Abstract
Alterations in the intracellular levels of calcium are a common response to cell death stimuli in animals and fungi and, particularly, in the Neurospora crassa response to staurosporine. We highlight the importance of the extracellular availability of Ca2+ for this response. Limitation of the ion in the culture medium further sensitizes cells to the drug and results in increased accumulation of reactive oxygen species (ROS). Conversely, an approximately 30-fold excess of external Ca2+ leads to increased drug tolerance and lower ROS generation. In line with this, distinct staurosporine-induced cytosolic Ca2+ signaling profiles were observed in the absence or presence of excessive external Ca2+. High-throughput RNA sequencing revealed that different concentrations of extracellular Ca2+ define distinct transcriptional programs. Our transcriptional profiling also pointed to two putative novel Ca2+-binding proteins, encoded by the NCU08524 and NCU06607 genes, and provides a reference dataset for future investigations on the role of Ca2+ in fungal biology.
Keywords: calcium, cell death, ROS, Ca2+-binding motif, Neurospora crassa
INTRODUCTION
Calcium (Ca2+) is crucial for diverse processes in fungi, including tip growth, virulence, circadian rhythms, nutrient sensing, cell wall regeneration, chemotropic interaction and cell death 1. Ca2+ and cell death are intimately connected, especially because of the vital role of Ca2+ as an intracellular messenger affecting numerous signaling pathways that determine cell fate. Therefore, it is not surprising that cells commonly respond to cell death stimuli with rises in the intracellular levels of this divalent ion. On one hand, these transient or stable modifications in the levels of Ca2+ can operate as survival signals, leading to the activity of anti-death proteins or stimulating transcriptional defence responses. On the other hand, the loss of Ca2+ homeostasis can behave as a pro-death signal, since in some systems overload or a simple perturbation of the distribution of the ion within storage organelles is sufficient to trigger cell death 2. In yeast species like Saccharomyces cerevisiae, Candida albicans and Cryptococcus neoformans, cytosolic Ca2+ increases are a common feature of the cellular response to a number of antifungal stresses, including pheromone 3, the plant essential oils carvacrol 4 and eugenol 5, amiodarone 3,6, and ER stress-inducing agents like tunicamycin and azole drugs 7,8. In N. crassa, a rise in the cytosolic levels of Ca2+ was associated with cell death induced by chitosan 9, Penicillium chrysogenum protein PAF 10, PAF26 11 and staurosporine 12.
We have been using N. crassa to investigate the molecular mechanisms underlying the cell death process induced by the bacterial alkaloid staurosporine. Treatment with staurosporine leads to a complex response that includes the efflux of reduced glutathione (GSH), with a concomitant change in the intracellular redox state to a more oxidative environment and the consequent accumulation of reactive oxygen species (ROS) 13,14. The drug also activates a drug resistance pathway that comprises the zinc binuclear cluster transcription factor CZT-1 15 and ABC-3, an ATP-binding cassette (ABC) transporter 16. A combined genetic and pharmacological strategy coupled to measurements of cytosolic alterations in Ca2+ levels with the photoreporter aequorin revealed a three-step signature response to the drug that involves mobilization of the ion from intracellular stores as well as uptake from the external medium. N. crassa is a good model for the study of Ca2+ dynamics as sequencing of its genome revealed a rich and versatile assortment of molecules involved in Ca2+ homeostasis, including channels, pumps and signaling transducers 17,18,19. A substantial fraction of the genome, however, remains to be characterized and additional unknown members of Ca2+ machinery may exist. The best-studied Ca2+-binding moiety is the classical EF-hand, in which a Ca2+-interacting loop is flanked by two helices 20. However, there are variations on this motif, especially at the structural level, and it is now accepted that more than a dozen different types of Ca2+-binding domains exist 21. In particular, several proteins classes were shown to carry convergently evolved Ca2+-binding Dx[DN]xDG motifs. The distinct evolutionary origins of the motif are evident from the variability of structural contexts flanking the Ca2+-binding region 22,23. Though initially identified in bacteria 24, it was later perceived that this motif is widespread across all kingdoms 22,23, including in a fungal caleosin 23,25 and lectin from Psathyrella velutina 22,26.
We have recently shown that staurosporine-induced cell death in N. crassa involves a precise sequence of Ca2+ signaling events which are completely abolished by the inhibition of extracellular Ca2+ uptake 12. Thus, we investigated the effects of the drug in N. crassa cells growing in limited or excessive amounts of the ion. The results presented here revealed that extracellular Ca2+ modulates cell death and the transcriptional alterations induced by staurosporine, and led to the identification of two novel putative Ca2+-binding proteins, encoded by the NCU08524 and NCU06607 genes.
RESULTS
The availability of extracellular Ca2+ impacts staurosporine-induced cell death and intracellular Ca2+ signaling
Intracellular Ca2+ dynamics play an important role during staurosporine-induced cell death in N. crassa, eliciting a complex response that includes Ca2+ release from internal stores as well as uptake from the extracellular space 12. Thus, we tested the response to staurosporine of cells growing with different Ca2+ concentrations. We prepared a culture medium, designated ‘no Ca2+’, by removing CaCl2 from Vogel’s minimal medium (MM) 27. Small amounts of Ca2+ as impurities from the other reagents in the solution allowed N. crassa to grow well under such conditions (Fig. 1A, middle panel). In contrasting experiments, we supplemented medium with 20 mM CaCl2 (’20 mM CaCl2’ medium), which represents an approximately 30 fold (but not toxic) increase as compared with the 0.68 mM CaCl2 present in standard Vogel’s medium (Fig. 1A, lower panel). We inoculated wild type cells on the centre of Petri dishes containing standard, no Ca2+ or 20 mM CaCl2 solid Vogel’s MM supplemented with staurosporine and measured radial growth during a 104 hour time course. The drug inhibited growth in all media (Fig. 1A-C). However, the staurosporine inhibitory effect was amplified in the absence of Ca2+ (e.g., ~39% of inhibition after 32 hours of treatment with 1 μM staurosporine in no Ca2+ versus ~14% in standard MM). On the other hand, growth inhibition was partially overcome in 20 mM CaCl2 medium (e.g., ~37% of inhibition after 32 hours of treatment with 2.5 μM staurosporine in 20 mM CaCl2 versus ~65% in standard MM). In GFS medium, which contains sorbose that promotes colonial growth 28, the outcome of modifying the extracellular concentration of Ca2+ was similar: the effects of staurosporine were exacerbated in no Ca2+ medium whereas growth inhibition was partially suppressed in the presence of 20 mM CaCl2, (Fig. 1D). Enhancement and suppression of inhibition of growth by absence of Ca2+ and 20 mM CaCl2, respectively, was not reproduced when cells are treated with phytosphingosine (data not shown), another cell death inducer in N. crassa, supporting previous observations that staurosporine and phytosphingosine act by distinct mechanisms 14,29,30.
Figure 1. FIGURE 1: Extracellular Ca2+ modulates the N. crassa sensitivity to staurosporine.
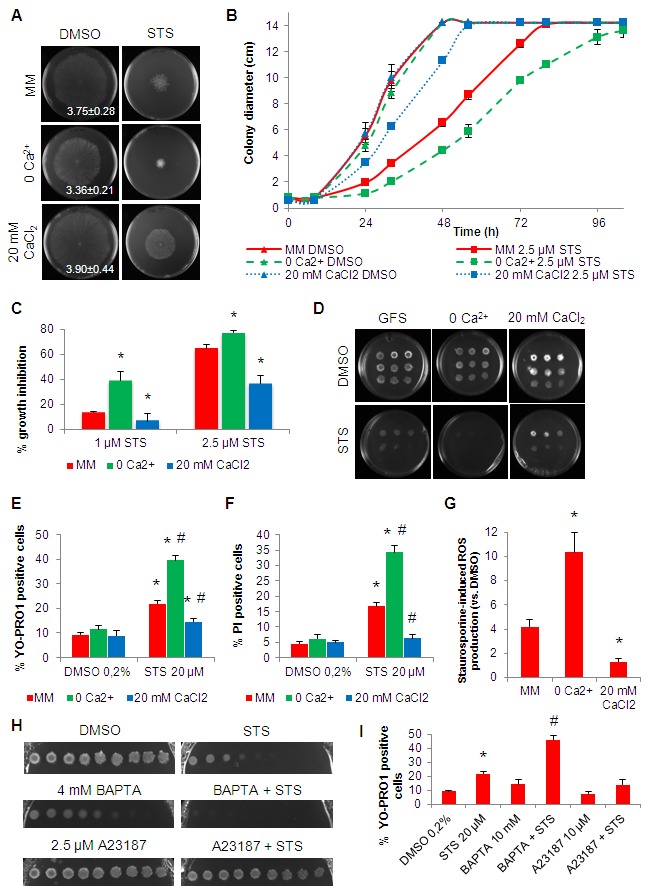
(A-C) Conidia were inoculated on the centre of a Petri dish containing Vogel’s MM with 2.5 μM staurosporine (STS) and different concentrations of Ca2+. Growth at 32h (A), over time (B) and the percentage of growth in STS-treated cells versus control (C) are shown. Growth rates (mm/h) are indicated in the left panels in (A). *, p-value <= 0.05.
(D) Serial dilutions (from left to right) of conidia spotted in GFS agar medium supplemented with 5 μM STS and different concentrations of Ca2+ were incubated for 3 days.
(E-F) Cell death following treatment of the cells with 20 μM staurosporine in the indicated liquid culture media was evaluated by flow cytometry quantification of positive cells for YOPRO-1 (E) or PI (F). *, p-value <= 0.05 for the comparison STS versus DMSO in each media; #, p-value <= 0.05 for the comparison between media after the treatment with STS.
(G) The fold increase in cellular ROS accumulation following treatment with 20 μM staurosporine in the indicated liquid culture media was evaluated by staining with DHR123. *, p-value <= 0.05.
(H-I) Growth inhibition and cell death in 2.5 μM STS-treated cells in the presence of BAPTA or A23187 was assessed by spots in GFS medium (H) or cell growth in liquid medium followed by staining with YOPRO-1 (I) *, p-value <= 0.05 for the comparison STS versus DMSO; #, p-value <= 0.05 for the comparison between STS alone and BAPTA+STS.
In order to check cell death, we analysed cell staining with YOPRO-1 (early apoptosis marker) or PI (late apoptosis/necrosis marker) by flow cytometry. A 2-hour treatment with staurosporine in standard MM resulted in ~22% and ~17% of YOPRO-1 and PI positive cells, respectively (Fig. 1E-F). In no Ca2+ medium, these levels were augmented to ~40% (p-value ~ 0) and ~34% (p-value = 0.029), respectively (Fig. 1E-F). In contrast, the percentage of YOPRO-1 and PI-positive cells was reduced to ~15% (p-value = 0.042) and ~6% (p-value = 0.050), respectively, in 20 mM CaCl2 medium.
The production of ROS is an essential event during the response of N. crassa to staurosporine, as the addition of antioxidants blocks cell death 13. We asked if the availability of Ca2+ in the culture medium influenced cellular ROS production induced by staurosporine. While in standard MM cells stressed for 30 minutes with staurosporine displayed a ~4.2 fold-increase in ROS accumulation, the equivalent accumulation was ~10.4 (p-value = 0.001) and ~1.3 (p-value = 0.014) in no Ca2+ and 20 mM CaCl2 media, respectively (Fig. 1G). Given the importance of ROS formation for cell death provoked by staurosporine, this Ca2+-ROS dependence is likely associated with the distinct cell death phenotypes in the different culture media.
To further stress the modulatory effect of altering the levels of Ca2+ in staurosporine-induced cell death, we combined the drug with the Ca2+ chelator BAPTA and the Ca2+ ionophore A23187 (calcimycin) in GFS plates with standard MM. As expected, extracellular Ca2+ blockage with BAPTA synergized whereas elevation of intracellular Ca2+ with A23187 antagonized the effects of staurosporine (Fig. 1H). In agreement, pre-incubation with BAPTA significantly enhanced apoptotic levels induced by staurosporine, whereas they were partially inhibited by A23187, as measured by YOPRO-1 staining (Fig. 1I). Altogether, these data consistently indicate that the absence of Ca2+ enhances staurosporine-induced cell death, whereas excess (but not toxic) concentrations of Ca2+ partially prevent it.
Using cells expressing the cytosolic Ca2+ reporter gene aequorin, we described recently that staurosporine induces a three-step Ca2+ response comprising three main signals called "A", "B" and "C". Peak "A" occurs immediately after the addition of the drug and lasts approximately 20 minutes; peak "B" appears after 35-40 minutes and lasts approximately 80 minutes; phase "C" is a prolonged and continuous elevation of cytosolic Ca2+ 12. In the absence of extracellular Ca2+, the Ca2+ response to staurosporine was compromised, with peaks "A" and "B" showing substantial reduction (Fig. 2A-B). In medium with 20 mM CaCl2 there was a strong boost in peak "A", while peak "B" remained similar to standard MM. These alterations in intracellular Ca2+ dynamics are in agreement with our findings that extracellular Ca2+ uptake is important for the Ca2+ signaling response to staurosporine 12.
Figure 2. FIGURE 2: Extracellular Ca2+ availability affects intracellular Ca2+ dynamics in response to staurosporine.
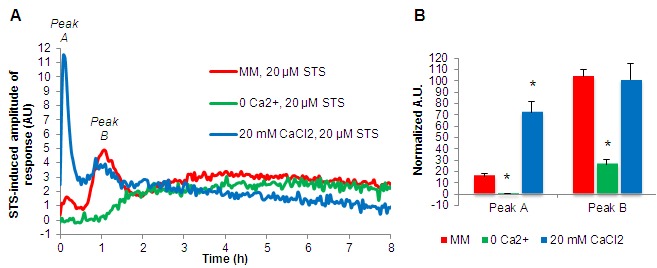
(A) Aequorin-expressing wild type cells were cultured for 6 hours in the indicated media and after the addition of 20 μM staurosporine (STS), luminescence was followed over time. The STS-induced amplitude of response is shown.
(B) Quantification (in arbitrary units) of the cytosolic Ca2+ peaks "A" and "B" (indicated in A). *, p-value < 0.05.
Distinct transcriptional programs in staurosporine-treated cells depend on the extracellular Ca2+ availability
Treatment with 20 μM staurosporine led to significant alterations in the expression of ~28% of the N. crassa genes (1921 genes) 15. The extent of this transcriptional response was amplified in the absence of Ca2+ (~36% of genes (2749 genes) were altered by the drug), and markedly suppressed by 20 mM of CaCl2 (~6% genes (454 genes) showed altered profiles) (Fig. 3A-B). In standard medium, approximately half of the genes were induced and approximately half were repressed by staurosporine 15. In no Ca2+ medium, 43.1% of the altered genes were upregulated, whereas 56.9% where downregulated. Remarkably, in the presence of excessive Ca2+ most of the altered genes were repressed (~68%). The complete dataset obtained from these RNA-seq experiments is presented as File S1.
Figure 3. FIGURE 3: Overview of the N. crassa transcriptional response to staurosporine in minimal media containing different concentrations of Ca2+.
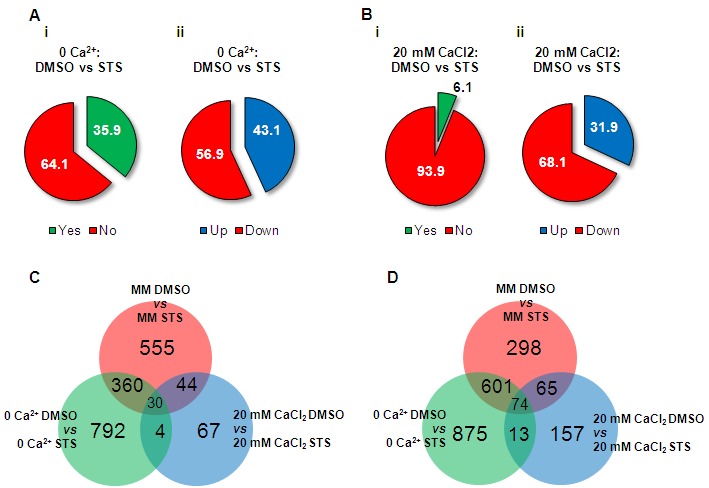
(A-B) The percentages of genes with altered expression upon treatment with staurosporine (i) and the fraction of induced and repressed genes (ii) were calculated for culture medium with no Ca2+ (A) or with 20 mM CaCl2 (B).
(C-D) Venn diagrams were used to assess the amount of Ca2+-specific staurosporine-induced (C) and -repressed genes (D). General statistics for Ca2+-specific transcriptional responses are included.
We analysed the distribution of the individual genes induced and repressed by staurosporine in the different culture media. In the absence of Ca2+, 792 (66.8%) of the induced genes were specific to this condition while in 20 mM CaCl2 this corresponded to 67 (46.2%) genes (Fig. 3C and File S1). Among repressed genes, 875 (56.0%) and 157 (50.8%) genes were specific to no Ca2+ and 20 mM CaCl2 media, respectively (Fig. 3D and File S1). Only minor fractions of the altered genes (30 and 74 genes, induced and repressed, respectively) were common to all conditions.
FunCat was used to examine the enrichment of functional categories in the different gene sets (File S2; Table S1-S2). The most interesting Ca2+ concentration-specific differences were found among the downregulated genes. In the absence of Ca2+, staurosporine led to the downregulation of genes included in numerous categories. These encompass various groups within the ‘Metabolism’ super-category such as ‘metabolism of the aspartate family’, ‘purine nucleotide/nucleoside/nucleobase metabolism’, ‘phosphate metabolism’ and ‘tetracyclic and pentacyclic triterpenes metabolism’. Also, there was a specific repression of genes involved in ‘Cell cycle and DNA processing’ and ‘Transcription’, namely ‘mRNA synthesis’ and ‘mRNA processing’. Categories related to stress responses like ‘Protein fate’, ‘unfolded protein response’, ‘cellular sensing and response to external stimulus’, ‘cell growth’, ‘anti-apoptosis’ were also repressed. Finally, while in standard MM there was a downregulation of ‘aerobic respiration’ genes, in no Ca2+ medium, other bioenergetic pathways were repressed: ‘glycolysis and gluconeogenesis’ and ‘pentose-phosphate pathway’. These data indicates that a highly complex transcriptional program is established by staurosporine-treated cells growing in the absence of Ca2+ resulting in the repression of several genes related with an anti-stress response. It seems that, when Ca2+ is limited, staurosporine causes the downregulation of cellular defences and this may be related to the high susceptibility of these cells to the drug.
Limited and excess of Ca2+ alters basal gene expression
Figure 4. FIGURE 4: Overview of the N. crassa transcriptional response to limited or excess Ca2+.
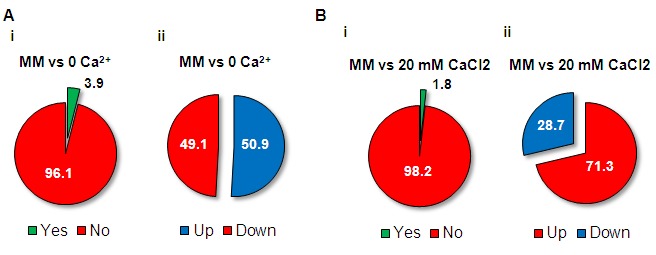
(A-B) The percentages of genes with altered expression (i) and the fraction of induced and repressed genes (ii) were calculated for no Ca2+ (A) and 20 mM CaCl2 medium (B) in comparison with standard MM.
The transcriptional response to the lack or excess of Ca2+ was analysed by comparing basal gene expression in no Ca2+, 20 mM CaCl2 and standard MM (File S3). The fraction of genes with altered expression in no Ca2+ and 20 mM CaCl2 compared with standard MM was 3.9% and 1.8%, respectively (Fig. 4A-B), corresponding to 281 and 129 genes, respectively. While half of the genes altered in no Ca2+ were induced and half were repressed, in 20 mM CaCl2 most of the altered genes were upregulated (71.3%). Most of the genes were specifically induced or repressed depending on the particular culture medium: 97.2% and 97.1%, respectively, for no Ca2+, and 95.7% and 89.2%, respectively, for 20 mM CaCl2 (Fig. 5A-B). Not surprisingly, categories related to ‘ion transport’, ‘homeostasis of cations’ and ‘homeostasis of anions’ were enriched in the set of genes induced in the absence of Ca2+ (Fig. 5A and File S4), which is perhaps linked to a cellular attempt to recover proper conditions of osmolarity. Consistently, these categories were also significantly enriched among the genes repressed in the presence of 20 mM CaCl2 (Fig. 5B). Lack of Ca2+ also seems to be associated to a metabolic adaptation of the cells, since there was enrichment in the induction of genes falling on the ‘metabolism’ super-category, like ‘amino acid metabolism’, ‘nitrogen, sulfur and selenium metabolism’ and ‘anaerobic respiration’.
Figure 5. FIGURE 5: Functional enrichment analysis of the transcriptional alterations caused by limited or excess Ca2+.

(A-B) The transcriptional response to the absence or excess of Ca2+ was investigated by building Venn diagrams showing the distribution of genes with induced (A) or repressed (B) expression in no Ca2+ and 20 mM CaCl2 medium. Lists of enriched categories are included.
We looked for alterations in the expression of known Ca2+ channels, Ca2+-ATPases, Ca2+-exchangers, Ca2+-dependent signaling molecules and other Ca2+ binding proteins 18,19,31,32 (Fig. S1 and File S5). NCU11680, NCU04736 and NCU07075, encoding the TRP channel YVC-1, the Ca2+-ATPase NCA-2 and the Ca2+/H+ antiporter CAX, respectively, were repressed in the absence of Ca2+. NCU04265, encoding the invertase enzyme (INV) was induced in the absence of Ca2+. The induction of INV, an enzyme that hydrolyses extracellular sucrose into glucose and fructose, is a marker of the process of carbon catabolite repression 33,34. NCU08147, encoding the stress-related Ca2+-ATPase ENA-2 35 was repressed in the presence of 20 mM CaCl2. NCU05046 and NCU07966, encoding ENA-1 and TRM-1, respectively, were induced by no Ca2+ and repressed in 20 mM CaCl2 medium. These results further suggest an intracellular metabolic remodeling in response to Ca2+ availability.
Two putative novel components of the N. crassa Ca2+ homeostasis machinery
We found that 188 genes with unknown function ("hypothetical proteins") had altered expression in no Ca2+ or in 20 mM CaCl2 medium. We screened the respective protein sequences for the presence of Ca2+-binding patterns using CaPS 20 and identified 6 candidates (NCU01697, NCU08524, NCU06116, NCU07582, NCU06607 and NCU03647). We further considered NCU08524 and NCU06607, because the others did not display convincing similarities with known proteins, even around the predicted Ca2+-binding motif. NCU08524 was repressed in Ca2+-free medium whereas NCU06607 was induced by 20 mM CaCl2 (Fig. 6A). Fig. 6B shows the alignment of the predicted Ca2+-binding motifs of NCU08524 and NCU06607 with those of proteins containing the Dx[DN]xDG motif 22,23. The NCU08524 motif sequence resembles that of the rhamnogalacturonan lyase YesW from Bacillus subtilis. YesW exemplifies β-propeller structures in which the Dx[DN]xDG motif can be embedded in one of the propeller blades, forming a ‘calcium blade’ structure 22. Such a calcium blade was recently described in a fungal lectin from Psathyrella velutina 22. NCU08524 is similar to a fucose-specific lectin from Macrophomina phaseolina (Table S3) and matches from HHpred suggest that it contains a 6-bladed propeller structure in which β-strands flank the putative Ca2+-binding motif. Taking advantage of the similarity between fucose-specific lectins and NCU08524, molecular models were built for the latter in a situation of Ca2+ binding (Fig. S2A-C). In proteins containing the Dx[DN]xDG motif, interactions with bound Ca2+ ions are supplemented by the interaction of side chains from at least one downstream acidic residue 22. Therefore, three molecular models were prepared, considering Glu502, Glu503 or Asp504 as the possible additional acidic residue. The models show that NCU08524 can accommodate a Ca2+-binding geometry by combining the Dx[DN]xDG motif with any of these downstream residues (Fig. S2A-C).
Figure 6. FIGURE 6: Identification of putative novel Ca2+-binding proteins.
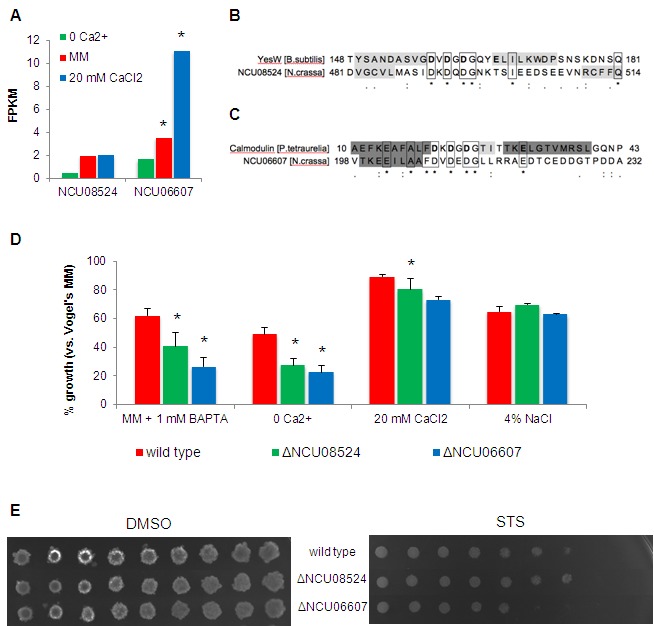
(A) Expression levels of NCU08524 and NCU06607 in no Ca2+, standard MM and 20 mM CaCl2 media. *, p-value <= 0.05.
(B-C) Sequence alignments of the EF-hand-like domain of NCU08524 and YesW from B. subtilis (accession number: O31526; PDB code: 2z8r) (B), and NCU06607 and calmodulin from P. tetraurelia (accession number: P07463; PDB code: 1exr) (C). Residues binding Ca2+ in YesW and calmodulin are in bold; β-strands and α-helices are shaded in light and dark grey, respectively (confirmed by crystallography for YesW 61 and calmodulin 62 and predicted with MEMSAT3 for NCU08524 and NCU06607).
(D) The percentages of growth of the different strains in the indicated media versus standard MM were determined by measuring absorbance of the respective liquid cultures at 450 nm. *, p-value <= 0.05.
(E) The spot assay was used to examine the sensitivity of ΔNCU08524 and ΔNCU06607 to 5 μM staurosporine (STS).
The predicted Ca2+-binding motif of NCU06607 (Fig. 6C) is comparable to the Dx[DN]xDG motif of a calmodulin from Paramecium tetraurelia 23. Molecular modeling of NCU06607 was not feasible because the Dx[DN]xDG motif is not located within a folded domain of known structure, but at the beginning of a predicted intrinsically disordered region. Anyway, the Dx[DN]xDG motif is sometimes found in highly disordered regions 36. NCU06607 has a predicted extracellular localization and there is a strong bias in the localization of proteins possessing the Dx[DN]xDG motif towards the cell surface or secretion 22. Plasma membrane localization together with a transmembrane domain was predicted for NCU08524 (Table S3).
The growth of ΔNCU08524 and ΔNCU06607 was compared with the wild type strain under different conditions of Ca2+ availability. The growth of both mutants was significantly reduced in no Ca2+ medium and in medium containing the Ca2+ chelator BAPTA (Fig. 6D). ΔNCU06607 also showed a significant reduction in growth in the presence of 20 mM CaCl2 as compared with wild type. The observed differences in growth were not merely due to non-specific osmotic stress, since all strains were similarly affected by the addition of 4% NaCl. ΔNCU06607 was slightly more sensitive to staurosporine than wild type, suggesting that the respective protein may be involved in the response to the drug (Fig. 6E). In summary, the RNA-seq dataset and further analysis revealed two putative novel components of the Ca2+-handling machinery in N. crassa.
Several members of the Ca2+ homeostasis machinery are involved in the fungal response to staurosporine
Figure 7. FIGURE 7: Several members of the Ca2+ handling machinery are involved in the fungal response to staurosporine.
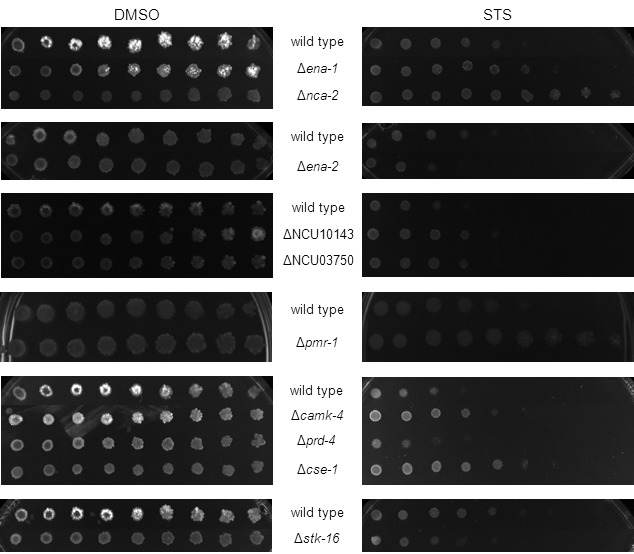
Serial dilutions (from left to right) of conidia from the indicated strains were spotted on GFS medium supplemented with 5 μM staurosporine (STS) and incubated for 3 days.
To further substantiate the crucial role of Ca2+ during the N. crassa response to staurosporine and to identify mediators of the process, we assayed the drug sensitivity profile of approximately 50 deletion strains encoding proteins involved in Ca2+ homeostasis. We showed recently that deletion of genes encoding phospholipase C enzymes, especially PLC-2, results in mutants resistant to staurosporine, while strains lacking the Ca2+ channels CCH-1 and MID-1 are hypersensitive 12. Several other deletion strains that lack Ca2+-permeable channels, Ca2+-ATPases or other Ca2+ binding molecules involved in cellular signaling showed a resistance phenotype to staurosporine that was dissimilar from the wild type strain (Fig. 7). A resistance phenotype was particularly strong in deletion mutants for genes encoding the Ca2+-ATPases NCA-2 and PMR-1, and deletion of other Ca2+-ATPases, namely ENA-1, ENA-2 and NCU10143 also led to increased resistance. The disruption of the signaling molecules calmodulin, CAMK-4, and CSE-1 augmented resistance, whereas strains carrying deletions in genes encoding STK-16 and PRD-4 resulted in increased susceptibility to staurosporine. Table 1 lists strains with altered sensitivity to staurosporine and Table S4 lists all strains tested. The deletion strains for NCU05360, NCU03804, NCU03833, NCU09871, NCU06347, NCU06617, NCU02411 and NCU09265 were not tested due to strain unavailability or their heterokaryotic nature. This data further stresses the importance and complexity of the fungal response to staurosporine, implicating numerous Ca2+-related proteins of different function and subcellular localization in the process.
Table 1.
Strains with Ca2+-related deleted genes that exhibit altered sensitivity to staurosporine when compared with wild type N. crassa.
SS: much more sensitive; S: slightly more sensitive; RR: much more resistant; R: slightly more resistant.
| Strain | Gene name | Classification 17,18,19 | Phenotype |
| ΔNCU02762 | cch-1 | Ca2+-permeable channel | SS 12 |
| ΔNCU06703 | mid-1 | Ca2+-permeable channel | SS 12 |
| ΔNCU04736 | nca-2 | Ca2+-ATPase | RR |
| ΔNCU05046 | ena-1 | Ca2+-ATPase | R |
| ΔNCU08147 | ena-2; ph-7 | Ca2+-ATPase | S |
| ΔNCU10143 | Ca2+-ATPase | R | |
| ΔNCU03292 | pmr-1 | Ca2+-ATPase | RR |
| ΔNCU06245 | plc-1 | Phospholipase C (δ-type) | R 12 |
| ΔNCU01266 | plc-2 | Phospholipase C (δ-type) | RR 12 |
| ΔNCU09655 | plc-3 | Phospholipase C (δ-type) | R 12 |
| ΔNCU03750 | Calmodulin | R | |
| ΔNCU04379 | cse-1 | Ca2+ and/or CaM binding protein | R |
| ΔNCU09212 | camk-4 | Ca2+ and/or CaM binding protein | R |
| ΔNCU00914 | stk-16 | Ca2+ and/or CaM binding protein | S |
| ΔNCU02814 | prd-4 | Ca2+ and/or CaM binding protein | S |
DISCUSSION
Here we show that controlling the amount of Ca2+ available in the extracellular milieu is enough for a robust modulation of the effects of staurosporine in N. crassa. Cell death is strongly enhanced by Ca2+ limitation whereas it is partially suppressed by a ~30-fold excess of external Ca2+. A similar behavior was observed in S. cerevisiae cells treated with miconazole or terbinafine, inhibitors of ergosterol biosynthesis 37, suggesting common stress-responsive pathways in the two fungi, even when the stimuli have apparently distinct targets. Supplementation of the medium with high Ca2+ concentrations (20 mM CaCl2, as used here) protected N. crassa cells from toxicity induced by PAF, an antifungal protein from Penicillium chrysogenum that also disturbs intracellular Ca2+ homeostasis 10. Our results substantiate the view that Ca2+ is an important player during cell death.
The effects of staurosporine in media containing different amounts of Ca2+ are remarkably paralleled by distinct cytosolic Ca2+ dynamics, in line with our previous findings that extracellular Ca2+ uptake is required for the development of the cytosolic Ca2+ signature induced by staurosporine 12. We have recently demonstrated that during the staurosporine-induced Ca2+ signature, peaks "A" and "B" precede cell death, suggesting that they represent signaling events. In the presence of 20 mM CaCl2 the peak "A" is enhanced, whereas in medium lacking Ca2+ peaks "A" and "B" of the staurosporine-induced cytosolic Ca2+ response are abolished. Thus, intracellular Ca2+ signaling that is triggered by staurosporine is markedly modulated by the concentration of Ca2+ in the external medium and might have a role in determining cell survival. However, the intracellular response to staurosporine is very complex and direct conclusions on the susceptibility of the cells based solely on the cytosolic Ca2+ profile, although tempting, are not possible. For instance, a Δplc-2 mutant shows increased resistance to the drug despite that its cytosolic Ca2+ response to staurosporine is nearly obliterated 12. Thus, it seems that events downstream of the cytosolic changes in Ca2+ levels ultimately define cell fate in response to staurosporine.
The screening for staurosporine sensitivity of Ca2+-handling mutant strains revealed a number of proteins likely involved in the cell death response to the drug. Further studies are needed to contextualize them functionally in the mechanism of action of the drug. NCA-2 is a Ca2+-ATPase which pumps excess of cytosolic Ca2+ to the vacuoles or extracellular space 38,39, and the respective knockout cells are very resistant to staurosporine. The knockout strain for PMR-1, a Golgi-localized Ca2+-ATPase 40, is also very resistant to staurosporine and yeast ΔPMR1 knockout cells accumulate much less Ca2+ in the ER 41. Lack of these functions in Δnca-2 and Δpmr-1 cells is likely related to the phenotype of the strains upon treatment with staurosporine, as both the ER and the vacuoles play an important role during the fungal response to staurosporine 12. It is plausible that the lack of NCA-2 or PMR-1 results in an increased accumulation of Ca2+ in the cytosol and this is consistent with the observations that high levels of Ca2+ (below a toxicity threshold) protect against the effects of staurosporine. The lack of NCA-2 has been recently associated with increased sensitivity to UV exposure, further implicating the molecule in cell survival 42. Other Ca2+-related deletion strains present altered resistance to staurosporine, including Δcse-1, Δcamk-4 and Δstk-16. Knockout of cse-1 has been shown to lead to impaired germling communication and fusion 43, as well as increased sensitivity to UV irradiation and Ca2+ 44. A previous study has shown that disruption of camk-4 results in benomyl resistance, while disruption of stk-16 leads to increased sensitivity to fludioxonil, sodium chloride and benomyl 45.
The N. crassa transcriptional response to staurosporine is robustly reduced in the presence of 20 mM CaCl2 when compared with standard MM, whereas limited extracellular Ca2+ causes alterations in an increased number of genes. Thus, different concentrations of extracellular Ca2+ not only lead to distinct intracellular Ca2+ dynamics, ROS accumulation and cell death levels, but also seem to be coupled to unique transcriptional profiles. It appears that, in the absence of Ca2+, cells treated with staurosporine turn off a diverse array of biological processes. We discovered several enriched functional categories in the set of no Ca2+ medium-specific repressed genes, including the repression of genes involved in cell cycle, signal transduction, cell growth, anti-apoptosis, cellular polarization, protein fate and the unfolded protein response. It is possible that the decreased activity of various intracellular pathways explains the inability of cells to cope with the staurosporine insult and the consequent increased cell death.
N. crassa clearly translates changes in extracellular Ca2+ into a transcriptional response, since alterations in gene expression are caused alone by extracellular Ca2+ limitation or overload. This likely results from the homeostatic adaptation to the abnormal Ca2+ environment, i.e., the observed differences in gene expression might be triggered after the uptake or release of Ca2+ to balance the intracellular levels of the ion. Alternatively, these alterations may be secondary to the activity of a surface-localized Ca2+ sensor, although a sequence homologue of the extracellular Ca2+-sensing receptor of animals and plants 46,47 has not been found so far in N. crassa. Given the predicted cell surface localization of the two putative novel Ca2+-binding proteins, NCU08524 and NCU06607, and the growth phenotype of the respective deletion strains under Ca2+ stress, it is tempting to speculate that they could play a role in Ca2+ sensing. The chemotropic interaction during conidial anastomosis tubes during cell fusion is Ca2+-dependent 43 and both NCU08524 and NCU06607 genes have been recently shown to be under the control of the PP-1 transcription factor, a regulator of cell fusion 48,49.
In summary, we show that the effects of the antifungal 13,14,15,16,50 and anticancer 51 agent staurosporine are robustly modulated by the accessibility to Ca2+ in the culture medium. The amount of extracellular Ca2+ affects staurosporine-triggered intracellular events like Ca2+ signaling, the production of ROS and cell death. Our results indicate that these differences are linked to the stimulation of unique transcriptional programs and further highlight the importance of Ca2+ during fungal cell death. In addition, our RNA-seq dataset will be a useful resource for investigations on the role of Ca2+ on different aspects of fungal biology.
MATERIALS AND METHODS
Strains, culture media and chemicals
Wild type 74-OR23-1VA strain and deletion mutants were obtained from the Fungal Genetics Stock Center 52 and handled with standard procedures 28. Minimal medium refers to Vogel’s medium containing 1.5% (w/v) sucrose 27 plus 1.5% (w/v) agar to obtain solid medium. The concentration of KH2PO4 in the 50x Vogel’s stock solution was limited to 10 mM to avoid precipitation with supplemental calcium 10. The following chemicals were used: staurosporine (from LC Laboratories), dimethyl sulfoxide (DMSO), A23187, calcium chloride and sodium chloride (Sigma-Aldrich), phytosphingosine (Avanti Polar Lipids), 1,2-bis(ortho-aminophenoxy)ethane-N,N,N’,N’-tetrasodium (BAPTA) (Merck Biochemicals).
Growth assays
Hyphal growth in solid medium at 26°C was obtained by measuring colony elongation after the inoculation of 20 μl containing 5x104 conidia on the centre of large Petri dishes (ϕ 14.2 cm). Growth in liquid medium was examined by incubating 1x104 conidia/ml at 26°C, 100 rpm, under constant light in 96-well plates (200 μl/well) and following absorbance at 450 nm 53 during 24 hours. The % growth versus the respective control and the growth rate were calculated for each condition. For the spot assays, 3-fold serial dilutions up to 1x104 cells/ml were prepared for each strain. 5 μl from each dilution were spotted on plates containing glucose-fructose-sorbose (GFS) 28 plus the relevant chemicals and incubated for 3 days at 26°C.
Flow cytometry measurements of cell death and reactive oxygen species (ROS)
For each experiment and strain, a control without staining was prepared to define autofluorescence. Samples were read in a BD FACS Calibur and data analyzed with FlowJo (Tree Star). The levels of cell death were determined by staining cells with YOPRO-1 (Life Technologies) or propidium iodide (PI; Sigma-Aldrich). An inoculum of 106 conidia/ml in the appropriate medium was incubated for 4 hours at 26°C (140 rpm, constant light). Staurosporine (20 μM) was added and growth resumed for further 2 hours. The cells were harvested by centrifugation, washed twice with PBS and incubated either with 0.1 μM YOPRO-1 (samples were kept 20 min on ice before reading) or 2 μg/ml PI (samples were immediately ready for cytometry). The production of ROS was measured using the fluorescent probe dihydrorhodamine 123 (Sigma-Aldrich). After growing 106 conidia/ml for 4 hours in the indicated medium at 26°C, 20 μg/ml dihydrorhodamine 123 and staurosporine were added for further 30 minutes. Samples were harvested by centrifugation and washed twice with PBS before being resuspended in PBS and read in the cytometer.
Cytosolic Ca2+ measurements
A bioluminescent method based on the reaction of cytosolic Ca2+ with the genetic probe aequorin in wild type cells was employed as previously described 12. Briefly, 100 μl of conidia at a concentration of 2x106 cells/ml, 5 μM coelenterazine (Santa Cruz Biotechnology) and minimal medium were added to white opaque 96-well plates and incubated for 6 hours at 26°C, in the dark, without agitation. Luminescence (in RLU, relative light units) was captured over time on a Bio-Tek Synergy HT microplate reader. Maximum levels of luminescence, determined in extra wells by measuring luminescence during 3 mins after the injection of 100 μl of 3M CaCl2 in 20% ethanol, were used to normalize each experiment (cytosolic Ca2+ levels (arbitrary units) = experimental RLU values / total emitted luminescence). Quantification was performed by summing the normalized values and data are expressed as mean ± SEM. The values from control DMSO-treated samples were subtracted from staurosporine-treated samples to obtain the "staurosporine-induced amplitude of response".
Statistical and protein sequence analyses
The non-parametric Mann-Whitney test was used for comparisons between two groups using SPSS 20 (SPSS Inc.) and p-values <= 0.05 were considered statistically significant. ClustalW2 54 was used to align protein sequences and PSI-BLAST 55 was used for iterative searches. Secondary structure was predicted with MEMSAT3 56, conserved domains with InterProScan 57, transmembrane domains with TMHMM 2.0 58, subcellular localization with WoLF PSORT 59 and Ca2+-binding motifs with CaPS 20.
High-throughput RNA sequencing (RNA-seq)
Conidial suspensions of 1x106 cells/ml were incubated in culture medium with the indicated concentration of Ca2+ for 6 hours (26°C, 140 rpm, constant light), staurosporine (or DMSO alone) was added and growth resumed for 1 more hour. Cells were harvested and immediately frozen in liquid nitrogen. RNA isolation, mRNA purification and fragmentation and cDNA synthesis was performed as previously described 15. The cDNA libraries were generated using an Illumina TruSeq kit and sequenced in an Illumina HiSeq2000 (single reads of 50 bp were obtained). Sequencing data was handled with Tophat, Cufflinks and Cuffdiff and expression levels are presented as Fragments Per Kilobase of transcript per Million mapped reads (FPKM). Functional enrichment of sets of genes was assessed with FunCat 60. The resulting dataset is available at the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo; series record: GSE53013).
SUPPLEMENTAL MATERIAL
All supplemental data for this article are also available online at http://microbialcell.com/researcharticles/extracellular-calcium-triggers-unique-transcriptional-programs-and-modulates-staurosporine-induced-cell-death-in-Neurospora-crassa/.
Funding Statement
We would like to thank Dr. Nick D Read (Manchester Fungal Infection Group, Institute of Inflammation and Repair, University of Manchester, UK) for providing the aequorin-containing plasmid. A.P.G. was recipient of a fellowship from Fundação Calouste Gulbenkian (104210) and a short-term fellowship from EMBO (329-2012). J.M.C. was hired under the scope of FCT Portugal CIÊNCIA 2008 Programme (FSE-POPH-QREN). This work was supported by FCT Portugal (PEst-C/SAU/LA0002/2013 and FCOMP-01-0124-FEDER-037277 to A.V. and PEst-OE/SAU/UI0215/2014 to P.C.S.), the European POCI program of QCAIII co-participated by FEDER (NORTE-07-0124-FEDER-000003), a grant from the University of Porto (PP_IJUP2011-20 to A.V.) and a grant from the National Institutes of Health (NIH R24 GM081597 to N.L.G. with Drs. JW Taylor and RB Brem).
References
- 1.Shaw BD, Hoch HC. In: Howard RJ, Gow NAR, editors. The Mycota VIII. Berlin: Springer-Verlag KG; 2001. Biology of the fungal cell. pp. 73–89. [DOI] [Google Scholar]
- 2.Zhivotovsky B, Orrenius S. Calcium and cell death mechanisms: a perspective from the cell death community. Cell Calcium. 2011;50(3):211–221. doi: 10.1016/j.ceca.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Pozniakovsky AI, Knorre DA, Markova OV, Hyman AA, Skulachev VP, Severin FF. Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J Cell Biol. 2005;168(2):257–269. doi: 10.1083/jcb.200408145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao A, Zhang Y, Muend S, Rao R. Mechanism of antifungal activity of terpenoid phenols resembles calcium stress and inhibition of the TOR pathway. Antimicrob Agents Chemother. 2010;54(12):5062–5069. doi: 10.1128/AAC.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts SK, McAinsh M, Widdicks L. Cch1p mediates Ca2+ influx to protect Saccharomyces cerevisiae against eugenol toxicity. PLoS One. 2012;7(9):e43989. doi: 10.1371/journal.pone.0043989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta SS, Ton VK, Beaudry V, Rulli S, Cunningham K, Rao R. Antifungal activity of amiodarone is mediated by disruption of calcium homeostasis. J Biol Chem. 2003;278(31):28831–28839. doi: 10.1074/jbc.M303300200. [DOI] [PubMed] [Google Scholar]
- 7.Bonilla M, Nastase KK, Cunningham KW. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 2002;21(10):2343–2353. doi: 10.1093/emboj/21.10.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin DC, Kim H, Mackin NA, Maldonado-Baez L, Evangelista Jr CC, Beaudry VG, Dudgeon DD, Naiman DQ, Erdman SE, Cunningham KW. New regulators of a high affinity Ca2+ influx system revealed through a genome-wide screen in yeast. J Biol Chem. 2011;286(12):10744–10754. doi: 10.1074/jbc.M110.177451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palma-Guerrero J, Huang IC, Jansson HB, Salinas J, Lopez-Llorca LV, Read ND. Chitosan permeabilizes the plasma membrane and kills cells of Neurospora crassa in an energy dependent manner. Fungal Genet Biol. 2009;46(8):585–594. doi: 10.1016/j.fgb.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Binder U, Chu M, Read ND, Marx F. The antifungal activity of the Penicillium chrysogenum protein PAF disrupts calcium homeostasis in Neurospora crassa. Eukaryot Cell. 2010;9(9):1374–1382. doi: 10.1128/EC.00050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munoz A, Marcos JF, Read ND. Concentration-dependent mechanisms of cell penetration and killing by the de novo designed antifungal hexapeptide PAF26. Mol Microbiol. 2012;85(1):89–106. doi: 10.1111/j.1365-2958.2012.08091.x. [DOI] [PubMed] [Google Scholar]
- 12. Gonçalves AP, Cordeiro JM, Monteiro J, Muñoz A, Correia-de-Sá P, Videira A. Activation of a TRP-like channel and intracellular calcium dynamics during phospholipase C-mediated cell death.. J Cell Sci . 2014;in press doi: 10.1242/jcs.152058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro A, Lemos C, Falcao A, Fernandes AS, Glass NL, Videira A. Rotenone enhances the antifungal properties of staurosporine. Eukaryot Cell. 2010;9(6):906–914. doi: 10.1128/EC.00003-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes AS, Castro A, Videira A. Reduced glutathione export during programmed cell death of Neurospora crassa. Apoptosis. 2013;18(8):940–948. doi: 10.1007/s10495-013-0858-y. [DOI] [PubMed] [Google Scholar]
- 15.Goncalves AP, Hall C, Kowbel DJ, Glass NL, Videira A. CZT-1 Is a Novel Transcription Factor Controlling Cell Death and Natural Drug Resistance in Neurospora crassa. G3 (Bethesda) 2014;4(6):1091–1102. doi: 10.1534/g3.114.011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandes AS, Goncalves AP, Castro A, Lopes TA, Gardner R, Glass NL, Videira A. Modulation of fungal sensitivity to staurosporine by targeting proteins identified by transcriptional profiling. Fungal Genet Biol. 2011;48(12):1130–1138. doi: 10.1016/j.fgb.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422(6934):859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 18.Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff C, Galagan JE, Dunlap JC, Loros JJ, Catcheside D, Inoue H, Aramayo R, Polymenis M, Selker EU, Sachs MS, Marzluf GA, Paulsen I, Davis R, Ebbole DJ. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev. 2004;68(1):1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zelter A, Bencina M, Bowman BJ, Yarden O, Read ND. A comparative genomic analysis of the calcium signaling machinery in Neurospora crassa, Magnaporthe grisea, and Saccharomyces cerevisiae. Fungal Genet Biol. 2004;41(9):827–841. doi: 10.1016/j.fgb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Yang W, Kirberger M, Lee HW, Ayalasomayajula G, Yang JJ. Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins. 2006;65(3):643–655. doi: 10.1002/prot.21139. [DOI] [PubMed] [Google Scholar]
- 21.Santamaria-Hernando S, Krell T, Ramos-Gonzalez MI. Identification of a novel calcium binding motif based on the detection of sequence insertions in the animal peroxidase domain of bacterial proteins. PLoS One. 2012;7(7):e40698. doi: 10.1371/journal.pone.0040698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigden DJ, Woodhead DD, Wong PW, Galperin MY. New structural and functional contexts of the Dx[DN]xDG linear motif: insights into evolution of calcium-binding proteins. PLoS One. 2011;6(6):e21507. doi: 10.1371/journal.pone.0021507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rigden DJ, Galperin MY. The DxDxDG motif for calcium binding: multiple structural contexts and implications for evolution. J Mol Biol. 2004;343(4):971–984. doi: 10.1016/j.jmb.2004.08.077. [DOI] [PubMed] [Google Scholar]
- 24.Rigden DJ, Jedrzejas MJ, Moroz OV, Galperin MY. Structural diversity of calcium-binding proteins in bacteria: single-handed EF-hands? Trends Microbiol. 2003;11(7):295–297. doi: 10.1016/S0966-842X(03)00153-7. [DOI] [PubMed] [Google Scholar]
- 25.Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J. Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol. 2000;44(4):463–476. doi: 10.1023/A:1026564411918. [DOI] [PubMed] [Google Scholar]
- 26.Cioci G, Mitchell EP, Chazalet V, Debray H, Oscarson S, Lahmann M, Gautier C, Breton C, Perez S, Imberty A. Beta-propeller crystal structure of Psathyrella velutina lectin: an integrin-like fungal protein interacting with monosaccharides and calcium. J Mol Biol. 2006;357(5):1575–1591. doi: 10.1016/j.jmb.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 27.Vogel HJ. A convenient growth medium for Neurospora (Medium N). Microbial Genet Bull. 1956;13:42–43. [Google Scholar]
- 28.Davis RH, de Serres FJ. Genetic and microbiological research techniques for Neurospora crassa. Meth Enzymol. 1970;17(79-143) doi: 10.1016/0076-6879(71)17168-6. [DOI] [Google Scholar]
- 29.Castro A, Lemos C, Falcao A, Glass NL, Videira A. Increased resistance of complex I mutants to phytosphingosine-induced programmed cell death. J Biol Chem. 2008;283(28):19314–19321. doi: 10.1074/jbc.M802112200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Videira A, Kasuga T, Tian C, Lemos C, Castro A, Glass NL. Transcriptional analysis of the response of Neurospora crassa to phytosphingosine reveals links to mitochondrial function. Microbiology. 2009;155(Pt 9):3134–3141. doi: 10.1099/mic.0.029710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bick AG, Calvo SE, Mootha VK. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336(6083):886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavinder B, Trail F. Role of Fig1, a component of the low-affinity calcium uptake system, in growth and sexual development of filamentous fungi. Eukaryot Cell. 2012;11(8):978–988. doi: 10.1128/EC.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herwig C, Doerries C, Marison I, von Stockar U. Quantitative analysis of the regulation scheme of invertase expression in Saccharomyces cerevisiae. Biotechnol Bioeng. 2001;76(3):247–258. doi: 10.1002/bit.10004. [DOI] [PubMed] [Google Scholar]
- 34.Ziv C, Gorovits R, Yarden O. Carbon source affects PKA-dependent polarity of Neurospora crassa in a CRE-1-dependent and independent manner. Fungal Genet Biol. 2008;45(2):103–116. doi: 10.1016/j.fgb.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Benito B, Garciadeblas B, Rodriguez-Navarro A. Molecular cloning of the calcium and sodium ATPases in Neurospora crassa. Mol Microbiol. 2000;35(5):1079–1088. doi: 10.1046/j.1365-2958.2000.01776.x. [DOI] [PubMed] [Google Scholar]
- 36.Rodrigues JJ, Ferreira HB, Farias SE, Zaha A. A protein with a novel calcium-binding domain associated with calcareous corpuscles in Echinococcus granulosus. Biochem Biophys Res Commun. 1997;237(2):451–456. doi: 10.1006/bbrc.1997.7025. [DOI] [PubMed] [Google Scholar]
- 37.Edlind T, Smith L, Henry K, Katiyar S, Nickels J. Antifungal activity in Saccharomyces cerevisiae is modulated by calcium signalling. Mol Microbiol. 2002;46(1):257–268. doi: 10.1046/j.1365-2958.2002.03165.x. [DOI] [PubMed] [Google Scholar]
- 38.Bowman BJ, Abreu S, Margolles-Clark E, Draskovic M, Bowman EJ. Role of four calcium transport proteins, encoded by nca-1, nca-2, nca-3, and cax, in maintaining intracellular calcium levels in Neurospora crassa. Eukaryot Cell. 2011;10(5):654–661. doi: 10.1128/EC.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowman BJ, Draskovic M, Freitag M, Bowman EJ. Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot Cell. 2009;8(12):1845–1855. doi: 10.1128/EC.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowman BJ, Abreu S, Johl JK, Bowman EJ. The pmr gene, encoding a Ca2+-ATPase, is required for calcium and manganese homeostasis and normal development of hyphae and conidia in Neurospora crassa. Eukaryot Cell. 2012;11(11):1362–1370. doi: 10.1128/EC.00105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strayle J, Pozzan T, Rudolph HK. Steady-state free Ca2+ in the yeast endoplasmic reticulum reaches only 10 microM and is mainly controlled by the secretory pathway pump pmr1. EMBO J. 1999;18(17):4733–4743. doi: 10.1093/emboj/18.17.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deka R, Tamuli R. Neurospora crassa ncs-1, mid-1 and nca-2 double-mutant phenotypes suggest diverse interaction among three Ca2+-regulating gene products. J Genet. 2013;92(3):559–563. doi: 10.1007/s12041-013-0270-y. [DOI] [PubMed] [Google Scholar]
- 43.Palma-Guerrero J, Hall CR, Kowbel D, Welch J, Taylor JW, Brem RB, Glass NL. Genome wide association identifies novel loci involved in fungal communication. PLoS Genet. 2013;9(8):e1003669. doi: 10.1371/journal.pgen.1003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deka R, Kumar R, Tamuli R. Neurospora crassa homologue of Neuronal Calcium Sensor-1 has a role in growth, calcium stress tolerance, and ultraviolet survival. Genetica. 2011;139(7):885–894. doi: 10.1007/s10709-011-9592-y. [DOI] [PubMed] [Google Scholar]
- 45.Park G, Servin JA, Turner GE, Altamirano L, Colot HV, Collopy P, Litvinkova L, Li L, Jones CA, Diala FG, Dunlap JC, Borkovich KA. Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot Cell. 2011;10(11):1553–1564. doi: 10.1128/EC.05140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown EM, Gamba G, Riccardi D, Lombardi M, Butters R, Kifor O, Sun A, Hediger MA, Lytton J, Hebert SC. Cloning and characterization of an extracellular Ca2+-sensing receptor from bovine parathyroid. Nature. 1993;366(6455):575–580. doi: 10.1038/366575a0. [DOI] [PubMed] [Google Scholar]
- 47.Han S, Tang R, Anderson LK, Woerner TE, Pei ZM. A cell surface receptor mediates extracellular Ca2+ sensing in guard cells. Nature. 2003;425(6954):196–200. doi: 10.1038/nature01932. [DOI] [PubMed] [Google Scholar]
- 48.Leeder AC, Jonkers W, Li J, Glass NL. Early colony establishment in Neurospora crassa requires a MAP kinase regulatory network. Genetics. 2013;195(3):883–898. doi: 10.1534/genetics.113.156984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palma-Guerrero J, Leeder AC, Welch J, Glass NL. Identification and characterization of LFD1, a novel protein involved in membrane merger during cell fusion in Neurospora crassa. Mol Microbiol. 2014;92(1):164–182. doi: 10.1111/mmi.12545. [DOI] [PubMed] [Google Scholar]
- 50.Gonçalves AP, Videira A. Programmed cell death in Neurospora crassa. New J Sci. 2014:479015. doi: 10.1155/2014/479015. [DOI] [Google Scholar]
- 51.Gescher A. Staurosporine analogues - pharmacological toys or useful antitumour agents? Crit Rev Oncol Hematol. 2000;34(2):127–135. doi: 10.1016/S1040-8428(00)00058-5. [DOI] [PubMed] [Google Scholar]
- 52.McCluskey K, Wiest A, Plamann M. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosci. 2010;35(1):119–126. doi: 10.1007/s12038-010-0014-6. [DOI] [PubMed] [Google Scholar]
- 53.Alberghina FA. Growth regulation in Neurospora crassa. Effects of nutrients and of temperature. . Arch Mikrobiol. 1973;89(2):83–94. doi: 10.1007/BF00425025. [DOI] [PubMed] [Google Scholar]
- 54.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 55.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones DT. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23(5):538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- 57.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. . Nucleic Acids Res . 2005;33(Web Server issue):W116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 59.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res . 2007;35(Web Server issue):W585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, Mewes HW. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32(18):5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ochiai A, Itoh T, Maruyama Y, Kawamata A, Mikami B, Hashimoto W, Murata K. A novel structural fold in polysaccharide lyases: Bacillus subtilis family 11 rhamnogalacturonan lyase YesW with an eight-bladed beta-propeller. J Biol Chem. 2007;282(51):37134–37145. doi: 10.1074/jbc.M704663200. [DOI] [PubMed] [Google Scholar]
- 62.Wilson MA, Brunger AT. The 1.0 A crystal structure of Ca2+-bound calmodulin: an analysis of disorder and implications for functionally relevant plasticity. . J Mol Biol. 2000;301(5):1237–1256. doi: 10.1006/jmbi.2000.4029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


