Structural analysis of two variant forms of truncated hemolysin A reveals additional proteolysis of the C-terminal 31 amino acids under high-salt crystallization conditions. This structural plasticity may support the implementation of stably exposed, on-edge β-strands during template-assisted folding.
Keywords: hemolysin A, two-partner secretion, alternate crystal forms, proteolysis, β-helix, Proteus mirabilis
Abstract
Wild-type and variant forms of HpmA265 (truncated hemolysin A) from Proteus mirabilis reveal a right-handed, parallel β-helix capped and flanked by segments of antiparallel β-strands. The low-salt crystal structures form a dimeric structure via the implementation of on-edge main-chain hydrogen bonds donated by residues 243–263 of adjacent monomers. Surprisingly, in the high-salt structures of two variants, Y134A and Q125A-Y134A, a new dimeric interface is formed via main-chain hydrogen bonds donated by residues 203–215 of adjacent monomers, and a previously unobserved tetramer is formed. In addition, an eight-stranded antiparallel β-sheet is formed from the flap regions of crystallographically related monomers in the high-salt structures. This new interface is possible owing to additional proteolysis of these variants after Tyr240. The interface formed in the high-salt crystal forms of hemolysin A variants may mimic the on-edge β-strand positioning used in template-assisted hemolytic activity.
1. Introduction
Hemolysin A from Proteus mirabilis (HpmA) is a secreted exoprotein capable of lysing red blood cells (Swihart & Welch, 1990 ▸). HpmA belongs to the type Vb or two-partner secretion pathway (TPS), the most widespread of the porin-type translocating systems (Jacob-Dubuisson et al., 2013 ▸). Similar to other type Vb exoproteins, HpmA is synthesized with an N-terminal Sec signal peptide that directs the protein to the periplasmic space, where the signal peptide is cleaved (Jacob-Dubuisson et al., 2001 ▸). HpmA then relies on its cognate partner, hemolysin B (HpmB), to assist in its activation and secretion to the extracellular matrix (Jacob-Dubuisson et al., 2004 ▸). Our studies focus on a truncated version of hemolysin A termed HpmA265 which, upon Sec signal cleavage, is composed of residues 30–265 (Weaver et al., 2009 ▸). Full-length HpmA is 1577 residues in length; however, the HpmA265 construct utilized in this study is amenable to crystallographic analysis and has been demonstrated to facilitate template-assisted hemolysis (Weaver et al., 2009 ▸). Structurally, HpmA265 harbors the so-called TPS domain and adopts a nonglobular right-handed, three-sided parallel β-helix similar to other two-partner secretion pathway A proteins (Weaver et al., 2009 ▸; Baelen et al., 2013 ▸; Yeo et al., 2007 ▸; Clantin et al., 2004 ▸).
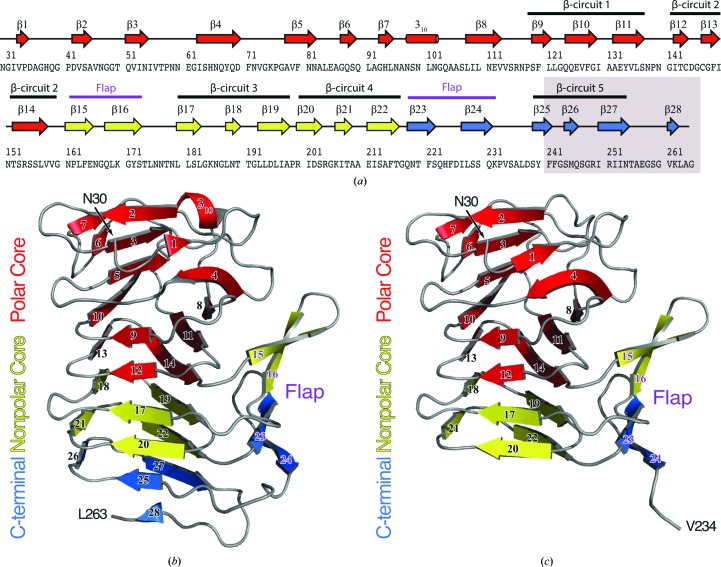
Structurally, the HpmA265 TPS domain can be divided into three subdomains: (i) a polar core subdomain composed of residues 30–160, (ii) a nonpolar core subdomain composed of residues 161–216 and (iii) a carboxy-terminal subdomain composed of residues 217–263 (Fig. 1 ▸ a; Wimmer et al., 2015 ▸). Interestingly, residues 111–265 produce the five complete β-circuits that form the HpmA265 β-helix. Therefore, the β-helix structure within HpmA265 is built from β-circuits donated from both the polar and nonpolar subdomains (Figs. 1 ▸ a and 1 ▸ b), where each β-circuit contains three parallel β-strands. Furthermore, a four-stranded antiparallel β-sheet, termed the flap, projects off the β-helix structure. The flap region forms from pairs of antiparallel β-strands positioned between β-circuits 2–3 and 4–5, respectively (Wimmer et al., 2015 ▸; Figs. 1 ▸ a and 1 ▸ b).
Figure 1.
(a) Secondary-structure diagram of HpmA265. The gray box indicates the region removed by proteolysis in the high-salt structure. The cartoon representations denote the subdomain architecture embedded within (b) the low-salt HpmA265 structure and (c) the high-salt HpmA265 structure. Despite proteolysis in the high-salt structure, both the low-salt and high-salt structures harbor polar core, nonpolar core and carboxy-terminal subdomains. The β-helix core of the HpmA265 structure comprises both the polar and nonpolar core subdomains. Additionally, a four-stranded antiparallel β-sheet (flap) frames one side of the β-helix core. The low-salt crystal form (b) includes residues Asn30–Leu263 and extends through β28 at the C-terminus. The high-salt form (c) includes residues Asn30–Val234 and terminates at β24, which is the final antiparallel β-strand within the flap region.
Past investigations using a template-assisted hemolytic assay (TAHA) have shown that HpmA265 is able to convert inactive, full-length HpmA (termed HpmA*) to active full-length HpmA (Weaver et al., 2009 ▸). The TAHA monitors lysis of red blood cells, which is only possible with folded, active full-length HpmA harboring the carboxy-terminal pore-forming domain. We previously postulated that HpmA265 acts as a template and assists the refolding of HpmA* into the hemolytically functional form (Weaver et al., 2009 ▸). ShlA, a Serritia marcescens homolog of HpmA, also exhibits template-assisted activity (Hertle et al., 1997 ▸). Furthermore, the TAHA mechanism has been proposed to rely upon β-helix activation similar to that implemented during filament growth (Margittai & Langen, 2008 ▸; Tsemekhman et al., 2007 ▸; Gurry & Stultz, 2014 ▸; Kajava et al., 2005 ▸). In this model of filament growth, an exposed layer of β-strands serves as a template onto which the unstructured protein becomes ordered through cooperative hydrogen bonding and the alignment of side chains. Currently, details of the HpmA265 interface responsible for providing the template for HpmA* activation are unknown, but the implementation of exposed on-edge β-strands is an attractive hypothesis.
During our recent protein-folding studies (Wimmer et al., 2015 ▸) we found that it was difficult to obtain crystals of several HpmA265 variants under the originally reported crystallization conditions (termed the low-salt condition; Weaver et al., 2009 ▸). In an effort to structurally characterize these HpmA265 variants, we identified an additional high-salt condition (see §2.3) that yielded well diffracting crystals. Here, we describe the structures of two variants, Y134A and Q125A-Y134A (termed AA), determined from crystals grown in both the original (low-salt) and high-salt conditions. We additionally report a higher resolution structure of native HpmA265 in the original low-salt condition. Molecular-replacement structure determination of Y134A and AA grown in the high-salt condition initially failed when using the low-salt HpmA265 crystal form as a search model. A complete molecular-replacement solution was only achieved when the carboxy-terminal ∼31 residues of the search model were removed. The first 231 residues in the high-salt variant structures are very similar to the respective low-salt structures, with both forms possessing the polar core, nonpolar core and carboxy-terminal subdomains (Figs. 1 ▸ b and 1 ▸ c). However, after residue 231, the backbones of the high-salt and low-salt variant structures diverge dramatically. Furthermore, for each of the variants crystallized in the high-salt condition, we were unable to find electron density in the maps corresponding to the ∼31 carboxy-terminal residues (Figs. 1 ▸ a and 1 ▸ c). We believe that the observed carboxy-terminal truncations within the high-salt crystal forms of Y134A and AA are caused by contaminating protease. These missing residues at the carboxy-terminus result in major structural changes: firstly, the newly exposed β-strand main-chain atoms form a novel dimerization interface; secondly, the subunit packing facilitates the formation of a previously unobserved tetramer; and thirdly, the main-chain atoms after residue 231 participate in an eight-stranded inter-subunit antiparallel β-sheet between crystallographically related flap regions. These missing residues also eliminate a major portion of the unstable carboxy-terminal subdomain, thereby exposing the more stable nonpolar core subdomain (Fig. 1 ▸). This stable subdomain may serve as a template for the folding of inactive HpmA*.
2. Methods
2.1. DNA plasmids and mutagenesis
Plasmid pHpmA265 expresses truncated hemolysin A encoding the first 265 amino acids of the HpmA protein and a carboxy-terminal six-histidine tag (Weaver et al., 2009 ▸). The first 29 residues of this variant contain the Sec signal peptide, which is proteolytically cleaved upon secretion into the periplasmic space. Thus, the fully processed HpmA265 protein consists of residues 30–265. The QuikChange mutagenesis kit (Stratagene, La Jolla, California, USA) was used to construct both single and double site-specific replacements: Y134A (pHpmA265_Y134A) and Q125A-Y134A (pHpmA265_Q125A-Y134A), respectively. DNA sequencing (Eton Biosciences, San Diego, California, USA or GenScript, Piscataway, New Jersey, USA) and X-ray crystallography verified the successful incorporation of the site-specific alterations.
2.2. Protein expression and purification
Briefly, Escherichia coli C41(DE3) (Lucigen Corporation, Middleton, Wisconsin, USA) cells were co-transformed with plasmids encoding HpmB and either HpmA265, Y134A or AA. Co-transformed cell cultures were grown overnight and used to inoculate 1.4 l Luria broth supplemented with the appropriate antibiotics. Protein expression was induced for 12 h via the adjustment of each flask to 1 mM IPTG at an OD600 of between 0.4 and 0.8. Each protein was purified via a previously published procedure (Weaver et al., 2009 ▸). Purified proteins were vacuum-concentrated to 10 mg ml−1 and dialyzed extensively against 10 mM sodium phosphate pH 7.4, 30 mM NaCl.
2.3. Crystallization
Y134A and AA were crystallized in two different solutions: (i) 100 mM citrate pH 5.0, 12–14% PEG 4000 and (ii) Crystal Screen condition No. 49 (Hampton Research, Aliso Viejo, California, USA), which consists of 1 M LiSO4, 2% PEG 8000, using the hanging-drop vapor-diffusion method. Native HpmA265 was crystallized in condition (i). Structures obtained from crystals grown from the PEG 4000 condition are termed the low-salt form, while those structures obtained from crystals grown from the 1 M LiSO4 condition are termed the high-salt form throughout this manuscript. The low-salt crystals appeared within two weeks, while the high-salt crystals appeared after five months of setting up the trays. All crystals were cryoprotected against mother liquor adjusted to 25% glycerol and cryocooled in liquid nitrogen. X-ray diffraction data were collected at 100 K on beamlines GM/CA-CAT 23-ID-B and LS-CAT 21-ID-F at the Advanced Photon Source (APS) (Table 1 ▸).
Table 1. X-ray diffraction data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Protein | Wild type | Y134A | Y134A | Q125A-Y134A | Q125A-Y134A |
|---|---|---|---|---|---|
| Condition | Low salt | Low salt | High salt | Low salt | High salt |
| PDB code | 4w8q | 5kf3 | 5kkd | 5keh | 5sz8 |
| Data collection | |||||
| Beamline | 23-ID-B, GM/CA-CAT, APS | 21-ID-F, LS-CAT, APS | 21-ID-F, LS-CAT, APS | 21-ID-F, LS-CAT, APS | 21-ID-F, LS-CAT, APS |
| Wavelength (Å) | 1.03 | 0.98 | 0.98 | 0.98 | 0.98 |
| Space group | P2 | P22121 | P41212 | P2 | P41212 |
| Unit-cell parameters | |||||
| a (Å) | 59.7 | 33.9 | 69.1 | 51.6 | 69.23 |
| b (Å) | 34.3 | 59.8 | 69.1 | 33.6 | 69.23 |
| c (Å) | 68.3 | 115.8 | 234.5 | 59.6 | 234.86 |
| β (°) | 99.2 | 100.7 | |||
| No. of unique reflections | 49625 | 12612 | 33020 | 29433 | 51639 |
| Resolution limit (Å) | 1.43 (1.46–1.43) | 2.20 (2.24–2.20) | 2.13 (2.17–2.13) | 1.55 (1.58–1.55) | 1.83 (1.86–1.83) |
| Completeness (%) | 97.2 (74.9) | 99.7 (98.4) | 100.0 (100.0) | 99.8 (100.0) | 99.9 (100) |
| Multiplicity | 6.9 (4.4) | 8.8 (7.5) | 24.1 (20.1) | 7.4 (7.1) | 22.0 (22.1) |
| 〈I/σ(I)〉 | 25.4 (2.2) | 23.0 (10.2) | 32.0 (3.2) | 18.6 (12.8) | 38.2 (3.3) |
| R merge (%) | 7.8 (81.4) | 9.4 (20.7) | 12.9 (78.8) | 7.4 (25.2) | 8.5 (80.3) |
| R p.i.m. (%) | 3.2 (43.0) | 3.2 (7.7) | 2.6 (16.8) | 3.0 (10.1) | 1.8 (16.7) |
| Refinement | |||||
| Resolution range (Å) | 29.7–1.43 | 32.5–2.20 | 47.8–2.13 | 35.2–1.55 | 44.8–1.83 |
| R free (%) | 16.3 | 20.7 | 19.5 | 18.0 | 20.6 |
| R work (%) | 13.9 | 17.3 | 17.0 | 14.7 | 17.8 |
| No. of atoms | |||||
| Total | 2062 | 1890 | 3135 | 2099 | 3239 |
| Protein | 1781 | 1725 | 2968 | 1874 | 2968 |
| Water | 281 | 165 | 162 | 225 | 261 |
| Ramachandran plot† | |||||
| Favoured (%) | 98.7 | 98.7 | 97.8 | 98.8 | 97.8 |
| Allowed (%) | 1.3 | 1.3 | 2.2 | 0.8 | 2.2 |
| Outliers (%) | 0 | 0 | 0 | 0.4 | 0 |
| Average B factors (Å2) | |||||
| Protein | 22.5 | 22.1 | 44.3 | 18.0 | 43.1 |
| Water | 38.9 | 33.1 | 47.4 | 34.1 | 49.7 |
| R.m.s.d.† | |||||
| Bond lengths (Å) | 0.016 | 0.004 | 0.007 | 0.016 | 0.017 |
| Bond angles (°) | 1.52 | 0.641 | 0.818 | 1.37 | 1.479 |
| Coordinate error (Å) | 0.11 | 0.20 | 0.17 | 0.12 | 0.21 |
Root-mean-square deviations, coordinate error estimates and Ramachandran plot data were obtained from PHENIX (Adams et al., 2010 ▸).
2.4. Structure determination and refinement
Diffraction data were indexed, integrated and scaled with HKL-3000 (Minor et al., 2006 ▸). The L-test for twinning yielded 〈|L|〉 values of 0.49 for all data sets, indicating that the crystals were untwinned. Molecular replacement was carried out with Phaser (McCoy et al., 2007 ▸) using PDB entry 3fy3 (with and without the carboxy-terminal 29 amino acids; Weaver et al., 2009 ▸) as the search model. Multiple rounds of model building and refinement were performed with Coot (Emsley et al., 2010 ▸) and PHENIX (Adams et al., 2010 ▸), respectively. Model validity was checked using the PDB_REDO (Joosten et al., 2014 ▸) and MolProbity (Chen et al., 2010 ▸) web servers. Coordinate and structure factors for HpmA265 and its variants were deposited in the Protein Data Bank. PDB codes and data-collection and refinement statistics are given in Table 1 ▸.
2.5. Mass spectrometry
Protein samples, harvested in crystalline and liquid forms, were analyzed via electrospray ionization-mass spectrometry (ESI-MS; University of Wisconsin-Madison Mass Spectrometry Facility, Madison, Wisconsin, USA). Liquid protein samples were subjected directly to ESI-MS analysis, while protein crystals were harvested, washed and precipitated using the following procedure. Firstly, protein crystals were grown and harvested from a droplet consisting of 1 M LiSO4, 2% PEG 8000. To remove traces of noncrystalline protein, 100 µl mother liquor was added to the harvested protein crystals. The majority of the mother liquor was removed via pipetting, while any residual solution was wicked away from the protein crystals using filter paper. The crystals were then washed with 100 µl ice-cold acetone followed by 100 µl ice-cold 80% methanol. Residual solvent was wicked away after each step using filter paper. Crystals were washed an additional two times with acetone and 80% methanol. Finally, crystals were suspended in ice-cold acetone, transferred to a microfuge tube and spun at 21 000g for 2 min. The solvent was removed and the sample was subjected to ESI-MS analysis.
3. Results and discussion
3.1. Tertiary and quaternary structures of HpmA265 and variants
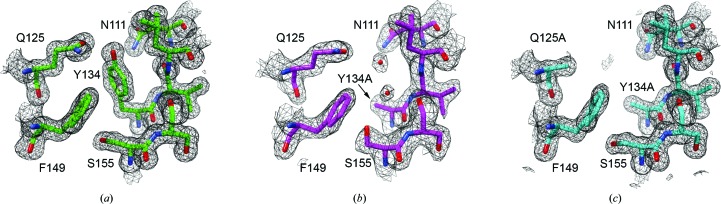
The native structure of HpmA265 was determined in the low-salt condition at 1.43 Å resolution (PDB entry 4w8q), yielding improved maps and water placement over the existing 1.8 Å resolution structure (PDB entry 3fy3; Weaver et al., 2009 ▸). As expected, the overall structure of HpmA265 remained largely unchanged; superposition of PDB entry 3fy3 onto PDB entry 4w8q results in an r.m.s.d. of only 0.31 Å over 234 Cα pairs. Superposition of the low-salt Y134A and AA structures onto the 1.43 Å resolution HpmA265 structure results in r.m.s.d. values of 0.53 and 0.45 Å, respectively, and indicates that the overall structure is not significantly perturbed by these mutations. Fig. 2 ▸ shows sample electron-density maps at the mutation site for the native HpmA265, Y134A and AA structures (Figs. 2 ▸ a, 2 ▸ b and 2 ▸ c, respectively). There is clear electron density for two ordered water molecules in place of the tyrosine in the Y134A mutant structure (Fig. 2 ▸ b); however, none are supported in the density for the AA mutant (Fig. 2 ▸ c). Given the overall similarity in the structures, and the large distance between these mutations and the cleavage site (>15 Å), it is not clear how these mutations may facilitate proteolysis in the high-salt condition (see below).
Figure 2.
2F o − F c electron-density maps contoured at 1σ near the mutation site. (a) shows the low-salt HpmA265 structure in green. (b) shows the low-salt Y134A structure in magenta and clear density for two ordered water molecules in place of the tyrosine side chain. (c) shows the low-salt AA double-mutant structure in cyan. Despite the loss of these two side chains the overall structure remains unaffected, and no electron density is observed that may account for ordered water molecules. Fig. 2 was created with the UCSF Chimera molecular visualization program (Pettersen et al., 2004 ▸).
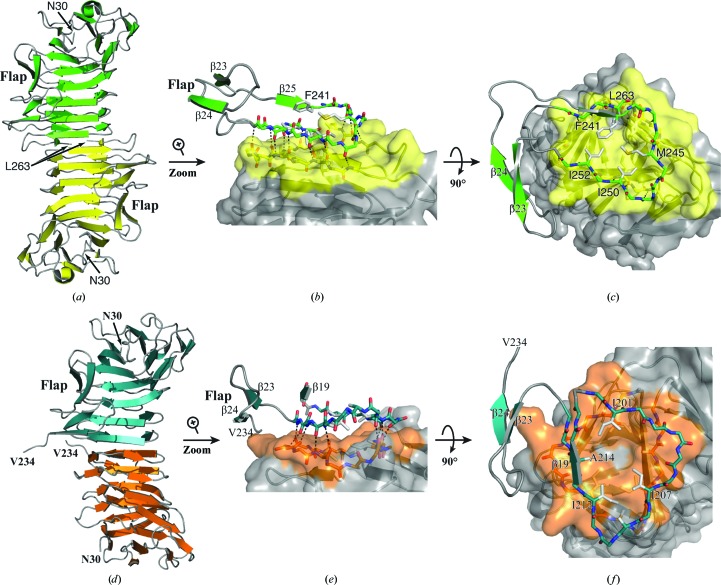
In the low-salt crystal form, all HpmA265 variants form a dimeric structure by implementing main-chain hydrogen bonds donated from exposed on-edge β-strands of each monomer. In this fashion, the last three β-strands (β26–β28; residues 244–262) of the protein lie in an antiparallel arrangement at the dimer interface (Fig. 3 ▸ a). Beyond the implementation of main-chain hydrogen bonds, van der Waals contacts between Leu263, Val261, Ile252, Ile250 and Met245 assist in maintaining the solvent-inaccessible dimer interface (Figs. 3 ▸ b and 3 ▸ c). Additionally, the antiparallel β-strands in the flap region do not participate in higher order antiparallel β-sheets with crystallographically related molecules.
Figure 3.
Dimerization interfaces in the low-salt and high-salt HpmA265 structures. (a) The HpmA265 low-salt crystal structure uses β26–β28 (residues Ser244–Leu263) at the dimerization interface. (b) Magnification of the side view showing hydrogen bonds donated by exposed on-edge β-strands (β26, β27 and β28 residues) from each monomer in the low-salt structure. (c) Top view emphasizing van der Waals interactions during the formation of the low-salt dimer interface. (d) In the AA high-salt crystal structure, proteolysis allows β20–β22 (residues Arg200–Phe215) to establish the dimer interface. (e) Side view emphasizing the hydrogen bonds donated by exposed on-edge β-strands (β20–β22, residues Arg200–Phe215) from each monomer in the high-salt AA structure. (f) A top view showing the van der Waals interactions of Ile201, Ile207, Ile212 and Ala214 during the formation of the high-salt dimer interface.
ESI-MS was used to verify the protein masses in both the original liquid AA sample (stored at 4°C) and a crystallized AA sample from the high-salt condition. The liquid AA protein sample (used to obtain the AA high-salt crystals) provided a mass of 25 308 Da, which closely matches the theoretical mass of 25 309 Da (Gasteiger et al., 2003 ▸). The harvested AA crystals yielded two species with masses of 25 308 and 21 905 Da. The higher mass species appears to be full-length AA carried over from the residual mother liquor during crystal harvesting. The AA construct consisting of residues Asn30–Tyr240 has a theoretical mass of 21 907 Da (Gasteiger et al., 2003 ▸). This mass closely agrees with the lower molecular-weight species, and suggests proteolytic cleavage after Tyr240. Therefore, we suspect that a small amount of contaminating protease cleaved the mutant proteins in the high-salt condition. Given the high purity of the protein samples, we are unable to identify the specific protease responsible for the cleavage.
In the high-salt condition, both the Y134A and AA variants are proteolyzed after residue Tyr240, and the structures reveal a new dimer interface formed by β20–β22 (residues 200–215; Fig. 3 ▸ d). Beyond the main-chain hydrogen bonds, a new set of van der Waals contacts are established between Ile201, Ser203, Ile207, Ile212 and Ala214 at the solvent-inaccessible interface (Figs. 3 ▸ e and 3 ▸ f).
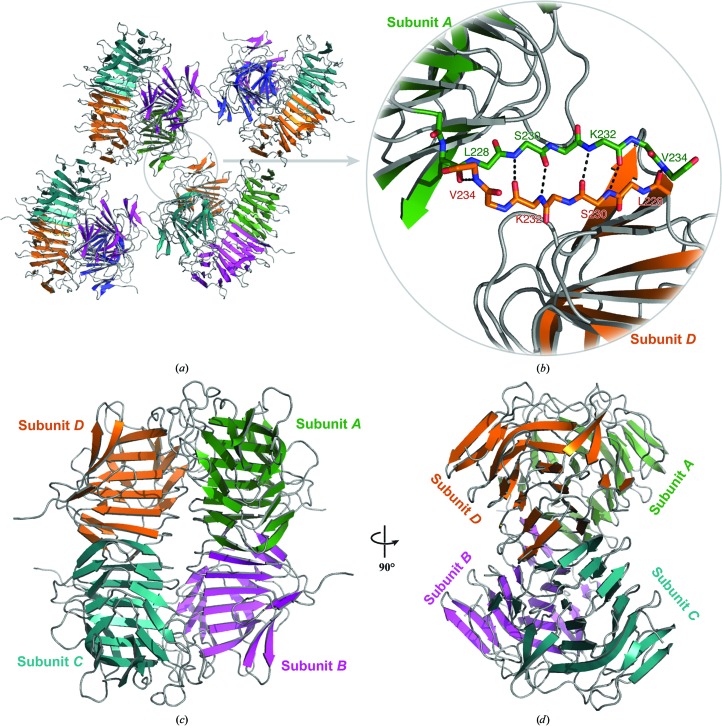
In addition to the establishment of a new dimer interface, the conformation of residues 230–234 of the flap region facilitates the formation of a higher order antiparallel β-sheet with a crystallographically related molecule (Figs. 4 ▸ a and 4 ▸ b). This β-sheet does not result in a substantial binding interface according to PISA (Krissinel & Henrick, 2007 ▸). However, the packing arrangement within the high-salt crystal form yields a previously unobserved tetramer (Figs. 4 ▸ c and 4 ▸ d). PISA calculations yield ΔG diss values of 2.2 and 8.4 kcal mol−1 for the Y134A and AA high-salt tetramers, respectively (Krissinel & Henrick, 2007 ▸). ΔG diss values greater than zero suggest that this interface is stable in solution. This newly resolved structural conformation might simulate the on-edge β-strands of HpmA265 implemented during template-assisted hemolysis. This observed structural plasticity and inherent stability is similar to that observed for other β-helix-type proteins (Choi et al., 2009 ▸; Kamen et al., 2000 ▸; Junker et al., 2006 ▸; Nelson et al., 2005 ▸).
Figure 4.
Tetramer formation in the high-salt structures. (a) Depiction of the eight-stranded antiparallel β-sheet formed from the flap regions of crystallographically related tetramers in the high-salt condition. The removal of β25–β28 owing to the proteolysis event has allowed β24 to expose on-edge main-chain hydrogen-bonding partners. (b) Magnified view of the main-chain atoms, including those donated from β24, used to assemble the inter-subunit antiparallel β-sheet. The newly discovered dimerization facilitates the formation of a stable tetramer (c, d). Interactions between adjacent N-terminal polar core subdomains form the majority of this interface.
3.2. Proteolysis of HpmA265 variants exposes a stable interface
Our recent unfolding studies conducted on a series of HpmA265 variants led to the discovery of three subdomains within HpmA265 (Wimmer et al., 2015 ▸). These subdomains, termed the polar core, nonpolar core and carboxy-terminal, have decreasing levels of stability as determined via chemical denaturation studies. An HpmA265 variant where Phe241 was replaced by a lysine aided assignment of the carboxy-terminal subdomain boundary. The F241K variant eliminated the first transition associated with the carboxy-terminal subdomain, while leaving the more stable nonpolar and polar core subdomains unaffected. In effect, this buried lysine at position 241 destabilized the carboxy-terminal subdomain to the point where it was completely unfolded even in the absence of denaturant. We utilized mass spectrometry to verify that the F241K variant was fully intact and that loss of the carboxy-terminal subdomain transition was not an artefact of proteolysis.
Interestingly, the F241K variant depicted an enhanced rate of template-assisted activity (Wimmer et al., 2015 ▸). Typically, the template-assisted activity presents as a biphasic profile, where the initial lag phase progresses to a rapid hemolytic phase (Weaver et al., 2009 ▸; Gurry & Stultz, 2014 ▸; Tsemekhman et al., 2007 ▸; Junker et al., 2006 ▸). However, the F241K variant of HpmA265 displayed a pronounced reduction in the initial lag phase and an enhanced hemolytic phase. We surmised that destabilization of the the F241K variant carboxy-terminal subdomain allowed the exposure of a more stable interface within the nonpolar core. Exposure of β-strands localized within a more stable subdomain would then act as better templates during on-edge template-assisted hemolytic activation of unfolded full-length hemolysin A. In further support of this idea, PISA calculations yielded ΔG diss values of 6.1 and 5.0 kcal mol−1 for the Y134A and AA high-salt dimers, respectively. These values suggest the proteolyzed dimers are more stable than the wild-type HpmA265 low-salt dimer, which possesses a ΔG diss value of −0.6 kcal mol−1, which falls into a ‘gray region of complex formation’ (Krissinel & Henrick, 2007 ▸).
Our earlier denaturation studies approximated the division between the carboxy-terminal and nonpolar subdomains very near the flap region ending at Gln231 (Wimmer et al., 2015 ▸). Remarkably, mass-spectrometric data and the new high-salt crystal forms of AA and Y134A indicate proteolysis of the β-helix after Tyr240, near the beginning of β-circuit 5. It appears that the high-salt condition encourages proteolysis of the least stable portion of the carboxy-terminal subdomain, revealing the stable nonpolar core subdomain residues 200–215 to form the new dimer interface (Fig. 3 ▸ f). This surface presented by the nonpolar core subdomain may be exposed during transport of HpmA to the extracellular matrix, and may act as a superior template during the on-edge template-assisted activation of HpmA*.
Supplementary Material
PDB reference: HpmA265, 4w8q
PDB reference: Y134A variant, low salt, 5kf3
PDB reference: high salt, 5kkd
PDB reference: Q125A-Y134A variant, low salt, 5keh
PDB reference: high salt, 5sz8
Acknowledgments
The authors would like to thank Megan Wimmer, Christopher Woods and Kyle Adamczak for assistance during crystallization trials. The authors would also like to thank Professor James Keck (University of Wisconsin-Madison Department of Biomolecular Chemistry) for graciously allowing access to the Advanced Photon Source. This research used the resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). GM/CA at the Advanced Photon Source has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The authors would also like to thank Gregory Barrett-Wilt PhD and Grzegorz Sabat MS from the Mass Spectrometry Facility in the Biotechnology Center at the University of Wisconsin-Madison for the mass-spectrometry results.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Baelen, S., Dewitte, F., Clantin, B. & Villeret, V. (2013). Acta Cryst. F69, 1322–1327. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Choi, J. H., May, B. C. H., Govaerts, C. & Cohen, F. E. (2009). Structure, 17, 1014–1023. [DOI] [PubMed]
- Clantin, B., Hodak, H., Willery, E., Locht, C., Jacob-Dubuisson, F. & Villeret, V. (2004). Proc. Natl Acad. Sci. USA, 101, 6194–6199. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D. & Bairoch, A. (2003). Nucleic Acids Res. 31, 3784–3788. [DOI] [PMC free article] [PubMed]
- Gurry, T. & Stultz, C. M. (2014). Biochemistry, 53, 6981–6991. [DOI] [PubMed]
- Hertle, R., Brutsche, S., Groeger, W., Hobbie, S., Koch, W., Könninger, U. & Braun, V. (1997). Mol. Microbiol. 26, 853–865. [DOI] [PubMed]
- Jacob-Dubuisson, F., Fernandez, R. & Coutte, L. (2004). Biochim. Biophys. Acta, 1694, 235–257. [DOI] [PubMed]
- Jacob-Dubuisson, F., Guérin, J., Baelen, S. & Clantin, B. (2013). Res. Microbiol. 164, 583–595. [DOI] [PubMed]
- Jacob-Dubuisson, F., Locht, C. & Antoine, R. (2001). Mol. Microbiol. 40, 306–313. [DOI] [PubMed]
- Joosten, R. P., Long, F., Murshudov, G. N. & Perrakis, A. (2014). IUCrJ, 1, 213–220. [DOI] [PMC free article] [PubMed]
- Junker, M., Schuster, C. C., McDonnell, A. V., Sorg, K. A., Finn, M. C., Berger, B. & Clark, P. L. (2006). Proc. Natl. Acad. Sci. USA, 103, 4918–4923. [DOI] [PMC free article] [PubMed]
- Kajava, A. V., Aebi, U. & Steven, A. C. (2005). J. Mol. Biol. 348, 247–252. [DOI] [PubMed]
- Kamen, D. E., Griko, Y. & Woody, R. W. (2000). Biochemistry, 39, 15932–15943. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Margittai, M. & Langen, R. (2008). Q. Rev. Biophys. 41, 265–297. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Nelson, R., Sawaya, M. R., Balbirnie, M., Madsen, A. Ø., Riekel, C., Grothe, R. & Eisenberg, D. (2005). Nature (London), 435, 773–778. [DOI] [PMC free article] [PubMed]
- Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C. & Ferrin, T.E. (2004). J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed]
- Swihart, K. G. & Welch, R. A. (1990). Infect. Immun. 58, 1861–1869. [DOI] [PMC free article] [PubMed]
- Tsemekhman, K., Goldschmidt, L., Eisenberg, D. & Baker, D. (2007). Protein Sci. 16, 761–764. [DOI] [PMC free article] [PubMed]
- Weaver, T. M., Smith, J. A., Hocking, J. M., Bailey, L. J., Wawrzyn, G. T., Howard, D. R., Sikkink, L. A., Ramirez-Alvarado, M. & Thompson, J. R. (2009). J. Biol. Chem. 284, 22297–22309. [DOI] [PMC free article] [PubMed]
- Wimmer, M. R., Woods, C. N., Adamczak, K. J., Glasgow, E. M., Novak, W. R. P., Grilley, D. P. & Weaver, T. M. (2015). Protein Sci. 24, 1841–1855. [DOI] [PMC free article] [PubMed]
- Yeo, H.-J., Yokoyama, T., Walkiewicz, K., Kim, Y., Grass, S. & Geme, J. W. S. (2007). J. Biol. Chem. 282, 31076–31084. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: HpmA265, 4w8q
PDB reference: Y134A variant, low salt, 5kf3
PDB reference: high salt, 5kkd
PDB reference: Q125A-Y134A variant, low salt, 5keh
PDB reference: high salt, 5sz8