Graphical abstract
Keywords: Alcohol, Green tea extract, Antioxidant, ROS, Mitochondrial DNA, D-loop
Abstract
The present study principally sought to investigate the effect of green tea extract (GTE) supplementation on hepatic mitochondrial DNA (mtDNA) damage in alcohol receiving rats. MtDNA was isolated from hepatic tissues of albino wistar rats after alcohol treatment with and without GTE supplementation. Entire displacement loop (D-loop) of mtDNA was screened by PCR-Sanger’s sequencing method. In addition, mtDNA deletions and antioxidant activity were measured in hepatic tissue of all rats. Results showed increased frequency of D-loop mutations in alcoholic rats (ALC). DNA mfold analysis predicted higher free energy for 15507C and 16116C alleles compared to their corresponding wild alleles which represents less stable secondary structures with negative impact on overall mtDNA function. Interestingly, D-loop mutations observed in ALC rats were successfully restored on GTE supplementation. MtDNA deletions were observed in ALC rats, but intact native mtDNA was found in ALC + GTE group suggesting alcohol induced oxidative damage of mtDNA and ameliorative effect of GTE. Furthermore, markedly decreased activities of glutathione peroxidise, superoxide dismutase, catalase and glutathione content were identified in ALC rats; however, GTE supplementation significantly (P < 0.05) restored these levels close to normal. In conclusion, green tea could be used as an effective nutraceutical against alcohol induced mitochondrial DNA damage.
Introduction
Alcohol (ethanol) is a commonly abused psychoactive drug affecting diverse cellular and molecular processes in the liver and other organs of the body with no exception [1] As per the reports of World health organization (2014) there are nearly three billion alcoholics worldwide now and chronic excessive alcohol consumption is the third leading cause of global deaths accounting for 6% of the total deaths. Harmful use of alcohol is an important cause of mortality and morbidity associated with a number of diseases with multiple pathologies, such as malnutrition, gastritis, chronic pancreatitis, cardiomyopathy, alcoholic liver disease (ALD) and cancers of all organs leading to death [2], [3]. Elevated oxidative stress due to the excessive liberation of reactive oxygen species (ROS) in ethanol metabolism affects the antioxidant defense system leading to various diseases including cancer [4], [5].
Mitochondria are highly dynamic and energy transducing cell organelles playing a key role in cellular ATP generation via oxidative phosphorylation [6]. In addition, mitochondria involved in antioxidant defense system, fat oxidation, intermediary metabolic processes which includes alcohol metabolism and bioenergetics of the hepatocytes [7]. Ethanol induced hepatotoxicity often exhibits mitochondrial dysfunction associated with mitochondrial DNA (mtDNA) damage [8]. Hepatic mitochondria are more susceptible for alcoholic damage as 90% of ingested alcohol is metabolized here [9] producing its metabolites and free radicals which in turn lead to damage of several biomolecules including mtDNA.
Mitochondrial genome is a double-stranded, closed-circular DNA molecule of 16.5 kb in size (16.313 kb in rats) and encodes for 13 essential subunits of the respiratory chain complexes along with 2 ribosomal and 22 transfer rRNAs [10]. The mutation rate of mtDNA is higher than nuclear DNA due to the presence of limited DNA repair mechanisms and lack of associated histones. Displacement loop (D-loop), the only regulatory site of mitochondrial genome, is a hot spot for mtDNA mutations providing a unique opportunity to investigate the ethanol-induced hepatic mtDNA damage for which therapeutic strategy is sought [11].
Polyphenols exert a broad spectrum of therapeutic health effects against various chronic pathological conditions and diseases associated with oxidative stress such as ALD, cancer, neurodegenerative diseases, diabetes, and cardiovascular diseases [12]. Green tea (Camellia sinensis L.), a widely used beverage is rich in polyphenols. As compared to conventional pharmaceutical drugs, the ‘biosafety’ of green tea constituents, in particular, catechins are considerably higher and can more easily be incorporated into lifestyle changes [12]. Hence, polyphenols of green tea have become a nucleus of scientific interest targeted for developing novel therapeutic agents. Earlier studies suggested the protective effect of green tea catechins as effective scavengers of ROS, a key factor of mtDNA damage [13], [14]. So far no information is available on the protective effect of green tea on alcohol induced mitochondrial DNA damage. The present study is an attempt to investigate the effect of green tea supplementation on hepatic mtDNA damage in alcohol receiving rats with a view to recommend the same for therapeutic purpose.
Material and methods
All the chemicals and reagents used in the current study were purchased from Sigma-Aldrich chemical Co. (St. Louis, MO, USA) and SRL chemicals (Mumbai, India). Aqueous green tea leaf extract dry powder (extract contains 75% catechins with 50% EGCG) was obtained from Guardian Biosciences, Phoenix, Arizona, USA.
Animals
Albino wistar rats weighing 120–140 g procured from Sri Venkateswara Agencies, Bangalore, India, were maintained on a standard pellet diet (M/s. Hindustan Lever Ltd., Mumbai, India) and water ad libitum with 24 h light-dark cycle in the university animal house. After acclimatization for a week, animals were divided into four groups (n = 8) viz., group-I control (C), group-II alcohol (ALC), group-III green tea extract supplemented (GTE) and group-IV alcoholic rats with green tea extract supplementation (ALC + GTE). Alcohol (20%) was administered at a dose of 5 g/kg b.wt/day and GTE was administered at a dose of 300 mg/kg b.wt/day for 60 days. Experimentation and animal maintenance were done with prior approval of institutional animal ethical committee (Registered No: 1889/GO/Re/S/16/CPCSEA; F.No: 25/30/2015-CPCSEA, dated 30-05-2016). Animals of all experimental groups were fasted overnight and sacrificed by cervical dislocation at the end of 60 days period. Livers were collected and used for experimentation.
Isolation of total DNA
Total DNA was extracted from frozen liver tissues by using proteinase K and sodium dodecyl sulfate (SDS) as per the methods described previously [15]. DNA was quantified by Biophotometer (Eppendorf) using absorbance at 260 nm. The extract containing both nuclear DNA and mtDNA, was used for PCR and sequencing analysis without further purification.
Comprehensive screening of mtDNA D-loop
The entire mitochondrial D-loop region (np15416-16313) was screened by PCR-Sanger’s sequencing analysis using specific primers (Table 1) as described earlier [16]. PCR amplicons of 432 bp (primer set 1) and 519 bp were subjected to gel-purification and sequences were obtained by direct sequencing technique using an automated DNA-sequencer (Applied BioSystems, USA).
Table 1.
Primers used for PCR-Sanger’s sequencing analysis of mtDNA D-loop.
| S. no. | Primer sequences | NT location | Amplicon (bp) |
|---|---|---|---|
| 1 | F: 5′-CACCATCAACACCCAAAGC-3′ | 15358-15376 | 432 bp |
| R: 5′-GGCCCTGAAGTAAGAACCA-3′ | 15771-15789 | ||
| 2 | F: 5′-GGTTCTTACTTCAGGGCCATC-3′ | 15772-15792 | 519 bp |
| R: 5′-GTGGAATTTTCTGAGGGTAGGC-3′ | 16269-16290 | ||
For mutational analysis, the mtDNA sequence of all experimental animals was compared with the reference mtDNA sequence (wistar rat strain BBDP/Rhw; Acc. No. FJ919760). Sequences were aligned using CLUSTAL-X software and mutations were scored as described earlier [17]. Impact of identified mutations on D-loop secondary structures was assessed by DNA mfold web server.
Determination of mtDNA deletions
MtDNA deletions were analyzed by PCR method as described earlier [18] using specific primers (Table 2). Whole mtDNA genome was amplified by long extension PCR using Expand Long Template PCR system (Roche). Whole mitochondrial genome was amplified using 25 cycles of primary PCR followed by nested PCR. The 1st primers set (primary PCR) amplifies mtDNA fragment of 16,007 bp size while the 2nd primers set (nested PCR) amplifies a 15,708 bp fragment. The quality of PCR amplification products was analyzed by agarose gel electrophoresis.
Table 2.
Primers and PCR conditions used for mtDNA deletion analysis.
| Primer set | Primer sequences | NT location | Amplicon size (bp) | PCR conditions |
|---|---|---|---|---|
| 1 | F: 5′-CCATCCTCCGTGAAATCAACAACCCG-3′ | 15671-15696 | 16,007 bp | 93 °C for 15 s, 62 °C for 30 s, 68 °C for 15 min, 25 cycles |
| R: 5′-CTTTGGGTGTTGATGGTGGGGAGGTAG-3′ | 15377-15350 | |||
| 2 | F: 5′-AAGACATCTCGATGGTAACGGGTC-3′ | 15826-15849 | 15,708 bp | |
| R: 5′-CCAGAGATTGGTATGAGAATGAGG-3′ | 15233-15209 |
Activity of liver antioxidants
Liver tissue was homogenized (10% w/v) in ice cold 0.1 M Tris buffer (pH 7.4), and supernatant was collected by centrifugation (10,000g for 20 min at 4 °C) and used to assess the activities of enzymatic and non-enzymatic antioxidants. Total glutathione (GSH) content was measured by Ellman’s method [19] and the activities of glutathione peroxidise (GPx) [20], catalase [21] and superoxide dismutase (SOD) [22] were determined. Protein concentration was estimated by standard protocols [23].
Results
Mitochondrial DNA D-loop mutations
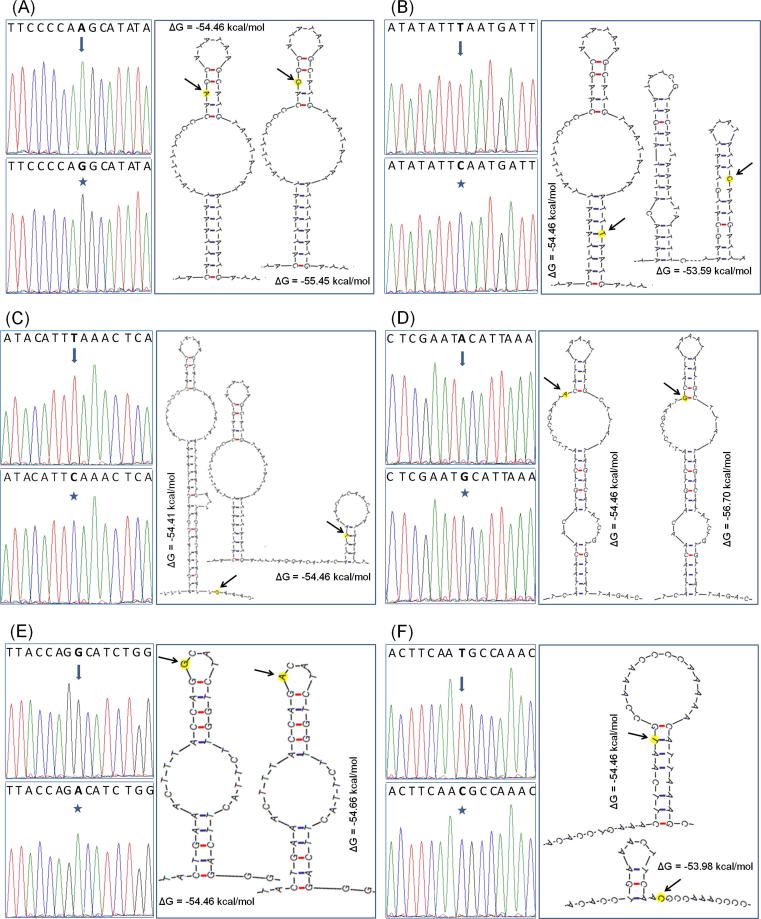
A total of 6 mutations were identified in the D-loop region of investigated groups (Table 3; Fig. 1). All the identified mutations were transition substitutions of purines (Y) or pyrimidines (R). Among them, 4 were present in alcoholic rats (ALC) while remaining 2 were present in all experimental groups viz., C, ALC, GTE and ALC + GTE groups. In overall, 4 mutations were present in the termination associated sequences (TAS, ETAS), 1 was in the central block (CB) and 1 was located in conserved sequence block 3 (MT-CSB3).
Table 3.
Mitochondrial DNA D-loop mutations observed in the present study.
| Locus (position in D-loop) | Nucleotide position | Ref sequence | Base change | IUPAC code | Status |
|||
|---|---|---|---|---|---|---|---|---|
| C | AL | GT | AG | |||||
| ETAS1 (15446-15503) | 15483 | A | G | R | ||||
| TAS-D (15497-15511) | 15507 | T | C | Y | × | × | × | |
| TAS-C (15520-15531) | 15529 | T | C | Y | × | × | × | |
| TAS-A (15571-15584) | 15572 | A | G | R | ||||
| CB (15673-15979) | 15779 | G | A | R | × | × | × | |
| MT-CSB3(16116-16133) | 16116 | T | C | Y | × | × | × | |
ETAS: Extended Termination-associated sequence; TAS: Termination associated sequence; CB: Central Block; MT-CSB: Conserved sequence block; IUPAC: International Union of Pure and Applied Chemistry; C: Control rats; AL: Alcoholic rats; GT: Green Tea Extract supplemented rats; and AG: Alcoholic rats with Green Tea Extract supplementation.
Fig. 1.
Mitochondrial DNA D-loop mutations identified in the present study: Chromatogram of sequence analysis and consequent secondary structure alterations are shown. (A) ETAS1 15483 A/G; (B) TAS-D 15507 T/C; (C) TAS-C 15529 T/C; (D) TAS-A 15572 A/G; (E) CB 15779 G/A; and (F) MT-CSB3 16116 T/C.
Effect of mutations on secondary structure of D-loop
To find out the impact of D-loop mutations on its secondary structure conformation, in silico analysis was performed using DNA mfold web server (Fig. 1). Results showed lesser free energy for 15483G (ETAS1), 15572G (TAS-A) alleles and higher free energy for 15507C (TAS-D), 16116C (MT-CSB3) alleles when compared to their corresponding wild alleles (Fig. 1). However, for 15529 T/C (TAS-C) and 15779 G/A (CB) variants no considerable difference was observed in free energy levels between wild and mutant alleles.
Mitochondrial DNA deletions
Whole mitochondrial genome from all the investigated groups was analyzed by Long-extension PCR technique. Large scale mtDNA deletions were observed only in alcoholic (ALC) rats while intact wild type mtDNA was observed in rats of C, GTE and ALC + GTE groups (Fig 2).
Fig. 2.
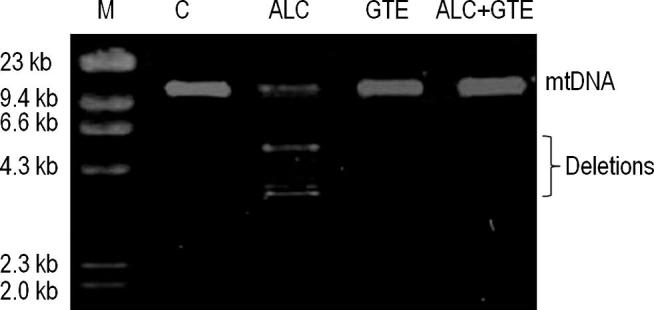
Long-extension PCR analysis of mtDNA deletions in hepatic tissue of experimental rats: M: DNA size marker; C: Controls; ALC: Alcohol; and GTE: Green tea extract.
Activity of liver antioxidants
The data on the effects of green tea extract on liver antioxidants in alcohol administered rats are summarized in Table 4. The activities of antioxidant enzymes viz., GPx, SOD, catalase and the content of GSH were markedly decreased in alcohol administered rats in comparison with the other experimental groups. Treatment of green tea extract to alcohol administered rats significantly (P < 0.05) restored these levels close to normal levels.
Table 4.
Effect of green tea extract on antioxidant enzymes and glutathione content of liver in alcohol administered rats.
| Parameter | C | ALC | GTE | ALC + GTE |
|---|---|---|---|---|
| GSH | 6.2 ± 0.29a | 3.4 ± 0.16b | 6.7 ± 0.43a | 5.9 ± 0.33a |
| GPx | 9.2 ± 0.24a | 5.6 ± 1.3b | 9.9 ± 0.4a | 8.6 ± 0.7a |
| SOD | 34 ± 3.8a | 23 ± 3.3b | 36 ± 2.1a | 31 ± 4.5b |
| CAT | 5.4 ± 0.19a | 3.6 ± 0.38b | 5.8 ± 0.21a | 5.1 ± 0.15a |
GSH is expressed as µg/mg protein and remaining values as µmole/min/mg protein. Values are mean ± SD of eight rats in each group. a,b Within a row, means not sharing a common superscript letter are significantly different at P < 0.05 (Tukey HSD method post hoc analysis for all groups, P < 0.01). C: Control rats; ALC: Alcohol fed rats; GTE: Green tea extract fed rats; and ALC + GTE: Alcohol and green tea extract fed rats.
Discussion
Green tea has many bioactive components, chiefly catechins viz., epigallocatechingallate (EGCG), epigallocatechin (EGC), epicatechingallate (ECG), and epicatechin (EC) along with other constituents such as caffeine, theobromine, theophylline, organic acids, free amino acids, carbohydrates, alkaloids and minerals [24]. The antioxidant activity of green tea polyphenols was primarily attributed to catechins. However, polyphenols are highly target specific with different efficacies and bio-availabilities. Earlier studies have shown that green tea catechins are effective scavengers of ROS including superoxide anions [14]. Thus, by lowering the levels of ROS and oxidative stress, green tea catechins may ameliorate mtDNA damage, and at the same time, the possibility of involvement of several other mechanisms related to beneficiary actions of catechins cannot be ruled out.
Mitochondrial DNA D-loop, the key regulating site of mtDNA function, is highly vulnerable to oxidative damage [25]. Thus, D-loop mutations might affect the overall mitochondrial function by altering mitochondrial replication, transcription and/or biogenesis. Numerous studies have reported association between D-loop mutations and risk of developing various complex diseases [26], [27], [28]. The present study reports increased frequency of D-loop mutations in ALC group rats (Table 3). Alcohol metabolism linked production of ROS might be responsible for this enhancement. However, alcoholic rats supplemented with green tea extract (ALC + GTE) showed no D-loop mutations that were observed in ALC group (Table 3). This could be due to the effective ROS scavenging nature of green tea catechins.
It is evident that DNA secondary structures can influence the molecular mechanisms of replication, transcription and recombination [29], [30]. In general, hairpin or cruciform structures serve as binding sites for several transacting elements [31], [32]. Hence, local intra-strand DNA secondary structures have a key role in replication and transcription processes. As key regulatory site of mtDNA replication and transcription, D-loop mutations can influence overall mtDNA stability. Therefore, impact of identified mutations on D-loop secondary structure was analyzed. Results showed higher free energy for 15507C (TAS-D) and 16116C (MT-CSB3) alleles compared to their corresponding wild alleles (Fig. 1). Higher free energy represents less stable secondary structures which may have negative impact on overall mtDNA function. The 15507C (TAS-D) and 16116C (MT-CSB3) variants observed in alcoholic rats were not present in alcoholic rats supplemented with GTE, indicating ameliorative effect of green tea. However, further studies are warranted to clarify the underlying molecular mechanisms involved in these findings.
DNA mfold analysis predicted lesser free energy for 15483G (ETAS1) and 15572G (TAS-A) alleles when compared to their corresponding wild alleles (Fig. 1). Lesser free energy represents more stable secondary structures. Interestingly, both of these variants were present in all groups of rats; hence, they can be considered as single nucleotide polymorphisms rather than mutations. The remaining 2 variants observed in alcoholic rats [15529 T/C (TAS-C) and 15779 G/A (CB)] showed no much difference in free energy levels (Fig. 1) and were restored by GTE treatment.
Oxidative stress can lead to the accumulation of mtDNA deletions [33], [34]. Large scale deletions of mitochondrial genome have been reported in several complex diseases including diabetes [26], [27]. Altered mtDNA replication and/or repair system could lead deletions in mtDNA [35], [36]. The present study identified mtDNA deletions in alcoholic rats while ALC + GTE group rats showed no detectable mtDNA deletions (Fig. 2). This could be attributed to elevated oxidative stress by alcohol induced ROS in ALC group and ameliorative effect of green tea catechins on mtDNA damage by ROS scavenging nature in ALC + GTE group. Although this is an interesting finding, further studies are warranted to clarify the underlying molecular mechanisms.
SOD, CAT and GPx are the major antioxidant enzymes that stand in the first-line of defense against oxidative damage [37]. These antioxidants play a key role in scavenging ROS, reduction in hydrogen peroxide and maintaining redox balances in biological system. GSH, an important non-enzymatic antioxidant biomolecule in tissues, is the substrate for GPx and GST. It plays a central role in the maintenance of membrane protein thiols and elimination of free oxygen species, such superoxide anions, alkoxy radicals including H2O2 [38]. The present study showed diminished activities of SOD, CAT and GPx and reduced GSH content in alcohol administered rats (Table 4). The lowered GSH content might be responsible for the reduced GPx activity. Decreased catalase activity accounts for less hydrogen peroxide decomposition, consequently the possible overproduction of hydroxyl radicals via fenton reaction. Decreased GSH content and lowered activity of catalase, SOD and GPx favor the environment for oxidative stress, which leads to mtDNA damage. Amelioration of mtDNA damage and restoration of antioxidant status in terms of GSH content and activities of defense enzymes to normal level in alcoholic rats receiving GTE supplementation are evident from the results of the study. This finding confirms the reports of Lodhi et al. [39] and others who reported such GTE induced restorative effect in antioxidant status in alcohol receiving rats.
Conclusions
The present study reports therapeutic effect of green tea extract against alcohol induced hepatic mitochondrial DNA damage in rats. To the best of our knowledge, this is the first report demonstrating the ameliorative effect of green tea extract on alcohol mediated mtDNA damage. However, further investigation is warranted to explore the molecular mechanisms involved in the reported findings.
Conflict of interest
The authors have declared no conflict of interest.
Acknowledgments
Dr. Suresh Govatati acknowledges the financial support from the University Grants Commission, New Delhi, under its Dr. D.S. Kothari postdoctoral scheme [No. F.4-2/2006 (BSR)/13-1014/2013 (BSR)].
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Wang Z., Su B., Fan S., Fei H., Zhao W. Protective effect of oligomeric proanthocyanidins against alcohol-induced liver steatosis and injury in mice. Biochem Biophys Res Commun. 2015;458:757–762. doi: 10.1016/j.bbrc.2015.01.153. [DOI] [PubMed] [Google Scholar]
- 2.Adjemian M.K., Volpe R.J., Adjemian J. relationships between diet, alcohol preference, and heart disease and type 2 diabetes among Americans. Plos One. 2015;11:e0124351. doi: 10.1371/journal.pone.0124351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louvet A., Mathurin P. Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat Rev Gastroenterol Hepatol. 2015;4:231–242. doi: 10.1038/nrgastro.2015.35. [DOI] [PubMed] [Google Scholar]
- 4.Pyun C.W., Mandal P.K., Hong G.E., Lee C.H. Effect of chronic alcohol consumption on phosphatidylcholine hydroperoxide content of rat liver and brain. Trop J Pharm Res. 2015;7:1225–1230. [Google Scholar]
- 5.Chuang S.C., Lee Y.C., Wu G.J., Straif K., Hashibe M. Alcohol consumption and liver cancer risk: a meta-analysis. Cancer Causes Control. 2015;26:1205–1231. doi: 10.1007/s10552-015-0615-3. [DOI] [PubMed] [Google Scholar]
- 6.Yin F., Cadenas E. Mitochondria: the cellular hub of the dynamic coordinated network. Antioxid Redox Signal. 2015;12:961–964. doi: 10.1089/ars.2015.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song B.J., Akbar M., Abdelmegeed M.A., Byun K., Lee B., Yoon S.K. Mitochondrial dysfunction and tissue injury by alcohol, high fat, nonalcoholic substances and pathological conditions through post-translational protein modifications. Redox Biol. 2014;3:109–123. doi: 10.1016/j.redox.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zelickson B.R., Benavides G.A., Johnson M.S., Chacko B.K., Venkatraman A., Landar A. Nitric oxide and hypoxia exacerbate alcohol-induced mitochondrial dysfunction in hepatocytes. Biochim Biophys Acta. 2011;12:1573–1582. doi: 10.1016/j.bbabio.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han D., Ybanez M.D., Johnson H.S., McDonald J.N., Mesropyan L., Sancheti H. Dynamic adaptation of liver mitochondria to chronic alcohol feeding in mice biogenesis, remodeling, and functional alterations. J Biol Chem. 2012;50:42165–42179. doi: 10.1074/jbc.M112.377374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson S., Bankier A.T., Barrell B.G., De Bruijn M.H., Coulson A.R., Drouin J. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 11.Nassir F., Ibdah J.A. Role of mitochondria in alcoholic liver disease. World J Gastroenterol. 2014;9:2136–2142. doi: 10.3748/wjg.v20.i9.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H.S., Quon M.J., Kim J.A. New insights into the mechanisms of polyphenols beyond antioxidant properties; lessons from the green tea polyphenol, epigallocatechin 3-gallate. Redox Biol. 2014;2:187–195. doi: 10.1016/j.redox.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Q., Zhao B., Shen S., Hou J., Hu J., Xin W. ESR study on the structure–antioxidant activity relationship of tea catechins and their epimers. Biochim Biophys Acta. 1999;1427:13–23. doi: 10.1016/s0304-4165(98)00168-8. [DOI] [PubMed] [Google Scholar]
- 14.Velayutham P., Babu A., Liu D. Green tea catechins and cardiovascular health: an update. Curr Med Chem. 2008;15:1840–1850. doi: 10.2174/092986708785132979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govatati S., Singamsetty G.K., Nallabelli N., Malempati S., Rao P.S., Madamchetty V.K.K. Contribution of cyclin D1 (CCND1) and E-cadherin (CDH1) alterations to colorectal cancer susceptibility: a case–control study. Tumor Biol. 2014;35:12059–12067. doi: 10.1007/s13277-014-2505-9. [DOI] [PubMed] [Google Scholar]
- 16.Govatati S., Challa K., Reddy S.B., Pramod K., Deenadayal M., Chakravarty B. BRCA1 alterations are associated with endometriosis, but BRCA2 alterations show no detectable endometriosis risk: a study in Indian population. J Assist Reprod Genet. 2015;2:277–285. doi: 10.1007/s10815-014-0379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govatati S., Tipirisetti N.R., Perugu S., Kodati V.L., Deenadayal M., Satti V. Mitochondrial genome variations in advanced stage endometriosis: a study in South Indian population. Plos One. 2012;7:e40668. doi: 10.1371/journal.pone.0040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govatati S., Malempati S., Saradamma B., Divyamaanasa D., Naidu B.P., Bramhachari P.V. Manganese-superoxide dismutase (Mn-SOD) overexpression is a common event in colorectal cancers with mitochondrial microsatellite instability. Tumor Biol. 2016;37:10357–10364. doi: 10.1007/s13277-016-4918-0. [DOI] [PubMed] [Google Scholar]
- 19.Ellman's Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 20.Rotruck J.T., Pope A.L., Ganther H.E., Swanson A.B., Hafeman D.G., Hoekstra W.G. Selenium: biochemical role as a component of glutathione peroxidase. Science. 1973;179:588–590. doi: 10.1126/science.179.4073.588. [DOI] [PubMed] [Google Scholar]
- 21.Aebi H. Catalase in vitro. Meth Enzymol. 1984;105:21–26. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 22.Marklund S., Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 23.Lowry O.H., Nira J.R., Farr L., Rose J.R. Protein measurement with the Follin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 24.Chaturvedula V.S., Prakash I. The aroma, taste, color and bioactive constituents of tea. J Med Plants Res. 2011;5:2110–2124. [Google Scholar]
- 25.Tan X., Lei Z., Jiang Y., Yang Y., Zhang W., Li Y. Post conditioning ameliorates mitochondrial DNA damage and deletion after renal ischemic injury. Nephrol Dial Transplant. 2013;28:2754–2765. doi: 10.1093/ndt/gft278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tipirisetti N.R., Govatati S., Pullari P., Malempati S., Thupurani M.K., Perugu S. Mitochondrial control region alterations and breast cancer risk: a study in South Indian population. Plos One. 2014;1:e85363. doi: 10.1371/journal.pone.0085363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Govatati S., Saradamma B., Malempati S., Dasi D., Thupurani M.K., Nagesh N. Association of mitochondrial displacement loop polymorphisms with risk of colorectal cancer in south Indian population. Mitochond DNA A DNA Mapp Seq Anal. 2016 doi: 10.3109/24701394.2016.1160076. [DOI] [PubMed] [Google Scholar]
- 28.Govatati S., Deenadayal M., Shivaji S., Bhanoori M. Mitochondrial displacement loop alterations are associated with endometriosis. Fertil Steril. 2013;7:1980–1986. doi: 10.1016/j.fertnstert.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Seffens W., Digby D. MRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucl Acids Res. 1999;27:1578–1584. doi: 10.1093/nar/27.7.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katz L., Burge C.B. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003;13:2042–2051. doi: 10.1101/gr.1257503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright B.E., Reschke D.K., Schmidt K.H., Reimers J.M., Knight W. Predicting mutation frequencies in stem-loop structures of derepressed genes: implications for evolution. Mol Microbiol. 2003;48:429–441. doi: 10.1046/j.1365-2958.2003.t01-1-03436.x. [DOI] [PubMed] [Google Scholar]
- 32.Hoede C., Denamur E., Tenaillon O. Selection acts on DNA secondary structures to decrease transcriptional mutagenesis. Plos Genet. 2006;2:e176. doi: 10.1371/journal.pgen.0020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikushima T., Andoh T., Kaikawa T., Hashiguchi K. Induction of a large deletion in mitochondrial genome of mouse cells induced by X-ray irradiation. Int Congr. 2002;1236:331–334. [Google Scholar]
- 34.Murphy J., Nugent S., Seymoura C., Mothersill C. Mitochondrial DNA point mutations and a novel deletion induced by direct low-LET radiation and by medium from irradiated cells. Mutat Res. 2005;12:127–136. doi: 10.1016/j.mrgentox.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Krishnan K.J., Reeve A.K., Samuels D.C., Chinnery P.F., Blackwood J.K., Taylor R.W. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;3:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 36.Sadikovic B., Wang J., El-Hattab A., Landsverk M., Douglas G., Brundage E.K. Sequence homology at the breakpoint and clinical phenotype of mitochondrial DNA deletion syndromes. Plos One. 2010;12:e15687. doi: 10.1371/journal.pone.0015687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maturu P., Reddy V.D., Padmavathi P., Varadacharyulu N. Ethanol induced adaptive changes in blood for the pathological and toxicological effects of chronic ethanol consumption in humans. Exp Toxicol Pathol. 2012;64:697–703. doi: 10.1016/j.etp.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 38.Padmavathi P., Reddy V.D., Varadacharyulu N. Influence of chronic cigarette smoking on serum biochemical profile in male human volunteers. J Health Sci. 2009;55:265–270. [Google Scholar]
- 39.Lodhi P., Tandan N., Singh N., Kumar D., Kumar M. Camellia sinensis (L.) Kuntze Extract Ameliorates Chronic Ethanol-Induced Hepatotoxicity in Albino Rats. Evid Based Complement Alternat Med. 2014 doi: 10.1155/2014/787153. ID:787153. [DOI] [PMC free article] [PubMed] [Google Scholar]