Abstract
The repair of DNA double strand breaks (DSBs) requires tight regulation with the DNA damage response that mediates apoptotic death of damaged cells. Multiple protein ubiquitylation events at the sites of DSBs regulate damage recognition, repair, and signalling processes. However, the spatiotemporal calibration of DNA repair and the apoptotic response remains poorly understood. We identified the E4 ubiquitin ligase UFD-2 in a genetic screen for apoptosis defects after ionizing radiation in Caenorhabditis elegans. Following the initiation of homologous recombination (HR) at DSBs, UFD-2 forms foci, which also contain processivity factors including the ubiquitin-selective segregase CDC-48, the deubiquitylation enzyme ATX-3/Ataxin-3, and the proteasome. UFD-2 foci formation requires the recombinase RAD-51 and UFD-2 foci are retained until recombination intermediates are removed by the Holliday junction resolvases GEN-1, MUS-81 or XPF-1. In the absence of UFD-2, the removal of RAD-51-marked DSB repair foci is delayed indicative of inefficient repair. Similarly to ufd-2 deletion or E4 ubiquitin ligase inactivation, elevated RAD-51 levels lead to defects in DNA damage-induced apoptosis. UFD-2 foci formation also depends on the pro-apoptotic C. elegans p53 tumour suppressor homolog CEP-1, suggesting an intricate coordination between DSB processing and the apoptotic response. We establish a central role for the UFD-2 ubiquitin ligase in the coordination between the DNA repair process and the apoptotic response.
Introduction
DNA double strand breaks (DSBs) are highly cytotoxic and require the assembly of DNA damage signalling complexes and DSB repair machinery at the DNA breaks 1. In the C. elegans germline DSBs are mainly removed through homologous recombination (HR) 2. RAD-51 accumulates at the site of DSBs and mediates the strand invasion into the undamaged template ultimately leading to the formation of cruciform recombination intermediates called Holliday junctions (HJ) 3. HJs can be processed by two major pathways: HJ dissolution via the combined action of Blooms helicase and Topoisomerase TopoIIIα 4, or by resolution of HJ by nucleases acting as resolving enzymes 5. While HJ dissolution predominates in most systems 6,7, in C. elegans the GEN-1 resolvase is needed for completion of HR repair of DSBs 8. The resolution of HR intermediates is important for the apoptotic response to DSBs as GEN-1 and HJ processing factors are required for the DNA damage-induced programmed cell death. While the mechanisms for such regulation are not known yet, the C-terminal non-catalytic domain of GEN-1 appears to be important for DNA damage signalling 8,9. The apoptotic response to persistent DSBs facilitates the removal of germ cells in C. elegans when DSBs or meiotic recombination intermediates are not repaired, which occurs in the meiotic pachytene zone of the nematode germline (Fig. 1a) 10. DNA damage checkpoint signalling leads to the activation of the C. elegans p53 homolog CEP-1 followed by apoptosis induction (Fig. 5a) 11,12. CEP-1/p53 protein becomes available in the late pachytene region of the germline, leading to apoptosis competency of these germ cells. CEP-1 expression in earlier stages of meiosis is translationally repressed by the conserved mRNA binding protein GLD-1 13. Thus, apoptosis is only initiated when aberrant meiotic recombination intermediates or ionizing radiation (IR)-induced DSBs persist in late pachytene cells. It remains, however, unclear how the active repair process coordinates with the apoptotic execution in order to allow sufficient timing for resolving HR intermediates.
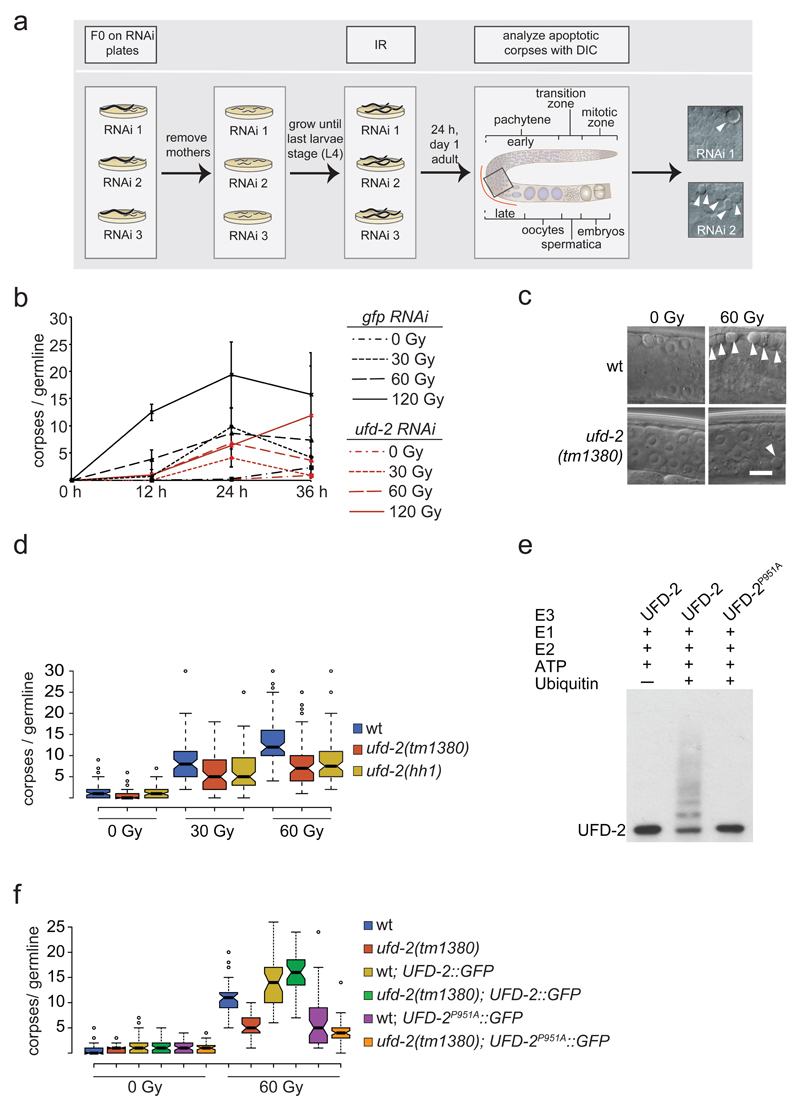
Figure 1.
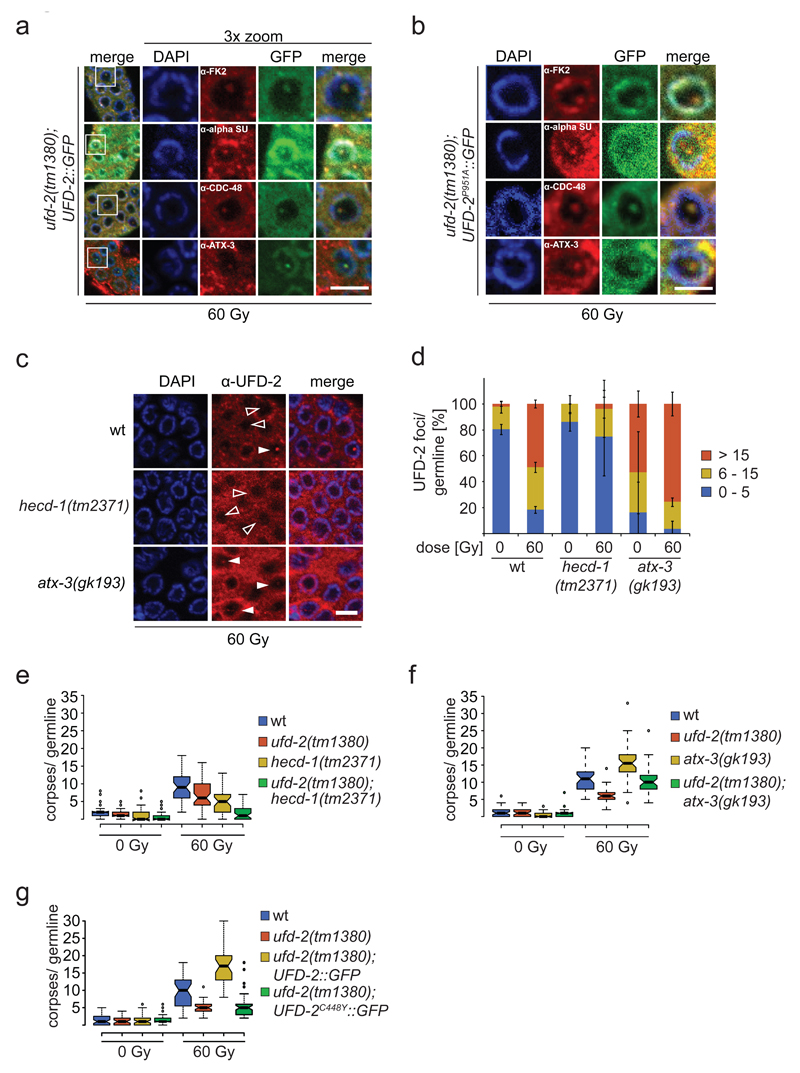
Ubiquitin ligase activity of UFD-2 is required for apoptosis execution. (a) Schematic illustration of RNAi screen for identification of DNA damage-induced apoptosis mediators. After RNAi treatment worms were subjected to IR and scored for apoptotic corpses (indicated by filled arrowheads) 24 hrs later by differential interference contrast (DIC) microscopy. (b) Worms treated with indicated RNAi constructs were exposed to IR of increasing dose and scored for apoptotic corpses 24 hrs after treatment. Data represent mean ± s.e.m. of selected data of RNAi screen. (c) Representative images of late pachytene cells of C. elegans germline 24 hrs after IR treatment. Filled arrowheads indicate an apoptotic corpse. Scale bar 5 µm. (d) Indicated genotypes were scored for DNA damage induced apoptosis 24 hrs after IR. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. The notches are defined as +/- 1.58*IQR/sqrt(n) and represent the 95% confidence interval for each median. Non-overlapping notches give roughly 95% confidence that two medians differ. Sample points of 5 independent experiments. (e) Auto-ubiquitylation of UFD-2. Ubiquitylation reactions were carried out as indicated using UFD-2 (wild-type) and UFD-2P951A as ubiquitin ligases. Representative immunoblot of 3 independent experiments. (f) Indicated genotypes were scored for DNA damage induced apoptosis 24 hrs after IR. Sample points of 3 independent experiments. For n-values see Supplementary Table 1.
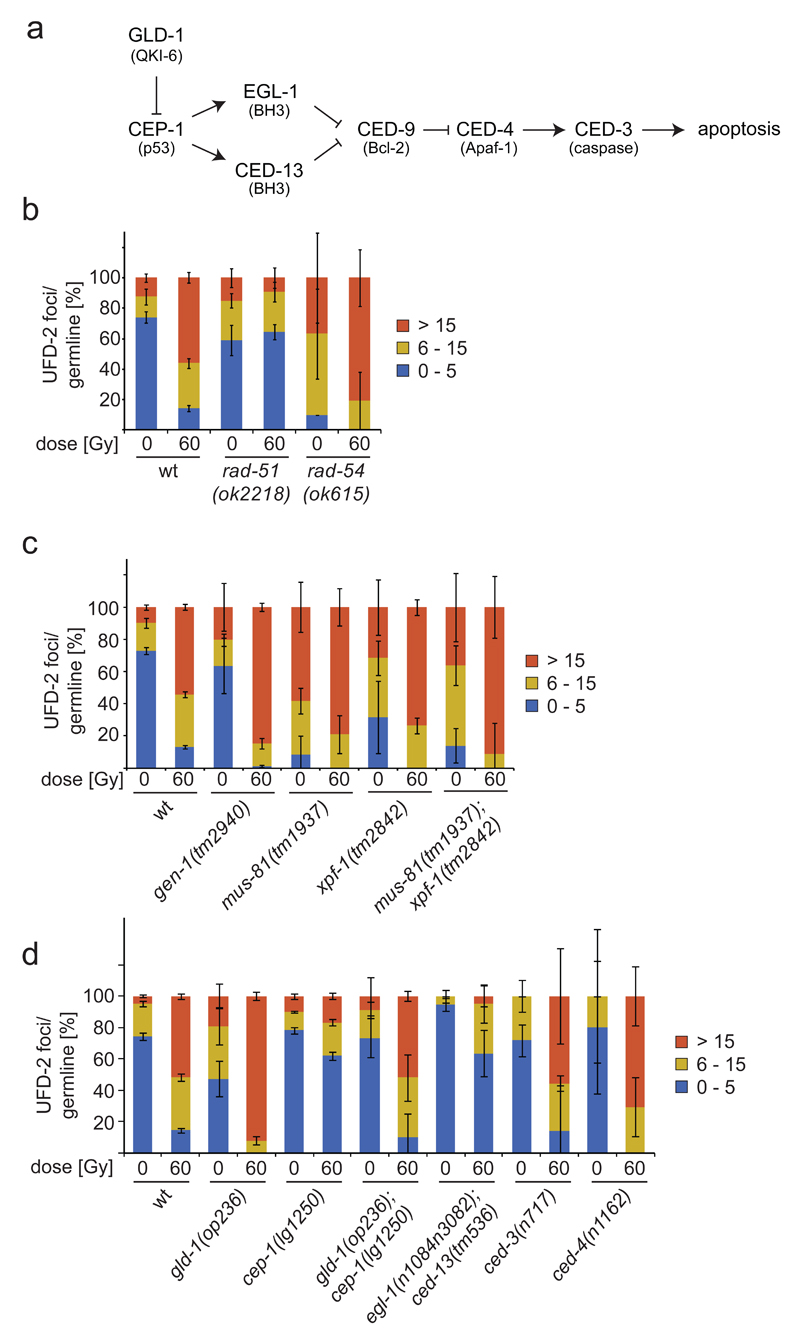
Figure 5.
UFD-2 foci in repair and apoptosis after DNA damage. (a) Schematic illustration of apoptosis pathway in C. elegans. Names in brackets indicate human homologues. (b, c and d) Quantification of UFD-2 foci in pachytene region of germlines of indicated genotypes isolated 24 hrs after irradiation with 60 Gy. Data show means ± s.e.m. of 3 independent experiments. For n-values see Supplementary Table 1.
Results
Ligase activity of UFD-2 mediates DNA damage induced cell death
To identify new regulators of the apoptotic response to DNA damage, we performed an RNA interference (RNAi) screen targeting 770 genes whose transcription is enriched in the C. elegans germline 14 (Fig. 1a). We focused on those genes because in C. elegans DNA damage induced apoptosis only occurs in germ cells 10,15. We identified the E4 ubiquitin ligase UFD-2 as to the most prominent candidate resulting from this screen. RNAi against ufd-2 led to a dose dependent reduction of IR induced apoptosis (Fig. 1b), a phenotype confirmed by analysing ufd-2(tm1380) and ufd-2(hh1) null alleles (Fig. 1c, d). In contrast, neither developmental apoptosis that occurs during the somatic development of the worm, nor physiological germ cell apoptosis, a background level of germ cell apoptosis that occurs independently of DNA damage, is defective in ufd-2 mutants (Supplementary Fig. 1a, b).
UFD-2 is a component of the ubiquitin fusion degradation (UFD) pathway first identified in budding yeast 16. Substrate ubiquitylation involves E1 ubiquitin activating, E2 ubiquitin conjugating, and E3 ubiquitin ligase enzymes. UFD-2 defines a class of so-called E4 enzymes, which act by further elongation of pre-existing ubiquitin chains to facilitate efficient substrate degradation 17–19. Ubiquitin forms chains of varying topology dependent on how molecules are linked to each other, thereby expanding its signalling capacity 20. UFD-2 plays an important role in the process of ubiquitin chain editing and supports proteasomal degradation 19,21. It preferentially makes use of lysine residues 29 and 48 of ubiquitin for autoubiquitylation (Supplementary Fig. 1e). A P951A point mutation in the U-box domain completely blocks the ligase activity of UFD-2 22 (Fig. 1e). To determine if UFD-2 catalytic activity is required for DNA damage-induced apoptosis, we transgenically expressed UFD-2::GFP or UFD-2P951A::GFP in the germline of wild-type or the ufd-2 deletion background. UFD-2::GFP expression fully restored the apoptotic DNA damage response in ufd-2(tm1380) mutant animals (Fig. 1f). In contrast, the catalytically dead mutant UFD-2P951A::GFP showed strongly reduced apoptosis after treatment with 60 Gy IR. The apoptosis defect caused by overexpressing UFD-2P951A::GFP, in a wild-type background indicates that the inactive U-box mutant acts dominant-negatively in response to DNA damage (Fig. 1f).
UFD-2 forms foci after DSB induction
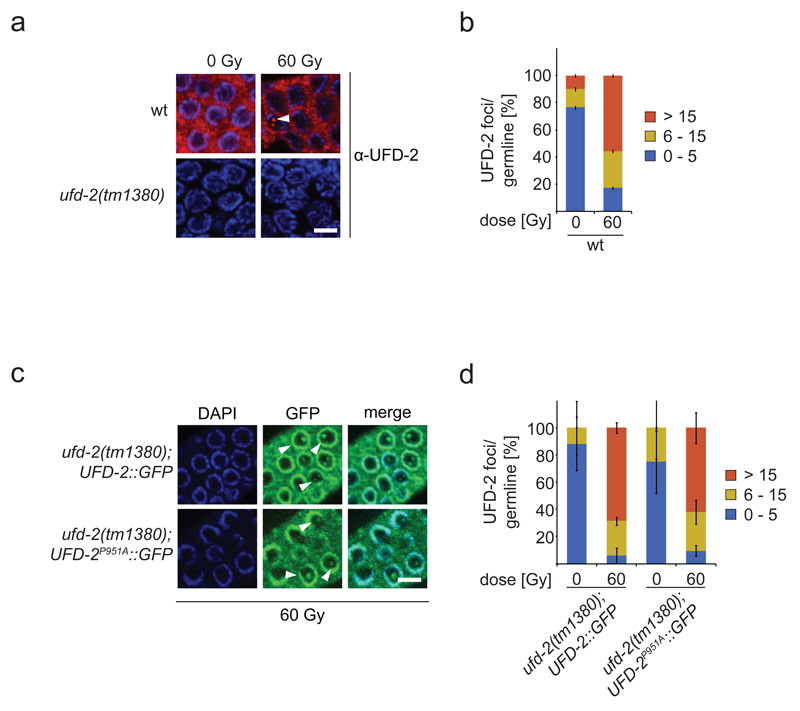
To determine UFD-2 localization, we raised polyclonal antibodies that specifically recognize UFD-2 both by western blot analysis and immunofluorescence staining (Fig. 2a and Supplementary Fig. 2a). Immunostaining revealed that under unperturbed conditions the protein is excluded from nucleoli but otherwise evenly distributed in the C. elegans germ line syncytium (Supplementary Fig. 2b). Commencing from late pachytene cells, UFD-2 accumulates at the nuclear periphery resulting into a ring-shaped staining pattern. After IR treatment UFD-2 foci of varying size and number became detectable within the nucleolus (Fig. 2a, b and Supplementary Fig. 2b). The pattern of antibody staining was confirmed by GFP-tagged UFD-2 transgenes (Fig. 2c, d). These UFD-2 foci occur in the mitotic zone as well as in the mid-late pachytene zone of the germline after IR. Given our interest in apoptosis we focussed on UFD-2 foci formation within nucleoli in the pachytene region. Pachytene cells elicit a DNA damage-induced apoptotic response upon DNA damage checkpoint activation, whereas mitotic nuclei in the distal germ line compartment are subjected to cell cycle arrest 10. In contrast to the IR-induced apoptosis defect, the cell cycle arrest, which can be monitored by scoring for enlarged mitotic nuclei due to continuous growth of cellular and nuclear compartments in the absence of cell division (Supplementary Fig. 2b) 10,23, was normally induced in ufd-2 mutants animals, indicating that the DNA damage checkpoint in general is functional (Supplementary Fig. 1c, d). Unlike IR-induced RAD-51 repair foci which are detectable immediately after damage induction (Fig. 4d) UFD-2 foci accumulated after 12 hrs following damage induction (Supplementary Fig. 2c). We therefore scored UFD-2 foci formation 24 hrs after IR, a time concomitant with full apoptosis activation 10, using both antibodies and GFP transgenes. The number of foci observed in pachytene cells increased from 0-5 foci per germline to more than 15 upon treatment with 60 Gy of IR (Fig. 2a, b and Supplementary Fig. 2b). Collectively, these data suggest that UFD-2 ligase function at first place is dispensable for foci formation (Fig. 2a) but is required to trigger the full apoptotic response.
Figure 2.
UFD-2 forms foci late after IR treatment. (a) Representative images of worm germlines of indicated genotypes irradiated with 60 Gy IR and stained with α-UFD-2 antibody and DAPI 24 hrs later. Filled arrowhead indicated nucleus with UFD-2 foci. Scale bar, 5 µm and (b) corresponding quantification of UFD-2 foci in pachytene region of germlines. Data show means ± s.e.m. of 12 independent experiments. (c) Representative images of worm germlines of indicated genotypes irradiated with 60 Gy IR. Germlines were isolated and stained with GFP-booster and DAPI 24 hrs after IR. Filled arrowheads indicate nuclei with UFD-2 foci. Scale bar, 5 µm and (d) corresponding quantification of UFD-2 foci in pachytene region of germlines. Data show means ± s.e.m. of 3 independent experiments. For n-values see Supplementary Table 1.
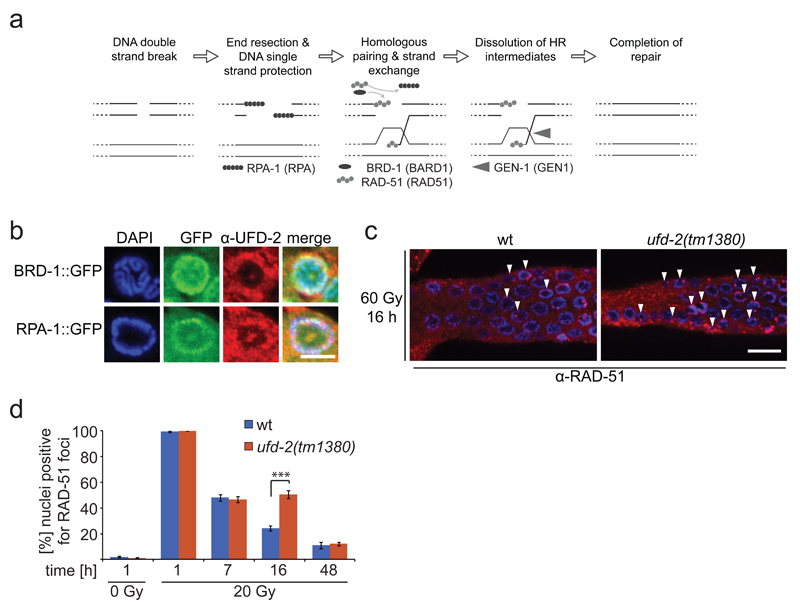
Figure 4.
Loss of ufd-2 delays DSB repair processing. (a) Schematic illustration of DNA DSB repair by HR in C. elegans. Upon DSB induction RPA binds resected single stranded DNA, BRD-1 acts together with BRCA-1 at DSB site, RPA is exchanged for RAD-51, which mediates strand invasion, Gen-1 resolves HJ resulting in repaired DSB. Names in brackets indicate human homologues. (b) Representative images of brd-1::gfp and rpa-1::gfp germlines isolated and stained with α-UFD-2 and DAPI 24 hrs after treatment with 60 Gy of IR. Scale bar, 5 µm. Representative images of 3 independent experiments. (c) Representative images of germlines isolated from wild-type and ufd-2(tm1380) worms 16 hrs after IR treatment with 20 Gy. Germlines were stained with α-RAD-51 and DAPI. Filled arrowheads indicate nuclei positive for RAD-51 staining. Scale bar, 10 µm. (d) Quantification of germ cells that were positive for RAD-51 staining. Wild-type and ufd-2(tm1380) worms were treated with 0 or 20 Gy of IR and isolated 1, 7, 16, 48 hrs after treatment (7, 16, 48 hrs only for 60 Gy treated worms) and immunostained with α-RAD-51 and DAPI to stain DNA. The last 50 nuclei of pachytene germ cells prior entering diakinesis were evaluated. Data show means ± s.e.m. of 3 independent experiments. The triple asterisk indicates P value of ≤ 0.001 in Student’s t-test. For n-values see Supplementary Table 1.
Ubiquitin-proteasome system factors fine-tune apoptosis response after DNA damage
Since yeast Ufd2 has been implicated in the degradation of the UFD substrates 16,17 and our evidence for UFD-2-mediated ubiquitylation having a role in DNA damage induced apoptosis, we examined if factors associated with the ubiquitin-proteasome system (UPS) accumulate at UFD-2 foci 17,24,25. Hence, we analysed ubiquitin localization 24 hrs after irradiation. In fact, an antibody that recognises conjugated mono- and polyubiquitin chains co-stained UFD-2 foci (Fig. 3a, Supplementary Fig. 3e). Additional staining experiments detected co-localization of the proteasome and the ubiquitin-selective segregase CDC-48/p97 with UFD-2 foci (Fig. 3a). Among other processes, CDC-48/p97 coordinates the degradation of chromatin-associated proteins during DNA replication or DNA repair by extracting ubiquitylated substrate proteins from higher order complexes 26–28. Transgenic over-expression of UFD-2C448Y::GFP, which is not able to interact with CDC-48, but importantly retains ligase activity forms high amount of UFD-2 foci before and after IR treatment (Supplementary Fig. 3f-j). However, UFD-2C448Y::GFP cannot rescue the apoptosis phenotype displayed by ufd-2 deletion worms (Fig. 3g). CDC-48 guides ubiquitin chain topology by coordinating different UPS-related substrate processing enzymes such as UFD-2 and the deubiquitylation enzyme ATX-3 21. Intriguingly, we also found ATX-3 localized to UFD-2 marked foci (Fig. 3a and Supplementary Fig. 3c, d), suggesting an orchestrated action of UFD-2, ATX-3, and CDC-48 at ubiquitylation hubs in the presence of DNA damage. Interestingly, ubiquitylation activity of UFD-2 is dispensable for the recruitment of ubiquitin processing enzymes. In contrast, apoptosis induction requires ligase activity of UFD-2 as well as its interaction with CDC-48 (Fig. 1e, f and 3b, g).
Figure 3.
UPS factors accumulate in UFD-2 hubs and balance apoptotic signalling. Representative images of (a) ufd-2(tm1380); ufd-2::gfp and (b) ufd-2(tm1380); ufd-2P951A::gfp immunostained with indicated antibodies. Germlines were isolated 24 hrs after treatment with 60 Gy. DNA stained with DAPI. The boxed area is three times magnified (3x zoom). α-alpha SU, α-Proteasome 20S alpha subunits. Scale bars, 5 µm. Representative images of 3 independent experiments. (c) Representative images of worm germlines of indicated genotypes irradiated with 60 Gy IR and stained with α-UFD-2 antibody and DAPI 24 hrs later. Empty and filled arrowhead indicated nuclei positive or negative for UFD-2 foci, respectively. Scale bar, 5 µm and (d) corresponding quantification of UFD-2 foci in pachytene region of germlines. Data show means ± s.e.m. of 3 independent experiments. (e, f and g) Indicated genotypes were scored for apoptosis 24 hrs after 60 Gy IR. Sample points of 3 independent experiments. For n-values see Supplementary Table 1.
Given that in yeast and humans, Ufd2/UBE4B mediates elongation of preformed ubiquitin chains, we tested whether UFD-2 collaborates with the E3 ligase HECD-1, the ortholog of budding yeast Ufd4 and human HECTD1 or TRIP12, to trigger DNA damage induced apoptosis 17,29–31. Importantly, loss of HECD-1 prevented UFD-2 foci formation, suggesting ubiquitin-dependent recruitment of UFD-2 (Fig. 3c, d). Supporting the role of UFD-2 focal accumulation in response to DNA damage, apoptosis was reduced in hecd-1 mutants (Fig. 3e). The apoptosis defect was even more pronounced in ufd-2; hecd-1 double mutants, indicating that the activity of both enzymes is required to achieve a full apoptotic response (Fig. 3e). In contrast, the deubiquitylation enzyme ATX-3 counteracts UFD-2 recruitment as both UFD-2 foci formation and apoptosis (Fig. 3d, f) was increased in atx-3 mutants (Fig. 3f). Accordingly, the excessive DNA damage-induced apoptosis occurring in atx-3 mutants was suppressed in ufd-2; atx-3 double mutant worms (Fig. 3f). In support of this notion, the number of ubiquitin foci per germline is decreased in hecd-1, whereas it is increased in atx-3 (Supplementary Fig. 3k). We therefore conclude that the apoptotic response to DNA damage is regulated by ubiquitylation signals defined by UFD-2 that cooperates in this response with HECD-1 and ATX-3.
UFD-2 supports RAD-51 dissociation from repair sites after DNA damage
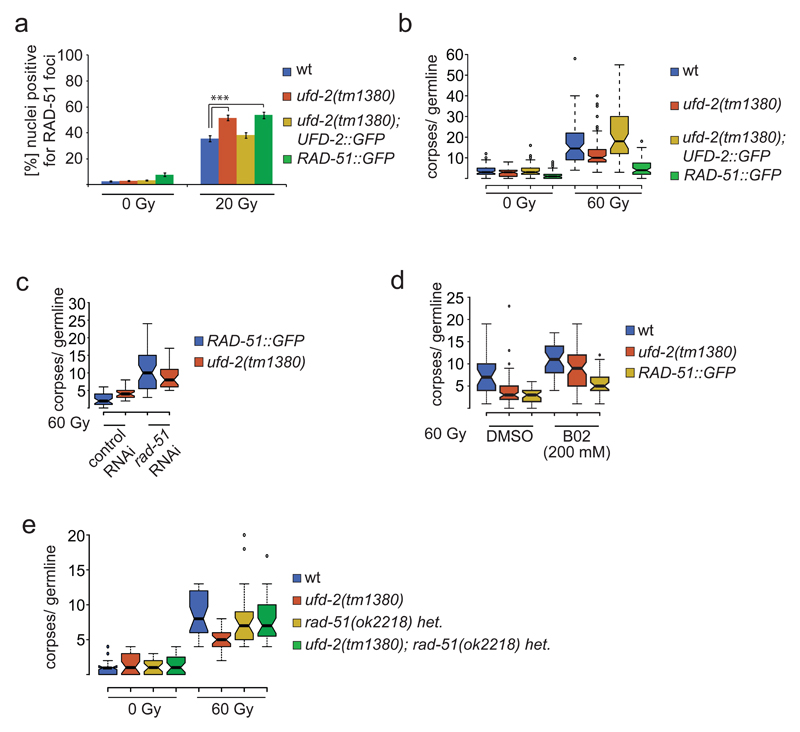
We next analysed if UFD-2 affects the DNA repair process in addition to apoptosis. In contrast to DSB induction by IR, UV irradiation did not result in formation of UFD-2 foci consistent with a specific role of UFD-2 in DSB repair (Supplementary Fig. 3a). In line with this observation we found that RPA-1::GFP and BRD-1::GFP HR proteins 32,33 accumulate in UFD-2 foci 24 hrs after IR treatment (Fig. 4b). Furthermore, IR of L4 staged ufd-2 mutant larvae resulted in reduced embryonic survival in the ensuing generation (Supplementary Fig. 3b). To establish whether ufd-2 promotes the processing of DNA repair intermediates we analysed the kinetics of RAD-51 foci. While both wild-type and ufd-2 mutants accumulated an equal amount of RAD-51 positive nuclei one hour after IR, twice as many RAD-51 stained nuclei persisted 16 hrs later in ufd-2 mutants (Fig. 4d). The delay in RAD-51 foci dissociation that temporally coincides with UFD-2 foci formation indicates that UFD-2 might contribute to resolution of repair intermediates.
UFD-2 coordinates DSB repair with apoptotic response
We further investigated the role of DSB repair in UFD-2 foci formation. Impairment of HR in rad-51 deletion worms blocked UFD-2 foci formation. Conversely, rad-54 deletion that inhibits removal of RAD-51 from DNA during HR repair 34, causes an accumulation of UFD-2 foci (Fig. 5b). Furthermore, deletion of the gen-1, mus-81and/or xpf-1 HJ resolvases led to the accumulation of high levels of UFD-2 foci (Fig. 5c and Supplementary Fig. 4a). These results indicate that HR needs to commence for UFD-2 foci to form and UFD-2 foci are only dissolved once HR is completed (Fig. 5b).
As ufd-2 mutant worms displayed reduced apoptosis, we assessed whether apoptotic signalling was affected in ufd-2 mutant worms. The apoptotic core machinery is conserved from C. elegans to the mammalian system. The p53 homologue CEP-1 induces transcription of the two BH3-only proteins EGL-1 and CED-13 13,35, which bind to the only Bcl2-like protein CED-9. As a consequence, the inhibitory effect of CED-9 on the Apaf1-like CED-4 is overruled and CED-4 activates the caspase CED-3, which executes the cell death (Fig. 5a) 36. In view of the ubiquitin ligase activity, we tested whether CEP-1 protein accumulates after damage induction in the absence of UFD-2. However, in wild-type and ufd-2 mutant worms CEP-1 protein was equally upregulated 2-fold upon 60 Gy irradiation (Supplementary Fig. 4b, c, d). Additional evaluation of mRNA transcripts of the CEP-1 target gene egl-1 showed a comparable transcriptional upregulation in both genotypes 4 and 24 hrs after damage infliction (Supplementary Fig. 4d). After having established that CEP-1 activation occurs independently of ufd-2, we wondered if UFD-2 foci formation might be dependent on CEP-1. Strikingly, loss of CEP-1 prevented UFD-2 foci induction after IR (Fig. 5d), whereas UFD-2 protein expression remained unaffected (Supplementary Fig. 5b). Consistently, a double mutant of the two pro-apoptotic CEP-1 effectors, egl-1; ced-13 phenocopied the cep-1 defect in UFD-2 foci formation after DNA damage (Fig. 5d and Supplementary Fig. 5a). To further correlate CEP-1 activity and UFD-2 foci formation, we enhanced CEP-1 activity by employing a gld-1 mutation, previously shown to lead to elevated CEP-1 levels and activity 13. gld-1 mutants indeed displayed strongly elevated number of UFD-2 foci, supporting the role of CEP-1 in promoting UFD-2 focal accumulation. The cep-1; gld-1 double mutant displayed a similar number of UFD-2 foci as wild-type germ cells (Fig. 5c and Supplementary Fig. 5a). One potential explanation for the failure of cep-1 to completely suppress the foci formation in gld-1 might be the numerous additional target mRNAs of GLD-1 37,38. Of note, the failure of cep-1 to initiate apoptosis is not correlated to its repair capacity. DNA repair assessed by embryonic survival is not perturbed in cep-1, but rather slightly increased in gld-1 mutants (Supplementary Fig. 5c). In ced-3 and ced-4 mutant worms UFD-2 foci generated normally (Fig. 5d), emphasising the necessity of CEP-1 rather than apoptotic signalling in general for UFD-2 foci formation. Taken together, UFD-2 foci formation depends on pro-apoptotic CEP-1 signalling but does not affect CEP-1 protein levels. Thus, UFD-2 seems to act downstream of pro-apoptotic signalling mediated by CEP-1 and EGL-1/CED-13.
We further tested a possible correlation between the effect of UFD-2 on DNA damage and apoptosis. Germline-specific expression of UFD-2::GFP in transgenic ufd-2 deletion mutants rescued the delay of RAD-51 removal from DNA. In contrast, increased RAD-51 retention occurred in moderately RAD-51::GFP overexpressing worms after 24 hrs of IR compared to wild-type (Fig. 6a). Nevertheless, this line possesses normal repair capacity as assessed by embryonic survival after IR (Supplementary Fig. 6c). Of note, loss of atx-3, important for restriction of UFD-2 foci and apoptosis execution after DNA damage, showed decreased RAD-51 retention 16 hrs after IR (Supplementary Fig. 4e). Obviously, the amount of retained RAD-51 foci negatively correlated with the apoptosis response (Fig. 6a, b). As control, ufd-2 mutants were crossed with rad-51. Similarly, RNAi depletion of RAD-51 in ufd-2 mutants or RAD-51::GFP expressing worms or treatment with the RAD-51 inhibitor B02 39reverted the apoptosis phenotype of ufd-2 deletion mutants or the RAD-51 overexpression line (Fig. 6c, d, Supplementary Fig. 6a, b), suggesting that RAD-51 accumulation directly blocks apoptosis signalling. In summary, these observations suggest that UFD-2 contributes to resolution of DNA repair sites, possibly by supporting the repair process directly or regulating the dynamics of repair proteins in specified degradation centres. Our data indicate that loss of E4 activity might cause the retention of RAD-51 on the DNA, which manifests in a transient block of apoptosis (Fig. 7).
Figure 6.
UFD-2 coordinates communication between repair and apoptosis after DNA damage. (a) Quantification of germ cells positive for RAD-51 staining. Worms of indicated genotypes were treated with 0 or 20 Gy of IR. Germlines were isolated 24 hrs after treatment and stained with α-RAD-51 and DAPI. The last 50 nuclei of pachytene germ cells prior entering diakinesis were evaluated. Data show means ± s.e.m. of 3 independent experiments. The triple asterisk indicates P value of ≤ 0.001 in Student’s t-test. (b) Indicated genotypes were scored for apoptosis 24 hrs after 60 Gy IR. Sample points of 3 independent experiments. (c) ufd-2 and RAD-51::GFP worms were treated with rad-51 or control RNAi and scored for apoptosis 24 hrs after 60 Gy IR. Sample points of 3 independent experiments. (d) wild-type, ufd-2 and RAD-51::GFP worms were treated with RAD51 inhibitor B02 (200mM) from L1 larvae on and scored for apoptosis 24 hrs after 60 Gy IR. Sample points of 3 independent experiments. (e) Indicated genotypes were scored for apoptosis 24 hrs after 60 Gy IR. Sample points of 3 independent experiments. For n-values see Supplementary Table 1.
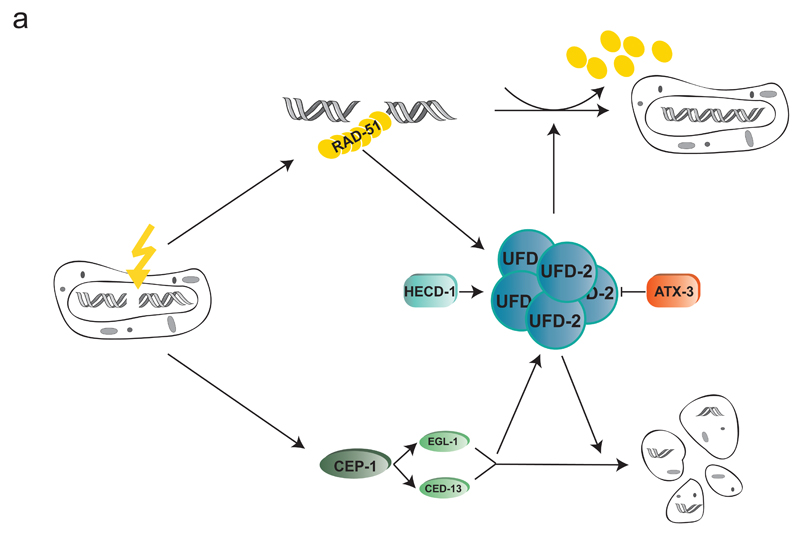
Figure 7.
Model of how UFD-2 integrates HR repair and apoptotic signalling. UFD-2 forms hubs late after IR (additionally containing proteolytic factors as CDC-48 and the proteasome (not shown)) that are dependent on active repair and apoptotic CEP-1 signalling. UFD-2 hub formation is balanced by the E3 ligase HECD-1 and the DUB ATX-3. In accordance with hub formation at later stages after DSB induction, UFD-2 supports RAD-51 dissociation from DSB site at advanced time points and mediates signal to apoptosis pathway.
Discussion
In this study we uncovered a ubiquitin dependent process that facilitates the communication between DNA repair and the apoptotic response. We identified the E4 ubiquitin ligase UFD-2 as a central regulator for the spatiotemporal coordination of both processes. Our data suggest that defects in timely proceeding of HR either by failure to resolve HJs as previously demonstrated 8,9 or by aberrant retention of RAD-51 at the chromatin caused by loss of UFD-2 as shown here, halt the apoptotic response. Conversely, RAD-51 filament assembly and pro-apoptotic signalling by the tumour suppressor CEP-1/p53 are both required for the formation of UFD-2-specific hubs that are defined by proteolytic factors of the UPS machinery. We propose that these degradation hubs calibrate the DNA repair status with apoptotic activity via modulation of ubiquitin signalling. Since the E3 ligase HECD-1 is required for UFD-2 hub formation and apoptosis execution, we further propose that E4 activity 17,30,40 is providing an additional layer of regulation by editing ubiquitin chain topology. The human E4 homolog UBE4B cooperates similarly with the HECT domain E3 ligase TRIP12 in substrate ubiquitylation, suggesting the existence of a conserved signalling pathway 29. In support of this idea, TRIP12 fine-tunes ubiquitin controlled events at DSBs 41 and recent reports linked UBE4B to different cancer types, highlighting the relevance of ubiquitin signalling in the decision between DNA damage and apoptosis response 42–45. Not only during meiotic recombination and DSB repair in germ cells but also during the maintenance of tissue integrity following DNA injury the apoptotic response requires timely adjustment to on-going activity of DNA repair processes particularly when they are as complex as HR. Defects in the both DNA repair and apoptosis are especially relevant in tumour formation. Thus, understanding the conserved role of UFD-2/UBE4B in response to IR induced DNA damage might open new therapeutic directions for drug development and cancer treatment.
Methods
C. elegans strains
C. elegans strains were cultured at 20 °C on nematode growth medium (NGM) and fed with Escherichia coli (E. coli) strain OP50 according to standard procedures 46. The Bristol strain N2 was used as wild-type. Mutants and transgenic animals used in this study are listed in the following: mus-81(tm1937) I, rad-54&snx-3(ok615) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III), cep-1(lg12501)I, ced-1(e1735)I, gld-1(op236)I, ufd-2(tm1380)II, ufd-2(hh1)II, xpf-1(tm2842) II, gen-1(tm2940)III, ced-4(n1162) III, hecd-1(tm2371)IV, rad-51(ok2218) IV/nT1[qIs51](IV;V), ced-3(n717) IV, atx-3(gk193)V, egl-1(n1084n3082)V; ced-13(tm536)X, , Is[rad-51::GFP:3xFLAG], gla-3(op216)I, hus-1(op241)I, unc-119(ed3)III; gtIs[unc-119(+), Ppie-1::GFP::rpa-1::pie-1-3'UTR], hhIs121[unc-119(+), Pmex-5::ufd-2::GFP::tbb-2 3'UTR], hhIs135[unc-119(+), Pmex-5 (w/o ATG)::ufd-2 (w/o TAA, P951A)::(Gly)5Ala::gfp F64LS65T(w introns/stop)::tbb-2 3'UTR], hhIs134[unc-119(+), Pmex-5::ufd-2 (C448Y)::GFP::tbb-2 3'UTR].
The transgenic lines hhIs121, hhIs134 and hhIs135 were generated for this study. Briefly, fosmid WRM0621dE05 was used as template to obtain the genomic sequence of ufd-2 that was cloned together with ppJA252, pJA257 into pCG150 containing the unc-119(+) marker for selection of transgenic worms 47. ufd-2 was modified by directed mutagenesis to create ufd-2P951A or ufd-2C448Y. The constructs were bombarded into unc-119(ed4)III mutants as described previously 48.
Ionizing radiation
Synchronized hermaphrodites were grown until L4 stage and irradiated with the corresponding dose (Radiation source: 120-kV X-rays (25 mA; 0.5mm Alu-filter; ISOVOLT 160 M1/10-55, GE Sensing & Inspection Technologies) or Biobeam 8000 using Cs137 as radiation source).
RNAi treatment
RNA interference was performed using the feeding method 49. Three P0 worms were placed on IPTG (isopropylthiogalactoside) and ampicillin-containing NGM-plates seeded with E. coli [HT115(DE3)] expressing double-stranded RNA (dsRNA) and incubated at 15°C for 72 hrs. Three single F1 worms were transferred each to a new, freshly seeded plate and allowed to lay eggs for approximately 20 hrs. F1 worms were removed and F2 worms were allowed to grow up to the L4 stage, treated with ionizing radiation and analyzed for radiation induced apoptosis. Clones in RNAi feeding vectors were provided by Marc Vidal of Dana Farber Cancer Center.
Apoptotic corpses
For physiological apoptosis analysis, synchronized L1 larvae were grown until L4 stage. Apoptotic corpses were scored 24 hrs later. For this, worms were mounted on 3% agar pads, paralyzed with 60 nM NaN3 and analysed via DIC microscopy 50. For DNA damage induced apoptosis worms were subjected to IR at L4 stage before apoptosis was evaluated 24 hrs later. Developmental apoptosis was assessed in L1 larvae. Therefore worms were grown until day one adulthood. 100 worms were transferred to a NGM-agar plate without E. coli and allowed to lay eggs until they were removed again after 1 h. Freshly hatched L1 larvae were scored for apoptotic corpses 51.
UFD-2 foci
Synchronized worms were grown until L4 larvae stage and irradiated with 0 and 60 Gy. 24 hrs later, germlines were isolated and immunostained. Number of UFD-2 foci was scored in all focal planes in pachytene germ cells. One germline per worm was scored.
Protein expression and purification
cDNAs encoding ufd-2b, ufd-2bC448Y and ufd-2bP951A were cloned into the pET-21d expression vector (Novagen) and pGex4T1 (GE Healthcare). Recombinant proteins were expressed in Escherichia coli strain BL21 Codon Plus (Novagen) and purified using the ÄKTA purifier system (GE Healthcare).
Antibody production
His-tagged purified proteins (UFD-2, ATX-321) were used for immunization of rabbits and anti-sera were affinity purified using respective GST-tagged recombinant proteins (BioGenes).
Preparation of worm lysates
Synchronized L1 larvae were grown on NGM-agar plates with OP50 bacteria until they reached adulthood. Worm lysates used for SDS-PAGE were either prepared from a distinct number of worms (n=150) or by washing worms from NGM-agar plates followed by multiple washing step with M9 buffer (3 g/l KH2P04, 6 g/l Na2 HPO4, 5 g/l NaCl, 1 mM Mg S04 (added after sterilization)), until bacteria were removed. The samples were heated to 95°C for 5 min and subsequently shock-frozen in liquid nitrogen. After thawing, samples were subjected to sonication (two times for 15 s, on ice; 50% power; Sonopuls UW 2200, Bandelin) and taken up in 4 x SDS sample buffer followed by centrifugation at 15,000 rpm for 10 min.
Immunotechniques
Immunostaining of isolated germlines was done according to the ‘freeze-crack’ protocol. Worms were dissected onto polylysine-coated slides (Thermo Scientific) in 60 nM NaN3 to isolate germlines and fixed in fixation buffer (3.7 % Formaldehyde, 0.2 % Tween 20) for 10 min with subsequent shock freezing in liquid nitrogen. This was followed by incubation in 1:1 mixture of methanol and acetone at -20 °C for 10 min. Germlines were permeabilized 3 times in 1 % PBS-Triton X-100 for 20 min followed by washing in 0.1 % PBS-Tween 20 (PBS-T) for 10 min and blocking in 10 % goat serum in 0.1 % PBS-T. A specific staining protocol was followed for GFP-expressing lines avoiding freezing. Isolated germlines were fixed with fixation buffer for 10 min in PCR tubes, directly followed by permeabilization and blocking as described above. Germlines were incubated with primary antibody overnight at 4 °C (anti-UFD-2 1:3,000, anti-CDC-48 1:12,000, anti-RAD-51 1:350 (Novus), anti-FK2-ubiquitin 1:100 (Millipore), anti-Proteasome 20S alpha 1+2+3+5+6+7 antibody 1:300 (abcam), anti-ATX-3 1:700). Incubation with the fluorescently labelled secondary antibodies (Life Technologies; 1:200) or GFP-booster (ChromoTek; 1:400) was done at room temperature for 1 h. Germlines were mounted in DAPI Fluoromount-G medium (SouthernBiotech). For western blotting, worm lysates were separated by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to nitrocellulose membranes (Whatman, Protran). Membranes were blocked in 1x Roti-Block (Roth) and incubated with the primary antibodies overnight at 4 °C in Roti-Block (Roth; anti-UFD-2 1:20,000, anti-ATX-3 1:10,000, anti-CEP 1:15,000, anti-tubulin 1:5000 (Sigma-Aldrich, clone DM1A). Incubation with fluorescently labelled secondary antibodies (1:10,000) was done at room temperature, before detection of signals using the Li-Cor Odyssey scanner. Quantification of signal intensities was done using the Odyssey V4.0 software (Li-Cor). The uncropped versions of western blots that have been used to assemble the main figures are collected in Supplementary Fig. 6.
Microscopy and image acquisition
Immunostained germlines were imaged with AxioImager.M1/Z1 microscope with Apoptome equipped with an AxioCam MRm camera (Carl Zeiss). To allow direct comparison of signal intensities, images were recorded under identical conditions. Processing of selected pictures was done in ZEN2011 and ImageJ.
In vitro ubiquitylation assay
UFD-2b::His, UFD-2bC448Y::GST and UFD-2bP951A::His fusion proteins were expressed in BL21-AI E. coli strain and lysed in buffer A (50 mM Tris pH 7.5, 250 mM NaCl, 5 mM DTT, 1% Triton X-100, 2 mM PMSF and protease inhibitor mix; Roche). 10 μg of the aforementioned bacterial lysate was mixed with E1 (25 ng), E2 (Let-70; 400 ng), 2 μg of FLAG::ubiquitin, energy regenerating solution (Boston Biochemicals) and ubiquitin conjugation reaction Buffer (Enzo Life Sciences). Samples were incubated at 30 °C for 1.5 h, terminated by boiling for 5 min with SDS-sample buffer, and resolved by SDS-PAGE followed by immunoblotting using anti-UFD-2 antibodies to monitor ubiquitylation of UFD-2.
Persistence of RAD-51 foci after IR
Synchronized worms were grown until L4 larvae stage and irradiated with 0 and 20 Gy. 1 to 48 hrs later, germlines were isolated and immunostained. Z-stacks were taken of late pachytene cells of the germline. Two focal planes covering the upper and lower part of the germline were subjected to analysis by scoring each plane for RAD-51 positive cells in the last 25 nuclei.
RNA isolation and real-time PCR
Total RNA was isolated using TRIzol (Invitrogen) and Qiagen RNeasy kit. Briefly, worms were washed off the plates using M9 buffer (3 g/l KH2P04, 6 g/l Na2 HPO4, 5 g/l NaCl, 1 mM Mg S04 (added after sterilization)) and 600 µl TRIzol, and silica beads (1 mm diameter) were added to the samples and homogenized by Precellys tissue homogeniser. Chloroform was added and samples were vortexed vigorously before phase separation through centrifugation. The aqueous phase was transferred on the Qiagen RNeasy Mini spin column and RNA was isolated according to manufacturer’s instructions. cDNA was synthesized using 200 ng total RNA and the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression levels were determined by real time PCR using Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies) and Biorad CFX96 Real-Time PCR Detection System. Relative gene expressions were normalized to tbg-1 (F58A4.8) mRNA levels. In the experiment three biological and three technical replicate samples were analyzed. The primer sequences used in the RT–PCR reactions are the following: tbg-1 forward: 5′-GTACACTCCACTGATCTCTGCTGACAAG-3′, tbg-1 reverse: 5′-CTCTGTACAAGAGGCAAACAGCCATG-3′ 52, egl-1 forward: 5′-TACTCCTCGTCTCAGGACTT-3′, egl-1 reverse: 5′- CATCGAAGTCATCGCACAT-3′.
Radiation sensitivity
To determine the radiation sensitivity, L4-stage hermaphrodites were irradiated with a single dose of IR as indicated. After 12 hrs, worms were transferred to fresh plates (three worms per plate, five plates in total) and allowed to lay eggs for 5 hrs. After this period, adults were removed and 24 hrs later the number of hatched and unhatched embryos was scored. As a control for DNA damage sensitivity, a heterozygous deletion mutant lacking rad-51 on one chromosome was used.
Mitotic germ cell cycle arrest upon IR
Worms were irradiated with 0 and 60 Gy at the late L4 larval stage as described previously 10. 16 hours post-irradiation, worms were mounted on 3% agar pads and paralyzed with 60 nM NaN3 for DIC microscopy and the distal region of the germline was scored for number of nuclei in all focal planes within a defined area of 2 μm x 6 μm.
Statistical analysis
Statistical analysis was performed using Excel (Microsoft). Statistical significance was calculated with two-tailed paired Student’s t-test. Box plots were generated using BoxPlotR 53. Centre lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. The notches are defined as +/-1.58*IQR/sqrt(n) and represent the 95% confidence interval for each median. Non-overlapping notches give roughly 95% confidence that two medians differ.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank Y. Kohara, M. Marr, the Caenorhabditis Genetics Center (funded by the NIH National Center for Research Resources), the Bloomington Stock Center, the Dana-Farber Cancer Institute, Addgene and Geneservice Ltd for antibodies, plasmids, cDNAs, and strains; Agata Lisowski and Ela Stellbrink for technical help. We thank André Franz and Aljona Gutschmidt for critical reading of the manuscript. We thank Kristijan Ramadan and Yossi Shiloh for insightful discussions on the project and exchange of unpublished results. This work was supported by a Wellcome Trust Senior Research award (090944/Z/09/Z) to AG, grants of the German-Israeli Foundation (GIF 1104-68.11/2010), the Deutsche Forschungsgemeinschaft (EXC 229, SFB 829, SFB 670, and KFO 286), the European Research Council (ERC Starting grant 260383), Marie Curie (FP7 ITN CodeAge 316354, aDDRess 316390, MARRIAGE 316964), and the Bundesministerium für Forschung und Bildung (Sybacol FKZ0315893A-B) to B.S., and the Deutsche Forschungsgemeinschaft (EXC 229, HO 2541/8-1, and KFO 286) and the European Research Council (consolidator grant 616499) to T.H. In addition, this work was supported by COST Action (PROTEOSTASIS BM1307), supported by COST (European Cooperation in Science and Technology).
Footnotes
Author Contributions L.A. and M.S. designed, performed and analysed the experiments. W.P. performed in vitro ubiquitylation assays. É.K. generated ATX-3 antibody. A.G. and B.S. designed and performed the RNAi screen. B.S. and T.H. supervised the design and data interpretation. L.A., B.S., and T.H. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Clejan I, Boerckel J, Ahmed S. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics. 2006;173:1301–1317. doi: 10.1534/genetics.106.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmens BB, Tijsterman M. DNA double-strand break repair in Caenorhabditis elegans. Chromosoma. 2011;120:1–21. doi: 10.1007/s00412-010-0296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bizard AH, Hickson ID. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol. 2014;6:a016477. doi: 10.1101/cshperspect.a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matos J, West SC. Holliday junction resolution: regulation in space and time. DNA Repair (Amst) 2014;19:176–181. doi: 10.1016/j.dnarep.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West SC, et al. Resolution of Recombination Intermediates: Mechanisms and Regulation. Cold Spring Harbor symposia on quantitative biology. 2015 doi: 10.1101/sqb.2015.80.027649. [DOI] [PubMed] [Google Scholar]
- 8.Bailly AP, et al. The Caenorhabditis elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling to DNA double-strand break repair. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva N, Adamo A, Santonicola P, Martinez-Perez E, Volpe LA. Pro-crossover factors regulate damage-dependent apoptosis in the Caenorhabditis elegans germ line. Cell Death & Differentiation. 2013;20:1209–1218. doi: 10.1038/cdd.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage--induced apoptosis and cell cycle arrest in C. elegans. Molecular cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 11.Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Current biology : CB. 2001;11:1722–1727. doi: 10.1016/S0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- 12.Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- 13.Schumacher B, et al. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Reinke V, et al. A global profile of germline gene expression in C. elegans. Molecular cell. 2000;6:605–616. doi: 10.1016/S1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 15.Vermezovic J, Stergiou L, Hengartner MO, d'Adda di Fagagna F. Differential regulation of DNA damage response activation between somatic and germline cells in Caenorhabditis elegans. Cell death and differentiation. 2012;19:1847–1855. doi: 10.1038/cdd.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson ES, Ma PCM, Ota IM, Varshavsky A. A Proteolytic Pathway That Recognizes Ubiquitin as a Degradation Signal. Journal of Biological Chemistry. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 17.Koegl M, et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 18.Saeki Y, Tayama Y, Toh-e A, Yokosawa H. Definitive evidence for Ufd2-catalyzed elongation of the ubiquitin chain through Lys48 linkage. Biochemical and biophysical research communications. 2004;320:840–845. doi: 10.1016/j.bbrc.2004.05.216. [DOI] [PubMed] [Google Scholar]
- 19.Hoppe T. Multiubiquitylation by E4 enzymes: 'one size' doesn't fit all. Trends Biochem Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. Journal of cell science. 2016;129:875–880. doi: 10.1242/jcs.183954. [DOI] [PubMed] [Google Scholar]
- 21.Kuhlbrodt K, et al. The Machado-Joseph disease deubiquitylase ATX-3 couples longevity and proteostasis. Nat Cell Biol. 2011;13:273–281. doi: 10.1038/ncb2200. [DOI] [PubMed] [Google Scholar]
- 22.Okumura F, Hatakeyama S, Matsumoto M, Kamura T, Nakayama KI. Functional regulation of FEZ1 by the U-box-type ubiquitin ligase E4B contributes to neuritogenesis. The Journal of biological chemistry. 2004;279:53533–53543. doi: 10.1074/jbc.M402916200. [DOI] [PubMed] [Google Scholar]
- 23.Hodgkin J, Horvitz HR, Brenner S. Nondisjunction Mutants of the Nematode CAENORHABDITIS ELEGANS. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rape M, et al. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 25.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Meerang M, et al. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol. 2011;13:1376–1382. doi: 10.1038/ncb2367. [DOI] [PubMed] [Google Scholar]
- 27.Acs K, et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- 28.Dantuma NP, Hoppe T. Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol. 2012;22:483–491. doi: 10.1016/j.tcb.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Park Y, Yoon SK, Yoon J-BB. TRIP12 functions as an E3 ubiquitin ligase of APP-BP1. Biochemical and biophysical research communications. 2008;374:294–298. doi: 10.1016/j.bbrc.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 30.Liu G, Rogers J, Murphy CT, Rongo C. EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. The EMBO journal. 2011;30:2990–3003. doi: 10.1038/emboj.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PloS one. 2011;6 doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marechal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell research. 2015;25:9–23. doi: 10.1038/cr.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boulton SJ, et al. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr Biol. 2004;14:33–39. doi: 10.1016/j.cub.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 34.Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 35.Hofmann ER, et al. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Current biology : CB. 2002;12:1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 36.Horvitz HR. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 1999;59:1701s–1706s. [PubMed] [Google Scholar]
- 37.Lee MH, Schedl T. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes & development. 2001;15:2408–2420. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doh JH, Jung Y, Reinke V, Lee M-HH. C. elegans RNA-binding protein GLD-1 recognizes its multiple targets using sequence, context, and structural information to repress translation. Worm. 2013;2 doi: 10.4161/worm.26548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang F, Mazin AV. A small molecule inhibitor of human RAD51 potentiates breast cancer cell killing by therapeutic agents in mouse xenografts. PloS one. 2014;9 doi: 10.1371/journal.pone.0100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park Y, Yoon SK, Yoon J-BB. The HECT domain of TRIP12 ubiquitinates substrates of the ubiquitin fusion degradation pathway. The Journal of biological chemistry. 2009;284:1540–1549. doi: 10.1074/jbc.M807554200. [DOI] [PubMed] [Google Scholar]
- 41.Gudjonsson T, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 42.Krona C, et al. Screening for gene mutations in a 500 kb neuroblastoma tumor suppressor candidate region in chromosome 1p; mutation and stage-specific expression in UBE4B/UFD2. Oncogene. 2003;22:2343–2351. doi: 10.1038/sj.onc.1206324. [DOI] [PubMed] [Google Scholar]
- 43.Carén H, Holmstrand A, Sjöberg RMM, Martinsson T. The two human homologues of yeast UFD2 ubiquitination factor, UBE4A and UBE4B, are located in common neuroblastoma deletion regions and are subject to mutations in tumours. European journal of cancer (Oxford, England : 1990) 2006;42:381–387. doi: 10.1016/j.ejca.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 44.Zage PE, et al. UBE4B levels are correlated with clinical outcomes in neuroblastoma patients and with altered neuroblastoma cell proliferation and sensitivity to epidermal growth factor receptor inhibitors. Cancer. 2013;119:915–923. doi: 10.1002/cncr.27785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kloppsteck P, Ewens CA, Forster A, Zhang X, Freemont PS. Regulation of p97 in the ubiquitin-proteasome system by the UBX protein-family. Biochim Biophys Acta. 2012;1823:125–129. doi: 10.1016/j.bbamcr.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeiser E, Frøkjær-Jensen C, Jorgensen E, Ahringer J. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PloS one. 2011;6 doi: 10.1371/journal.pone.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 50.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development (Cambridge, England) 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 51.Hengartner MO, Ellis RE, Horvitz HR. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature. 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 52.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC molecular biology. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spitzer M, Wildenhain J, Rappsilber J, Tyers M. BoxPlotR: a web tool for generation of box plots. Nature methods. 2014;11:121–122. doi: 10.1038/nmeth.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.