Abstract
Autophagy, a form of lysosomal degradation capable of eliminating dysfunctional proteins and organelles, is a cellular process associated with homeostasis. Autophagy functions in cell survival by breaking down proteins and organelles and recycling them to meet metabolic demands. However, aberrant up regulation of autophagy can function as an alternative to apoptosis. The duality of autophagy, and its regulation over cell survival/death, intimately links it with human disease. Non-coding RNAs regulate mRNA levels and elicit diverse effects on mammalian protein expression. The most studied non-coding RNAs to-date are microRNAs (miRNA). MicroRNAs function in post-transcriptional regulation, causing profound changes in protein levels, and affect many biological processes and diseases. The role and regulation of autophagy, whether it is beneficial or harmful, is a controversial topic in cardiovascular disease. A number of recent studies have identified miRNAs that target autophagy-related proteins and influence the development, progression, or treatment of cardiovascular disease. Understanding the mechanisms by which these miRNAs work can provide promising insight and potential progress towards the development of therapeutic treatments in cardiovascular disease.
Keywords: Autophagy, microRNAs, Cardiovascular Diseases, Post-Transcriptional Regulation, Review
2. INTRODUCTION
Adaptive and maladaptive autophagy has been linked to a wide variety of human processes and disease. Studies have found autophagy to be associated with development, aging, neurological disorders, liver and muscle disorders, infection, immunity, and cancer (1,2). The controversy regarding autophagy lies in the role it plays in pathology, and whether it serves as a protective response to insult or a means of exacerbating cell death.
The homeostasis between life and death is pertinent to human physiology. Maladaptive regulation of cell death and cell survival is an underlying cause of many human diseases. The balance between death and survival is maintained predominantly via three cellular pathways: apoptosis, necrosis, and autophagy. An intimate crosstalk exists between these pathways, however, the apoptosis and necrosis pathways have been reviewed extensively elsewhere and, thus, autophagy will remain the focus of this review (3–5). Autophagy is an intracellular vesicular process by which protein aggregates and organelles are eliminated via lysosomal degradation (2). The etymology of autophagy is Greek, derived from words that translate to “eating of self.” This finely tuned process is capable of mediating cell survival at basal levels by breaking down proteins and organelles into their building blocks, which are then capable of being reused by the cell. In this respect, autophagy functions as a cellular form of recycling. It is believed that decreased levels of autophagy explain protein aggregates often seen as the cause of neurological disorders such as Alzheimer’s disease (6,7). Excessive levels of autophagy, on the other hand, can be detrimental and provide an alternative route to cell death, often termed non-apoptotic type II cell death (2).
Protein regulation is important for the maintenance of cellular homeostasis. A multitude of regulatory mechanisms responsible for controlling protein expression and turnover transcend evolutionary biology (8). Research spanning the last several decades has shed light on the role of non-coding RNA in regulating protein expression. Several non-coding RNA protein regulatory pathways exist, including: microRNA (miRNA), short-interfering RNA (siRNA), repeat-associated small interfering RNA (rasiRNA), piwi-interacting RNA (piRNA), and long non-coding RNA (lncRNA) (9–11). Of these, miRNAs are the most evolutionarily conserved (12), and, thus, are the most commonly researched non-coding RNA. MicroRNAs are a small chain of nucleotides, roughly 22 nucleotides on average, that regulate specific messenger RNAs (mRNA), allowing them to suppress post-transcriptional gene expression and protein production (9). To date, over 1000 miRNAs have been identified within the human genome (8) and are expressed throughout various, yet specific, tissues. It is believed that the vast number of miRNAs control a plethora of cellular processes and regulate nearly half of all protein coding genes within humans (13–15).
2.1. Autophagy
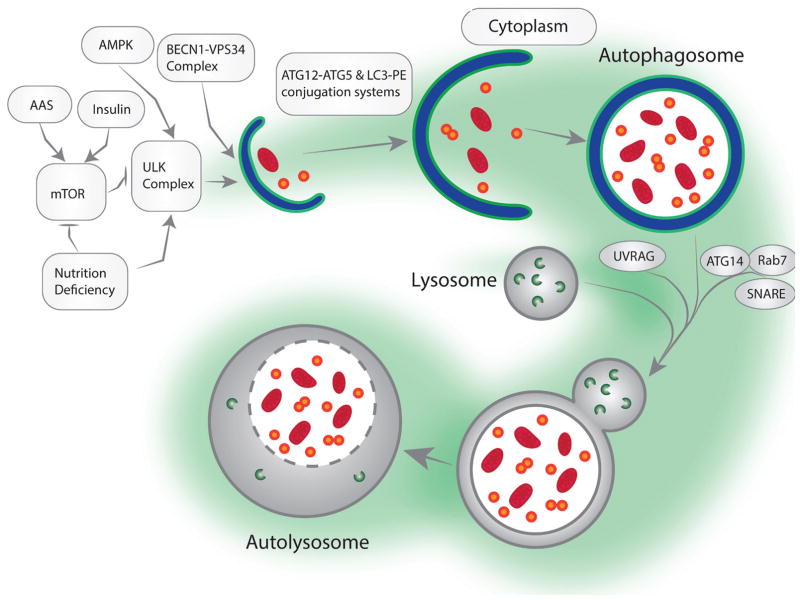
Three main subsets of autophagy have been identified: macroautophagy, microautophagy, and chaperone-mediated autophagy, which differ in cellular function and the means by which targets are delivered to lysosomes. The most commonly studied form is macroautophagy, and hence will be the emphasis of this review. Macroautophagy is distinguished by the formation of a double membrane vesicle known as an autophagosome that is used to deliver cargo to lysosomes (16). Five steps characterize the process: induction, vesicle nucleation, elongation/completion, docking/fusing with the lysosome, and cargo degradation (Figure 1) (17). Upon stimulus, such as stress, lipid nucleation begins. These lipids are believed to derive from membranes such as the mitochondria, plasma membrane, endoplasmic reticulum, or golgi apparatus and will eventually become the autophagic vacuole (18–23). After nucleation, the lipid membrane will elongate around target cargo and enclose it, completing the formation of an autophagosome. This vacuole will then fuse with a lysosome, forming a mature autolysosome, and undergo hydrolysis in order to degrade the vesicle’s contents (2). Many proteins are involved in the regulation and execution of autophagy. However, it is predominantly regulated by mammalian target of rapamycin (mTOR) and AMP-responsive protein kinase (AMPK) pathways, two processes responsible for nutrient sensing and the regulation of unc-51-like kinase 1(ULK1), a protein important for the induction of autophagy. In times of nutrient abundance, mTOR is active and inhibits the autophagic process via phosphorylation of ULK1. Alternatively nutrient deprivation stimulates AMPK to activate suppressors of mTOR, Tuberous Sclerosis Complex (TSC1/2), and stimulate autophagy (24).
Figure 1.
A schematic figure of key proteins that are involved in formation of the whole process of autophagy.
Early genetic studies done in yeast revealed a set of genes responsible for autophagosome formation, together known as autophagy-related genes (ATG) (25,26). Many of the initial genes discovered in yeast were later found to be conserved in mammals and have human homologs (27). Induction of autophagy is mediated through the activation of the ULK complex, composed of ULK1/2, mATG13, Family interacting protein of 200 kD (FIP200), and ATG101 (28). Under starvation conditions, ULK1 is dephosphorylated, disassociating it from mTOR, where it can proceed to autophosphorylate itself as well as the ULK complex. It then proceeds to activate vesicle nucleation by means of phosphorylating Beclin-1 (BECN1), a key component of the nucleation complex (29). Nucleation is dependent on a group of proteins including: class III phosphotidylinositol-3 kinase (PI3K) vacuolar protein sorting (VPS) 15, Beclin-1, activating molecule in BECN1 regulated autophagy protein 1 (AMBRA1), ultraviolet irradiation resistance-associated gene (UVRAG), ATG14L, and bax-interacting factor 1 (BIF-1). This complex forms to activate the class III PI3K VPS34 and generate PI3 (16).
After vesicle nucleation occurs, elongation of the membrane/phagophore takes place via ubiquitin-like conjugations with multiple ATG proteins (ATG12, ATG5, ATG 10, ATG 16L, ATG8, ATG4, ATG7, and ATG3) as well as the microtubule-associated protein 1 light chain (LC3). LC3 is capable of coupling to phosphotidylethanolamine (PE) to form LC3II and become a membrane bound conjugate (30). LC3-II is important in both membrane expansion as well as cargo recognition by means of its interaction with Sequestosome 1, which recognizes ubiquitinated target proteins (1). Sequestosome 1 is regulated by miR-17/20/93/106 (31). Upon completion of autophagosome formation many ATG and fellow proteins disassociate from the vesicle and into the cytoplasm for reuse; the rare exception being LC3II. LC3II bound to the outer membrane of autophagosomes is cleaved and returned to the cytoplasm by ATG4; however inner-membrane bound LC3II remains anchored to autophagosomes up until lysosomal fusion occurs, where it is degraded within the lysosome (1,2,16,17,25–27).
An autophagosome is fully matured once it has fused to a lysosome, creating the autolysosome. This fusion process is accomplished through Rab proteins, soluble N-ehtylmaleimide sensitive factor attachment proteins (SNAREs), the homotypic fusion and vacuole protein sorting (HOPS) complex, and tethering proteins (1). Rab7 GTPase is the most studied Rab and has an important role in autophagosome-to-lysosome trafficking through its interactions with effecter proteins such as FYCO1, RILP, and Csn8 (32). Recently Syntaxin 17 has been identified as a SNARE capable of interacting with VPS33A and VPS16 to mitigate membrane fusion between autophagic vacuoles and lysosomes (33). Upon fusion the inner membrane of the autophagosome and its cargo are degraded via lysosomal cathepsins and rendered back into usable amino and fatty acids (1,2,16,22,27).
Autophagy can be regulated by proteins aside from mTOR, AMPK, and the ATG family. For example, the B-cell lymphoma family of proteins are known to regulate apoptosis via mitochondrial outer membrane permeabilization. These proteins can either be pro-apoptotic or anti-apoptotic in function, but all contain at least one of four Bcl-2 homology domains (BH1-4) and all contain a BH3 domain which allow them to form homodimers or heterodimers and control cell death or cell survival (34). Beclin-1 contains a BH3 domain, and this allows the anti-apoptotic protein Bcl-2 to bind to and inhibit Beclin-1’s autophagic function (35). Apoptosis repressor with caspase recruitment domain (ARC) has been reported to negatively regulate autophagy (36). Meanwhile master gene transcription regulators such as Histone acetyltransferaces (HATs) and deacetylases (HDAC) can influence autophagy levels (28,37). Transcription factors, such as FOX1/3 and the RB1-E2F1 pathway, regulate autophagy as well (1).
It is safe to say that autophagy can certainly function as an adaptive and maladaptive response, depending on the level of regulation and cellular milieu. Beclin-1 can function in both apoptosis, as a BH3-only protein capable of sequestering pro-apoptotic proteins away from mitochondria, via its BH3 domain, as well as autophagosome nucleation (5,35). The dual functionality of this protein has led to it being one of the most well studied autophagy proteins. Deletion of the beclin1 gene in murine models leads to embryonic death as a result of failed pro-amniotic canal closure (38). In utero lethality is commonly associated with proteins involved in the induction and nucleation processes of autophagy (1). This same protein, Beclin-1, is also a tumor suppressor. Mice heterozygous for the BECN1 display haplo-insufficiency and significantly elevated levels of tumor development throughout various tissues (38,39).
Similarly, accumulation of misfolded proteins is commonly seen in liver diseases such as alpha-1-antitrypsin deficiency, where autophagy is regarded as the primary means of clearing the misfolded protein (40–42). More and more it is becoming evident that autophagy provides an essential link to disease.
2.2. MicroRNAs
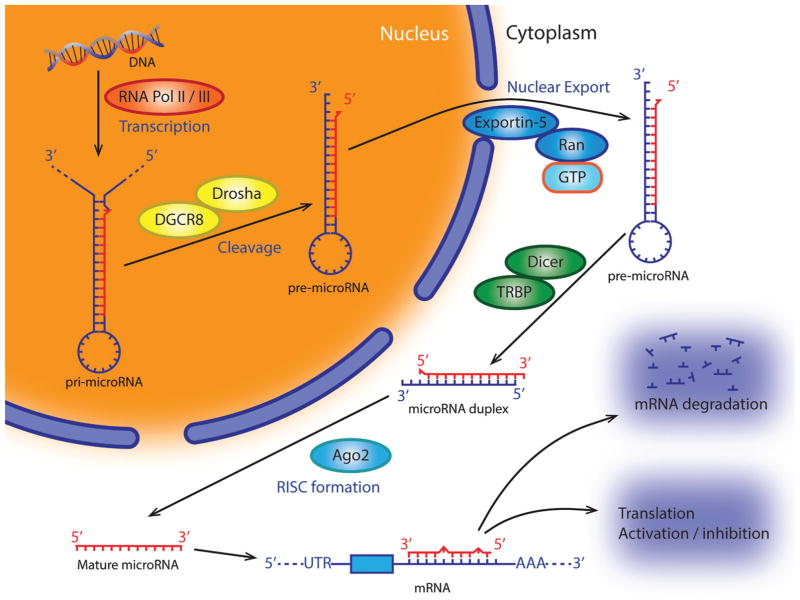
MicroRNAs are predominantly transcribed in the nucleus under the control of RNA polymerase II. These RNAs are much longer than their final 22 nucleotide form, and thus require processing within the nucleus by Drosha, a class two ribonuclease III enzyme. After processing by Drosha, the RNAs are deemed precursor miRNA (pre-miRNA) and contain a hairpin structure of close to 70 nucleotides (43). Precursor miRNA then leaves the nucleus by means of Exportin- 5, which recognizes a two-nucleotide overhang at the 3′ end of the RNA, and transports it to the cytoplasm (44). Once outside the nucleus, the RNA is further processed into a 22-nucleotide double-stranded RNA (dsRNA) by means of the RNase III family enzyme Dicer (8,44). Once in small dsRNA form the miRNA can be incorporated into the RNA-induced silencing complex (RISC). Incorporation of dsRNA into RISC is carried out by the complex of Dicer, transactivating response RNA-binding protein (TRBP), and Argonaut-2 (Ago2) (Figure 2) (45). The dsRNA is then unwound into two single-stranded RNAs. One strand will remain bound to Ago2, thus becoming mature miRNA, while the other strand gets degraded (44).
Figure 2.
A schematic figure shows biogenesis of microRNAs and their functions.
Mature miRNA will then bind to complimentary nucleotides along mRNA. Often complementarity is found between the “seed” region of miRNA (nucleotides 2–8) and the 3′ untranslated region (UTR) of mRNA, prompting Ago2 to bind GW182 and activate translational suppression (44). Once the ribonucleoprotein complex has bound its target mRNA it can go about suppressing it in several manners. Canonical miRNA/RISC mRNA inhibition is mediated through mRNA decay. This process consists of perfect complementarity between the “seed” region of miRNA and mRNA and leads to Argonaut proteins (Ago 1–4) within the RISC complex cleaving mRNA at cut sites that line up with nucleotides 10 and 11 of the miRNA (46). MicroRNA/RISC can also lead to mRNA decay by recruiting decapping factors (DCP1/2), deadenylase, and exonucleases (3′ or 5′) as well as endonucleases such as PMR1 (47–50). It is believed that the protein GW182 to essential for some of these alternative methods of mRNA degradation.
MicroRNA can suppress protein expression in an alternative fashion to simple mRNA decay; miRNAs can also interfere with the process of translation. This interference can occur both prior to and after translation activation. It has been hypothesized that miRNA can silence mRNA by preventing translation initiation by means of ribonucleoproteins, specifically, Ago. Ago competes with cap binding proteins (CBPs) and eukaryotic translation initiation factor 4E (EIF4E) to perturb cap structure and closed-loop formation essential for translation initiation; as well as sequestering mRNA in processing bodies, mRNA, Ago, and GW182 dense regions of the cytoplasm, for degradation or delayed expression (51–55). It is also possible for miRNA to inhibit mRNA once the initiation of translation has already taken place and elongation is in progress. This can happen in the form of: RISC inhibiting ribosomal subunit joining or prompting the subunits to disassociate from mRNA, competing with elongation factors and thus slowing the process, or recruiting decay factors, to degrade proteins mid translation (56–59).
It is of note that miRNA is not always associated with inhibitory or down regulatory effects. In rare circumstances, dependent on cell cycle and co-factors, miRNA has been shown to activate translation, thus up regulating protein levels (46). While miRNA mediated up regulation is far less common, multiple miRNAs have been shown to have this effect. Often miRNA up regulation occurs phase) (60). In this phase, when cells are quiescent (G0 GW182 is often down regulated. While the mere absence of GW182 is enough to abrogate mRNA degradation, its absence also allows Ago2 to bind with other proteins, such as fragile X mental retardation protein 1 (FXR1), which leads to gene activation (61). The cleavage activity of Ago is also absent in quiescent cells (62). This means it is possible for the RISC complex, with its capacity to bind EIF4E, previously deemed a means to block translation, could actually assist mRNA in forming a closed loop structure and initiate translation (51).
MicroRNAs have been shown to have vast effects on translational regulation. The inherent complexity of this regulatory system allows it to range from managing fine changes to grand alterations and functions both as a down regulator as well as an up regulator. The intricacies and implications of miRNAs make it abundantly clear how large a role they play in physiology, and thus present themselves as a wonderful tool in which to address the treatment of disease.
2.3. MiRNA regulation of autophagy and vice-versa
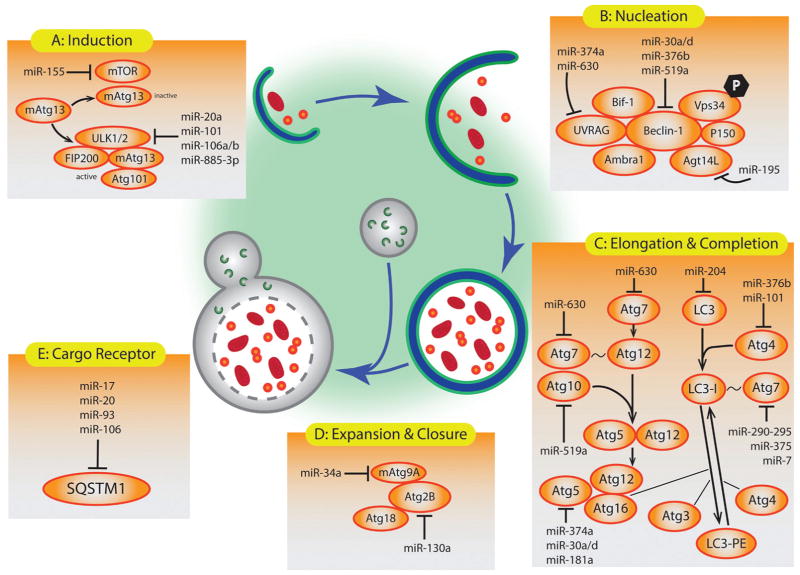
Regulation of pathways such as apoptosis and autophagy are pertinent to health. As mentioned before, autophagy is a complex process with many levels of regulation built in. It was not until 2009 however that a link was made between autophagy and miRNA. Zhu et al. discovered that miR-30a could negatively regulate Beclin-1, and, thus, autophagy, in cancer cells (63). Since that study, a plethora of others have followed revealing that miRNAs can influence all stages of autophagy (Figure 3).
Figure 3.
Summary of identified microRNAs that target key proteins of autophagic pathway for regulation of autophagy.
The master regulators of autophagy, mTOR and AMPK, are subject to miRNA regulation. RHEB and RICTOR, components of mTOR signaling, are both affected by miR-155 (64). Likewise AMPK is directly targeted by miR-148b (65).
The ULK complex, responsible for autophagy induction, is targeted by multiple miRNAs. ULK1/2 is regulated by miR-20a, miR-101, and miR-106a/b (66–68). Mammalian ATG13, another component of the ULK complex, contains binding sites for miR-885-3p, as do ATG9a and ATG2b (69).
The class III PI3K/Beclin-1 nucleation complex is under the control of miR-30a/d, miR-216a, miR-376b, and miR-519a (30,66,70–72). Vps34 is silenced by miR-338-5p (30). UVRAG is affected by miR-374a and miR-630 (72). MiR-195 is capable of inhibiting ATG14L (73). PI3K itself, an important factor of nucleation, is negatively regulated by phosphatase and tensin homolog (PTEN) which is the target of miR-18a, miR-21, miR-26a, miR-214, miR-216a, miR-17-92, miR221 and miR-222 (30,74).
The ubiquitin-like conjugation system required for phagophore elongation is made up of a multitude of proteins, each a subject of miRNA regulation. ATG4 is targeted by miR-101 and miR-376b, while LC3 is silenced by miR-204 (30,75). ATG12 is regulated by miR-23b and miR-630, while its conjugation partner ATG5 is targeted by miR-30a/d, miR-181a, and miR-374a (30,72,76). ATG7 is suppressed by miR-17, mir188-3p, miR-290-295, and miR-375 (77–80). Meanwhile microRNA-519a targets ATG10 (30,72).
The fusion of mature autophagosomes with lysosomes is regulated by miR-34a and miR-130a (81,82). It is important to emphasize the diverse impact and multiple functions individual miRNAs have. Each miRNA is capable of binding to and altering multiple mRNAs, thus altering more than just one isolated process or system. Take for instance miR-30a; it targets autophagy at two levels, Beclin-1 and ATG5, nucleation and elongation respectively, but also p53, an important player in DNA repair and apoptosis (30). It is fitting that a miRNA exists, capable of mediating apoptosis and autophagy, for it is believed that Beclin-1 serves in a similar function. However, this goes to show that more research needs to be done to understand the complexity of these systems.
Much as miRNAs can serve to regulate elements of the autophagic process, autophagy itself can also regulate miRNAs and their production. As reported by Gibbings et al., Dicer and Ago2, two main components of miRNA biogenesis, are selectively degraded via NDP52-mediated autophagy (83). MicroRNA production is dependent upon proper autophagy levels. For example, during states of low autophagic turnover, higher levels of inactive Dicer and Ago2 accumulate. These inactive proteins are incapable of binding to miRNA, but can bind to active RISC components, rendering them useless, and suppressing miRNA/RISC activity (84). Autophagy is also a capable means of degrading some miRNAs such as miR-224 (28).
3. AUTOPHAGY AND CARDIOVASCULAR DISEASE
Since 2001, it has become apparent that autophagy is intimately associated with human heart disease. The pioneering study examined human tissue samples from patients with dilated cardiomyopathies and revealed that dying or degrading cardiomyocytes often contained higher level of autophagic vacuoles (85). This study, along with others soon after, showed that autophagy might function as a method of cellular death in cardiomyocytes; spawning an ongoing controversy: is autophagy beneficial or detrimental to cardiovascular biology?
As with any other biological process, autophagy is not inherently all good or all bad in the context of cardiovascular physiology. Rather, autophagy is regarded as a double-edged sword in the context of heart and vascular biology. The heart is an energy demanding environment, one in which cardiomyocytes are continuously beating to accommodate the metabolic demands of the body. As a result, cardiomyocytes must ensure that their mitochondria sufficiently supply adequate amounts of ATP in an efficient manner. Failure of cardiomyocytes to degrade old mitochondria can lead to inefficient energy production and a buildup of radical oxygen species (ROS) (86). Excessive ROS can induce apoptosis within cardiomyocytes and jeopardize heart function. Because of this, it is crucial for cardiomyocytes to properly execute mitophagy, a specific form of macroautophagy targeting mitochondrial degradation, as a means of preventing cellular death (86). Thus, it is clear that perturbations that lower basal autophagy can be just as harmful as those seen in over activation of the pathway. A proper balance of autophagy must be struck to maintain proper cardiovascular homeostasis.
3.1. Autophagic miRNAs and cardiac remodeling
The hemodynamic load associated with cardiovascular stresses often leads to the heart remodeling (87). Remodeling is viewed as a structural change within the heart such as alterations in chamber dimensions, wall thickness, mass, etc. Depending on the stress and other factors, the heart can remodel in multiple ways: a compensatory growth of the heart in an effort to preserve contractility, known as hypertrophy, a non-compensatory increase in chamber volume associated with thinning of the cardiac walls, known as dilation, or, in rare circumstances, a loss of heart mass, known as atrophy. Much as autophagy is viewed as both friend and foe, hypertrophy can also be viewed as adaptive and maladaptive in response to cardiac stress (88). After an acute stress, such as myocardial infarction (MI), hypertrophy is a compensatory mechanism in which cardiomyocytes undergo structural, molecular, metabolic, and biochemical changes that enhance cell size in an effort to preserve cardiac function (89). However, in cases of chronic stress, compensatory hypertrophy can lead to left ventricular dilation, apoptosis, excessive fibrosis, and cardiac dysfunction (89,90). Cardiac hypertrophy results in an enhanced cardiac workload and an elevated demand for energy. In the circumstance of hypertrophy, cardiomyocytes switch from fatty acid (FA) oxidation to glucose as the primary source of ATP production. This switch is believed to be more favorable because glucose-derived ATP requires less oxygen than FAs, and the thickened walls of a hypertrophic heart alter the diffusion rate and availability of oxygen to cardiomyocytes (89).
Protein synthesis and efficient energy production are elemental to cardiac remodeling, and under acute stresses it is believed that autophagy is beneficial to both of these processes by: 1) clearing old or detrimental protein aggregates, preventing protein toxicity, 2) providing the necessary amino acids to accommodate new protein production, and 3) regulating mitophagy to optimize ATP production, while minimizing ROS and the risk of mitochondrial mediated cell death (89–91). Amelioration of autophagy within the heart, specifically ATG5 and ATG7 genes, has been shown to cause rapid cardiac hypertrophy in mice (89,91). Danon disease derives from defective autophagic flux due to improper autophagosome-lysosomal fusion, and is marked by cardiomyopathy (92). Alternatively it is hypothesized that once autophagy becomes overly activated, it becomes maladaptive, leading to cell death. Inhibition of autophagy has resulted in the restoration of systolic function and reversal of hypertrophy in mice that underwent transverse aortic constriction (TAC) (87).
MicroRNAs have also been found to play a significant role in the heart (Table 1). Mice deficient of Dicer develop heart failure and die within 4 days of being born (93). MiRNAs are important for cardiac remodeling. In 2008, miR-208 was identified as the first cardiac miRNA regulating myosin heavy chain gene expression and left ventricular hypertrophy (94). Since then a host of other miRNAs, not necessarily cardiac specific, have been identified in cardiac remodeling, several of which function through regulation of autophagy.
Table 1.
Known miRs that target autophagy in cardiovascular diseases
| miR (s) | Autophagy Target | Associated Cardiovascular Disease | Implication in Disease | References |
|---|---|---|---|---|
|
| ||||
| Regulation: Anti-autophagic | ||||
|
| ||||
| MiR-30a | Beclin-1 | Cardiac Remodeling | Down regulated in pressure overload and angiotensinII-mediated hypertrophy. | (95,96) |
| Stroke | Inhibition of miR alleviates neuronal death associated with I/R | (125) | ||
|
| ||||
| miR-34 | Atg9 | Cardiac Remodeling | Down regulated in angiotensinII-mediated hypertrophy. | (82,97) |
| Diabetic Cardiomyopathy* | Elevated during cardiac aging. | |||
| Up-regulated in Type 2 diabetes patients with ischemic HF | (117,118) | |||
|
| ||||
| miRs-212/132 | FOXO3a | Cardiac Remodeling | Over-expression results in hypertrophy and HF. | (98) |
| Suppression rescues pressure over-load induced hypertrophy | ||||
|
| ||||
| miR-221 | P27 | Cardiac Remodeling | Over-expression results in hypertrophy and HF | (99) |
|
| ||||
| miR-451 | TSC1 | Cardiac Remodeling | Lower levels seen in HCM patients compared to healthy hearts and deletion elevates rat cardiomyocyte surface area | (100) |
|
| ||||
| miR-199a | GSK3-b | Cardiac Remodeling | Induces hypertrophy | (101) |
|
| ||||
| miR-497 | LC3 and Bcl-2 | Ischemic/Reperfusion | Decreased in MI hearts and hypoxia treated cardiomyocytes. | (114) |
| Diabetic Cardiomyopathy* | Suppression reduces apoptosis and infarct area. | (119) | ||
| Down regulated in diabetic hearts | ||||
|
| ||||
| miR-204 | LC3II | Ischemic/Reperfusion | Down regulated during I/R | (115) |
|
| ||||
| miR-188-3p | Atg7 | Ischemic/Reperfusion | Administration of miR prevents death and minimizes infarct size | (78) |
|
| ||||
| miR-101 | Rab5a | Ischemic/Reperfusion | Down regulated during H/R. | (107) |
| Inhibition of miR prevents apoptosis | ||||
|
| ||||
| miR-221 | Not defined in study | Diabetic Cardiomyopathy | Up-regulated in STZ-treated mice hearts | (118) |
|
| ||||
| miR-133a | Not defined in study | Diabetic Cardiomyopathy | Down-regulation in DCM correlates with increase in beclin1, LC3, Atg3, and suppression of mTOR. | (119,120) |
| miR133a mimics prevent glucose mediated hypertrophy | ||||
|
| ||||
| miR-195 | Not defined in study | Diabetic Cardiomyopathy | Up-regulated in STZ-treated mice hearts | (118) |
|
| ||||
| miR-20a | Not defined in study | Diabetic Cardiomyopathy | Down regulated in STZ-treated mice hearts | (118) |
|
| ||||
| miR-216a | Beclin1 | Atherosclerosis | Over expression causes ox-LDL accumulation. | (70) |
| Suppression protects against ox-LDL | ||||
|
| ||||
| miR-207 | LAMP2 | Stroke | Down regulated after ischemia. | (126) |
| Mimics reduce infarct area | ||||
|
| ||||
| miR-352 | LAMP2 | Stroke | Down regulated after ischemia | (126) |
|
| ||||
| miR-99a | mTOR | Cardiac Remodeling | Attenuates remodeling post MI | (102) |
| Ischemic/Reperfusion | Over-expression improves cardiac function and prevents cell death in rodent MI model | |||
|
| ||||
| Regulation: Pro-autophagic | ||||
|
| ||||
| miR-144 | mTOR | Ischemic/Reperfusion | Administration and pre-conditioning reduce apoptosis and infarct size in mice. | (113) |
|
| ||||
| miR-325 | ARC | Ischemic/Reperfusion | Up-regulated in I/R injury. | (36) |
| Over-expression enlarges infarct size, while knockdown reduces cell death. | ||||
autophagic gene target not directly identified in study
Just as the genes responsible for autophagy and hypertrophy are controversial, so too are the miRNAs that regulate their transcripts. The well-studied miR-30a, known to target and suppress Beclin-1, is reportedly down regulated in murine models of pressure-overload (TAC) and Angiotensin-II-mediated cardiac hypertrophy (95,96). The down regulation of miR-30a was met with enhanced levels of Beclin-1, LC3II, autophagy, and hypertrophic markers such as atrial naturietic peptide (ANP) and beta-MHC. Administration of miR-30a mimics and autophagy inhibitors ameliorated autophagy and the hypertrophic phenotype (66).
Another miRNA that targets autophagy is miR-34, and it is also decreased in Angiotensin-II models of cardiac hypertrophy (82). This miRNA interferes with autophagosome elongation by down regulating ATG9. MiR-34 levels have been shown to increase in response to cardiac aging, and that suppression of miR-34 reduced age-associated cell death (97). The process of aging is generally associated with decreases in basal autophagy, suggesting that miR-34, and its anti-autophagic function, might be responsible for cardiac aging phenotypes.
While miR-30a and miR-34 directly regulate autophagic machinery, several studies have found miRNAs that control transcriptional regulators of autophagy, and function as anti-autophagic miRNAs that promote hypertrophy. Fox03a is a pro-autophagic transcription factor, as well as anti-hypertrophic, that is negatively regulated by miR-212/132. Over expression of miR-212/132 significantly perturbs autophagy and results in drastic cardiac hypertrophy and heart failure (98). Furthermore, deletion of miR-212/132 yields enhanced autophagy and the capacity to rescue pressure-overload induced hypertrophy.
The master regulator of metabolism and autophagy, mTOR, is regulated by cyclin-dependent kinase 2 (Cdk2), which is, in turn, inhibited by p27 (also known as cyclin-dependent kinase inhibitor 1B). MiR-221 negatively regulates p27, and, therefore, is anti-autophagic. Similar to miR-212/132, cardiac over expression of miR-221 results in diminished autophagy, cardiac hypertrophy, and heart failure (99). Song et al. analyzed miRNA differences between patients with hypertrophic cardiomyopathy (HCM) and healthy donors and reported that miR-451 was the most drastically down regulated miRNA (100). They went on to show that miR-451 silences TSC1, a negative regulator of mTOR. When miR-451 was deleted, rat neonatal cardiomyocyte cell surface area increased, while over expression of miR-451 resulted in inhibited formation of autophagosomes. Similarly, tissue samples from HCM patients, deficient of miR-451, consistently displayed higher levels of LC3II and Beclin-1 compared to healthy donor samples (100). Mir-199a is capable of inhibiting autophagy via its suppression of GSK3-b (96). GSK3-b promotes autophagy through its phosphorylation and activation of TSC2, thereby inhibiting mTOR. As seen with miRs-212/132 and miR-221, miR-199a also induces cardiac hypertrophy (101).
Li et al. reported that miR-99a is an anti-apoptotic/pro-autophagic miRNA capable of improving heart function and attenuating remodeling post MI (102). They show miR-99a is able to lower protein expression of mTOR while elevating autophagy proteins such as ATG5, ATG12, and LC3II/LC3I and increasing autophagic flux (as shown by p62 turnover). Interestingly their data revealed that mRNA for mTOR did not decrease in miR-99a’s presence, suggesting that perhaps the miRNA blocks translation of the mTOR transcript, rather than degrading it.
Fibrosis is a phenotype seen in many diseases, and multiple studies have shown that autophagy can help protect against cardiac fibrosis (103,104). Inhibition of autophagy, with either chloroquine or ATG5 targeted siRNA, was shown to aggravate Angiotensin II-mediated cardiac fibrosis in rat cardiac fibroblasts (103). Singh et al. reported that loss of ATG7 leads endothelial cells to undergo endothelial-mesenchymal transition and enhance organ fibrosis via TGF-beta signaling pathways (104). While these two studies examined autophagy as a regulator of fibrosis, Duisters et al. observed that miR-30 is highly expressed in cardiac fibroblast under normal conditions, however, it becomes down regulated during heart failure in both humans and rodents (105). They identified connective tissue growth factor (CTGF) as the gene target responsible for fibrosis regulation. It is worth noting that miR-30 also regulates autophagy via the BECN1 gene (66,105). Rat cardiac fibroblasts express miR-101, a regulator of ATG4, and expression is decreased under hypertrophic conditions as well as in the peri-infarct area 4 weeks after MI, however, TGF-β1 was identified as the target in this study (30, 75, 106). While they did not identify an autophagic target, this does not rule out autophagy’s involvement, and aligns with Wu et al. findings in hypoxia/reoxygenation (H/R) studies with H9c2 cells (106,107).
3.2. Autophagic miRNAs and ischemic cardiomyopathy
Globally, ischemic heart disease is often the leading cause of death (108). Ischemia is characterized by diminished blood supply, resulting in reduced access to oxygen and nutrients in peripheral tissues and, in this circumstance, the heart. Ischemic heart disease can present itself in an acute or chronic fashion. MIs, the most common form of acute ischemic cardiomyopathy, occur when plaques in the vasculature become dislodged and cause thrombotic occlusions that completely block coronary vessels, depriving the myocardium of oxygen and nutrients.
It is evident that ischemic events that endure over a prolonged period of time will result in myocardial death. It is also commonly observed that reperfusion following nutrient deprivation, despite being capable of rescuing cells from the direct threat of death, elicits its own deleterious effects (109). Myocardium directly occluded from blood supply quickly die from necrosis, meanwhile surrounding tissue, also known as the border area, becomes ischemic. Longer durations of ischemia result in larger areas of damage. This ischemic phase is characterized by cells transitioning from primarily oxidative phosphorylation to glucose and lactic acid fermentation as a means of ATP production (89). This alteration in respiration leads to an imbalance in ion concentrations within the cell, specifically sodium. With elevated intracellular levels of Na, the sodium-calcium exchanger (NCX) can no longer expel calcium, and intracellular calcium becomes overloaded. This abundance in calcium allows for elevated mitochondrial calcium concentration, which initiates both apoptotic and necrotic pathways (110). The acidic buildup within cells during ischemia, derived from elevated intracellular sodium as a result of lactic acid fermentation and the sodium-hydrogen exchanger (NHE), is restored upon reperfusion. However, this restoration is believed to reactivate the NHE as well as NaHCO3 exchangers and, again, cause buildup of both sodium and calcium (111). The increase in intracellular calcium elicits several effects such as calpain activity and excessive contractility. However, again, elevation in mitochondrial calcium is seen. Mitochondrial calcium, coupled with elevated ROS, leads to activation of mitochondrial permeability transition pores (MPTP), initiating apoptosis (112).
While research has pointed towards apoptosis and necrosis as the predominant mediators of cell death associated with ischemia and reperfusion, studies have been done to look at whether targeting autophagy can help protect against ischemic/reperfusion (I/R) related injuries (109). Several pro-autophagy microRNAs have been associated with cardiac I/R injury. As mentioned earlier, Li et al. found that over expression of miR-99a protected against maladaptive remodeling, but also is beneficial during I/R injuries (102). MiR-99a targets mTOR, promoting autophagy. MiR-99a over expression improved cardiac function in mice subjected to MI, while also diminishing levels of apoptosis. MTOR is also targeted by miR-144 and was shown to be beneficial during preconditioning stages to ameliorate apoptosis and infarct size during MI in mice (113). MiR-325 functions to negatively regulate ARC and, therefore, serves as a pro-autophagic miRNA (36). During myocardial I/R injury miR-325 is up regulated. However, unlike MiR-99a, over expression of miR-325 managed to increase autophagy and exacerbate infarct size in MI models. Likewise, knockdown of miR-325 reduced autophagy and cell death within the heart (36).
There are also anti-autophagic miRNAs that play a role in I/R injury. It is reported that miR-497 is decreased in hearts that undergo MI as well as cardiomyocytes subjected to hypoxia (114). This reduction yields an increase in the target genes LC3B and BCL2, and is believed to be beneficial to cells. Further suppression of miR-497 results in elevated autophagy and reduction in both apoptosis and infarct size. Light-chain microtubule associated protein LC3 is targeted by miR-204 and altered during I/R injury (115). Another miRNA down regulated by I/R injury is miR-188-3p. This is an anti-autophagic miR that negatively regulates the elongation phase of autophagosome formation via ATG7. It is believed that this miR prevents deleterious autophagy. Compensatory administration of miR-188-3p prevents death and minimizes infarct size in murine models of MI (78). It was reported by Wu et al. that miR-101 is down regulated when H9c2 cells are treated under hypoxia/reoxygenation conditions and undergo apoptosis (107). They looked at the role of RAB5a in regulating autophagy and found that inhibition of miR-101 prevented apoptosis and elevated levels of autophagy (107). It is of note, however, that miR-101 has also been shown to target other elements of the autophagy machinery such as ATG4 and ULK1/2 (67,75).
3.3. Autophagic miRNAs and diabetic cardiomyopathy
The American Heart Association reports that individuals suffering from diabetes are two to four times more likely to suffer from heart disease or stroke. The comorbidity that exists between diabetes and cardiovascular disease is thought to stem from common risk factors associated with diabetes such as obesity, high blood pressure, and high LDL as well as triglyceride levels (116). Due to the systemic nature of diabetes, diabetic cardiomyopathies have different levels of miR expression than their non-diabetic counterparts (117–120).
Interestingly, miR-34 is up regulated in type II diabetes patients with ischemic heart failure (HF) and miR-221 is similarly up regulated in STZ-induced diabetic mouse hearts, a model of type I diabetes (117, 118). MiR-34 also regulates the p53 pathway (121,122). This supports the findings that cardiomyocytes derived from OVE26 mice display reduced levels of autophagy (123). In a clinical study, Nandi et al. report that left ventricle samples from diabetic left ventricular assist device (LVAD) patients consistently showed a down regulation of miR-133a, a muscle specific miRNA, when compared to non-diabetic LVAD counterparts (120). This reduction corresponded with induction of autophagy, shown by increases in Beclin-1 and LC3II, suppression of mTOR, and hypertrophy; however, the direct target of miR-133a was not identified. Feng et al. performed a study in which they identified a number of miRNAs that are altered between diabetic and non-diabetic mouse hearts (119). In their study, they identified miR-133a as being significantly down regulated in diabetic hearts and showed neonatal rat cardiomyocytes decrease expression of miR-133a in response to high glucose treatment. In addition, treatment with a miR-133a mimic has been shown to prevent glucose-mediated cardiomyocyte hypertrophy (119). Feng et al. also noted that miR-497, capable of inhibiting LC3, was reduced in diabetic hearts (119, 114). MiR-195 has been shown to inhibit ATG14L and was reported as up regulated in STZ-induced diabetic mouse hearts (73, 118). From Diao et al.’s study they reported miR-20a, an effecter of ULK1/2 (68), as down regulated in STZ mouse hearts as well (118, 68).
3.4. Autophagic miRNAs and other cardiovascular diseases
Ischemic events are significant beyond the heart. Atherosclerosis is a disease afflicting vasculature, and is capable of causing systemic injury. As was alluded to before, chronic ischemic heart disease is often caused by atherosclerosis, specifically within the coronary arteries, that leads to undernourishment of myocardium over time. This relatively modern disease is characterized by the formation of plaques within the inner wall of blood vessels. These plaques are made up of cholesterol, lipids, cellular debris from immune cells such as monocytes/macrophages, calcium, etc. and are believed to form as a result of cellular damage to the vascular endothelium. These plaques pose the threat of forming a blood clot either at the initial site of formation or breaking off and lodging elsewhere in the blood vessels of the body. Atherosclerosis leads to reduced blood flow and ischemia and, therefore, is often the cause of heart attacks, strokes, and gangrene (124).
It was hypothesized and shown by Menghini et al. that miR-216a can regulate ox-LDL induced autophagy in human endothelial cells by directly inhibiting Beclin-1 (70). They also showed that ATG5 levels diminished, however luciferase assays could not verify ATG5 as a direct target of miR-216a. Over expression of the miR suppresses autophagy, leading to ox-LDL accumulation, as well as elevated monocyte adherence; suggesting a potential role in the early plaque formation of atherosclerosis. Alternatively, down regulation of miR-216a induces autophagy in human endothelial cells and provides protection against ox-LDL treatment (70).
Atherosclerosis is intimately associated with cerebrovascular accidents such as strokes. Wang et al. report that inhibition of miR-30a, and subsequent rescue of Beclin-1, can alleviate death/injury associated with cerebral ischemia as well as protect against neurological deficits in mice (125). Two other miRNAs play an important role in regulation of autophagy post-cerebral ischemia: miR-207 and miR-352 (126). It was reported that both miRNAs are down regulated after ischemia and suppresses lysosome associated membrane protein (LAMP) 2. Tao et al.’s data reveal that up regulation of miR-207 can reduce the number of lysosomes and autolysosomes, while increasing the number of autophagic vacuoles, suggesting miR-207 prevents full autophagosome maturation and autophagic degradation. MiR-207 mimics can reduce infarct size of ischemic insults, indicating that miR-207 protects against maladaptive autophagic cell death (126).
Currently, there is no literature supporting the role of autophagic-miRNA in arrhythmic or valvular cardiovascular disease. That said, each pathology has evidence that points towards autophagy being involved. Clinical work by Yuan et al. revealed that patients suffering from atrial fibrillation express higher levels of LC3II as well as increased levels of AMPK phosphorylation (127). In the same study, they were able to recapitulate these findings using cardiac pacing in canines. Meanwhile, a recent study using clinical samples revealed LC3 elevations as a biomarker for myxomatous mitral valves (128). Therefore, it is certainly within the realm of possibility that miRNAs might be responsible for the alterations in autophagic machinery associated with both pathologies. It could prove insightful to see whether some of the miRNAs previously discussed, such as miRs-497/204, which directly targets LC3, or miR-30a451/99a, which directly target upstream mRNAs and generally regulate autophagy levels, are altered in these diseases.
4. CONCLUSION
Autophagy and microRNA play a prevalent role in the development, progression, and protection against cardiovascular disease. Studies that implement miRNA agonist and antagonist mimics provide promising hope for potential clinical application. However, nuances regarding numerous gene targets for each miRNA, as well as establishing tissue specific regulation, still remain to be elucidated. While advances have been made in the fields of heart failure, cardiac remodeling, ischemia, atherosclerosis, and cerebrovascular accidents, more research is required to see whether miRNA is associated with autophagic regulation of arrhythmias and valvular disease. Additionally, studies have indicated that some miRs may be responsible for diabetic cardiomyopathy, however, the direct autophagic targets of the miRs involved with diabetic cardiomyopathy have not been identified. Fibrosis is another major player in cardiovascular disease, and as of now there is little known regarding the role of autophagic miRNAs in cardiovascular fibrosis. Non-coding RNAs, such as miRNA and lcRNA, are burgeoning fields of science capable of revealing many new insights in human health.
Acknowledgments
The Zhu laboratory is supported by National Institute of Health Grant #HL124122 and #AR067766, and American Heart Association Grant 12SDG12070174.
References
- 1.Hale AN, Ledbetter DJ, Gawriluk TR, Rucker EB., 3rd Autophagy: regulation and role in development. Autophagy. 2013;9:951–972. doi: 10.4161/auto.24273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: The Release of Damage-Associated Molecular Patterns and Its Physiological Relevance. Immunity. 2013;38:209–223. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead? Protein Cell. 2012;3:17–27. doi: 10.1007/s13238-011-1127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang DS, Stavrides P, Mohan PS, Kaushik S, Kumar A, Ohno M, Schmidt SD, Wesson D, Bandyopadhyay U, Jiang Y, Pawlik M, Peterhoff CM, Yang AJ, Wilson DA, St George-Hyslop P, Westaway D, Matthews PM, Levy E, Cuervo AM, Nixon RA. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer’s disease ameliorates amyloid pathologies and memory deficits. Brain. 2011;134:258–277. doi: 10.1093/brain/awq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitoh Y, Fujikake N, Okamoto Y, Popiel HA, Hatanaka Y, Ueyama M, Suzuki M, Gaumer S, Murata M, Wada K, Nagi Y. P62 Plays a Protective Role in the Autophagic Degradation of Polyglutamine Protein Oligomers in Polyglutamine Disease Model Flies. J Biol Chem. 2015;290:1442–1453. doi: 10.1074/jbc.M114.590281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Aravin A, Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 11.Ounzain S, Burdet F, Ibberson M, Pedrazzini T. Discovery and functional characterization of cardiovascular long noncoding RNAs. J Mol Cell Cardiol. 2015;89:17–26. doi: 10.1016/j.yjmcc.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Pasquinelli A, Reinhart B, Slack F, Martindales M, Kuroda M, Maller B, Hayward D, Ball E, Degnan B, Müller P, Spring J, Srinivasan A, Fishamn M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 13.Zhai H, Fesler A, Ju J. MicroRNA A third dimension in autophagy. Cell Cycle. 2013;12:246–250. doi: 10.4161/cc.23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Shen X, Zou Q, Wang S, Tang S, Zhang G. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 16.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang CW, Klionsky DJ. The Molecular Mechanism of Autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- 18.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria Supply Membranes for Autophagosome Biogenesis during Starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre- autophagosomal structures. Cell. 2011;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse Autophagosome Membrane Sources Coalesce in Recycling Endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine B, Klionsky DJ. Development by Self-Digestion Molecular Mechanisms and Biological Functions of Autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 23.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Komatsu M. Autophagy: Renovation of Cells and Tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Jing Z, Han W, Sui X, Xie J, Pan H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015;356:332–338. doi: 10.1016/j.canlet.2014.09.039. [DOI] [PubMed] [Google Scholar]
- 29.Nazarko VY, Zhong Q. ULK1 targets Beclin-1 in autophagy. Nat Cell Biol. 2013;15:727–728. doi: 10.1038/ncb2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6 doi: 10.18632/oncotarget.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, Sun SM, Taskesen E, Stern P, de Ru AH, van Adrichem AJ, Demmers J, Jongen-Lavrencic M, Löwenberg B, Touw IP, Sharp PA, Erkeland SJ. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 2011;118:916–925. doi: 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2013;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 35.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bo L, Su-Ling D, Fang L, Lu-Yu Z, Tao A, Stefan D, Kun W, Pei-Feng L. Autophagic program is regulated by miR-325. Cell Death Differ. 2014;21:967–977. doi: 10.1038/cdd.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sui X, Zhu J, Zhou J, Wang X, Li D, Han W, Fang Y, Pan H. Epigenetic modifications as regulatory elements of autophagy in cancer. Cancer Lett. 2015;360:106–113. doi: 10.1016/j.canlet.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Embryonic Development, Is a Haploinsufficient Tumor Suppressor. Proc Natl Acad Sci USA. 2003;100 doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perlmutter D. The role of autophagy in alpha-1-antitrypsin deficiency: a specific cellular response in genetic diseases associated with aggregation-prone proteins. Autophagy. 2006;2:258–263. doi: 10.4161/auto.2882. [DOI] [PubMed] [Google Scholar]
- 41.Yorimitsu T, Klionsky DJ. Eating the endoplasmic reticulum: quality control by autophagy. Trends Cell Biol. 2007;17:279–285. doi: 10.1016/j.tcb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Kamimoto T. Intracellular Inclusions Containing Mutant 1-Antitrypsin Z Are Propagated in the Absence of Autophagic Activity. J Biol Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 44.Leung AKL. The Whereabouts of microRNA Actions: Cytoplasm and Beyond. Trends Cell Biol. 2015;25:601–610. doi: 10.1016/j.tcb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chendrimada TP, Gregory RI, Kumaraswamy E, Cooch N, Nishikura K, Shiekhattar R. TRBP recruites the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 48.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 49.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 50.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs Results in Target mRNA Degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 51.Iwasaki S, Tomari Y. Argonaute-mediated translational repression (and activation) Fly (Austin) 2009;3:204–206. doi: 10.4161/fly.3.3.9025. [DOI] [PubMed] [Google Scholar]
- 52.Eulalio A, Huntzinger EE. Izaurralde Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 53.Eystathioy T, Jakymiw A, Chan EKL, Séraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 56.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 57.Ding XC, Großhans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathonnet G. MicroRNA Inhibition of Translation. Science (80-) 2007;317:1764. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 59.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 60.Vasudevan S, Tong Y, Steitz JA. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 61.Yang Z. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 62.Lund E, Sheets MD, Imboden SB, Dahlberg JE. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev. 2011;25:1121–1131. doi: 10.1101/gad.2038811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu CG, Yang JM. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816–823. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Yang K, Zhou L, Minhaowu, Wu Y, Zhu M, Lai X, Chen T, Feng L, Li M, Huang C, Zhong Q, Huang X. MicroRNA-155 Promotes Autophagy to Eliminate Intracellular Mycobacteria by Targeting Rheb. PLoS Pathog. 2013;9:e1003697. doi: 10.1371/journal.ppat.1003697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao G, Zhang JG, Liu Y, Qin Q, Wang B, Tian K, Liu L, Li X, Niu Y, Deng SC, Wang CY. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKα1. Mol Cancer Ther. 2013;12:83–93. doi: 10.1158/1535-7163.MCT-12-0534-T. [DOI] [PubMed] [Google Scholar]
- 66.Pan B, Yi J, Song H. MicroRNA-mediated autophagic signaling networks and cancer chemoresistance. Cancer Biother Radiopharm. 2013;28:573–578. doi: 10.1089/cbr.2012.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ciuffreda L, Di Sanza C, Incani U, Milella M. The mTOR pathway: a new target in cancer therapy. Curr Cancer Drug Targets. 2010;10:484–495. doi: 10.2174/156800910791517172. [DOI] [PubMed] [Google Scholar]
- 68.Wu H, Wang F, Hu S, Yin C, Li X, Zhao S, Wang J, Yan X. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal. 2012;24:2179–2186. doi: 10.1016/j.cellsig.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 69.Huang Y, Chuang AY, Ratovitski EA. Phospho-ΔNp63α/miR-885-3p axis in tumor cell life and cell death upon cisplatin exposure. Cell Cycle. 2011;10:3938–3947. doi: 10.4161/cc.10.22.18107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Menghini R, Casagrande V, Marino A, Marchetti V, Cardellini M, Stoehr R, Rizza S, Martelli E, Greco S, Mauriello A, Ippoliti A, Martelli E, Lauro R, Federici M. MiR-216a: a link between endothelial dysfunction and autophagy. Cell Death Dis. 2014;5:e1029. doi: 10.1038/cddis.2013.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8:165–176. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 72.Huang Y, Guerrero-Preston R, Ratovitski EA. Phospho-ΔNp63α-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle. 2012;11:1247–1259. doi: 10.4161/cc.11.6.19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi G, Shi J, Liu K, Liu N, Wang Y, Fu Z, Ding J, Jia L, Yuan W. Increased miR-195 aggravates neuropathic pain by inhibiting autophagy following peripheral nerve injury. Glia. 2013;61:504–512. doi: 10.1002/glia.22451. [DOI] [PubMed] [Google Scholar]
- 74.Dai X, Tan C. Combination of microRNA therapeutics with small-molecule anticancer drugs: Mechanism of action and co-delivery nanocarriers. Adv Drug Deliv Rev. 2014;81:184–197. doi: 10.1016/j.addr.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 75.Xu Y, An Y, Wang Y, Zhang C, Zhang H, Huang C, Jiang H, Wang X, Li X. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;29:2019–2024. doi: 10.3892/or.2013.2338. [DOI] [PubMed] [Google Scholar]
- 76.Wang P, Zhang J, Zhang L, Zhu Z, Fan J, Chen L, Zhuang L, Luo J, Chen H, Liu L, Chen Z, Meng Z. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–1143.e12. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 77.Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F, Pirtoli L, Miracco C. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 2013;14:574–586. doi: 10.4161/cbt.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang K, Liu C, Zhou L, Wang J, Wang M, Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, Feng C, Wang CQ, Zhao YF, Li PF. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:1–11. doi: 10.1038/nature14444. [DOI] [PubMed] [Google Scholar]
- 79.Chen Y, Liersch R, Detmar M. The miR-290-295 cluster suppresses autophagic cell death of melanoma cells. Sci Rep. 2012;2:808. doi: 10.1038/srep00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang Y, Yan W, He X, Zhang L, Li C, Huang H, Nace G, Geller DA, Lin J, Tsung A. MiR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143:177–187. doi: 10.1053/j.gastro.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 81.Huang J, Sun W, Huang H, Ye J, Pan W, Zhong Y, Cheng C, You X, Liu B, Xiong L, Liu S. miR-34a modulates angiotensin II-induced myocardial hypertrophy by direct inhibition of ATG9A expression and autophagic activity. PLoS One. 2014;9:e94382. doi: 10.1371/journal.pone.0094382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kovaleva V, Mora R, Park YJ, Plass C, Chiramel AI, Bartenschlager R, Döhner H, Stilgenbauer S, Pscherer A, Lichter P, Seiffert M. miRNA-130a Targets ATG2B and DICER1 to Inhibit Autophagy and Trigger Killing of Chronic Lymphocytic Leukemia Cells. Cancer Res. 2012;72:1763–1772. doi: 10.1158/0008-5472.CAN-11-3671. [DOI] [PubMed] [Google Scholar]
- 83.Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gibbings D, Mostowy S, Voinnet O. Autophagy selectively regulates miRNA homeostasis. Autophagy. 2013;9:781–783. doi: 10.4161/auto.23694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shimomura H, Terasaki F, Hayashi T, Kitaura Y, Isomura T, Suma H. Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy. Jpn Circ J. 2001;65:965–968. doi: 10.1253/jcj.65.965. [DOI] [PubMed] [Google Scholar]
- 86.Bartz RR, Suliman HB, Piantadosi CA. Redox mechanisms of cardiomyocyte mitochondrial protection. Front Physiol. 2015;6:1–8. doi: 10.3389/fphys.2015.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nemchenko A, Chiong M, Turer A, Lavandero S, Hill JA. Autophagy as a therapeutic target in cardiovascular disease. J Mol Cell Cardiol. 2011;51:584–593. doi: 10.1016/j.yjmcc.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Selvetella G. Adaptive and maladaptive hypertrophic pathways: points of convergence and divergence. Cardiovasc Res. 2004;63:373–380. doi: 10.1016/j.cardiores.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 89.Tham YK, Bernardo BC, Ooi JYY, Weeks KL, McMullen JR. Pathophysiology of cardiac hypertrophy and heart failure: signaling pathways and novel therapeutic targets. Arch Toxicol. 2015:1401–1438. doi: 10.1007/s00204-015-1477-x. [DOI] [PubMed] [Google Scholar]
- 90.Schiattarella GG, Hill JA. Therapeutic targeting of autophagy in cardiovascular disease. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martinet W, Knaapen MWM, Kockx MM, De Meyer GRY. Autophagy in cardiovascular disease. Trends Mol Med. 2007;13:482–491. doi: 10.1016/j.molmed.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 92.Xie M, Morales CR, Lavandero S, Hill JA. Tuning flux: autophagy as a target of heart disease therapy. Curr Opin Cardiol. 2011;26:216–222. doi: 10.1097/HCO.0b013e328345980a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang DZ. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 95.Yin X, Peng C, Ning W, Li C, Ren Z, Zhang J, Gao H, Zhao K. miR-30a downregulation aggravates pressure overload-induced cardiomyocyte hypertrophy. Mol Cell Biochem. 2013;379:1–6. doi: 10.1007/s11010-012-1552-z. [DOI] [PubMed] [Google Scholar]
- 96.Pan W, Zhong Y, Cheng C, Liu B, Wang L, Li A, Xiong L, Liu S. MiR-30-Regulated Autophagy Mediates Angiotensin II-Induced Myocardial Hypertrophy. PLoS One. 2013;8:1–14. doi: 10.1371/journal.pone.0053950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Tréguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Billiczki P, Ehrlich JR, Katus HA, Müller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 98.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A, Remke J, Caprio M, Jentzsch C, Engelhardt S, Geisendorf S, Glas C, Hofmann TG, Nessling M, Richter K, Schiffer M, Carrier L, Napp LC, Bauersachs J, Chowdhury K, Thum T. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Su M, Wang J, Wang C, Wang X, Dong W, Qiu W, Wang Y, Zhao X, Zou Y, Song L, Zhang L, Hui R. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ. 2015;22:986–999. doi: 10.1038/cdd.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Song L, Su M, Wang S, Zou Y, Wang X, Cui H, Zhao P, Hui R, Wang J. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J Cell Mol Med. 2014;18(11):1–9. doi: 10.1111/jcmm.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Buller C, Loberg R, Fan M, Zhu Q, Park JL, Vesely E, Inoki K, Guan KL, Brosius FC., 3rd A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. Am J Phsyiol Cell Phsyiol. 2008;295(3):836–843. doi: 10.1152/ajpcell.00554.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li Q, Xie J, Li R, Shi J, Sun J, Gu R, Ding L, Wang L, Xu B. Overexpression of microRNA-99a attenuates heart remodelling and improves cardiac performance after myocardial infarction. J Cell Mol Med. 2014;18:919–928. doi: 10.1111/jcmm.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu S, Chen S, Li M, Zhang B, Shen P, Liu P, Zheng D, Chen Y, Jiang J. Autophagy activation attenuates angiotensin II-induced cardiac fibrosis. Arch Biochem Biophys. 2016;590:37–47. doi: 10.1016/j.abb.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 104.Singh KK, Lovren F, Pan Y, Quan A, Ramadan A, Matkar PN, Ehsan M, Sandhu P, Mantella LE, Gupta N, Teoh H, Parotto M, Tabuchi A, Kuebler WM, Al-Omran M, Finkel T, Verma S. The essential autophagy gene ATG7 modulates organ fibrosis via regulation of endothelial-to-mesenchymal transition. J Biol Chem. 2015;290:2547–2559. doi: 10.1074/jbc.M114.604603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 106.Pan Z, Sun X, Shan H, Wang N, Wang J, Ren J, Feng S, Xie L, Lu C, Yuan Y, Zhang Y, Wang Y, Lu Y, Yang B. miR-101 Inhibited Post-Infarct Cardiac Fibrosis and Improved Left Ventricular Compliance via FOS/TGFβ1 Pathway. Circulation. 2012:840–850. doi: 10.1161/CIRCULATIONAHA.112.094524. [DOI] [PubMed] [Google Scholar]
- 107.Wu D, Jiang H, Chen S, Zhang H. Inhibition of microRNA-101 attenuates hypoxia/reoxygenation-induced apoptosis through induction of autophagy in H9c2 cardiomyocytes. Mol Med Rep. 2015;11:3988–3994. doi: 10.3892/mmr.2015.3215. [DOI] [PubMed] [Google Scholar]
- 108.Pagidipati NJ, Gaziano TA. Estimating Deaths From Cardiovascular Disease: A Review of Global Methodologies of Mortality Measurement. Circulation. 2013;127:749–756. doi: 10.1161/CIRCULATIONAHA.112.128413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao Z, Vinten-Johansen J. Myocardial apoptosis and ischemic preconditioning. Cardiovasc Res. 2002;55:438–455. doi: 10.1016/S0008-6363(02)00442-X. [DOI] [PubMed] [Google Scholar]
- 110.Pierce GN, Czubryt MP. The Contribution of Ionic Imbalance to Ischemia Reperfusion-Induced Injury. J Mol Cell Cardiol. 1995;27:53–63. doi: 10.1016/S0022-2828(08)80007-7. [DOI] [PubMed] [Google Scholar]
- 111.Schafer C, Ladilov YV, Siegmund B, Piper HM. Importance of bicarbonate transport for protection of cardiomyocytes against reoxygenation injury. Am J Hear Circ Physiol. 2000;278:1457–1463. doi: 10.1152/ajpheart.2000.278.5.H1457. [DOI] [PubMed] [Google Scholar]
- 112.Bonora M, Pinton P. The mitochondrial permeability transition pore and cancer: molecular mechanisms involved in cell death. Front Oncol. 2014;4:302. doi: 10.3389/fonc.2014.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li J, Sagar R, Gelber N, Rutka J, Sabah N, Gladstone RA, Wei C, Hu P, Kharbanda RK, Redington AN. MicroRNA-144 is a circulating effector of remote ischemic preconditioning. Basic Res Cardiol. 2014;109:1–15. doi: 10.1007/s00395-014-0423-z. [DOI] [PubMed] [Google Scholar]
- 114.Li X, Zeng Z, Li Q, Xu Q, Xie J, Hao H, Luo G, Liao W, Bin J, Huang X, Liao Y. Inhibition of microRNA-497 ameliorates anoxia/reoxygenation injury in cardiomyocytes by suppressing cell apoptosis and enhancing autophagy. Oncotarget. 2015;6:18829–18844. doi: 10.18632/oncotarget.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xiao J, Zhu X, He B, Zhang Y, Kang B, Wang Z, Ni X. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J Biomed Sci. 2011;18:35. doi: 10.1186/1423-0127-18-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cardiovascular Disease and Diabetes. Am Hear Assoc. 2015 [Google Scholar]
- 117.Greco S, Fasanaro P, Castelvecchio S, D’Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC, Menicanti L, Martelli F. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes. 2012;61:1633–1641. doi: 10.2337/db11-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Diao X, Shen E, Wang X, Hu B. Differentially expressed microRNAs and their target genes in the hearts of streptozotocin-induced diabetic mice. Mol Med Rep. 2011;4:633–640. doi: 10.3892/mmr.2011.489. [DOI] [PubMed] [Google Scholar]
- 119.Feng B, Chen S, George B, Feng Q, Chakrabarti S. miR133a regulates cardiomyocyte hypertrophy in diabetes. Diabetes Metab Res Rev. 2010;26:40–49. doi: 10.1002/dmrr.1054. [DOI] [PubMed] [Google Scholar]
- 120.Nandi SS, Duryee MJ, Shahshahan HR, Thiele GM, Daniel R. Induction of autophagy markers is associated with attenuation of miR-133a in diabetic heart failure patients undergoing mechanical unloading. Am J Transl Res. 2015;7:683–696. [PMC free article] [PubMed] [Google Scholar]
- 121.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 122.Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D’Andrea A, Vindigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-Dependent Responses in miR-34–Deficient Mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xie Z, Lau K, Eby B, Lozano P, He C, Pennington B, Li H, Rathi S, Dong Y, Tian R, Kem D, Zou MH. Improvement of Cardiac Functions by Chronic Metformin Treatment Is Associated With Enhanced Cardiac Autophagy in Diabetic OVE26 Mice. Diabetes. 2011;60:1770–1778. doi: 10.2337/db10-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ross R. The pathogenesis of atherosclerosis: A perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 125.Wang P, Liang J, Li Y, Li J, Yang X, Zhang X, Han S, Li S, Li J. Down-regulation of miRNA-30a alleviates cerebral ischemic injury through enhancing beclin 1-mediated autophagy. Neurochem Res. 2014;39:1279–1291. doi: 10.1007/s11064-014-1310-6. [DOI] [PubMed] [Google Scholar]
- 126.Tao J, Liu W, Shang G, Zheng Y, Huang J, Lin R, Chen L. MiR-207/352 regulate lysosomal-associated membrane proteins and enzymes following ischemic stroke. Neuroscience. 2015;305:1–14. doi: 10.1016/j.neuroscience.2015.07.064. [DOI] [PubMed] [Google Scholar]
- 127.Yuan Y, Zhao J, Yan S, Wang D, Zhang S, Yun F, Zhao H, Sun L, Liu G, Ding X, Liu L, Li Y. Autophagy: A potential novel mechanistic contributor to atrial fibrillation. Int J Cardiol. 2014;172:492–494. doi: 10.1016/j.ijcard.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 128.Deroyer C, Magne J, Moonen M, Le Goff C, Dupont L, Hulin A, Radermecker M, Colige A, Cavalier E, Kolh P, Pierard L, Lancellotti P, Merville MP, Fillet M. New biomarkers for primary mitral regurgitation. Clin Proteomics. 2015;12:25. doi: 10.1186/s12014-015-9097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]