 and their roles in regulating the yeast salt stress response. FEBS J. 2013;280:1991–2001. doi: 10.1111/febs.12219. [DOI] [PubMed] [Google Scholar]
- 19.Downie JA, Stewart JW, Brockman N, Schweingruber AM, Sherman F. Structural Gene for Yeast Iso-2-cytochrome c. J. Mol. Biol. 1977;113:369–384. doi: 10.1016/0022-2836(77)90147-4. [DOI] [PubMed] [Google Scholar]
- 20.Barrientos A, Pierre D, Lee J, Tzagoloff A. Cytochrome oxidase assembly does not require catalytically active cytochrome C. J Biol Chem. 2003;278:8881–8887. doi: 10.1074/jbc.M212427200. [DOI] [PubMed] [Google Scholar]
- 21.Lawson JE1, Douglas MG. Separate genes encode functionally equivalent ADP/ATP carrier proteins in Saccharomyces cerevisiae. Isolation and analysis of AAC2. . J Biol Chem. 1988;263:14812–14818. [PubMed] [Google Scholar]
- 22.Drgon T, Sabová L, Nelson N, Kolarov J. ADP/ATP translocator is essential only for anaerobic growth of yeast Saccharomyces cerevisiae. FEBS Lett. 1991;289:159–162. doi: 10.1016/0014-5793(91)81059-H. [DOI] [PubMed] [Google Scholar]
- 23.Koonin EV. How many genes can make a cell: the minimalgene-set concept. Annu. Rev. Genomics Hum. Genet. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA III, Smith HO, Venter JC. Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci U S A. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saccharomyces Genome Database . GO analysis is based on the analysis tools at Saccharomyces Genome Database. Go Term Finder available at: http://www.yeastgenome.org/cgi-bin/GO/goTermFinder.pl . Go Slim Mapper available at: http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl . [Google Scholar]
- 27. Yeast Deletion Project . The essential gene list is based on the Yeast Deletion Project available at: http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html . [Google Scholar]
- 28.Madeo F, Zimmermann A, Maiuri MC, Kroemer G. Essential role for autophagy in life span extension. J Clin Invest. 2015;125:85–93. doi: 10.1172/JCI73946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 30.Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/S0960-9822(00)00854-X. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Ren Q, Yang H, Conrad MN, Guacci V, Kateneva A, Dresser ME. Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion. Mol Microbiol. 2005;56:670–680. doi: 10.1111/j.1365-2958.2005.04582.x. [DOI] [PubMed] [Google Scholar]
- 33.Euskirchen GM. Nnf1p, Dsn1p, Mtw1p, and Nsl1p: a new group of proteins important for chromosome segregation in Saccharomyces cerevisiae. Eukaryot Cell. 2002;1:229–240. doi: 10.1128/EC.1.2.229-240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar S, Shenoy RT, Dalgaard JZ, Newnham L, Hoffmann E, Millar JB, Arumugam P. Monopolin subunit Csm1 associates with MIND complex to establish monopolar attachment of sister kinetochores at meiosis I. PLoS Genet. 2013;9:e1003610. doi: 10.1371/journal.pgen.1003610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams A, Pringle J. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 37.Pereira C, Camougrand N, Manon S, Sousa MJ, Côrte-Real M. ADP/ATP carrier is required for mitochondrial outer membrane permeabilization and cytochrome c release in yeast apoptosis. Mol Microbiol. 2007;66:571–582. doi: 10.1111/j.1365-2958.2007.05926.x. [DOI] [PubMed] [Google Scholar]
- 38.Uren AG, Beilharz T, O'Connell MJ, Bugg SJ, van Driel R, Vaux DL, Lithgow T. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc Natl Acad Sci U S A. 1999;96:10170–10175. doi: 10.1073/pnas.96.18.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon HJ, Carbon J. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA 96, 1999;96:13208–13213. doi: 10.1073/pnas.96.23.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter D, Wissing S, Madeo F, Fahrenkrog B. The inhibitor-of-apoptosis protein Bir1p protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. . J Cell Sci. 2006;119(Pt 9):1843–1851. doi: 10.1242/jcs.02902. [DOI] [PubMed] [Google Scholar]
- 41.Ren Q, Liou LC, Gao Q, Bao X, Zhang Z. Bir1 deletion causes malfunction of the spindle assembly checkpoint and apoptosis in yeast. Front Oncol. 2012;2:93. doi: 10.3389/fonc.2012.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/S0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 43.Yang H, Ren Q, Zhang Z. Cleavage of Mcd1 by caspase-like protease Esp1 promotes apoptosis in budding yeast. Mol Biol Cell. 2008;19:2127–2134. doi: 10.1091/mbc.E07-11-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madeo F, Fröhlich E, Fröhlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren Q, Yang H, Rosinski M, Conrad MN, Dresser ME, Guacci V, Zhang Z. Mutation of the cohesin related gene PDS5 causes cell death with predominant apoptotic features in Saccharomyces cerevisiae during early meiosis. Mutat Res. 2005;570:163–173. doi: 10.1016/j.mrfmmm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Ren Q, Yang H, Gao B, Zhang Z. Global transcriptional analysis of yeast cell death induced by mutation of sister chromatid cohesin. Comparative and Functional Genomics . 2008;2008:634283. doi: 10.1155/2008/634283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Botstein D, Amberg D, Molholland J, Huffaker T, Adams A, Drubin D, Stearns T. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology, edited by Pringle JR, Broach JR and Jones EW . New York: Cold Spring Harbor Laboratory Press ; 1997. The yeast cytoskeleton. ; pp. 1–90. [Google Scholar]
- 48.Gourlay CW, Carpp LN, Timpson P, Winder SJ, Ayscough KR. A role for the actin cytoskeleton in cell death and aging in yeast. J Cell Biol. 2004;164:803–809. doi: 10.1083/jcb.200310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 50.Borrás C, Monleón D, López-Grueso R, Gambini J, Orlando L, Pallardó FV, Santos E, Viña J, Font de Mora J. RasGrf1 deficiency delays aging in mice. Aging (Albany NY). 2011;3:262–276. doi: 10.18632/aging.100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones S, Vignais ML, Broach JR. The CDC25 protein of Saccharomyces cerevisiae promotes exchange of guanine nucleotides bound to ras. Mol Cell Biol. 1991;11:2641–2646. doi: 10.1128/MCB.11.5.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altmann K, Westermann B. Role of Essential Genes in Mitochondrial Morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5410–5417. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moretti-Almeida G, Netto LE, Monteiro G. The essential gene YMR134W from Saccharomyces cerevisiae is important for appropriate mitochondrial iron utilization and the ergosterol biosynthetic pathway. FEBS Lett. 2013;587:3008–3013. doi: 10.1016/j.febslet.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 54.Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janáky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, Lill R. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005;24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/S0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 56.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 57.Eisenberg T, Büttner S, Kroemer G, Madeo F. The mitochondrial pathway in yeast apoptosis. Apoptosis. 2007;12:1011–1023. doi: 10.1007/s10495-007-0758-0. [DOI] [PubMed] [Google Scholar]
- 58.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13(9):837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 59.Davydenko SG, Juselius JK, Munder T, Bogengruber E, Jäntti J, Keränen S. Screening for novel essential genes of Saccharomyces cerevisiae involved in protein secretion. Yeast. 2004;21:463–471. doi: 10.1002/yea.1063. [DOI] [PubMed] [Google Scholar]
- 60.Cappell SD, Baker R, Skowyra D, Dohlman HG. Systematic analysis of essential genes reveals important regulators of G protein signaling. Mol Cell. 2010;38:746–757. doi: 10.1016/j.molcel.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flower TR1, Clark-Dixon C, Metoyer C, Yang H, Shi R, Zhang Z, Witt SN. YGR198w (YPP1) targets A30P alpha-synuclein to the vacuole for degradation. J Cell Biol. 2007;177:1091–1104. doi: 10.1083/jcb.200610071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baird D, Stefan C, Audhya A, Weys S, Emr SD. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhai C, Li K, Markaki V, Phelan JP, Bowers K, Cooke FT, Panaretou B. Ypp1/YGR198w plays an essential role in phosphoinositide signaling at the plasma membrane. Biochem J. 2008;415:455–466. doi: 10.1042/BJ20080209. [DOI] [PubMed] [Google Scholar]


