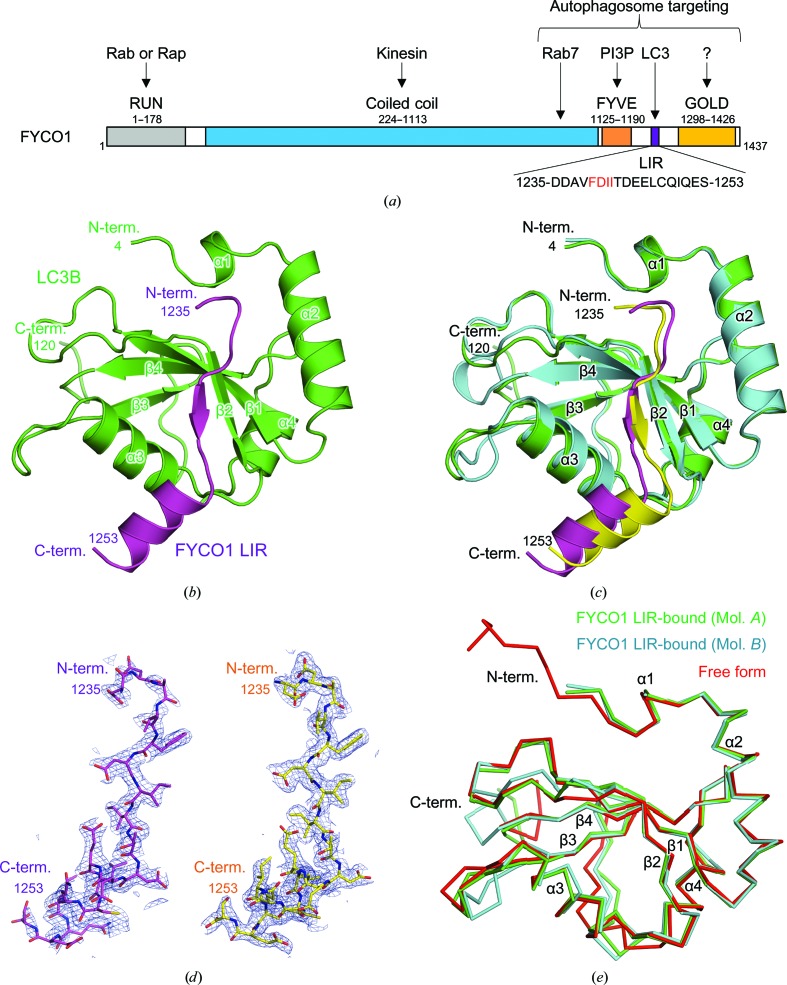
FYCO1 mediates microtubule plus-end-directed autophagosome transport by interacting with LC3. Here, the crystal structure of mouse LC3B in complex with the FYCO1 LC3-interacting region (LIR) motif peptide was determined.
Keywords: autophagy, FYCO1, LC3, LC3-interacting region (LIR) motif
Abstract
FYVE and coiled-coil domain-containing protein 1 (FYCO1), a multidomain autophagy adaptor protein, mediates microtubule plus-end-directed autophagosome transport by interacting with kinesin motor proteins and with the autophagosomal membrane components microtubule-associated protein 1 light chain 3 (LC3), Rab7 and phosphatidylinositol 3-phosphate (PI3P). To establish the structural basis for the recognition of FYCO1 by LC3, the crystal structure of mouse LC3B in complex with the FYCO1 LC3-interacting region (LIR) motif peptide was determined. Structural analysis showed that the flanking sequences N-terminal and C-terminal to the LIR core sequence of FYCO1, as well as the tetrapeptide core sequence, were specifically recognized by LC3B and contributed to the binding. Moreover, comparisons of related structures revealed a conserved mechanism of FYCO1 recognition by different LC3 isoforms among different species.
1. Introduction
Autophagy is a crucial pathway for maintaining intracellular homeostasis and is conserved among eukaryotes from yeast to humans. Nonessential or damaged cytoplasmic components, including proteins and organelles, as well as pathogens invading cells, are degraded during autophagy (Mizushima, 2007 ▸; Klionsky & Emr, 2000 ▸; Mizushima & Komatsu, 2011 ▸). Dysfunction of autophagy is related to various human diseases, including cancer and neurodegenerative diseases (Mizushima & Komatsu, 2011 ▸; Jiang & Mizushima, 2014 ▸). Macroautophagy (hereafter referred to as autophagy) is mediated by unique double-membrane vesicles called autophagosomes, in which the targets of autophagy are sequestered. Autophagosomes are transported along microtubules and then fuse with lysosomes to become autolysosomes, which are the sites of degradation (Mizushima, 2007 ▸; Klionsky & Emr, 2000 ▸; Mizushima & Komatsu, 2011 ▸; Mackeh et al., 2013 ▸).
Microtubule-associated protein 1 light chain 3 (LC3), a mammalian orthologue of Saccharomyces cerevisiae autophagy-related protein 8 (Atg8), plays important roles in autophagosome formation and transport (Mizushima & Komatsu, 2011 ▸; Shpilka et al., 2011 ▸; Nakatogawa et al., 2007 ▸). Mammalian Atg8 orthologues can be subdivided into the LC3, GABA type A receptor-associated protein (GABARAP) and Golgi-associated ATPase enhancer of 16 kDa (GATE16) subfamilies, which include LC3A, LC3B, LC3B2, LC3C, GABARAP, GABARAPL1 and GABARAPL2 (Shpilka et al., 2011 ▸). Atg8 proteins are cleaved at their C-terminal tail immediately after translation, exposing the conserved glycine residues. Phosphatidylethanolamine (PE) is then conjugated to the glycine residue. PE-conjugated Atg8 proteins are anchored to the surface of phagophores or autophagosomes and serve as markers of autophagy for protein–protein interactions (Ichimura et al., 2000 ▸). Atg8 proteins selectively recruit autophagic cargo by interacting with cargo receptors such as p62 and neighbour of BRCA1 gene 1 (NBR1) (Mizushima & Komatsu, 2011 ▸; Boyle & Randow, 2013 ▸; Johansen & Lamark, 2011 ▸; Ichimura & Komatsu, 2010 ▸). Moreover, Atg8 proteins contribute to the proper transport of autophagosomes by interacting via autophagy adaptor proteins, which interact with microtubule motor proteins (Pankiv et al., 2010 ▸; Fu et al., 2014 ▸). The specificity and selectivity of interactions with Atg8 proteins are defined by the LC3-interacting region (LIR) motif, which contains the tetrapeptide core sequence W/Y/FxxL/I/V; the hydrophobic residues of this sequence interact with the hydrophobic pockets of Atg8 family proteins (Birgisdottir et al., 2013 ▸; Noda et al., 2008 ▸, 2010 ▸).
FYVE and coiled-coil protein 1 (FYCO1) is an autophagy adaptor protein responsible for microtubule plus-end-directed autophagosome transport by mediating interactions between kinesin motor proteins and autophagosomes (Pankiv et al., 2010 ▸; Fig. 1 ▸ a). FYCO1 is a multidomain protein containing an N-terminal RUN domain, a central coiled-coil region and a C-terminal FYVE domain, LIR motif and Golgi dynamics (GOLD) domain. The RUN domain is thought to interact with Rab and Rap family proteins, whereas the central coiled-coil region is responsible for FYCO1 dimerization and interaction with kinesin and Rab7. The FYVE domain is defined as a phosphatidylinositol 3-phosphate (PI3P)-binding module (Gaullier et al., 1998 ▸) and is required for membrane targeting of FYCO1. The FYCO1 LIR motif is also important for membrane targeting through interaction with the membrane-anchored Atg8 protein LC3. Although the GOLD domain has not yet been characterized, it is thought to be involved in membrane association (Anantharaman & Aravind, 2002 ▸).
Figure 1.
Crystal structure of mouse LC3B in complex with FYCO1 LIR. (a) Schematic representation of the domain structure of mouse FYCO1. (b) Crystal structure of LC3B in complex with FYCO1 LIR. LC3B and FYCO1 LIR are shown in green and purple, respectively. The N- and C-termini and structural elements are labelled. (c) Superposition of the two LC3B–FYCO1 LIR complexes in the crystal asymmetric unit. LC3B structures (molecules A and B) are shown in green and cyan, respectively, and FYCO1 LIR structures (molecules C and D) are shown in purple and yellow, respectively. The two complexes consist of molecules A and C and molecules B and D. (d) Electron densities of FYCO1 LIR. The 2F o − F c difference electron-density map is contoured at the 1.0σ level with a blue mesh (left, molecule C; right, molecule D). (e) Superposition of LC3B proteins. The mouse LC3B in complex with FYCO1 LIR (molecules A and B in this study) and the free form of human LC3B (PDB entry 3vtu; Rogov et al., 2013 ▸) are superposed and are shown in green, cyan and red, respectively.
Therefore, in this study, in order to gain structural insights into the autophagosome targeting of FYCO1, we determined the crystal structure of LC3B in complex with the FYCO1 LIR peptide. In addition to the LIR core sequence, N-terminal and C-terminal extension sequences of FYCO1 LIR were involved in the interaction with LC3B and were important for high-affinity binding. Comparisons with related structures (Cheng et al., 2016 ▸; Olsvik et al., 2015 ▸) revealed a conserved mechanism of FYCO1 recognition by different LC3 isoforms among different species, providing important insights into the functional mechanisms of these proteins.
2. Materials and methods
2.1. Macromolecule production
The gene encoding mouse LC3B (residues 1–120) was inserted into the expression vector pGEX6P-1 (GE Healthcare) between the BamHI and EcoRI sites. Escherichia coli BL21(DE3)pLysS cells were transformed with the vector and cultured at 310 K to a suitable cell density (OD600 of 0.6–0.7); protein expression was then induced by addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and cultivation was continued for 14 h at 291 K. The cells were collected by centrifugation and lysed by sonication in phosphate-buffered saline (PBS) containing 2 mM dithiothreitol (DTT). The proteins were purified from the cleared lysate using Glutathione Sepharose 4B (GE Healthcare) followed by GST-tag cleavage with PreScission protease. Further purification was performed using a Superdex 200 gel-filtration column (GE Healthcare). The proteins were concentrated to about 37 mg ml−1 in a buffer consisting of 10 mM Tris–HCl pH 8.0, 150 mM NaCl. Mouse FYCO1 LIR peptide (residues 1235–1253, DDAVFDIITDEELCQIQES; Eurofins Genomics) was dissolved to about 9 mg ml−1 in a buffer consisting of 10 mM Tris–HCl pH 8.0, 70 mM HEPES–NaOH pH 7.0, 30 mM NaOH, 120 mM NaCl. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule information.
Additional residues after GST-tag cleavage with PreScission protease are underlined.
| LC3B | |
| Source organism | Mus musculus |
| DNA source | cDNA |
| Expression vector | pGEX6P-1 |
| Expression host | E. coli BL21(DE3)pLysS |
| Complete amino-acid sequence of the construct | GPLGSMPSEKTFKQRRSFEQRVEDVRLIREQHPTKIPVIIERYKGEKQLPVLDKTKFLVPDHVNMSELIKIIRRRLQLNANQAFFLLVNGHSMVSVSTPISEVYESERDEDGFLYMVYASQETFG |
| Formula weight of entity (Da) | 14554.7 |
| FYCO1 (synthetic peptide) | |
| Source organism | Mus musculus |
| Complete amino-acid sequence of the construct | DDAVFDIITDEELCQIQES |
| Formula weight of entity (Da) | 2183.3 |
2.2. Crystallization, data collection and structure determination
The protein solution for the cocrystallization of LC3B and the FYCO1 LIR peptide contained LC3B at 19 mg ml−1 and a 1.5-fold excess of the FYCO1 peptide. Crystals of the LC3B–FYCO1 LIR complex were grown at 293 K by the sitting-drop vapour-diffusion method by mixing the protein solution with an equal volume of reservoir solution (2 M ammonium sulfate, 0.1 M sodium citrate tribasic dihydrate pH 5.6, 0.2 M potassium sodium tartrate tetrahydrate). Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Sitting-drop vapour diffusion |
| Plate type | Intelli-Plate 96-2 shallow well (Hampton Research) |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 19 |
| Buffer composition of protein solution | 10 mM Tris–HCl pH 8.0, 150 mM NaCl |
| Composition of reservoir solution | 2 M ammonium sulfate, 0.1 M sodium citrate tribasic dihydrate pH 5.6, 0.2 M potassium sodium tartrate tetrahydrate |
| Volume and ratio of drop | 300 nl protein solution, 300 nl reservoir solution |
| Volume of reservoir (µl) | 50 |
X-ray diffraction data were collected (λ = 1.0000 Å) on beamline BL41XU at SPring-8, Hyogo, Japan under cryogenic conditions at 100 K. Crystals were equilibrated in a cryoprotectant solution consisting of reservoir solution supplemented with 25% glycerol and 0.15 M NaCl prior to flash-cooling. X-ray diffraction data were processed with HKL-2000 (Otwinowski & Minor, 1997 ▸). Data-collection and processing statistics are summarized in Table 3 ▸.
Table 3. Data-collection and processing statistics.
Values in parentheses are for the outer shell.
| Diffraction source | BL41XU, SPring-8 |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Detector | PILATUS 6M |
| Crystal-to-detector distance (mm) | 380 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 180 |
| Exposure time per image (s) | 0.5 |
| Space group | C2 |
| a, b, c (Å) | 113.4, 44.6, 63.6 |
| α, β, γ (°) | 90, 120.3, 90 |
| Matthews coefficient (V M) (Å3 Da−1) | 2.07 |
| Mosaicity (°) | 0.34 |
| Resolution range (Å) | 50.00–1.90 (1.93–1.90) |
| Total No. of reflections | 71783 |
| No. of unique reflections | 21371 |
| Completeness (%) | 98.1 (87.6) |
| Multiplicity | 3.4 (2.8) |
| 〈I/σ(I)〉 | 20.2 (2.0) |
| R meas † | 0.060 (0.436) |
| Overall B factor from Wilson plot (Å2) | 28.7 |
R
meas = 
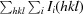 .
.
The crystal structure of the LC3B–FYCO1 LIR complex was solved by molecular replacement using MOLREP (Vagin & Teplyakov, 2010 ▸) using the free form of the human LC3B structure (PDB entry 3vtu; Rogov et al., 2013 ▸) as a search model. The models were subjected to iterative cycles of manual model building using Coot (Ensley et al., 2010 ▸) and restrained refinement using REFMAC (Murshudov et al., 2011 ▸) (Table 4 ▸). The quality of the refined model was evaluated with MolProbity (Chen et al., 2010 ▸). The structural figures were prepared with PyMOL (DeLano, 2002 ▸). The coordinate and structure-factor data of mouse LC3B in complex with the FYCO1 LIR peptide have been deposited in the Protein Data Bank (PDB) as PDB entry 5wrd.
Table 4. Refinement statistics.
| Resolution range (Å) | 50.00–1.90 |
| Completeness (%) | 98.0 |
| No. of reflections, working set | 20284 |
| No. of reflections, test set | 1087 |
| Final R cryst (%) | 18.8 |
| Final R free (%) | 24.4 |
| No. of non-H atoms | |
| LC3B | 1950 |
| FYCO1 | 304 |
| Other | 62 |
| Average B factors (Å2) | |
| LC3B | 39.7 |
| FYCO1 | 50.5 |
| Other | 38.5 |
| R.m.s. deviations | |
| Bonds (Å) | 0.019 |
| Angles (°) | 2.00 |
| Ramachandran plot | |
| Most favoured (%) | 98.1 |
| Allowed (%) | 1.1 |
2.3. Pull-down assay
The genes encoding mouse FYCO1 LIR shown in Fig. 2 ▸(g) were inserted into the expression vector pGEX6P-1 between the BamHI and EcoRI sites. E. coli BL21(DE3)RIPL cells were transformed with the vectors and cultured at 310 K to a suitable cell density; protein expression was then induced by addition of 0.5 mM IPTG and cultivation was continued for 15 h at 291 K. The cells were collected by centrifugation and lysed by sonication in PBS containing 2 mM DTT. The proteins from the cleared lysate were incubated with Glutathione Sepharose 4B and purified LC3B for 1 h at 277 K. After washing, proteins were eluted from Glutathione Sepharose 4B and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE).
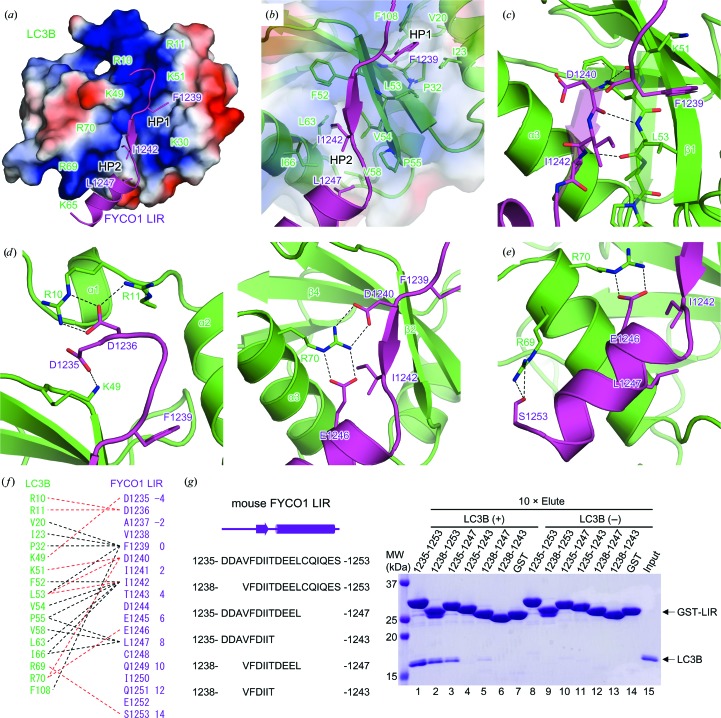
Figure 2.
Interaction of LC3B and FYCO1 LIR. (a) Electrostatic surface potential of LC3B. The positively charged residues of LC3B are labelled. The bound FYCO1 LIR peptide is shown as a cartoon model with side chains of hydrophobic residues at positions 0, 3 and 8. Positive and negative electrostatic potential is shown in blue and red, respectively. The hydrophobic pockets, HP1 and HP2, are indicated. (b) Detailed view of the interactions between the hydrophobic pockets of LC3B and the hydrophobic residues of FYCO1 LIR. (c) Intermolecular main-chain interactions between LC3B and FYCO1 LIR. Hydrogen bonds are indicated by dashed lines. (d) Detailed view of the electrostatic interactions at the N-terminal (left) and C-terminal (right) extensions of the FYCO1 LIR motif. (e) Detailed view of the interaction of the C-terminal α-helix of FYCO1 LIR with LC3B. (f) Schematic summary of the interactions between LC3B and FYCO1 LIR. Hydrogen bonds and hydrophobic interactions are indicated by red and black dashed lines, respectively. The numbering scheme used in this paper is shown on the right. (g) GST pull-down assay using FYCO1 LIR fused to GST and LC3B. The input and eluted proteins were subjected to SDS–PAGE and stained with Coomassie Brilliant Blue.
3. Results
3.1. Crystal structure of LC3B in complex with FYCO1 LIR
We conducted the cocrystallization of mouse LC3B (residues 1–120) with a 19-residue peptide corresponding to mouse FYCO1 LIR (residues 1235–1253, DDAVFDIITDEELCQIQES; Fig. 1 ▸ a) and determined the crystal structure of the complex at 1.9 Å resolution (Fig. 1 ▸ b, Tables 1 ▸, 2 ▸, 3 ▸ and 4 ▸). The final model contained two molecules of LC3B (molecules A and B, residues 4–120) and FYCO1 LIR (molecules C and D, residues 1235–1253) in the crystal asymmetric unit and formed two LC3B–FYCO1 LIR complexes with 1:1 stoichiometry (molecules A and C and molecules B and D; Figs. 1 ▸ b and 1 ▸ c). The electron density of the FYCO1 LIR peptide was clearly visible in both complexes (Fig. 1 ▸ d).
The structure of LC3B was a compact globular shape with dimensions of approximately 25 × 30 × 40 Å containing two N-terminal α-helices (α1 and α2) and a C-terminal ubiquitin-like fold, as reported previously (Noda et al., 2008 ▸; Cheng et al., 2016 ▸; Olsvik et al., 2015 ▸; Rogov et al., 2013 ▸; McEwan et al., 2015 ▸; Suzuki et al., 2014 ▸; von Muhlinen et al., 2012 ▸; Sugawara et al., 2004 ▸; Fig. 1 ▸ b). The two molecules of LC3B in the crystal asymmetric unit were essentially the same, with a root-mean-square deviation (r.m.s.d.) of 0.6 Å (Fig. 1 ▸ c). The structure of LC3B in this study was similar to previously reported free forms of LC3B (PDB entry 3vtu; Rogov et al., 2013 ▸), with an r.m.s.d. of between 0.6 and 0.7 Å (Fig. 1 ▸ e). Large structural deviations among the three LC3B molecules were observed at the loop regions between α3 and β3 and between β3 and α4.
The FYCO1 peptide used in this study contained a central tetrapeptide LIR core motif (FDII) and flanking sequences N- and C-terminal to the LIR core motif (Fig. 1 ▸ a). The core motif was extended to form a β-strand, and the C-terminal flanking region exhibited a helical conformation (Fig. 1 ▸ b).
3.2. LC3B–FYCO1 LIR interactions
FYCO1 LIR was recognized not only by the core motif-mediated interactions that are conserved among most LIR–Atg8 family protein complexes but also by flanking region-mediated interactions (Figs. 2 ▸ a–2 ▸ f). The FYCO1 LIR peptide bound to the surface groove of LC3B formed by the α1, α2, α3 and β2 structures (Figs. 1 ▸ b and 2 ▸ a). The groove was highly positively charged with basic residues (Arg10, Arg11, Lys30, Lys49, Lys51, Lys65, Arg69 and Arg70) suitable for facilitating interactions with the negatively charged FYCO1 LIR (Figs. 2 ▸ a and 2 ▸ f). The groove had two hydrophobic pockets, HP1 and HP2; HP1 was formed by residues Val20, Ile23, Pro32, Leu53 and Phe108, and HP2 was formed by residues Phe52, Val54, Pro55, Val58, Leu63 and Ile66 (Figs. 2 ▸ a and 2 ▸ b). HP1 and HP2 are conserved among Atg8 family proteins and accommodate aromatic W/Y/F and hydrophobic L/I/V moieties in the LIR core sequence (W/Y/FxxL/I/V), respectively (Birgisdottir et al., 2013 ▸; Noda et al., 2008 ▸, 2010 ▸; Cheng et al., 2016 ▸; Olsvik et al., 2015 ▸; Rogov et al., 2013 ▸; McEwan et al., 2015 ▸; Suzuki et al., 2014 ▸; von Muhlinen et al., 2012 ▸; Wu et al., 2015 ▸; Stadel et al., 2015 ▸; Khaminets et al., 2015 ▸; Genau et al., 2015 ▸; Rogov et al., 2014 ▸; Lystad et al., 2014 ▸; Rozenknop et al., 2011 ▸; Ichimura et al., 2008 ▸; Satoo et al., 2009 ▸). In the structure of the LC3B–FYCO1 LIR complex, HP1 and HP2 interacted with Phe1239 and Ile1242 of FYCO1, respectively (Figs. 2 ▸ a, 2 ▸ b and 2 ▸ f). Moreover, residues Lys51 and Leu53 of LC3B formed main-chain hydrogen bonds with residues Asp1240 and Ile1242 of FYCO1 LIR (positions 1 and 3) to form an intermolecular β-sheet, as observed in other LC3–LIR structures (Birgisdottir et al., 2013 ▸; Noda et al., 2008 ▸, 2010 ▸; Cheng et al., 2016 ▸; Olsvik et al., 2015 ▸; Rogov et al., 2013 ▸; McEwan et al., 2015 ▸; Suzuki et al., 2014 ▸; von Muhlinen et al., 2012 ▸; Wu et al., 2015 ▸; Stadel et al., 2015 ▸; Khaminets et al., 2015 ▸; Genau et al., 2015 ▸; Rogov et al., 2014 ▸; Lystad et al., 2014 ▸; Rozenknop et al., 2011 ▸; Ichimura et al., 2008 ▸; Satoo et al., 2009 ▸; Figs. 2 ▸ c and 2 ▸ f).
In addition to the conserved interactions, the flanking sequences N-terminal and C-terminal to the FYCO1 LIR core motif also contributed to interactions with LC3B. We observed electrostatic interactions between charged residues, some of which formed salt bridges, i.e. Arg10, Arg11 and Lys49 of LC3B with Asp1235 and Asp1236 of FYCO1 (positions −4 and −3; Figs. 2 ▸ d and 2f), and Arg70 of LC3B with Asp1240 and Glu1246 of FYCO1 (positions 1 and 7; Figs. 2 ▸ d and 2f). Leu1247 in the C-terminal α-helix of FYCO1 LIR (position 8) also contributed to hydrophobic interactions with HP2 of LC3B (Figs. 2 ▸ a, 2 ▸ b and 2 ▸ f). Additional hydrogen bonding was observed between Arg69 of LC3B and Ser1253 of FYCO1 (position 14) in the C-terminal α-helix of the FYCO1 peptide (Figs. 2 ▸ e and 2 ▸ f). Interactions involving the N-terminal acidic residues of the LIR are observed in most LIR–Atg8 family protein complexes (Birgisdottir et al., 2013 ▸; Noda et al., 2008 ▸, 2010 ▸; Cheng et al., 2016 ▸; Olsvik et al., 2015 ▸; Rogov et al., 2013 ▸; McEwan et al., 2015 ▸; Suzuki et al., 2014 ▸; von Muhlinen et al., 2012 ▸; Wu et al., 2015 ▸; Stadel et al., 2015 ▸; Khaminets et al., 2015 ▸; Genau et al., 2015 ▸; Rogov et al., 2014 ▸; Lystad et al., 2014 ▸; Rozenknop et al., 2011 ▸; Ichimura et al., 2008 ▸), whereas interactions in the C-terminal flanking region are observed only in the Atg4B LIR–LC3 (Glu and Phe at positions 7 and 8) and FYCO1 LIR–LC3 structures (Cheng et al., 2016 ▸; Olsvik et al., 2015 ▸; Satoo et al., 2009 ▸).
3.3. GST pull-down assay of the interactions between FYCO1 LIR and LC3B
To confirm the interactions mediated by the flanking sequences N-terminal and C-terminal to the FYCO1 LIR core motif observed in the crystal structure, we conducted GST pull-down assays using FYCO1 LIR fused to GST and LC3B (Fig. 2 ▸ g). Consistent with the structural analysis, deletion of the N-terminal and/or C-terminal sequences of FYCO1 LIR resulted in decreased binding to LC3B. The binding was only marginally reduced by deletions of the most N- and C-terminal moieties of the LIR (lanes 2 and 3), but was markedly reduced by deletion of the whole C-terminal α-helix of the LIR (from residue 1244; lanes 4 and 6), indicating that interactions mediated by Glu1246 and Leu1247 (positions 7 and 8, respectively) were important for LC3B–FYCO1 LIR binding (Figs. 2 ▸ a, 2 ▸ b, 2 ▸ d and 2 ▸ f).
3.4. Structural comparisons with related LC3–FYCO1 LIR structures
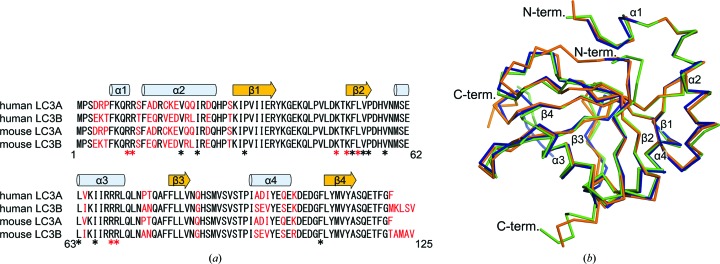
LC3A and LC3B from humans and mice were found to have a high sequence identity ranging from 79 to 100%, and the residues participating in the interaction with FYCO1 LIR were completely conserved (Fig. 3 ▸ a). In addition, the sequences of the LIR motif were perfectly conserved between human (residues 1276–1294) and mouse (residues 1235–1253) FYCO1. Therefore, the interactions between LC3 and FYCO1 LIR were expected to be conserved among different LC3 isoforms from different species. Recently, two groups have reported structures of LC3 in complex with FYCO1 LIR: human LC3A–FYCO1 LIR (PDB entry 5cx3; Cheng et al., 2016 ▸) and human LC3B–FYCO1 LIR (PDB entry 5d94; Olsvik et al., 2015 ▸). The structure of mouse LC3B–FYCO1 LIR in this study was similar to these structures, with r.m.s.d.s ranging from 0.4 to 0.6 Å (Fig. 3 ▸ b). Similar to the conserved interactions among Atg8 family protein–LIR complexes mediated by the LIR core motif, the additional electrostatic and hydrophobic interactions in the flanking sequence of the core motif (positions −3, 7 and 8) were commonly observed among the three structures.
Figure 3.
Sequence and structural comparison of LC3 proteins. (a) Sequence alignment of LC3A and LC3B from humans and mice. Secondary-structure elements are indicated above the alignment. The nonconserved residues are highlighted in red. Residues forming hydrogen bonds and hydrophobic interactions with FYCO1 LIR are indicated by red and black asterisks, respectively. (b) Superposition of LC3B proteins in complex with FYCO1 LIR. Mouse LC3B (this study), human LC3A (PDB entry 5cx3; Cheng et al., 2016 ▸) and human LC3B (PDB entry 5d94; Olsvik et al., 2015 ▸) complexed with FYCO1 LIR are shown in green, orange and blue, respectively.
4. Discussion
In this study, we determined the crystal structure of mouse LC3B in complex with the FYCO1 LIR peptide and established the structural basis for the recognition of the FYCO1 LIR motif by mouse LC3B. To date, many proteins containing the LIR motif have been reported (Birgisdottir et al., 2013 ▸; Noda et al., 2008 ▸, 2010 ▸; Cheng et al., 2016 ▸; Olsvik et al., 2015 ▸; Rogov et al., 2013 ▸; McEwan et al., 2015 ▸; Suzuki et al., 2014 ▸; von Muhlinen et al., 2012 ▸; Wu et al., 2015 ▸; Stadel et al., 2015 ▸; Khaminets et al., 2015 ▸; Genau et al., 2015 ▸; Rogov et al., 2014 ▸; Lystad et al., 2014 ▸; Rozenknop et al., 2011 ▸; Ichimura et al., 2008 ▸; Satoo et al., 2009 ▸). Although the tetrapeptide LIR core sequence of W/Y/FxxL/I/V has been characterized structurally based on its mode of interaction with Atg8 family proteins, the importance of the flanking sequences N-terminal and C-terminal to the LIR core motif have not been elucidated. In this study, we showed that LC3B recognizes not only the FYCO1 LIR core sequence but also the flanking sequences. In particular, Glu1246 and Leu1247 (positions 7 and 8) of the C-terminal flanking region of FYCO1 LIR were found to be important for the high-affinity interaction between LC3 and FYCO1. Consistent with this, the importance of these residues has also been independently demonstrated by other groups (Cheng et al., 2016 ▸; Olsvik et al., 2015 ▸); point mutations at each position abrogate the high-affinity binding of the protein to LC3, and positions 7 and 8 are interchangeable with the acidic residue Asp and the hydrophobic residues Phe, Ile, Leu and Val, respectively. Moreover, a structural comparison of the three LC3 structures in complex with the FYCO1 LIR peptide revealed the conserved recognition mechanism of FYCO1 among human LC3A and LC3B and mouse LC3B. A considerable number of LIR sequences contain several acidic residues in the flanking region of the LIR core motif (Olsvik et al., 2015 ▸). These acidic residues form electrostatic interactions with basic residues of Atg8 family proteins, as observed in our study; these interactions play an auxiliary role in fine-tuning the interactions between Atg8 family proteins and LIR motif-containing proteins.
The FYCO1–autophagosome interaction is mediated by multiple sites at the C-terminal region of FYCO1 (Fig. 1 ▸ a). In addition to LC3–LIR interactions, the C-terminal part of the coiled-coil region and the FYVE domain of FYCO1 interact with the autophagosomal membrane through their interactions with Rab7 and PI3P, respectively. Moreover, the GOLD domain may be involved in membrane binding through an unknown mechanism. The flexible loop region containing the LIR motif interferes with the lipid-binding surface of the FYVE domain in the absence of LC3, and the LC3–LIR interaction relieves this autoinhibition (Pankiv & Johansen, 2010 ▸). Additional studies are required to elucidate the detailed mechanisms of autophagosome transport by FYCO1.
Supplementary Material
PDB reference: LC3B in complex with FYCO1 LIR, 5wrd
Acknowledgments
We thank Professor Noboru Mizushima for providing the cDNA for mouse LC3B. We thank the beamline staff members at SPring-8 for their assistance with data collection. This work was supported by a Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science and Technology (UO and TS), the Takeda Science Foundation (UO and TS), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (UO), the Daiichi Sankyo Foundation of Life Science (UO) and the Naito Foundation (UO). We declare that none of the authors have financial interests related to this work.
References
- Anantharaman, V. & Aravind, L. (2002). Genome Biol. 3, research0023. [DOI] [PMC free article] [PubMed]
- Birgisdottir, Å. B., Lamark, T. & Johansen, T. (2013). J. Cell Sci. 126, 3237–3247. [DOI] [PubMed]
- Boyle, K. B. & Randow, F. (2013). Curr. Opin. Microbiol. 16, 339–348. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cheng, X. F., Wang, Y. L., Gong, Y. K., Li, F. X., Guo, Y. J., Hu, S. C., Liu, J. P. & Pan, L. F. (2016). Autophagy, 12, 1330–1339. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fu, M.-M., Nirschl, J. J. & Holzbaur, E. L. F. (2014). Dev. Cell, 29, 577–590. [DOI] [PMC free article] [PubMed]
- Gaullier, J. M., Simonsen, A., D’Arrigo, A., Bremnes, B., Stenmark, H. & Aasland, R. (1998). Nature (London), 394, 432–433. [DOI] [PubMed]
- Genau, H. M., Huber, J., Baschieri, F., Akutsu, M., Dötsch, V., Farhan, H., Rogov, V. & Behrends, C. (2015). Mol. Cell, 57, 995–1010. [DOI] [PubMed]
- Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., Mizushima, N., Tanida, I., Kominami, E., Ohsumi, M., Noda, T. & Ohsumi, Y. (2000). Nature (London), 408, 488–492. [DOI] [PubMed]
- Ichimura, Y. & Komatsu, M. (2010). Semin. Immunopathol. 32, 431–436. [DOI] [PubMed]
- Ichimura, Y., Kumanomidou, T., Sou, Y.-S., Mizushima, T., Ezaki, J., Ueno, T., Kominami, E., Yamane, T., Tanaka, K. & Komatsu, M. (2008). J. Biol. Chem. 283, 22847–22857. [DOI] [PubMed]
- Jiang, P. D. & Mizushima, N. (2014). Cell Res. 24, 69–79. [DOI] [PMC free article] [PubMed]
- Johansen, T. & Lamark, T. (2011). Autophagy, 7, 279–296. [DOI] [PMC free article] [PubMed]
- Khaminets, A. et al. (2015). Nature (London), 522, 354–358.
- Klionsky, D. J. & Emr, S. D. (2000). Science, 290, 1717–1721. [DOI] [PMC free article] [PubMed]
- Lystad, A. H., Ichimura, Y., Takagi, K., Yang, Y. J., Pankiv, S., Kanegae, Y., Kageyama, S., Suzuki, M., Saito, I., Mizushima, T., Komatsu, M. & Simonsen, A. (2014). EMBO Rep. 15, 557–565. [DOI] [PMC free article] [PubMed]
- Mackeh, R., Perdiz, D., Lorin, S., Codogno, P. & Poüs, C. (2013). J. Cell Sci. 126, 1071–1080. [DOI] [PubMed]
- McEwan, D. G. et al. (2015). Mol. Cell, 57, 39–54. [DOI] [PubMed]
- Mizushima, N. (2007). Genes Dev. 21, 2861–2873. [DOI] [PubMed]
- Mizushima, N. & Komatsu, M. (2011). Cell, 147, 728–741. [DOI] [PubMed]
- Muhlinen, N. von, Akutsu, M., Ravenhill, B. J., Foeglein, Á., Bloor, S., Rutherford, T., Freund, S. V., Komander, D. & Randow, F. (2012). Mol. Cell, 48, 329–342. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nakatogawa, H., Ichimura, Y. & Ohsumi, Y. (2007). Cell, 130, 165–178. [DOI] [PubMed]
- Noda, N. N., Kumeta, H., Nakatogawa, H., Satoo, K., Adachi, W., Ishii, J., Fujioka, Y., Ohsumi, Y. & Inagaki, F. (2008). Genes Cells, 13, 1211–1218. [DOI] [PubMed]
- Noda, N. N., Ohsumi, Y. & Inagaki, F. (2010). FEBS Lett. 584, 1379–1385. [DOI] [PubMed]
- Olsvik, H. L., Lamark, T., Takagi, K., Larsen, K. B., Evjen, G., Øvervatn, A., Mizushima, T. & Johansen, T. (2015). J. Biol. Chem. 290, 29361–29374. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pankiv, S., Alemu, E. A., Brech, A., Bruun, J. A., Lamark, T., Overvatn, A., Bjørkøy, G. & Johansen, T. (2010). J. Cell Biol. 188, 253–269. [DOI] [PMC free article] [PubMed]
- Pankiv, S. & Johansen, T. (2010). Autophagy, 6, 550–552. [DOI] [PubMed]
- Rogov, V., Dötsch, V., Johansen, T. & Kirkin, V. (2014). Mol. Cell, 53, 167–178. [DOI] [PubMed]
- Rogov, V. V., Suzuki, H., Fiskin, E., Wild, P., Kniss, A., Rozenknop, A., Kato, R., Kawasaki, M., McEwan, D. G., Löhr, F., Güntert, P., Dikic, I., Wakatsuki, S. & Dötsch, V. (2013). Biochem. J. 454, 459–466. [DOI] [PubMed]
- Rozenknop, A., Rogov, V. V., Rogova, N. Y., Löhr, F., Güntert, P., Dikic, I. & Dötsch, V. (2011). J. Mol. Biol. 410, 477–487. [DOI] [PubMed]
- Satoo, K., Noda, N. N., Kumeta, H., Fujioka, Y., Mizushima, N., Ohsumi, Y. & Inagaki, F. (2009). EMBO J. 28, 1341–1350. [DOI] [PMC free article] [PubMed]
- Shpilka, T., Weidberg, H., Pietrokovski, S. & Elazar, Z. (2011). Genome Biol. 12, 226. [DOI] [PMC free article] [PubMed]
- Stadel, D., Millarte, V., Tillmann, K. D., Huber, J., Tamin-Yecheskel, B. C., Akutsu, M., Demishtein, A., Ben-Zeev, B., Anikster, Y., Perez, F., Dötsch, V., Elazar, Z., Rogov, V., Farhan, H. & Behrends, C. (2015). Mol. Cell, 60, 89–104. [DOI] [PubMed]
- Sugawara, K., Suzuki, N. N., Fujioka, Y., Mizushima, N., Ohsumi, Y. & Inagaki, F. (2004). Genes Cells, 9, 611–618. [DOI] [PubMed]
- Suzuki, H., Tabata, K., Morita, E., Kawasaki, M., Kato, R., Dobson, R. J., Yoshimori, T. & Wakatsuki, S. (2014). Structure, 22, 47–58. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wu, F. et al. (2015). Mol. Cell, 60, 914–929. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: LC3B in complex with FYCO1 LIR, 5wrd