Abstract
Cathepsin D has garnered increased attention in recent years, mainly since it has been associated with several human pathologies. In particular, cathepsin D is often overexpressed and hypersecreted in cancer cells, implying it may constitute a therapeutic target. However, cathepsin D can have both anti- and pro-survival functions depending on its proteolytic activity, cellular context and stress stimulus. Therefore, a more detailed understanding of cathepsin D regulation and how to modulate its apoptotic functions is clearly needed. In this review, we provide an overview of the role of cathepsin D in physiological and pathological scenarios. We then focus on the opposing functions of cathepsin D in apoptosis, particularly relevant in cancer research. Emphasis is given to the role of the yeast protease Pep4p, the vacuolar counterpart of cathepsin D, in life and death. Finally, we discuss how insights from yeast cathepsin D and its role in regulated cell death can unveil novel functions of mammalian cathepsin D in apoptosis and cancer.
Keywords: cathepsin D, cancer, apoptosis, yeast cathepsin D, yeast model
CATHEPSINS
Cathepsins are members of a large protease family, which can be subdivided according to their structure and active-site amino acid into cysteine (cathepsins B, C, F, H, K, L, O, S, V, W, and X), serine (cathepsins A and G), and aspartic cathepsins (cathepsins D and E). While cathepsins B, L, H, C and D are ubiquitously expressed in human tissues, expression of cathepsins A, G, K, S, V, X and W is tissue and cell type specific 1,2,3,4. In general, cathepsins are found in acidic cellular organelles, lysosomes and endosomes. Initially, their function was thought to be limited to bulk degradation of proteins delivered to the lysosome by endocytosis or autophagocytosis. However, it was later demonstrated that cathepsins possess highly specific and directed proteolytic activity, and that they can be found in other cellular compartments 5,6,7,8,9,10. Numerous physiological functions of cathepsins have been uncovered, including a role in hormone and antigen processing, bone and tissue remodeling, growth factor and proenzyme activation and, more recently, in the immune response 5,6,11,12,13. Cathepsins also participate in apoptosis and are translocated from the lysosomal lumen to the cytosol of mammalian cells through lysosomal membrane permeabilization (LMP) in response to a variety of apoptotic signals 14,15,16. These lysosomal proteases can also be secreted from the cell and degrade extracellular matrix proteins such as collagen, fibronectin, proteoglycans and laminin 17.
In addition to their physiological function, cathepsins have also been associated with several pathologies such as cardiovascular diseases, osteoporosis, rheumatoid arthritis, atherosclerosis and cancer 6,11,17,18,19. Elucidating the mechanisms underlying the involvement of cathepsins in the pathogenesis of these diseases, and how they can be modulated to develop new prevention and therapeutic strategies, has therefore taken center stage. Among cathepsins, cathepsin D (CatD) has attracted increased attention in recent years due to its importance in the mediation of lysosomal cell death pathways and in cancer. In this review, we will concentrate on both physiological and pathological functions of CatD, as well as on yeast as a model system to study CatD pathophysiology.
ROLE OF CATHEPSIN D IN CELLULAR PHYSIOLOGY AND PATHOLOGY
CatD is a soluble aspartic endopeptidase found in the lysosomes of most mammalian cells. Like other cathepsins, CatD is activated by proteolytic cleavage of the synthetized inactive zymogen (preproCatD), which is composed of an N-terminal signal peptide, a propeptide, and a catalytic domain 20,21,22. The signal peptide directs the nascent chain to the endoplasmic reticulum, where it is cleaved in the lumen. ProCatD is then N-glycosylated and transported to the Golgi, where the N-glycan structures acquire mannose-6phosphate (Man-6P) residues that can bind to Man-6P receptor(s) (Man-6PR), and the complex is directed to the lysosomal compartment 23. In the acidic milieu, proCatD (52 kDa) undergoes further proteolytic processing by cleavage of the proregion, resulting in the 48 kDa single chain intermediate active form. Finally, this chain is processed into mature active CatD, composed of heavy (34 kDa) and light (14 kDa) chains linked by non-covalent interactions 24,25,26. It has been shown that CatD processing involves cysteine cathepsins 26,27 and, more recently, that it is independent of its own catalytic function and auto-activation but requires CatL and CatB 28. Although proCatD and CatD are mostly intracellular, they can also localize in the extracellular matrix and synovial fluid of cartilage 29,30,31. ProCatD/CatD are also found in human, bovine and rat milk 32,33,34, serum, sweat and urine 35,36, and extracellularly in macrophage-rich regions of atherosclerotic lesions 37. ProCatD secretion by human keranocytes 38, mammalian epithelial cells 39 and different types of cancer cells 18,40 was also demonstrated.
It is widely accepted that the major function of CatD is its involvement in general protein degradation and turnover within the lysosomal compartment. However, CatD has also emerged as an important regulator and signaling molecule with numerous physiological functions. These include activation of enzymatic precursors, prohormones and growth factors, processing of brain-specific antigens, tissue homeostasis, and participation in apoptosis 18,41. CatD has also been associated with different pathological scenarios such as cancer progression and metastasis, Alzheimer’s disease, atherosclerosis and inflammatory disorders 11,12,40,42, and found to be a specific biomarker for several pathologies. The involvement of CatD in both physiological and pathological processes has been addressed in multiple studies, some of which are summarized in Table 1 38,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62. A more detailed description of the role of CatD in cancer is given below.
Table 1.
Cellular roles of cathepsin D in physiological and pathological processes.
| Role | Model | References |
| Limited proteolysis of proteins regulating cell growth and/or tissue homeostasis | In vivo: CatD-deficient mice | 43 |
| Postnatal tissue homeostasis including tissue renewal, remodeling, aging and RCD | In vivo: CatD-deficient mice; CatD-mutant mice | 44,45,246,47 |
| Neuronal ceroid lipofuscinosis in both animals and humans characterized by severe neurodegeneration, developmental regression, visual loss and epilepsy | In vivo: CatD-deficient mice; CatD-mutant mice Human pathologies Animal diseases | 45 48 49 50 51 |
| Wound healing, epidermal differentiation and pathological conditions such as psoriasis | In vivo: CatD-deficient mice In vitro: normal and psoriatic keranocytes Human patients | 52 53 54 |
| Proliferation and regeneration in keratinocytes and possibly in skin regeneration | In vitro: keratinocyte cell line HaCaT | 38 |
| Processing of proteins involved in Alzheimer disease pathogenesis, such as apolipoprotein E (apoE) and Tau protein | Human patients Recombinant protein | 55 56 |
| Post-partum cardiomyopathy resulting in heart failure | In vivo: mutant mice | 57 |
| Autism pathogenesis | Autistic subjects | 58 |
| Innate immune responses and Parkinson disease | In vivo: CatD-deficient mice Human patients | 59 60 |
| Intracellular metabolism, transport of phospholipids and cholesterol | Human patients | 61 |
| Atherosclerotic lesions associated with proCatD release from monocyte-derived macrophages | Atherosclerosis patients In vitro: cultured atherosclerotic plaques | 62 |
THE ROLE OF CATHEPSIN D IN CANCER
Numerous reports have demonstrated that CatD is overexpressed in several cancer types 18,40,42,63,64,65, often correlating with poor prognosis. In particular, CatD is considered an independent prognostic marker in breast cancer associated with metastatic risk 66,67,68 and in colorectal cancer (CRC) 69,70. Mechanistically, the majority of reports attribute its role in cancer to overexpression of proCatD. As an example, transfection of rat tumor cells with human proCatD cDNA leads to increased proliferation, invasion and metastasis in vitro and in vivo 71. Accordingly, anti-proCatD antibodies can inhibit tumor growth both in vitro and in vivo 72,73,74. Overexpressed proCatD escapes normal targeting routes and is hypersecreted to the extracellular milieu, where it can act in multiple fashions. On one hand, it can exert an autocrine effect, inducing cancer cell growth by interacting with cell surface receptors 72,75,76,77. This autocrine role has so far been observed in breast, prostate, ovarian and lung cancer cells 72,73,74,78. In addition, proCatD can play a crucial paracrine role in the tumor microenvironment by stimulating fibroblast outgrowth and tumor angiogenesis 71,79, as well as inhibiting anti-tumor responses 80. When in the tumor microenvironment, proCatD may also affect stromal cell behavior and/or degrade components from the extracellular matrix 81,82, including the release of growth factors 83. Although it has been suggested that proCatD can be processed in the acidic extracellular space to catalytically active CatD 84, the enzymatic activity of CatD is reportedly not required for its mitogenic role. Indeed, a proteolytically inactive mutant of CatD (D231N) is still mitogenic for fibroblasts 85, as well as for cancer cells both in vitro, in three-dimensional matrices, and in athymic nude mice 71,86. Similarly, proCatD stimulates angiogenesis in tumor xenografts of athymic nude mice independently of its catalytic activity 85, also suggesting that CatD can signal through protein-protein interactions.
Though less extensive, there are also examples of CatD roles in cancer cells that are not attributed to proCatD. For instance, intracellular CatD can stimulate cancer cell growth by inactivating secreted growth inhibitors 87,88. Moreover, mature CatD released into the cytosol as a consequence of the reportedly higher susceptibility of cancer cells to LMP 15,89 may interact with and/or degrade pro- and anti-apoptotic proteins, modulating cell death 41.
Targeting CatD is a promising strategy in the clinic, but requires further detailed elucidation of its mechanisms of action. In the following section, we focus on the role of CatD in the apoptotic process, which is of particular relevance for cancer research. These studies may however also offer clues into the function of CatD in other physiological and pathological scenarios.
OPPOSING FUNCTIONS OF CATHEPSIN D IN APOPTOSIS
In recent years, multiple studies have shown that CatD is a central player in the apoptotic response, both under physiological and pathological conditions. In fact, depending on the cell type and context, CatD can induce or inhibit apoptosis, acting through different mechanisms 41. On one hand, CatD can directly induce apoptosis triggered by several stimuli such as staurosporine 90, etoposide, 5-fluorouracil and cisplatin 91, as well as resveratrol 92 and others, possibly mediated by intrinsic or extrinsic pathways 41. In the intrinsic pathway, the role of CatD is linked to the release of mature 34 kDa CatD into the cytosol and cleavage of Bid to form tBid, triggering insertion of the pro-apoptotic protein Bax into the mitochondrial membrane 15. Subsequent mitochondrial outer membrane permeabilization leads to the release of pro-apoptotic molecules such as cytochrome c and apoptosis inducing factor (AIF) to the cytosol 15. For instance, it has been shown that CatD mediates cytochrome c release and caspase activation in human fibroblasts undergoing staurosporine-induced apoptosis 90, and cleaves Bid and promotes apoptosis via oxidative stress-induced LMP in human neutrophils 93. In addition, Pepstatin A and/or knockdown of CatD expression by RNA interference prevent resveratrol toxicity, impeding Bax oligomerization, mitochondrial membrane permeabilization, cytochrome c release and caspase 3 activation in DLD1 and HT29 CRC cell lines 92. One study also reports that CatD mediates selective release of AIF in T lymphocytes entering the apoptosis early commitment phase through activation of Bax in a Bid-independent manner 94. This shows that CatD can be involved in caspase-independent apoptosis by activating Bax independently of Bid cleavage. Other studies strongly suggest that cytosolic CatD may have an additional role involving protein-protein interactions. As examples, it has been shown that overexpression of either catalytically active or inactive CatD by cancer cells enhances apoptosis-dependent chemo-sensitivity 95, and that stress-induced apoptosis is not affected in fibroblasts synthesizing a catalytically inactive CatD 96. Additionally, microinjection of inactive proCatD into the cytosol of both human fibroblasts and HeLa cells induces apoptosis 97. Interestingly, one report also indicates that cytosolic mature CatD may reach the nucleus during cell death 98.
In contrast with the multiple studies showing CatD is pro-apoptotic, other studies describe an anti-apoptotic function of CatD. Most of these suggest it plays an anti-apoptotic role in cancer cells. For example, CatD downregulation sensitizes human neuroblastoma cells to doxorubicin-induced apoptosis, while CatD overexpression has the opposite effect 99. Accordingly, inhibition of CatD with pepstatin A induces caspase-dependent apoptosis in neuroblastoma cell lines 100. Moreover, overexpression of intracellular CatD in mouse xenografs using rat-derived cell lines inhibits apoptosis 71, and expression of wild type or a catalytic mutant of CatD promotes survival and invasive growth of CatD‐deficient fibroblasts 85. Another study in glioblastoma cells proposes that CatD stimulates autophagy induction, inhibiting apoptotic cell death under genotoxic conditions 101. More recently, we showed that inhibition of CatD in CRC cells with small interfering RNA (siRNA) or pepstatin A enhances acetate-induced apoptosis associated with a decrease in mitochondria degradation independently of autophagy 102,103. An anti-apoptotic role of CatD has also been described under physiological conditions using CatD-deficient mice 43,44,45. Indeed, mutant mice developed apoptosis in the thymus, thalamus and retina.
In summary, it is well documented that CatD plays an important role in apoptosis regulation, both with and without involvement of its proteolytic activity. However, the exact role of CatD in apoptosis, particularly what determines whether this protease plays an anti- or pro-apoptotic function remains poorly understood. In this regard, a simpler model system would be particularly useful to offer additional clues into this dichotomy.
YEAST VACUOLAR PROTEASES
The versatility of the yeast Saccharomyces cerevisiae to study several conserved cellular functions such as cell metabolism, cell cycle, cell death and organelle biogenesis has justified the attractiveness of this system to study more complex mammalian physiological and pathological processes 104,105,106,107,108. Like other organelles, the yeast vacuole is functionally similar to its higher eukaryote counterpart, the lysosome. It harbors seven characterized proteases, namely three aminopeptidases, three serine proteases and one aspartyl protease. Among these, two are endopeptidases: proteinase A (Pep4p), ortholog to human CatD, and proteinase B (Prb1p). Five are exopeptidases: carboxypeptidase Y (CPY), ortholog to human CatA, carboxypeptidase S (CPS1), aminopeptidase I (Ape1) and Y (Ape3), and dipeptidylaminopeptidase B (Dap2).
More recently, Hecht et al. reported an eighth vacuolar protease, a transmembrane metalloprotease (Pff1) 109, but although evidence of Pff1 vacuolar localization was shown, its proteolytic activity has yet to be demonstrated.
The endopeptidases are responsible for the majority of bulk protein degradation, including of plasma membrane proteins. They are also fundamental for activation of the vacuolar proteolytic cascade, particularly Pep4p, since it is involved in proteolytic activation of Prb1p, CPY and Ape1 110,111. Prb1p, in turn, participates in the activation of Pep4p, CPY, CPS1, Ape1 and Ape3. Both carboxypeptidases and Ape1 are involved in peptide and glutathione degradation, respectively, but are not required for zymogen activation 111,112.
Substrates for the vacuolar proteases are mostly imported via endocytosis (extracellular and cell surface proteins) or autophagy (cytoplasmic material and organelles). Autophagy is activated under nutrient deprivation conditions, and both Pep4p and Prb1p are implicated in the dissolution of autophagic bodies 113,114.
In addition, vacuolar proteases play a role in sporulation. While absence of Prb1p activity alone results in partial reduction of sporulation, absence of Prb1p activity in a mutant lacking both CPY and CPS1 leads to almost complete loss of sporulation ability 115. In addition to ensuring protein homeostasis under physiological conditions, vacuolar proteolysis therefore also appears to be a stress-responsive process, particularly under nutrient stress conditions and during sporulation. However, additional roles for vacuolar proteases have emerged in recent years, in particular for Pep4p.
Pep4p PROTEASE - THE YEAST CATHEPSIN D
Yeast CatD (Pep4p), like its lysosomal counterpart, is synthesized as an inactive zymogen, traveling via the endoplasmic reticulum and Golgi to the acidic vacuoles, where it is activated through proteolytic removal of the inhibitory propeptide 116. Although Pep4p is mainly located in the vacuole, different cell death stimuli can lead to its release to the cytosol, involving a selective vacuolar membrane permeabilization (VMP) typical of apoptotic death.
Mason et al. were the first to report that Pep4p translocates from the vacuole to the cytosol 117. These authors observed an increase in nuclear permeability associated with increased accumulation of reactive oxygen species (ROS) during H2O2-induced cell death, and found that Pep4p is released into the cytosol and degrades nucleoporins during this process. However, Pep4p did not affect resistance to H2O2-induced cell death, probably because it migrates out of vacuoles after cells are effectively unviable. They further showed that the release of a Pep4p-EGFP (Enhanced Green Fluorescent Protein) fusion from the vacuole in H2O2-treated cells was not associated with major rupture of the vacuolar membrane, as cells maintained a vacuolar lumen morphologically distinct from the cytosol. Other authors reported that Pep4p is involved in protein degradation and removal of oxidized proteins during H2O2-induced oxidative stress, but also did not ascribe a role for this protease in cell death induced by H2O2 118.
Another study showed that stabilization of the actin cytoskeleton caused by lack of the actin regulatory protein End3p leads to loss of mitochondrial membrane potential, accumulation of ROS, increase in VMP and consequent migration of Pep4p to the cytosol, as well as apoptotic cell death 119. In that study, Pep4p-EGFP was visualized exclusively in the vacuole lumen in wild type cells, but distributed throughout the entire cell in an END3-deficient strain. Again, no role was attributed to this protease in actin-stabilized dying cells.
Pep4p is also involved in programmed nuclear destruction during yeast gametogenesis 120. Using cells co-expressing Pep4p-mCherry and Vma1-GFP, a GFP-tagged vacuolar membrane protein, Pep4p was shown to translocate from the vacuole into the ascal compartment of early postmeiotic cells during sporulation, with preservation of vacuolar integrity.
These observations show that VMP seems to mimic LMP in human cells. However, they do not indicate whether yeast vacuolar proteases play a role in cell survival and regulated death.
In this regard, it has been shown that Pep4p has a pro-survival role during chronological aging, since a Pep4p-deficient mutant has a shortened lifespan associated with higher levels of carbonylated proteins 118. Carmona-Gutiérrez et al. further showed that deletion of PEP4 results in both apoptotic and necrotic cell death during chronological aging 121. Using a panel of Pep4p mutants, they conclude that Pep4p plays a dual pro-survival role composed of both anti-apoptotic and anti-necrotic functions, conferred by its proteolytic activity and its proteolytically inactive propeptide, respectively. We also previously found that Pep4p-EGFP translocates to the cytosol during acetic acid-induced apoptosis involving selective VMP in S. cerevisiae W303 cells, with preservation of both vacuolar and plasma membrane integrity 122. Moreover, we demonstrated that Pep4p is required for increased cell survival and for efficient autophagy-independent mitochondrial degradation in response to this acid in a manner depending on its catalytic activity 122,123. This suggests that VMP associated with Pep4p release may act as an alternative mitochondrial degradation process, delaying cell death. In contrast, we recently demonstrated that absence of PEP4 resulted in increased resistance to acetic acid in S. cerevisiae BY4741 cells 124. This prompted the hypothesis that Pep4p plays a dual function in acetic acid-induced cell death depending on the genetic background, providing an interesting tool to explore the molecular determinants of CatD function.
YEAST AS A TOOL TO EXPLORE THE ROLE OF CATHEPSIN D IN APOPTOSIS AND CANCER
It is widely established that the process of regulated cell death (RCD) involves a genetically encoded molecular machinery 125. Core components of this machinery are conserved in yeast, which can undergo RCD exhibiting typical markers of apoptosis, autophagy and necrosis 126,127,128. Thus, this eukaryotic organism has been used extensively to study the molecular mechanisms of RCD pathways, reviewed elsewhere 126,127,128,129. These studies encompass not only analysis of yeast endogenous death pathways but also heterologous expression of human proteins involved in apoptosis, such as caspases, Bcl-2 family proteins, PKC isoforms and the p53 tumor suppressor protein 130,131.
As discussed above, the role of the lysosome-like vacuole in the regulation of RCD has been investigated in yeast, where it has been shown to play a role similar to lysosomes 132,133. However, the use of this model organism to study lysosomal cell death pathways in general and cathepsin function in particular is still underexplored. So far, only translocation of Pep4p to the cytosol during yeast apoptosis has been clearly demonstrated by different authors 117,119,122. One other study shows that the RNase T2 family member Rny1p is also released from the vacuole into the cytosol during oxidative stress, with preservation of vacuolar membrane integrity, directly promoting cell death 134. The need for a comprehensive analysis of the VMP process and the vacuolar proteins released in response to different stimuli is therefore evident.
Another approach that has not been sufficiently exploited is the heterologous expression of cathepsins in yeast. Two studies have shown that rat cathepsin L and D precursor polypeptides are recognized by mechanisms similar to those involved in the intracellular sorting of vacuolar proteins in yeast cells 135,136. We therefore sought to further explore this tool to understand the function of human CatD. As mentioned above, we previously showed the parallel between the role of human and yeast CatD in acetate/acetic acid-induced apoptosis and in the degradation of damaged mitochondria, which render CRC/yeast cells more resistant to apoptosis induced by acetate/acetic acid 102,122. We now found that heterologous expression of human CatD in yeast PEP4-deficient cells reverts their sensitivity to acetic acid-induced apoptosis and delays mitochondrial degradation 103, as previously observed for wild type Pep4p 122,123. These results provide evidence that the role of CatD in both apoptosis and mitochondrial degradation is conserved through evolution. Further elucidation of the molecular mechanisms underlying the involvement of CatD in apoptosis and in mitochondrial degradation will now be crucial to develop novel strategies to specifically inhibit this protease in apoptosis deficiency-associated diseases, such as cancer.
Taking into account the multiple functions of CatD, one caveat of using CatD inhibitors could be a negative effect on Bax activation, release of cytochrome c and downstream caspase activation. To address this question, we exploited the well-established system of heterologous expression of Bax in yeast, which lacks obvious orthologs of the Bcl-2 family, and allows studying how absence of yeast CatD affects Bax activity without interference from other Bcl-2 family members. Using yeast cells heterologously expressing a cytosolic inactive form of human Bax, which was activated by exposure to acetic acid, we could discard this hypothesis since absence of Pep4p enhanced Bax-induced cell death (Figure 1). It will be interesting to further exploit this system with heterologous co-expression of Bax and human CatD, in order to dissect the role of this lysosomal protease in the regulation of Bax activity independently of Bid.
Figure 1. FIGURE 1: Survival of S. cerevisiae cells expressing Bax during acetic acid treatment.
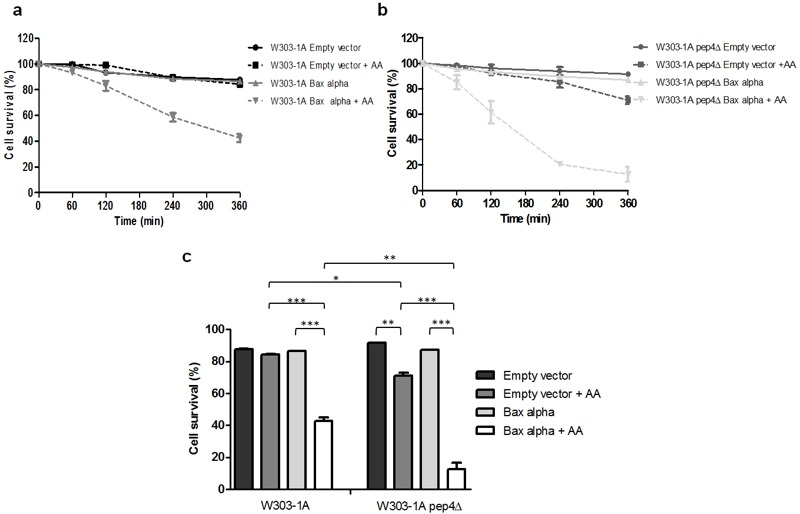
The wild type W303-1A and pep4Δ mutant strains transformed with the empty vector (PYES2) and PYES2-Bax alpha were incubated with 120 mM acetic acid for up to 360 min. (A) Cell survival of W303-1A strain and (B) W303-1A pep4Δ strain for up to 360 min was determined by standard dilution plate counts and expressed as a percentage of c.f.u. in relation to time 0. Data represents means ± S.D. (n=2). (C) Cell survival at time 360 min was determined by standard dilution plate counts and expressed as a percentage of c.f.u. in relation to time 0. Data represents means ± S.D. (n=2). *P < 0.05, **P < 0.01, ***P < 0.001.
As a final conclusion, it becomes apparent that the approaches with yeast have already provided and can further offer new perspectives for an increased understanding of the role of CatD in mammalian apoptosis, and its implications in cancer. Indeed, studies with yeast further reinforce the use of this eukaryotic organism as a valuable model to identify and characterize novel RCD processes, and open the door to new clinical opportunities, with a substantial impact in public health.
Funding Statement
This work was supported by FEDER through POFC – COMPETE and by Fundacao para a Ciencia e Tecnologia through projects PEst-OE/BIA/UI4050/2014 and FCTANR/BEX-BCM/0175/2012, as well as fellowships to H. Pereira (SFRH/BD/73139/2010), C.S.F. Oliveira (SFRH/BD/77449/2011), L. Castro (SFRH/BD/93589/2013) and S. Chaves (SFRH/ BPD/89980/2012).
References
- 1.Lecaille F, Kaleta J, Brömme D. Human and Parasitic Papain-Like Cysteine Proteases: Their Role in Physiology and Pathology and Recent Developments in Inhibitor Design. Chem Rev. 2002;102(12):4459–4488. doi: 10.1021/cr0101656. [DOI] [PubMed] [Google Scholar]
- 2.Turk B, Turk D, Turk V. Lysosomal cysteine proteases: more than scavengers. Biochim Biophys Acta. 2000;1477(1-2):98–111. doi: 10.1016/s0167-4838(99)00263-0. [DOI] [PubMed] [Google Scholar]
- 3.Wex T, Levy B, Smeekens SP, Ansorge S, Desnick RJ, Bromme D. Genomic structure, chromosomal localization, and expression of human cathepsin W. Biochem Biophys Res Commun. 1998;248(2):255–261. doi: 10.1006/bbrc.1998.8954. [DOI] [PubMed] [Google Scholar]
- 4.Brömme D, Li Z, Barnes M, Mehler E. Human cathepsin V functional expression, tissue distribution, electrostatic surface potential, enzymatic characterization, and chromosomal localization. Biochemistry (Mosc) 1999;38(8):2377–2385. doi: 10.1021/bi982175f. [DOI] [PubMed] [Google Scholar]
- 5.Brix K, Dunkhorst A, Mayer K, Jordans S. Cysteine cathepsins: cellular roadmap to different functions. Biochimie. 2008;90(2):194–207. doi: 10.1016/j.biochi.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824(1):68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pratt MR, Sekedat MD, Chiang KP, Muir TW. Direct measurement of cathepsin B activity in the cytosol of apoptotic cells by an activity-based probe. Chem Biol. 2009;16(9):1001–1012. doi: 10.1016/j.chembiol.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hook V, Funkelstein L, Wegrzyn J, Bark S, Kindy M, Hook G. Cysteine Cathepsins in the secretory vesicle produce active peptides: Cathepsin L generates peptide neurotransmitters and cathepsin B produces beta-amyloid of Alzheimer’s disease. Proteolysis 50 Years Discov Lysosome. 2012;1824(1):89–104. doi: 10.1016/j.bbapap.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulet B, Baruch A, Moon N-S, Poirier M, Sansregret LL, Erickson A, Bogyo M, Nepveu A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell. 2004;14(2):207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- 10.Duncan EM, Muratore-Schroeder TL, Cook RG, Garcia BA, Shabanowitz J, Hunt DF, Allis CD. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135(2):284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiser J, Adair B, Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest. 2010;120(10):3421–3431. doi: 10.1172/JCI42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conus S, Simon H-U. Cathepsins and their involvement in immune responses. Swiss Med Wkly. 2010;140:w13042. doi: 10.4414/smw.2010.13042. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson LS, Lima H, Goldberg MF, Gocheva V, Tsiperson V, Sutterwala FS, Joyce JA, Gapp BV, Blomen VA, Chandran K, Brummelkamp TR, Diaz-Griffero F, Brojatsch J. Cathepsin-mediated necrosis controls the adaptive immune response by Th2 (T helper type 2)-associated adjuvants. J Biol Chem. 2013;288(11):7481–7491. doi: 10.1074/jbc.M112.400655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Repnik U, Hafner Česen M, Turk B. Lysosomal membrane permeabilization in cell death: Concepts and challenges. Mitochondrion 19 Pt. 2014;A:49–57. doi: 10.1016/j.mito.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27(50):6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 16. Česen MH, Pegan K, Spes A, Turk B. Lysosomal pathways to cell death and their therapeutic applications. . Exp Cell Res . 2012;318(11): 1245-1251(11):1245–1251. doi: 10.1016/j.yexcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Fonović M, Turk B. Cysteine cathepsins and extracellular matrix degradation. Biochim Biophys Acta. 2014;1840(8):2560–2570. doi: 10.1016/j.bbagen.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Benes P, Vetvicka V, Fusek M. Cathepsin D--many functions of one aspartic protease. Crit Rev Oncol Hematol. 2008;68(1):12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saftig P, Hunziker E, Wehmeyer O, Jones S, Boyde A, Rommerskirch W, Moritz JD, Schu P, von Figura K. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc Natl Acad Sci U S A. 1998;95(23):13453–13458. doi: 10.1073/pnas.95.23.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasilik A, Neufeld EF. Biosynthesis of lysosomal enzymes in fibroblasts. Synthesis as precursors of higher molecular weight. . J Biol Chem. 1980;255(10):4937–4945. [PubMed] [Google Scholar]
- 21.Erickson AH. Biosynthesis of lysosomal endopeptidases. J Cell Biochem. 1989;40(1):31–41. doi: 10.1002/jcb.240400104. [DOI] [PubMed] [Google Scholar]
- 22.Gieselmann V, Pohlmann R, Hasilik A, Von Figura K. Biosynthesis and transport of cathepsin D in cultured human fibroblasts. J Cell Biol. 1983;97(1):1–5. doi: 10.1083/jcb.97.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranski TJ, Koelsch G, Hartsuck JA, Kornfeld S. Mapping and molecular modeling of a recognition domain for lysosomal enzyme targeting. J Biol Chem. 1991;266(34):23365–23372. [PubMed] [Google Scholar]
- 24.Erickson AH, Conner GE, Blobel G. Biosynthesis of a lysosomal enzyme. Partial structure of two transient and functionally distinct NH2-terminal sequences in cathepsin D. . J Biol Chem. 1981;256(21):11224–11231. [PubMed] [Google Scholar]
- 25.Conner GE, Richo G. Isolation and characterization of a stable activation intermediate of the lysosomal aspartyl protease cathepsin D. Biochemistry (Mosc) 1992;31(4):1142–1147. doi: 10.1021/bi00119a024. [DOI] [PubMed] [Google Scholar]
- 26.Gieselmann V, Hasilik A, von Figura K. Processing of human cathepsin D in lysosomes in vitro. J Biol Chem. 1985;260(5):3215–3220. [PubMed] [Google Scholar]
- 27. Samarel AM, Ferguson AG, Decker RS, Lesch M. Effects of cysteine protease inhibitors on rabbit cathepsin D maturation. . Am J Physiol . 1989;257(6Pt1): C1069– C1079. doi: 10.1152/ajpcell.1989.257.6.C1069. [DOI] [PubMed] [Google Scholar]
- 28.Laurent-Matha V, Derocq D, Prébois C, Katunuma N, Liaudet-Coopman E. Processing of human cathepsin D is independent of its catalytic function and auto-activation: involvement of cathepsins L and B. J Biochem (Tokyo) 2006;139(3):363–371. doi: 10.1093/jb/mvj037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole AR, Hembry RM, Dingle JT. Cathepsin D in cartilage: the immunohistochemical demonstration of extracellular enzyme in normal and pathological conditions. J Cell Sci. 1974;14(1):139–161. doi: 10.1242/jcs.14.1.139. [DOI] [PubMed] [Google Scholar]
- 30.Bjelle A, Osterlin S. Cathepsin D activity in bovine articular cartilage, synovial membrane and fluid: degradation of cartilage proteoglycans from same joint. J Rheumatol. 1976;3(4):400–408. [PubMed] [Google Scholar]
- 31.Vittorio N, Crissman JD, Hopson CN, Herman JH. Histologic assessment of cathepsin D in osteoarthritic cartilage. Clin Exp Rheumatol. 1986;4(3):221–230. [PubMed] [Google Scholar]
- 32.Vĕtvicka V, Vágner J, Baudys M, Tang J, Foundling SI, Fusek M. Human breast milk contains procathepsin D--detection by specific antibodies. Biochem Mol Biol Int. 1993;30(5):921–928. [PubMed] [Google Scholar]
- 33.Larsen LB, Petersen TE. Identification of five molecular forms of cathepsin D in bovine milk. Adv Exp Med Biol. 1995;362:279–283. doi: 10.1007/978-1-4615-1871-6_32. [DOI] [PubMed] [Google Scholar]
- 34.Benes P, Koelsch G, Dvorak B, Fusek M, Vetvicka V. Detection of procathepsin D in rat milk. Comp Biochem Physiol B Biochem Mol Biol. 2002;133(1):113–118. doi: 10.1016/s1096-4959(02)00112-4. [DOI] [PubMed] [Google Scholar]
- 35.Zühlsdorf M, Imort M, Hasilik A, von Figura K. Molecular forms of beta-hexosaminidase and cathepsin D in serum and urine of healthy subjects and patients with elevated activity of lysosomal enzymes. Biochem J. 1983;213(3):733–740. doi: 10.1042/bj2130733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baechle D, Flad T, Cansier A, Steffen H, Schittek B, Tolson J, Herrmann T, Dihazi H, Beck A, Mueller GA, Mueller M, Stevanovic S, Garbe C, Mueller CA, Kalbacher H. Cathepsin D is present in human eccrine sweat and involved in the postsecretory processing of the antimicrobial peptide DCD-1L. J Biol Chem. 2006;281(9):5406–5415. doi: 10.1074/jbc.M504670200. [DOI] [PubMed] [Google Scholar]
- 37.Hakala JK, Oksjoki R, Laine P, Du H, Grabowski GA, Kovanen PT, Pentikäinen MO. Lysosomal enzymes are released from cultured human macrophages, hydrolyze LDL in vitro, and are present extracellularly in human atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2003;23(8):1430–1436. doi: 10.1161/01.ATV.0000077207.49221.06. [DOI] [PubMed] [Google Scholar]
- 38. Vashishta A, Saraswat Ohri S, Vetvickova J, Fusek M, Ulrichova J, Vetvicka V. Procathepsin D secreted by HaCaT keratinocyte cells - A novel regulator of keratinocyte growth. . Eur J Cell Biol. 2007;86(6):303–313. doi: 10.1016/j.ejcb.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lkhider M, Castino R, Bouguyon E, Isidoro C, Ollivier-Bousquet M. Cathepsin D released by lactating rat mammary epithelial cells is involved in prolactin cleavage under physiological conditions. J Cell Sci. 2004;117(Pt 21):5155–5164. doi: 10.1242/jcs.01396. [DOI] [PubMed] [Google Scholar]
- 40.Masson O, Bach A-S, Derocq D, Prébois C, Laurent-Matha V, Pattingre S, Liaudet-Coopman E. Pathophysiological functions of cathepsin D: Targeting its catalytic activity versus its protein binding activity? Biochimie. 2010;92(11):1635–1643. doi: 10.1016/j.biochi.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Minarowska A, Minarowski L, Karwowska A, Gacko M. Regulatory role of cathepsin D in apoptosis. Folia Histochem Cytobiol Pol Acad Sci Pol Histochem Cytochem Soc. 2007;45(3):159–163. [PubMed] [Google Scholar]
- 42.Tan G-J, Peng Z-K, Lu J-P, Tang F-Q. Cathepsins mediate tumor metastasis. World J Biol Chem. 2013;4(4):91–101. doi: 10.4331/wjbc.v4.i4.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saftig P, Hetman M, Schmahl W, Weber K, Heine L, Mossmann H, Köster A, Hess B, Evers M, von Figura K. Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J. 1995;14(15):3599–3608. doi: 10.1002/j.1460-2075.1995.tb00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koike M, Shibata M, Ohsawa Y, Nakanishi H, Koga T, Kametaka S, Waguri S, Momoi T, Kominami E, Peters C, Figura K von, Saftig P, Uchiyama Y. Involvement of two different cell death pathways in retinal atrophy of cathepsin D-deficient mice. Mol Cell Neurosci. 2003;22(2):146–161. doi: 10.1016/s1044-7431(03)00035-6. [DOI] [PubMed] [Google Scholar]
- 45.Koike M, Nakanishi H, Saftig P, Ezaki J, Isahara K, Ohsawa Y, Schulz-Schaeffer W, Watanabe T, Waguri S, Kametaka S, Shibata M, Yamamoto K, Kominami E, Peters C, von Figura K, Uchiyama Y. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci Off J Soc Neurosci. 2000;20(18):6898–6906. doi: 10.1016/s0168-0102(00)81020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, Gotow T, Peters C, von Figura K, Mizushima N, Saftig P, Uchiyama Y. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Am J Pathol. 2005;167(6):1713–1728. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang D, Brankov M, Makhija MT, Robertson T, Helmerhorst E, Papadimitriou JM, Rakoczy PE. Correlation between inactive cathepsin D expression and retinal changes in mcd2/mcd2 transgenic mice. Invest Ophthalmol Vis Sci. 2005;46(9):3031–3038. doi: 10.1167/iovs.04-1510. [DOI] [PubMed] [Google Scholar]
- 48.Steinfeld R, Reinhardt K, Schreiber K, Hillebrand M, Kraetzner R, Bruck W, Saftig P, Gartner J. Cathepsin D deficiency is associated with a human neurodegenerative disorder. Am J Hum Genet. 2006;78(6):988–998. doi: 10.1086/504159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siintola E, Partanen S, Strömme P, Haapanen A, Haltia M, Maehlen J, Lehesjoki A-E, Tyynelä J. Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain J Neurol. 2006;129(Pt 6):1438–1445. doi: 10.1093/brain/awl107. [DOI] [PubMed] [Google Scholar]
- 50.Tyynelä J, Sohar I, Sleat DE, Gin RM, Donnelly RJ, Baumann M, Haltia M, Lobel P. Congenital ovine neuronal ceroid lipofuscinosis--a cathepsin D deficiency with increased levels of the inactive enzyme. Eur J Paediatr Neurol EJPN Off J Eur Paediatr Neurol Soc. 2001;5(Suppl A):43–45. doi: 10.1053/ejpn.2000.0433. [DOI] [PubMed] [Google Scholar]
- 51.Awano T, Katz ML, O’Brien DP, Taylor JF, Evans J, Khan S, Sohar I, Lobel P, Johnson GS. A mutation in the cathepsin D gene (CTSD) in American Bulldogs with neuronal ceroid lipofuscinosis. Mol Genet Metab. 2006;87(4):341–348. doi: 10.1016/j.ymgme.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Egberts F, Heinrich M, Jensen J-M, Winoto-Morbach S, Pfeiffer S, Wickel M, Schunck M, Steude J, Saftig P, Proksch E, Schütze S. Cathepsin D is involved in the regulation of transglutaminase 1 and epidermal differentiation. J Cell Sci. 2004;117(Pt 11):2295–2307. doi: 10.1242/jcs.01075. [DOI] [PubMed] [Google Scholar]
- 53.Chen SH, Arany I, Apisarnthanarax N, Rajaraman S, Tyring SK, Horikoshi T, Brysk H, Brysk MM. Response of keratinocytes from normal and psoriatic epidermis to interferon-gamma differs in the expression of zinc-alpha(2)-glycoprotein and cathepsin D. FASEB J Off Publ Fed Am Soc Exp Biol. 2000;14(3):565–571. doi: 10.1096/fasebj.14.3.565. [DOI] [PubMed] [Google Scholar]
- 54.Kawada A, Hara K, Kominami E, Hiruma M, Noguchi H, Ishibashi A. Processing of cathepsins L, B and D in psoriatic epidermis. Arch Dermatol Res. 1997;289(2):87–93. doi: 10.1007/s004030050160. [DOI] [PubMed] [Google Scholar]
- 55.Zhou W, Scott SA, Shelton SB, Crutcher KA. Cathepsin D-mediated proteolysis of apolipoprotein E: possible role in Alzheimer’s disease. Neuroscience. 2006;143(3):689–701. doi: 10.1016/j.neuroscience.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 56.Kenessey A, Nacharaju P, Ko LW, Yen SH. Degradation of tau by lysosomal enzyme cathepsin D: implication for Alzheimer neurofibrillary degeneration. J Neurochem. 1997;69(5):2026–2038. doi: 10.1046/j.1471-4159.1997.69052026.x. [DOI] [PubMed] [Google Scholar]
- 57.Hilfiker-Kleiner D, Kaminski K, Podewski E, Bonda T, Schaefer A, Sliwa K, Forster O, Quint A, Landmesser U, Doerries C, Luchtefeld M, Poli V, Schneider MD, Balligand J-L, Desjardins F, Ansari A, Struman I, Nguyen NQN, Zschemisch NH, Klein G, Heusch G, Schulz R, Hilfiker A, Drexler H. A cathepsin D-cleaved 16 kDa form of prolactin mediates postpartum cardiomyopathy. Cell. 2007;128(3):589–600. doi: 10.1016/j.cell.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 58.Sheikh AM, Li X, Wen G, Tauqeer Z, Brown WT, Malik M. Cathepsin D and apoptosis related proteins are elevated in the brain of autistic subjects. Neuroscience. 2010;165(2):363–370. doi: 10.1016/j.neuroscience.2009.10.035. [DOI] [PubMed] [Google Scholar]
- 59.Conus S, Perozzo R, Reinheckel T, Peters C, Scapozza L, Yousefi S, Simon H-U. Caspase-8 is activated by cathepsin D initiating neutrophil apoptosis during the resolution of inflammation. J Exp Med. 2008;205(3):685–698. doi: 10.1084/jem.20072152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cullen V, Lindfors M, Ng J, Paetau A, Swinton E, Kolodziej P, Boston H, Saftig P, Woulfe J, Feany MB, Myllykangas L, Schlossmacher MG, Tyynelä J. Cathepsin D expression level affects alpha-synuclein processing, aggregation, and toxicity in vivo. Mol Brain. 2009;2:5. doi: 10.1186/1756-6606-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haidar B, Kiss RS, Sarov-Blat L, Brunet R, Harder C, McPherson R, Marcel YL. Cathepsin D, a lysosomal protease, regulates ABCA1-mediated lipid efflux. J Biol Chem. 2006;281(52):39971–39981. doi: 10.1074/jbc.M605095200. [DOI] [PubMed] [Google Scholar]
- 62.Durán MC, Martín-Ventura JL, Mohammed S, Barderas MG, Blanco-Colio LM, Mas S, Moral V, Ortega L, Tuñón J, Jensen ON, Vivanco F, Egido J. Atorvastatin modulates the profile of proteins released by human atherosclerotic plaques. Eur J Pharmacol. 2007;562(1-2):119–129. doi: 10.1016/j.ejphar.2007.01.077. [DOI] [PubMed] [Google Scholar]
- 63.Gyrd-Hansen M, Nylandsted J, Jäättelä M. Heat shock protein 70 promotes cancer cell viability by safeguarding lysosomal integrity. Cell Cycle Georget Tex. 2004;3(12):1484–1485. doi: 10.4161/cc.3.12.1287. [DOI] [PubMed] [Google Scholar]
- 64.Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29(1):22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 65.Leto G, Tumminello FM, Crescimanno M, Flandina C, Gebbia N. Cathepsin D expression levels in nongynecological solid tumors: clinical and therapeutic implications. Clin Exp Metastasis. 2004;21(2):91–106. doi: 10.1023/b:clin.0000024740.44602.b7. [DOI] [PubMed] [Google Scholar]
- 66.Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, Prébois C, Rochefort H, Vignon F. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006;237(2):167–179. doi: 10.1016/j.canlet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 67.Fitzgibbons PL, Page DL, Weaver D, Thor AD, Allred DC, Clark GM, Ruby SG, O’Malley F, Simpson JF, Connolly JL, Hayes DF, Edge SB, Lichter A, Schnitt SJ. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. . Arch Pathol Lab Med. 2000;124(7):966–978. doi: 10.5858/2000-124-0966-PFIBC. [DOI] [PubMed] [Google Scholar]
- 68.Bossard N, Descotes F, Bremond AG, Bobin Y, De Saint Hilaire P, Golfier F, Awada A, Mathevet PM, Berrerd L, Barbier Y, Estève J. Keeping data continuous when analyzing the prognostic impact of a tumor marker: an example with cathepsin D in breast cancer. Breast Cancer Res Treat. 2003;82(1):47–59. doi: 10.1023/B:BREA.0000003919.75055.e8. [DOI] [PubMed] [Google Scholar]
- 69.Kirana C, Shi H, Laing E, Hood K, Miller R, Bethwaite P, Keating J, Jordan TW, Hayes M, Stubbs R. Cathepsin D Expression in Colorectal Cancer: From Proteomic Discovery through Validation Using Western Blotting, Immunohistochemistry, and Tissue Microarrays. Int J Proteomics. 2012;2012:245819. doi: 10.1155/2012/245819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shin IY, Sung NY, Lee YS, Kwon TS, Si Y, Lee YS, Oh ST, Lee IK. The expression of multiple proteins as prognostic factors in colorectal cancer: cathepsin D, p53, COX-2, epidermal growth factor receptor, C-erbB-2, and Ki-67. Gut Liver. 2014;8(1):13–23. doi: 10.5009/gnl.2014.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berchem G, Glondu M, Gleizes M, Brouillet J-P, Vignon F, Garcia M, Liaudet-Coopman E. Cathepsin-D affects multiple tumor progression steps in vivo: proliferation, angiogenesis and apoptosis. Oncogene. 2002;21(38):5951–5955. doi: 10.1038/sj.onc.1205745. [DOI] [PubMed] [Google Scholar]
- 72.Fusek M, Vetvicka V. Mitogenic function of human procathepsin D: the role of the propeptide. Biochem J . 1994;303( Pt 3):775–780. doi: 10.1042/bj3030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vetvicka V, Vetvickova J, Fusek M. Role of procathepsin D activation peptide in prostate cancer growth. The Prostate. 2000;44(1):1–7. doi: 10.1002/1097-0045(20000615)44:1<1::aid-pros1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 74.Bazzett LB, Watkins CS, Gercel-Taylor C, Taylor DD. Modulation of proliferation and chemosensitivity by procathepsin D and its peptides in ovarian cancer. Gynecol Oncol. 1999;74(2):181–187. doi: 10.1006/gyno.1999.5426. [DOI] [PubMed] [Google Scholar]
- 75.Mathieu M, Rochefort H, Barenton B, Prebois C, Vignon F. Interactions of cathepsin-D and insulin-like growth factor-II (IGF-II) on the IGF-II/mannose-6-phosphate receptor in human breast cancer cells and possible consequences on mitogenic activity of IGF-II. Mol Endocrinol Baltim Md. 1990;4(9):1327–1335. doi: 10.1210/mend-4-9-1327. [DOI] [PubMed] [Google Scholar]
- 76.Vetvicka V, Vetvickova J, Benes P. Role of enzymatically inactive procathepsin D in lung cancer. Anticancer Res. 2004;24(5A):2739–2743. [PubMed] [Google Scholar]
- 77.Vetvicka V, Benes P, Fusek M. Procathepsin D in breast cancer: what do we know? Effects of ribozymes and other inhibitors. . Cancer Gene Ther. 2002;9(10):854–863. doi: 10.1038/sj.cgt.7700508. [DOI] [PubMed] [Google Scholar]
- 78.Vashishta A, Ohri SS, Proctor M, Fusek M, Vetvicka V. Role of activation peptide of procathepsin D in proliferation and invasion of lung cancer cells. Anticancer Res. 2006;26(6B):4163–4170. [PubMed] [Google Scholar]
- 79.Hu L, Roth JM, Brooks P, Luty J, Karpatkin S. Thrombin up-regulates cathepsin D which enhances angiogenesis, growth, and metastasis. Cancer Res. 2008;68(12):4666–4673. doi: 10.1158/0008-5472.CAN-07-6276. [DOI] [PubMed] [Google Scholar]
- 80.Wolf M, Clark-Lewis I, Buri C, Langen H, Lis M, Mazzucchelli L. Cathepsin D specifically cleaves the chemokines macrophage inflammatory protein-1 alpha, macrophage inflammatory protein-1 beta, and SLC that are expressed in human breast cancer. Am J Pathol. 2003;162(4):1183–1190. doi: 10.1016/s0002-9440(10)63914-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vetvicka V, Vashishta A, Saraswat-Ohri S, Vetvickova J. Procathepsin D and cancer: From molecular biology to clinical applications. World J Clin Oncol. 2010;1(1):35–40. doi: 10.5306/wjco.v1.i1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khalkhali-Ellis Z, Hendrix MJ. Two Faces of Cathepsin D: Physiological Guardian Angel and Pathological Demon. Biol Med. 2014;6(206):2. doi: 10.4172/0974-8369.1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Briozzo P, Badet J, Capony F, Pieri I, Montcourrier P, Barritault D, Rochefort H. MCF7 mammary cancer cells respond to bFGF and internalize it following its release from extracellular matrix: a permissive role of cathepsin D. Exp Cell Res. 1991;194(2):252–259. doi: 10.1016/0014-4827(91)90362-x. [DOI] [PubMed] [Google Scholar]
- 84.Achour O, Bridiau N, Kacem M, Delatouche R, Bordenave-Juchereau S, Sannier F, Thiéry V, Piot J-M, Maugard T, Arnaudin I. Cathepsin D activity and selectivity in the acidic conditions of a tumor microenvironment: Utilization in the development of a novel Cathepsin D substrate for simultaneous cancer diagnosis and therapy. Biochimie. 2013;95(11):2010–2017. doi: 10.1016/j.biochi.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 85.Laurent-Matha V, Maruani-Herrmann S, Prébois C, Beaujouin M, Glondu M, Noël A, Alvarez-Gonzalez ML, Blacher S, Coopman P, Baghdiguian S, Gilles C, Loncarek J, Freiss G, Vignon F, Liaudet-Coopman E. Catalytically inactive human cathepsin D triggers fibroblast invasive growth. J Cell Biol. 2005;168(3):489–499. doi: 10.1083/jcb.200403078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Glondu M, Coopman P, Laurent-Matha V, Garcia M, Rochefort H, Liaudet-Coopman E. A mutated cathepsin-D devoid of its catalytic activity stimulates the growth of cancer cells. Oncogene. 2001;20(47):6920–6929. doi: 10.1038/sj.onc.1204843. [DOI] [PubMed] [Google Scholar]
- 87.Liaudet E, Derocq D, Rochefort H, Garcia M. Transfected cathepsin D stimulates high density cancer cell growth by inactivating secreted growth inhibitors. Cell Growth Differ Mol Biol J Am Assoc Cancer Res. 1995;6(9):1045–1052. [PubMed] [Google Scholar]
- 88.Nirdé P, Derocq D, Maynadier M, Chambon M, Basile I, Gary-Bobo M, Garcia M. Heat shock cognate 70 protein secretion as a new growth arrest signal for cancer cells. Oncogene. 2010;29(1):117–127. doi: 10.1038/onc.2009.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fehrenbacher N, Gyrd-Hansen M, Poulsen B, Felbor U, Kallunki T, Boes M, Weber E, Leist M, Jäättelä M. Sensitization to the lysosomal cell death pathway upon immortalization and transformation. Cancer Res. 2004;64(15):5301–5310. doi: 10.1158/0008-5472.CAN-04-1427. [DOI] [PubMed] [Google Scholar]
- 90.Johansson A-C, Steen H, Ollinger K, Roberg K. Cathepsin D mediates cytochrome c release and caspase activation in human fibroblast apoptosis induced by staurosporine. Cell Death Differ. 2003;10(11):1253–1259. doi: 10.1038/sj.cdd.4401290. [DOI] [PubMed] [Google Scholar]
- 91.Emert-Sedlak L, Shangary S, Rabinovitz A, Miranda MB, Delach SM, Johnson DE. Involvement of cathepsin D in chemotherapy-induced cytochrome c release, caspase activation, and cell death. Mol Cancer Ther. 2005;4(5):733–742. doi: 10.1158/1535-7163.MCT-04-0301. [DOI] [PubMed] [Google Scholar]
- 92.Trincheri NF, Nicotra G, Follo C, Castino R, Isidoro C. Resveratrol induces cell death in colorectal cancer cells by a novel pathway involving lysosomal cathepsin D. Carcinogenesis. 2007;28(5):922–931. doi: 10.1093/carcin/bgl223. [DOI] [PubMed] [Google Scholar]
- 93.Blomgran R, Zheng L, Stendahl O. Cathepsin-cleaved Bid promotes apoptosis in human neutrophils via oxidative stress-induced lysosomal membrane permeabilization. J Leukoc Biol. 2007;81(5):1213–1223. doi: 10.1189/jlb.0506359. [DOI] [PubMed] [Google Scholar]
- 94.Bidère N, Lorenzo HK, Carmona S, Laforge M, Harper F, Dumont C, Senik A. Cathepsin D triggers Bax activation, resulting in selective apoptosis-inducing factor (AIF) relocation in T lymphocytes entering the early commitment phase to apoptosis. J Biol Chem. 2003;278(33):31401–31411. doi: 10.1074/jbc.M301911200. [DOI] [PubMed] [Google Scholar]
- 95.Beaujouin M, Baghdiguian S, Glondu-Lassis M, Berchem G, Liaudet-Coopman E. Overexpression of both catalytically active and -inactive cathepsin D by cancer cells enhances apoptosis-dependent chemo-sensitivity. Oncogene. 2006;25(13):1967–1973. doi: 10.1038/sj.onc.1209221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tardy C, Tyynelä J, Hasilik A, Levade T, Andrieu-Abadie N. Stress-induced apoptosis is impaired in cells with a lysosomal targeting defect but is not affected in cells synthesizing a catalytically inactive cathepsin D. Cell Death Differ. 2003;10(9):1090–1100. doi: 10.1038/sj.cdd.4401272. [DOI] [PubMed] [Google Scholar]
- 97.Schestkowa O, Geisel D, Jacob R, Hasilik A. The catalytically inactive precursor of cathepsin D induces apoptosis in human fibroblasts and HeLa cells. J Cell Biochem. 2007;101(6):1558–1566. doi: 10.1002/jcb.21269. [DOI] [PubMed] [Google Scholar]
- 98.Zhao S, Aviles ER, Fujikawa DG. Nuclear translocation of mitochondrial cytochrome c, lysosomal cathepsins B and D, and three other death-promoting proteins within the first 60 minutes of generalized seizures. J Neurosci Res. 2010;88(8):1727–1737. doi: 10.1002/jnr.22338. [DOI] [PubMed] [Google Scholar]
- 99.Sagulenko V, Muth D, Sagulenko E, Paffhausen T, Schwab M, Westermann F. Cathepsin D protects human neuroblastoma cells from doxorubicin-induced cell death. Carcinogenesis. 2008;29(10):1869–1877. doi: 10.1093/carcin/bgn147. [DOI] [PubMed] [Google Scholar]
- 100.Castino R, Bellio N, Nicotra G, Follo C, Trincheri NF, Isidoro C. Cathepsin D-Bax death pathway in oxidative stressed neuroblastoma cells. Free Radic Biol Med. 2007;42(9):1305–1316. doi: 10.1016/j.freeradbiomed.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 101.Hah Y-S, Noh HS, Ha JH, Ahn JS, Hahm JR, Cho HY, Kim DR. Cathepsin D inhibits oxidative stress-induced cell death via activation of autophagy in cancer cells. Cancer Lett. 2012;323(2):208–214. doi: 10.1016/j.canlet.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 102.Marques C, Oliveira CSF, Alves S, Chaves SR, Coutinho OP, Côrte-Real M, Preto A. Acetate-induced apoptosis in colorectal carcinoma cells involves lysosomal membrane permeabilization and cathepsin D release. Cell Death Dis. 2013;4:e507. doi: 10.1038/cddis.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oliveira CSF, Pereira H, Alves S, Castro L, Baltazar F, Chaves SR, Preto A, Côrte-Real M. Cathepsin D protects colorectal cancer cells from acetate-induced apoptosis through autophagy-independent degradation of damaged mitochondria. Cell Death Dis. 2015;"In press” doi: 10.1038/cddis.2015.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hartwell LH. Nobel Lecture. Yeast and cancer. Biosci Rep. 2002;22(3-4):373–394. doi: 10.1023/a:1020918107706. [DOI] [PubMed] [Google Scholar]
- 105.Diaz-Ruiz R, Uribe-Carvajal S, Devin A, Rigoulet M. Tumor cell energy metabolism and its common features with yeast metabolism. Biochim Biophys Acta. 2009;1796(2):252–265. doi: 10.1016/j.bbcan.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 106.Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker’s yeast? Nat Rev Neurosci. 2010;11(6):436–449. doi: 10.1038/nrn2809. [DOI] [PubMed] [Google Scholar]
- 107.Smith MG, Snyder M. Yeast as a model for human disease. . Curr Protoc Hum Genet. 2006;Chapter 15: Unit 15.6. doi: 10.1002/0471142905.hg1506s48. [DOI] [PubMed] [Google Scholar]
- 108.Carmona-Gutierrez D, Ruckenstuhl C, Bauer MA, Eisenberg T, Büttner S, Madeo F. Cell death in yeast: growing applications of a dying buddy. Cell Death Differ. 2010;17(5):733–734. doi: 10.1038/cdd.2010.10. [DOI] [PubMed] [Google Scholar]
- 109.Hecht KA, Wytiaz VA, Ast T, Schuldiner M, Brodsky JL. Characterization of an M28 metalloprotease family member residing in the yeast vacuole. FEMS Yeast Res. 2013;13(5):471–484. doi: 10.1111/1567-1364.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rupp S, Hirsch HH, Wolf DH. Biogenesis of the yeast vacuole (lysosome). Active site mutation in the vacuolar aspartate proteinase yscA blocks maturation of vacuolar proteinases. . FEBS Lett. 1991;293(1-2):62–66. doi: 10.1016/0014-5793(91)81153-y. [DOI] [PubMed] [Google Scholar]
- 111.Hecht KA, O’Donnell AF, Brodsky JL. The proteolytic landscape of the yeast vacuole. Cell Logist. 2014;4(1):e28023. doi: 10.4161/cl.28023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Adamis PDB, Mannarino SC, Riger CJ, Duarte G, Cruz A, Pereira MD, Eleutherio ECA. Lap4, a vacuolar aminopeptidase I, is involved in cadmium-glutathione metabolism. Biometals Int J Role Met Ions Biol Biochem Med. 2009;22(2):243–249. doi: 10.1007/s10534-008-9160-9. [DOI] [PubMed] [Google Scholar]
- 113.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147(2):435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wolf DH, Ehmann C. Carboxypeptidase S- and carboxypeptidase Y-deficient mutants of Saccharomyces cerevisiae. J Bacteriol. 1981;147(2):418–426. doi: 10.1128/jb.147.2.418-426.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Van Den Hazel HB, Kielland-Brandt MC, Winther JR. Review: biosynthesis and function of yeast vacuolar proteases. Yeast Chichester Engl. 1996;12(1):1–16. doi: 10.1002/(sici)1097-0061(199601)12:1<1::aid-yea902>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 117.Mason DA, Shulga N, Undavai S, Ferrando-May E, Rexach MF, Goldfarb DS. Increased nuclear envelope permeability and Pep4p-dependent degradation of nucleoporins during hydrogen peroxide-induced cell death. FEMS Yeast Res. 2005;5(12):1237–1251. doi: 10.1016/j.femsyr.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 118.Marques M, Mojzita D, Amorim MA, Almeida T, Hohmann S, Moradas-Ferreira P, Costa V. The Pep4p vacuolar proteinase contributes to the turnover of oxidized proteins but PEP4 overexpression is not sufficient to increase chronological lifespan in Saccharomyces cerevisiae. Microbiol Read Engl. 2006;152(Pt 12):3595–3605. doi: 10.1099/mic.0.29040-0. [DOI] [PubMed] [Google Scholar]
- 119.Gourlay CW, Ayscough KR. Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26(17):6487–6501. doi: 10.1128/MCB.00117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eastwood MD, Cheung SWT, Lee KY, Moffat J, Meneghini MD. Developmentally programmed nuclear destruction during yeast gametogenesis. Dev Cell. 2012;23(1):35–44. doi: 10.1016/j.devcel.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 121.Carmona-Gutiérrez D, Bauer MA, Ring J, Knauer H, Eisenberg T, Büttner S, Ruckenstuhl C, Reisenbichler A, Magnes C, Rechberger GN, Birner-Gruenberger R, Jungwirth H, Fröhlich K-U, Sinner F, Kroemer G, Madeo F. The propeptide of yeast cathepsin D inhibits programmed necrosis. Cell Death Dis. 2011;2:e161. doi: 10.1038/cddis.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pereira C, Chaves S, Alves S, Salin B, Camougrand N, Manon S, Sousa MJ, Côrte-Real M. Mitochondrial degradation in acetic acid-induced yeast apoptosis: the role of Pep4 and the ADP/ATP carrier. Mol Microbiol. 2010;76(6):1398–1410. doi: 10.1111/j.1365-2958.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 123.Pereira H, Azevedo F, Rego A, Sousa MJ, Chaves SR, Côrte-Real M. The protective role of yeast cathepsin D in acetic acid-induced apoptosis depends on ANT (Aac2p) but not on the voltage-dependent channel (Por1p). FEBS Lett. 2013;587(2):200–205. doi: 10.1016/j.febslet.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 124.Sousa M, Duarte AM, Fernandes TR, Chaves SR, Pacheco A, Leão C, Côrte-Real M, Sousa MJ. Genome-wide identification of genes involved in the positive and negative regulation of acetic acid-induced programmed cell death in Saccharomyces cerevisiae. BMC Genomics. 2013;14:838. doi: 10.1186/1471-2164-14-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Galluzzi L, Bravo-San Pedro JM, Vitale I, Aaronson SA, Abrams JM, Adam D, Alnemri ES, Altucci L, Andrews D, Annicchiarico-Petruzzelli M, Baehrecke EH, Bazan NG, Bertrand MJ, Bianchi K, Blagosklonny MV, Blomgren K, Borner C, Bredesen DE, Brenner C, Campanella M, Candi E, Cecconi F, Chan FK, Chandel NS, Cheng EH, Chipuk JE, Cidlowski JA, Ciechanover A, Dawson TM, Dawson VL. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 2015;22(1):58–73. doi: 10.1038/cdd.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carmona-Gutierrez D, Eisenberg T, Büttner S, Meisinger C, Kroemer G, Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17(5):763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 127.Eisenberg T, Carmona-Gutierrez D, Büttner S, Tavernarakis N, Madeo F. Necrosis in yeast. Apoptosis Int J Program Cell. 2010;Death doi: 10.1007/s10495-009-0453-4. [DOI] [PubMed] [Google Scholar]
- 128.Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194(2):341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pereira C, Silva RD, Saraiva L, Johansson B, Sousa MJ, Côrte-Real M. Mitochondria-dependent apoptosis in yeast. Biochim Biophys Acta. 2008;1783(7):1286–1302. doi: 10.1016/j.bbamcr.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 130.Pereira C, Coutinho I, Soares J, Bessa C, Leão M, Saraiva L. New insights into cancer-related proteins provided by the yeast model. FEBS J. 2012;279(5):697–712. doi: 10.1111/j.1742-4658.2012.08477.x. [DOI] [PubMed] [Google Scholar]
- 131.Silva RD, Manon S, Gonçalves J, Saraiva L, Côrte-Real M. The importance of humanized yeast to better understand the role of bcl-2 family in apoptosis: finding of novel therapeutic opportunities. Curr Pharm Des. 2011;17(3):246–255. doi: 10.2174/138161211795049651. [DOI] [PubMed] [Google Scholar]
- 132.Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta. 2009;1793(4):650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sousa MJ, Azevedo F, Pedras A, Marques C, Coutinho OP, Preto A, Gerós H, Chaves SR, Côrte-Real M. Vacuole-mitochondrial cross-talk during apoptosis in yeast: a model for understanding lysosome-mitochondria-mediated apoptosis in mammals. Biochem Soc Trans. 2011;39(5):1533–1537. doi: 10.1042/BST0391533. [DOI] [PubMed] [Google Scholar]
- 134.Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185(1):43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nishimura Y, Kato K. Expression of mouse cathepsin L cDNA in Saccharomyces cerevisiae: evidence that cathepsin L is sorted for targeting to yeast vacuole. Arch Biochem Biophys. 1992;298(2):318–324. doi: 10.1016/0003-9861(92)90417-u. [DOI] [PubMed] [Google Scholar]
- 136.Nishimura Y, Takeshima H, Sakaguchi M, Mihara K, Omura T, Kato K, Himeno M. Expression of rat cathepsin D cDNA in Saccharomyces cerevisiae: implications for intracellular targeting of cathepsin D to vacuoles. J Biochem (Tokyo) 1995;118(1):168–177. doi: 10.1007/978-1-4615-1871-6_34. [DOI] [PubMed] [Google Scholar]


