INTRODUCTION
Autophagy, the main lysosomal degradative machinery, plays a major role in maintaining cellular homeostasis and thus a healthy state in an organism. This process recycles unnecessary or damaged material, therefore, not only providing nutrients to maintain vital cellular functions in times of starvation but also eliminating potentially harmful cellular material 1. Importantly, the autophagic rate declines with increasing age 2,3, suggesting a functional correlation between aging and autophagy. Indeed, the deregulation of autophagy is involved in the onset of various age-related diseases such as cancer, cardiomyopathy, type II diabetes, and neurodegeneration 4. Until recently, aging was regarded as an unregulated and inescapable consequence of the accumulation of incidental damage in macromolecules and/or organelles. However, the discovery of multiple ways to extend the lifespan in a variety of different model organisms, e.g., by genetic and pharmacological means, developed the formulation of alternative aging theories that consider aging as a molecular program 5. Indeed, the last years have provided important insights into the networks that control aging and have thus highlighted the interconnected nature of aging and various cellular processes. For instance, the process of aging is intimately coupled to metabolic processes 6, in particular to energy metabolism and nutrient availability. Nevertheless, specific metabolites that affect aging and autophagy remain poorly described.
NUTRIENT AVAILABILITY CONTROLS AUTOPHAGY AND AGING VIA ENERGY METABOLITES
As sensors of the current environmental status, nutrient signaling pathways represent central aging regulators. For instance, individual interventions in the insulin/insulin-like growth factor 1 (IGF-1), Ras, protein kinase A (PKA), target of rapamycin (Tor), or protein kinase B (SCH9/Akt) pathways have been shown to extend lifespan in various organisms, including mammals 7,8,9. Caloric restriction (CR) requires autophagy for lifespan extension 10,11,12 and CR-mediated autophagy induction follows molecular pathways that are shared with those known to affect aging, such as Tor, SCH9/Akt, or IGF-1 13. Furthermore, the AMP-activated protein kinase (AMPK) serves as a metabolic radar sensing changes in the AMP/ATP ratio and is conserved in the majority of eukaryotic species, and has also been established as a checkpoint for growth control and autophagy regulation 14. Consistently, several studies have revealed a connection between the AMP/ATP ratio, autophagic flux rates, senescence, and disease 15,16. Noteworthy, early studies on rat hepatocytes also suggested that the execution of autophagy depends on energy availability since inhibition of ATP production stalls autophagic flux 17. Other pivotal energy sources like butyrate, an essential energy component in the colon, and second messengers such as cAMP, which might also be implicated in Ras/PKA-mediated lifespan modulation in various organisms, were identified as potential autophagy mediators 18,19. This argues for a decisive function of nutrient signaling and energy metabolites during aging and its associated processes.
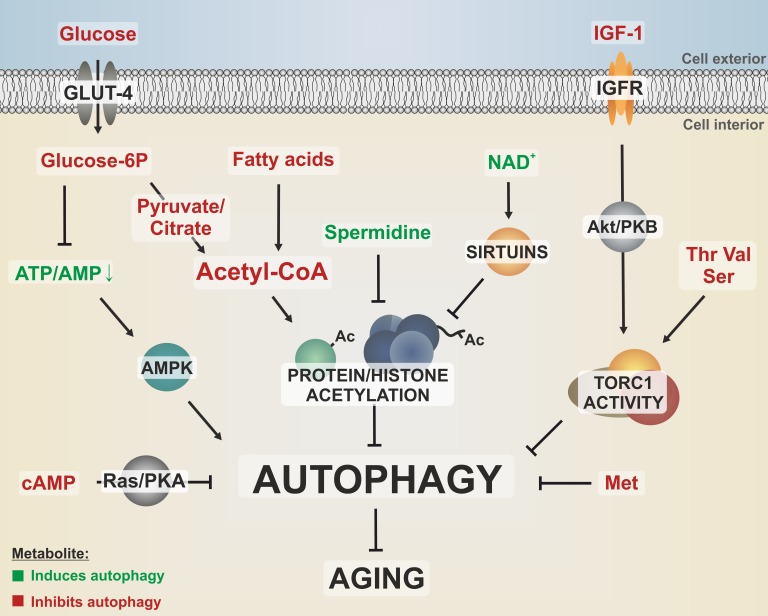
Figure 1. FIGURE 1: Different metabolites converge on pathways that regulate autophagy and aging.
Dietary nutrients like glucose, amino acids and fatty acids as well as growth signaling by IGF-1 activate nutrient-sensing kinases, like the target of rapamycin complex 1 (TORC1), which stalls autophagy via phosphorylation of downstream targets. Furthermore, autophagy is negatively regulated by the Ras/PKA pathway, which responds to nutrient availability by sensing intracellular cAMP levels. The cellular energy status is reflected by the ATP/AMP ratio, which is sensed by the autophagy activator AMPK. Methionine downregulates autophagy during aging in a yet to be elucidated fashion. The central energy intermediate acetyl-CoA integrates metabolites from glycolysis, β-oxidation or respiration and fuels acetylation of proteins such as histones, resulting in decreased autophagic flux. All these autophagy-limiting metabolic pathways have been linked to an accelerated aging phenotype. In contrast, polyamines, like spermidine, reduce protein acetylation, thereby promoting autophagy and longevity. Potential crosstalks between protein acetylation and nutrient sensing kinase signaling are yet to be elucidated. GLUT-4, glucose transporter 4; IGFR, IGF-1 receptor; Ac, acetyl-group; amino acids are indicated by three-letter code.
THE ENERGY METABOLITE ACETYL-CoA SUPPRESSES AUTOPHAGY VIA PROTEIN ACETYLATION
In the yeast S cerevisiae, glucose is the preferred carbon source for fueling energy metabolism and its fermentation produces acetate and ethanol that is used for subsequent respiration after the diauxic shift. Both intermediates are released into the medium and have been attributed a role in limiting yeast chronological life span 20, but considered a pro-aging factor specific for yeast 21. However, acetate is an important metabolite that is involved in central processes such as acetyl-Coenzyme A (acetyl-CoA) production. Thus, its metabolic fate rather than a simply extracellular toxicity may (at least in part) be responsible for its impact on lifespan. Consequently, acetate metabolism may also contribute to aging in higher eukaryotes, possibly via its impact on protein acetylation through acetyl-CoA generation and subsequent control of cellular function. Energy metabolites that can derive in the production of acetyl-CoA, such as citrate, pyruvate and fatty acids, were shown to be deregulated in senescence-accelerated mice 22. We could recently show that (nucleo-)cytosolic acetyl-CoA, in fact, serves as a modulator of longevity, suppressing starvation and age-associated autophagy in a variety of phyla 23,24. This function might be due to the fact that acetyl-CoA is the only donor for acetylation reactions and both, protein acetylation and epigenetic chromatin modifications, have repeatedly been linked to the regulation of aging and autophagy 23,25,26. For instance, the highly conserved protein family of NAD+-dependent histone deacetylases and ADP ribosylases (sirtuins) has been connected to aging modulation 27,28 and lifespan extension upon CR 29. Activation of histone deacetylases, such as sirtuins, has therefore been extensively studied for its capability to combat aging or age-associated pathologies. Interestingly, the lifespan-extending effects of sirtuin activation by CR or pharmacological interventions depend on the induction of autophagy 11. The dependency on NAD+ as a cofactor and the tight connections between sirtuins, longevity and autophagy induction have led to the hypothesis that sirtuins act as metabolic sensors that promote mitochondrial maintenance 30. Notably, nicotinamide metabolism has become an intensively investigated target for drug discovery against a variety of human diseases, including age-associated pathologies such as cancer or neurodegeneration 31.
POLYAMINE METABOLISM LEAVES A REGULATORY FINGERPRINT AT HISTONE MODIFICATION SITES
The levels of polyamines, a class of ubiquitously occurring small basic polycations, decline with progressing age in various organisms, including humans 32, yeast 25 and plants 33. Intriguingly, external application of a specific polyamine (spermidine) counteracts cell death during aging and improves the lifespan of yeast, flies, worms, and human immune cells in an autophagy-dependent manner 25. Furthermore, it causes a reduction of oxidative stress in mice. In addition, more recent studies also suggest longevity-promoting effects in mammals 34. Importantly, spermidine treatment appears to be associated with both histone hypoacetylation caused by inhibition of histone acetyl transferases 25 and deacetylation of cytosolic proteins 35. However, it has also been hypothesized that polyamines influence histone acetylation in dependence of the histones’ own acetylation status 36 and massive polyamine catabolism has even been shown to deplete the acetylation co-factor acetyl-CoA 37. Possibly, the involvement of spermidine in the biosynthesis of the methyl-group donor S-adenosyl-methionine (SAM) 38 could also culminate in regulatory methylation reactions, such as chromatin silencing. Thus, polyamines may influence chromatin structure and protein acetylation via multiple mechanisms. Given the aforementioned contribution of acetyl-CoA to aging and autophagy modulation, it is tempting to speculate that spermidine functions include a down-titration of the intracellular acetyl-CoA pool and thus a rearrangement of the metabolic state.
SPECIFIC AMINO ACID STARVATION INDUCES AUTOPHAGY AND PROLONGS LIFESPAN
In line with the vast importance of nutrient signaling during aging, amino acid metabolism has an important impact on eukaryotic aging and its related diseases. The levels of specific amino acids like tryptophane, methionine, arginine, or leucine has often been suggested to (positively or negatively) influence the autophagy pathway and impact aging in different eukaryotes 39,40,41,42. Indeed, we could recently demonstrate that limitation of the amino acid methionine enhances yeast chronological lifespan. Intriguingly, this lifespan extension requires autophagy-dependent vacuolar acidification 43. In the same line, vacuolar acidification also elongates replicative lifespan of yeast, where it protects mitochondria most probably via an improvement of the vacuolar amino acid storage function 44. In yeast, serine, threonine and valine promote cellular senescence probably via activation of the Sch9/TOR pathway and subsequent inhibition of the protein kinase Rim15p, which guides anti-aging stress response pathways 45. Altogether, these examples suggest that the intake or limitation of specific amino acids is a determining factor during aging, though the mechanistic specificities are expected to be complexly regulated, especially at the organismal level.
CONCLUSION
A variety of potential metabolic controllers of autophagy and health span have already been proposed. However, precise strategies to target the correlating pathways (e.g., by nutrition patterns) remain to be elucidated in more detail. For example, it would be of great interest to determine if special diets that include the limitation of (defined) amino acids or the uptake of certain polyamines, like spermidine, influence the metabolism towards improved cellular conditions during aging. It also remains elusive how certain diets may affect the microbiome and in turn impact the levels of certain metabolites that have been shown to regulate cellular fitness, such as citrate, pyruvate, butyrate, or acetate. The investigation of metabolites as powerful rheostats in aging and autophagy is supported by the improvement of technologies that have opened up new possibilities to detect and trace even small molecules in vitro and in vivo. This might bring up metabolomics as a future trend for aging analyses 46. Hopefully, the findings on the impact of metabolism on aging will culminate in amended dietary guidelines that would make eating the tastiest of all medicines.
References
- 1.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuervo AM. Autophagy and aging. Trends Genet TIG. 2008;24(12):604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Trends Genet TIG. 2008;4(2):176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autoph-agy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldsmith TC. On the pro-grammed/non-programmed aging controversy. Biochem Mosc. 2012;77(7):729–732. doi: 10.1134/S000629791207005X. [DOI] [PubMed] [Google Scholar]
- 6.Houtkooper RH, Williams RW, Auwerx J. Metabolic networks of longevity. Cell. 2010;142(1):9–14. doi: 10.1016/j.cell.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Longo VD. Mutations in signal transduction proteins increase stress resistance and longevity in yeast, nematodes, fruit flies, and mammalian neuronal cells. Neurobiol Ag-ing. 1999;20(5):479–486. doi: 10.1016/S0197-4580(99)00089-5. [DOI] [PubMed] [Google Scholar]
- 8.Tatar M, Kopelman A, Epstein D, Tu M-P, Yin C-M, Garofalo RS. A Mutant Drosophila Insulin Receptor Homolog That Extends Life-Span and Impairs Neuroendo-crine Function. Science. 2001;292(5514):107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 9.Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310(5751):1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 10.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3(6):597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 11.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, Michaud M, Madeo F, Tavernarakis N, Kroemer G. Caloric restriction and resveratrol pro-mote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minina EA, Sanchez-Vera V, Moschou PN, Suarez MF, Sundberg E, Weih M, Bozhkov PV. Autophagy mediates ca-loric restriction-induced lifespan extension in Arabidopsis. Aging Cell. 2013;12(2):327–329. doi: 10.1111/acel.12048. [DOI] [PubMed] [Google Scholar]
- 13.Rubinsztein DC, Mariño G, Kroemer G. Autophagy and Aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 14.Mihaylova MM, Shaw RJ. The AMP-activated protein kinase (AMPK) signaling pathway coordinates cell growth, au-tophagy, & metabolism. Nat Cell Biol. 2011;13(9):1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Fujii H, Mohan SV, Goronzy JJ, Weyand CM. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med. 2013;210(10):2119–2134. doi: 10.1084/jem.20130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubli DA, Gustafsson ÅB. Cardiomyocyte health: adapting to metabolic changes through autophagy. Trends Endo-crinol Metab. 2014;25(3):156–164. doi: 10.1016/j.tem.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plomp PJAM, Wolvetang EJ, Groen AK, Meijer AJ, Gordon PB, Seglen PO. Energy dependence of autophagic protein deg-radation in isolated rat hepatocytes. Eur J Bio-chem. 1987;164(1):197–203. doi: 10.1111/j.1432-1033.1987.tb11011.x. [DOI] [PubMed] [Google Scholar]
- 18.Holen I, Gordon PB, Strømhaug PE, Seglen PO. Role of cAMP in the regula-tion of hepatocytic autophagy. Eur J Biochem FEBS. 1996;236(1):163–170. doi: 10.1111/j.1432-1033.1996.00163.x. [DOI] [PubMed] [Google Scholar]
- 19.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, Bultman SJ. The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon. Cell Metab. 2011;13(5):517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirisola MG, Longo VD. Acetic acid and acidification accelerate chrono-logical and replicative aging in yeast. Cell Cycle. 2012;11(19):3532–3533. doi: 10.4161/cc.22042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burhans WC, Weinberger M. Acetic acid effects on aging in budding yeast: Are they relevant to aging in higher eukary-otes? Cell Cycle. 2009;8(14):2300–2302. doi: 10.4161/cc.8.14.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang N, Yan X, Zhou W, Zhang Q, Chen H, Zhang Y, Zhang X. NMR-Based Metabonomic Investigations into the Metabolic Profile of the Senescence-Accelerated Mouse. J Proteome Res. 2008;7(9):3678–3686. doi: 10.1021/pr800439b. [DOI] [PubMed] [Google Scholar]
- 23.Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Küttner V, Bhukel A, Mariño G, Pietrocola F, Harger A, Zimmermann A, Moustafa T, Sprenger A, Jany E, Büttner S, Carmona-Gutierrez D, Ruckenstuhl C, Ring J, Reichelt W, Schimmel K, Leeb T, Moser C, Schatz S, Kamolz L-P, Magnes C, Sinner F, Sedej S, Fröhlich K-U, Juhasz G, Pieber TR, Dengjel J, Sigrist SJ, Kroemer G, Madeo F. Nucleocytosolic Depletion of the Ener-gy Metabolite Acetyl-Coenzyme A Stimulates Autophagy and Prolongs Lifespan. Cell Metab. 2014;19(3):431–444. doi: 10.1016/j.cmet.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, Zamzami N, Scoazec M, Durand S, Enot DP, Fernández AF, Martins I, Kepp O, Senovilla L, Bauvy C, Morselli F, Vacchelli E, Bennetzen M, Magnes C, Sinner F, Pieber T, López-Otin C, Maiuri MC, Codogno P, Andersen JS, Hill JA, Madeo F, Kroemer G. Regulation of Autophagy by Cytosolic Acetyl-Coenzyme A . Mol Cell. 2014;53(5):710–25. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Fröhlich K-U, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11(11):1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 26.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silenc-ing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 27.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123(4):655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 28.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13(19):2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S-J, Defossez P-A, Guarente L. Requirement of NAD and SIR2 for Life-Span Extension by Calorie Restriction in Saccharomyces cerevisiae. Science. 2000;289(5487):2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 30.Sack MN, Finkel T. Mitochondrial Metabolism, Sirtuins, and Aging. Cold Spring Harb Perspect Biol. 2012;4(12):a013102. doi: 10.1101/cshperspect.a013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan JA, Forouhar F, Tao X, Tong L. Nicotinamide adenine dinucleotide me-tabolism as an attractive target for drug discov-ery. Expert Opin Ther Targets. 2007;11(5):695–705. doi: 10.1517/14728222.11.5.695. [DOI] [PubMed] [Google Scholar]
- 32.Scalabrino G, Ferioli ME. Polyamines in mammalian ageing: an oncolog-ical problem, too? A review. . Mech Ageing Dev. 1984;26(2-3):149–164. doi: 10.1016/0047-6374(84)90090-3. [DOI] [PubMed] [Google Scholar]
- 33.Kaur-Sawhney R, Shih L-M, Flores HE, Galston AW. Relation of Polyamine Synthesis and Titer to Aging and Senescence in Oat Leaves 1. Plant Physiol. 1982;69(2):405–410. doi: 10.1104/pp.69.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soda K, Dobashi Y, Kano Y, Tsujinaka S, Konishi F. Polyamine-rich food decreases age-associated pathology and mortality in aged mice. Exp Gerontol. 2009;44(11):727–732. doi: 10.1016/j.exger.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Morselli E, Marino G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Benit P, Rustin P, Criollo A, Kepp O, Gal-luzzi L, Shen S, Malik SA, Maiuri MC, Horio Y, Lopez-Otin C, Andersen JS, Tavernarakis N, Madeo F, Kroemer G. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192(4):615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Sutton A, Sternglanz R. A Yeast Polyamine Acetyltransferase. J Biol Chem. 2005;280(17):16659–16664. doi: 10.1074/jbc.M414008200. [DOI] [PubMed] [Google Scholar]
- 37.Kee K, Foster BA, Merali S, Kramer DL, Hensen ML, Diegelman P, Kisiel N, Vujcic S, Mazurchuk RV, Porter CW. Acti-vated polyamine catabolism depletes acetyl-CoA pools and suppresses prostate tumor growth in TRAMP mice. J Biol Chem. 2004;279(38):40076–40083. doi: 10.1074/jbc.M406002200. [DOI] [PubMed] [Google Scholar]
- 38.Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M. Methionine salvage and S-adenosylmethionine: essential links be-tween sulfur, ethylene and polyamine biosyn-thesis. Biochem J. 2013;451(2):145–154. doi: 10.1042/BJ20121744. [DOI] [PubMed] [Google Scholar]
- 39.Dillon EL. Nutritionally essential amino acids and metabolic signaling in aging. Amino Acids. 2013;45(3):431–441. doi: 10.1007/s00726-012-1438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low Methionine Ingestion by Rats Extends Life Span. J Nutr. 1993;123(2):269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 41.Van der Goot AT, Nollen EAA. Tryptophan metabolism: entering the field of aging and age-related pathologies. Trends Mol Med. 2013;19(6):336–344. doi: 10.1016/j.molmed.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Aris JP, Alvers AL, Ferraiuolo RA, Fishwick LK, Hanvivatpong A, Hu D, Kirlew C, Leonard MT, Losin KJ, Marraffini M, Seo AY, Swanberg V, Westcott JL, Wood MS, Leeuwenburgh C, Dunn Jr WA. Autophagy and leucine promote chronological longevity and respiration proficiency during calorie restriction in yeast. Exp Gerontol. 2013;48(10):1107–1119. doi: 10.1016/j.exger.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruckenstuhl C, Netzberger C, Entfellner I, Carmona-Gutiérrez D, Kickenweiz T, Stekovic S, Gleixner C, Smidt C, Klug L, Sor-go AG, Eisenberg T, Büttner S, Marino G, Ra-fal K, Jansen-Dürr P, Fröhlich K-U, Kroemer G, Madeo F. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification: in press. 2014 doi: 10.1371/journal.pgen.1004347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mi-tochondrial function and lifespan in yeast. Nature. 2012;492(7428):261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirisola MG, Taormina G, Fabrizio P, Wei M, Hu J, Longo VD. Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging. PLoS Genet. 2014;10(2):e1004113. doi: 10.1371/journal.pgen.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kristal BS, Shurubor YI. Metabolomics: Opening Another Window into Aging. Sci Aging Knowl Environ. 2005;2005(26):pe19. doi: 10.1126/sageke.2005.26.pe19. [DOI] [PubMed] [Google Scholar]