Abstract
Key flowering genes, FD and FD PARALOGUE (FDP) encoding bZIP transcription factors that interact with a FLOWERING LOCUS T (FT) in Arabidopsis were ectopically expressed in rice since we found AtFD and AtFDP also interact with HEADING DATE 3a (Hd3a) and RICE FLOWERING LOCUS T 1 (RFT1). Transgenic rice plants overexpressing AtFD and AtFDP caused reduction in plant height and spikelet size with decreased expression of genes involved in cell elongation without significant flowering time alteration in spite of increased expression of OsMADS14 and OsMADS15, rice homologues of APETALA1 (AP1) in the leaves. Simultaneous overexpression of AtFD and AtFDP enhanced phenotypes seen with overexpression of either single gene while transgenic rice plants expressing AtFD or AtFDP under the control of phloem-specific Hd3a promoter were indistinguishable from wild-type rice. Candidate genes responsible for the phenotypes were identified by comparison of microarray hybridization and their expression pattern was also examined in WT and transgenic rice plants. It has so far not been reported that AtFD and AtFDP affect cell elongation in plants, and our findings provide novel insight into the possible roles of AtFD and AtFDP in the mesophyll cells of plants, and potential genetic tools for manipulation of crop architecture.
Formation of a hexameric florigen activation complex (FAC) composed of a rice florigen Hd3a, GF14 proteins (14-3-3 s) and phosphorylated transcription factor OsFD1 has been reported to activate OsMADS15, a homologue of Arabidopsis APETALA1 (AP1) resulting in flowering1. Since interactions between certain isoforms of 14-3-3 and Arabidopsis FT have also been shown2,3 and AtFD is phosphorylated by calcium-dependent protein kinases4, without direct evidence it has been assumed that AtFT-AtFD interaction in Arabidopsis occurs in a manner similar to rice Hd3a-OsFD1. Recently, OsFD2, a homologue of OsFD1 was proposed to control leaf development through interaction with the same components of a FAC5, and this supports the notion that distinct transcription factors may confer the formation of various FAC-like complexes to control programs in plant growth and development.
Hd3a and RFT1 are two florigen genes in rice. The expression of both genes is controlled by a complicated genetic network integrating photoperiodic signaling and circadian clock information6. Hd3a is a leaf-borne florigenic signal moving to the shoot apex to induce flowering7. Among 13 homologues of Hd3a in the rice genome8, RICE FLOWERING LOCUS T 1 (RFT1) and FT-like 1 (FT-L1) share extensive sequence similarity with Hd3a and induce early flowering when overexpressed9,10 indicating that they have the features of floral activators. RFT1 is also expressed in the leaf vascular tissues, and simultaneous knockdown of Hd3a and RFT1 by RNAi approaches prevents flowering for at least 300 days under inductive short days11. Besides promoting flowering, FT homologues are known to play various roles in plant developmental processes although it remains unknown whether formation of FAC-like complexes is necessarily required for their function. Reported roles include controlling tuberization in potato12, growth cessation and bud set in Populus13, leaf shape and inflorescence architecture in tomato14, stomatal opening in Arabidopsis15 and bulb formation in onion16.
AtFD, an Arabidopsis bZIP transcription factor is preferentially expressed in the shoot apex and required for AtFT to promote flowering through the formation of a complex between them in the shoot apex. Phosphorylation of AtFD at the 282th threonine residue is essential for AtFT-AtFD complex formation and functional complementation of fd-117. In the case of OsFD1, phosphorylation of the 192th serine residue has been proposed to be critical for the formation of FAC and activation of OsMADS15 in the protoplasts1. Overexpression of OsFD1 for the phosphomimetic form (OsFD1 S192E) has been reported to promote flowering although there were no significant flowering time alterations in overexpressors or dsRNAi lines for OsFD11. Remarkably, OsFD genes are pretty strongly expressed in the leaves and other vegetative organs distinct from the expression pattern of AtFD in Arabidopsis5.
14-3-3s are being found to play key roles in an expanding variety of plant health, growth and developmental processes. Genetic verification of the roles of GF14 proteins as florigen receptors is complicated by gene redundancy and by many processes in which these molecules have been involved18. Eight isoforms of GF14 are available in rice and six of them are expressed in the shoot apex19. Simultaneous reduction of the expression of four GF14s only caused mild reduction of target gene transcription of the FAC indicating that other isoforms might play a compensatory role1. Additionally, GF14c knock-out mutant rice is seedling-lethal and transgenic rice with GF14c overexpression show delayed flowering19.
Even though it is still unknown whether all interactions between FT homologues and bZIP proteins are dependent on 14-3-3s, published data demonstrated that a FAC-like complex containing OsFD2 possesses the same components required for the formation of FAC, but is implicated in rice leaf development.
In this work, we observed distinct molecular characteristics in two homologous proteins, AtFT and Hd3a: in binding to bZIP proteins and flowering promotion in rice. We also found that transgenic rice overexpressing AtFD and AtFDP exhibited dwarfism with reduced organ size without significant alteration of flowering time. Transgenic Arabidopsis overexpressing AtFD also showed reduced body size as well as early flowering. Both AtFD and AtFDP are able to interact with two rice florigenic proteins, Hd3a and RFT1 in yeast and rice protoplast systems. Transgenic dwarf rice caused by AtFD/AtFDP overexpression showed reduced expression of genes involved in cell elongation but still responded to gibberellin (GA3). Candidate rice genes affected by AtFD and AtFDP which are likely responsible for the phenotypes were identified. The majority of these candidate genes were found to encode various types of kinases. In view of these results we propose that AtFD and AtFDP likely influence cell elongation or proliferation through GA biosynthesis control and/or various phosphorylation-mediated signal transduction cascades in rice although it is not clear whether the action of AtFD and/or AtFDP is mediated by the FAC-like complex formation or not. Functional evaluation of the candidate genes identified will be the next step for their potential use in manipulation of crop architecture.
Results
Arabidopsis FD and FDP interact with Hd3a and RFT1
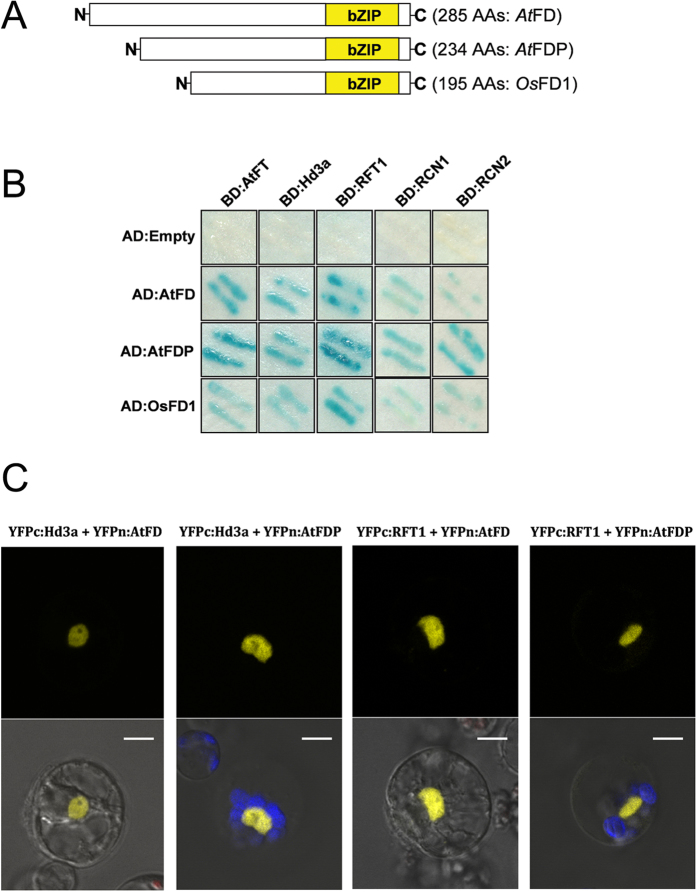
To test whether Arabidopsis FD and FDP are able to interact with rice Hd3a or RFT1, rice FT homologues, we utilized yeast two-hybrid systems and got positive results for Hd3a, RFT1, AtFD and AtFDP. First, AtFD and AtFDP proteins were co-localized with Hd3a and RFT1 in the nucleus of rice protoplasts derived from mesophyll cells (Supplementary Fig. S1). Although all the bZIP domain proteins, AtFD, AtFDP and rice OsFD1 interacted with phosphatidylethanolamine binding protein (PEBP) family proteins including AtFT, Hd3a, RICE CENTRORADIALIS1 (RCN1) and RCN2 in the yeast systems (Fig. 1), MBP pull-down assays demonstrated that Hd3a does not interact directly with the bZIP proteins such as AtFD, AtFDP and OsFD1. AtFT, however, was able interact with the bZIP proteins in the same system when the MBP:Hd3a was used as a bait protein (Supplementary Fig. S2). This result implies a molecular discrepancy between AtFT and Hd3a in terms of interaction with other proteins including bZIP domain proteins. In the case of Hd3a-OsFD1 interaction, for example, eukaryotic-specific 14-3-3 proteins are required1 and in eukaryotic cells including yeast and plant cells those proteins are believed to act as mediators for the Hd3a-OsFD1 interaction. The phenotypes of transgenic rice plants overexpressing AtFT and Hd3a were also obviously different: rice plants overexpressing Hd3a produced panicles from the callus during tissue culture period whereas none of the transgenic rice overexpressing AtFT exhibited such extreme early flowering (Supplementary Fig. S3). Results of BiFC in vivo assays using rice protoplasts also supported the AtFD/AtFDP-Hd3a and AtFD/AtFDP-RFT1 interactions, although it is unclear whether 14-3-3s are associated for the interactions in the rice cell. Yellow florescence from the reconstructed YFP was observed in the nucleus of rice protoplasts (Fig. 1).
Figure 1. Protein interactions among FTs and FDs.
(A) Structure of AtFD, AtFDP from Arabidopsis and OsFD1 from rice. bZIP domains are marked with yellow boxes. (B) AtFD and AtFDP are able to interact with rice phosphatidylethanolamine-binding protein (PEBP) family proteins as well as AtFT in yeast cells. (C) Positive interactions in the yeast system between AtFD/AtFDP and rice proteins Hd3a and RFT1 were verified in rice protoplasts by BiFC assays. Scale bars = 5 μm.
Overexpression of AtFD and AtFDP produces sheathed panicles with reduced height in rice
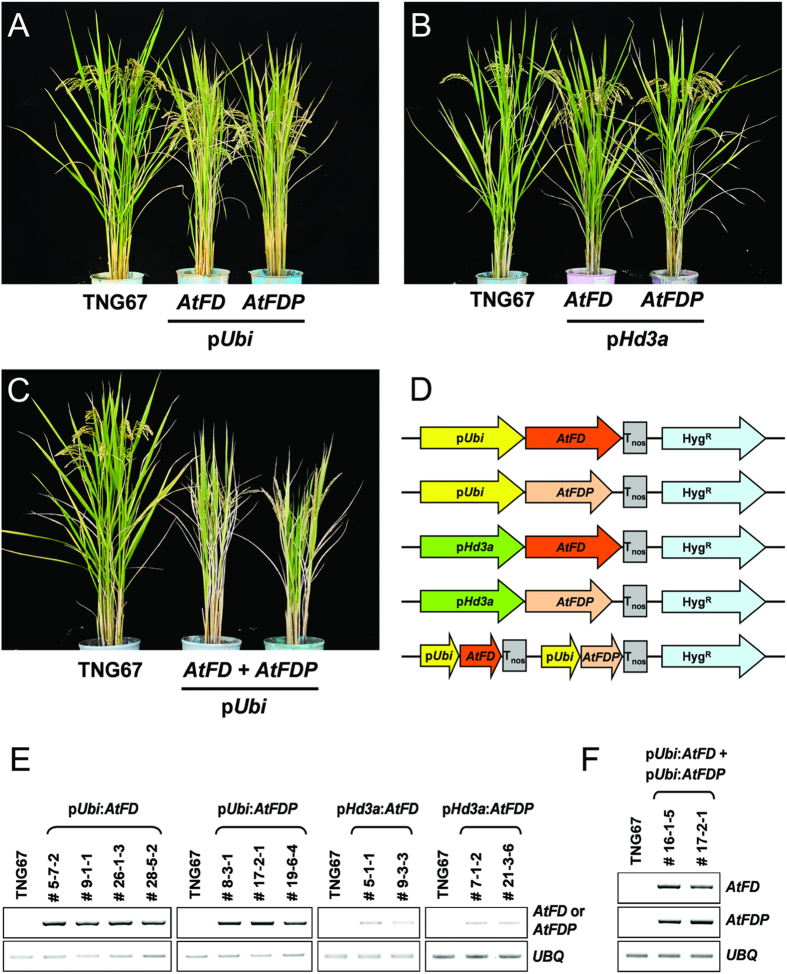
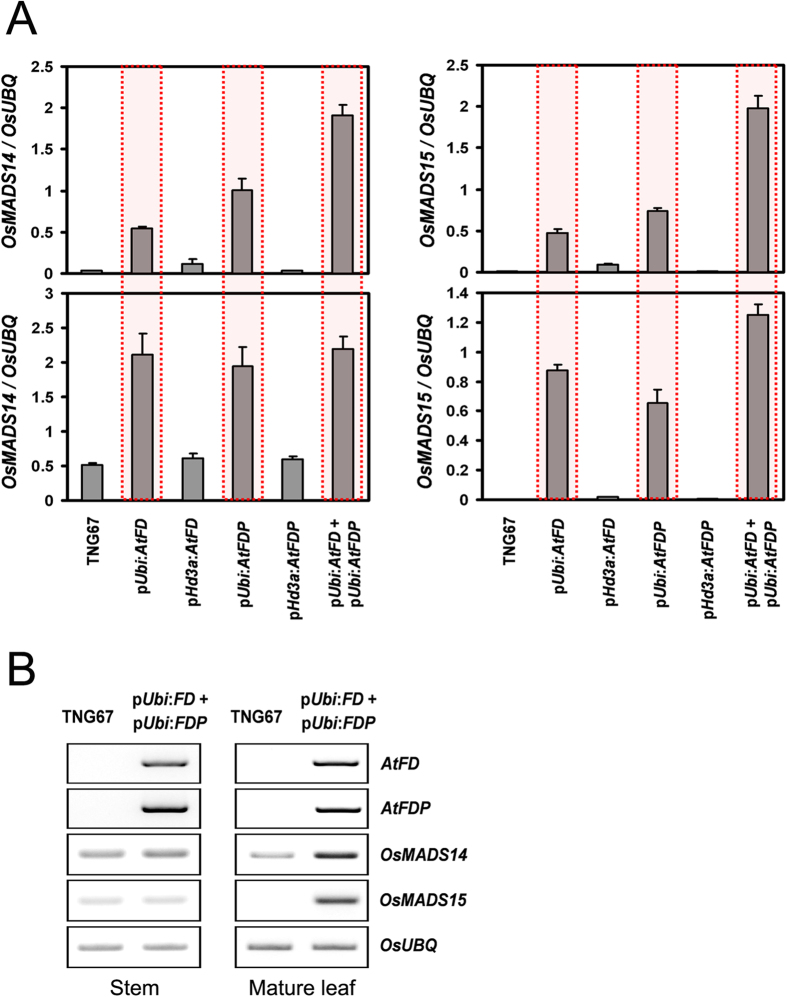
Since AtFD and AtFDP interact with Hd3a and RFT1, the effect of AtFD and AtFDP on rice development was investigated by transgenic approaches. Expression level of each transgene was investigated by RT-PCR using total RNA extracts from leaves of each transgenic rice plant. Compared with the wild-type control, TNG67, transgenic rice plants containing pUbi:AtFD or pUbi:AtFDP construct showed reduced plant height and grain size without significant alteration in heading date (Figs 2,3 and 4). Of particular note, sheathed panicles resulted in reduced fertility and consequently caused loss of yield. Measuring node numbers and the length of internodes in each transgenic plant revealed that the elongation of lower internodes was severely retarded (Fig. 3). Smaller size spikelets and grains were also observed in the transgenic rice plants indicating that overexpression of AtFD and/or AtFDP may affect cell elongation during rice development (Fig. 4). Notably, transcripts of OsMADS14 and OsMADS15 were accumulated in the leaves but not in the stems of transgenic plants although no significant changes in heading date were observed (Fig. 5).
Figure 2. Transgenic rice plants expressing AtFD and/or AtFDP.
(A) Transgenic rice plants expressing AtFD or AtFDP under the control of maize ubiquitin promoter show reduced height. (B) Phloem-specific expression of AtFD or AtFDP driven by Hd3a promoter does not affect rice architecture. (C) Simultaneous overexpression of AtFD and AtFDP in rice causes more severe dwarfism than either of the single overexpressors. (D) Simplified schema of each plasmid used for rice transformation in (A,B) and (C). (E,F) RT-PCR analyses for expression of introduced AtFD and/or AtFDP in the leaves of independent T3 transgenic lines. Original gel images are available in Supplementary Fig. S11A.
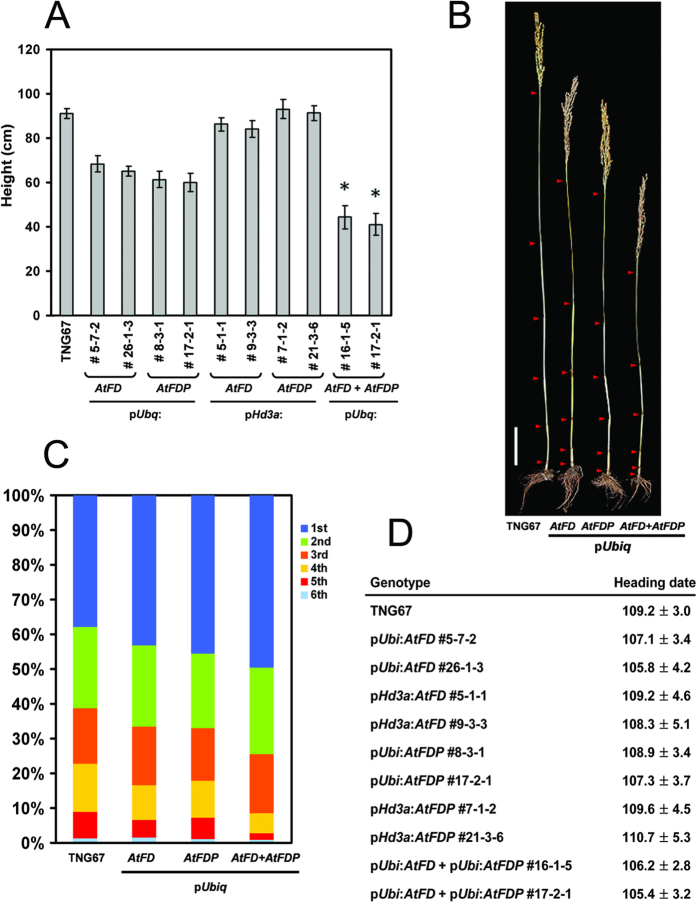
Figure 3. Characterization of transgenic rice plants.
(A) Measurement of plant height. Main culms of six to twelve plants per line were used for height measurement except panicle length. Values are means ± SD (n = 8–12, *P < 0.0001, Student’s t test). (B) Reduced plant height in transgenic rice plants. Arrowheads present each node. Scale bar = 10 cm (C) Proportion of each internode in transgenic rice plants. A representative plant from each line of pUbi:AtFD (#5-7-2), pUbi:AtFDP (#17-2-1) and pUbi:AtFD-pUbi:AtFDP (#16-1-5) was used together with TNG67. (D) Measurement of heading date from eight to twenty individual plants per line. Heading date is recorded based on number of days from sowing to the emergence of the first panicle. Values are means ± SD (n = 8–12).
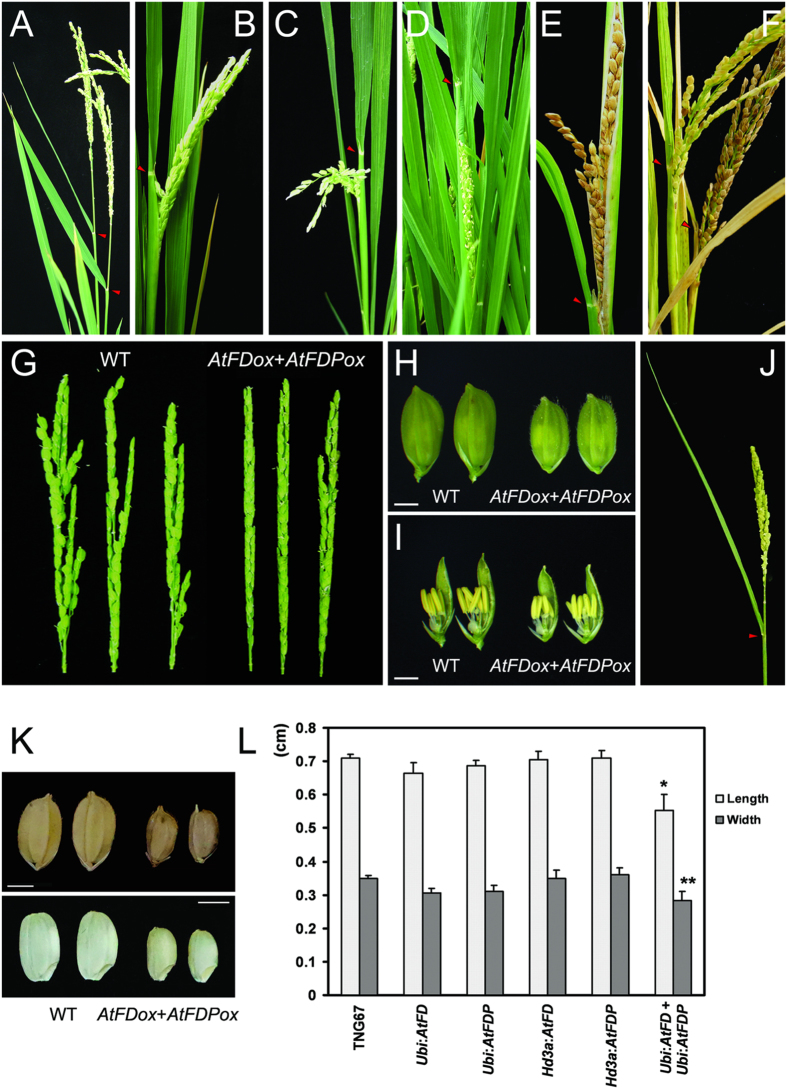
Figure 4. Transgenic rice plants exhibit sheathed panicles, small spikelets and grains.
(A) Panicles of wild-type TNG67. (B,D,E) Panicles of AtFD overexpressors. Sheathed panicles were observed and sometimes grains became mature in the sheath (E). (C) A panicle of a AtFDP overexpressor. (F) Panicles of AtFD-AtFDP dual overexpressor showing sheathed panicles with reduced fertility. The first three panicles from each independent homozygous transgenic and control plants were carefully checked for the sheathed panicle phenotype. All the panicles examined were sheathed in all AtFD, AtFDP and AtFD-AtFDP dual overexpressing transgenic rice plants; this was not observed in TNG67 or transgenic rice containing the empty vector. (G) Panicles of TNG67 (left three panicles) and AtFD-AtFDP dual overexpressors. Each spikelet of transgenic panicles is smaller than WT and panicles are also more compact (G,H,I). (H,I) Transgenic rice plants for AtFD-AtFDP dual overexpression produce smaller spikelets containing tiny floral organs (right two spikelets). (J) A panicle of transgenic rice overexpressing OsbZIP23/OsbZIP72 is indistinguishable from WT. (K,L) Grain size is also reduced in AtFD-AtFDP dual overexpressors. More than 20 grains randomly picked up from each genotype were used for size measurement and similar results were obtained from T3 and T4 generation plants. Values are means ± SD (n > 20, *P < 0.005; **P < 0.05, Student’s t test).
Figure 5. Differential expression of two rice AP1 homologues, OsMADS14 and OsMADS15 between WT and transgenic rice plants.
(A) Expression of OsMADS14 and OsMADS15 is up-regulated only in pUbi:AtFD, pUbi:AtFDP and pUbi:AtFD-pUbi:AtFDP double plants: in the aboveground parts of 50-day old plants (top) and in mature leaves of 100-day old plants. Values are means ± SD of three biological replicates. (B) Expression of OsMADS14 and OsMADS15 is increased in the mature leaves (right panel) but not in the stems of transgenic rice plants at the heading stage. Original gel images are available in Supplementary Fig. S11B.
Simultaneous overexpression of AtFD and AtFDP enhanced the phenotypes observed in pUbi:AtFD or pUbi:AtFDP transgenic rice plants
To investigate whether AtFD and AtFDP act in an additive manner in rice, a genetic construct for simultaneous expression of the two genes, AtFD and AtFDP was generated and introduced into rice plants through Agrobacterium-mediated transformation methods. The insertion of the construct into the rice genome was confirmed by genomic PCR (Supplementary Fig. S4) and the simultaneous expression of AtFD and AtFDP in the leaves of each transgenic plant was also verified by RT-PCR (Fig. 2). The transgenic plants exhibited dwarfism with severe reduction in grain size; lower internodes, in particular, showed heavily retarded elongation (Fig. 3) and the size of epidermal cells in the stem was also reduced (Supplementary Fig. S5). However, the transgenic plants responded to gibberellin (GA3) in cell elongation (Supplementary Fig. S6). The size of spikelets and the epidermal cells of floral organs such as palea were smaller than those of control wild-type plants as well (Supplementary Fig. S5). The length and width of grains from the transgenic rice plants were decreased by an average of 22.5% and 19%, respectively (Fig. 4). All the panicles were sheathed and fertility was also severely affected; only a few grains were properly matured in each plant (Fig. 4). Compared with wild-type pollen, the viability as well as the size of pollen from transgenic rice plants was decreased (Supplementary Fig. S5). Remarkably, the expression of OsMADS14 and OsMADS15, rice AP1 homologues was up-regulated only in the leaves but not in the stems of AtFD-AtFDP dual overexpressors controlled by ubiquitin promoter suggesting a distinct effect of AtFD and/or AtFDP on different cell types (Fig. 5). Since mesophyll cells are abundant in the leaves but not in the stems of rice20 and also phloem-specific expression of AtFD and/or AtFDP did not show any obvious phenotypes such as dwarfism or the reduced spikelet size shown in AtFD/AtFDP overexpressors driven by ubiquitin promoter, the effect of AtFD/AtFDP responsible for those phenotypes seems to mainly occur in mesophyll cells. Otherwise, leaf-preferentially expressed Hd3a/RFT1 may be responsible for the induction of OsMADS14 and OsMADS15 only in the leaves of AtFD/AtFDP overexpressors7,11.
Phloem-specific expression of AtFD and AtFDP by the Hd3a promoter in rice
AtFD and AtFDP were specifically expressed in the phloem tissue of rice by using Hd3a promoter. Expression of the transgenes was checked by RT-PCR using total RNAs extracted from the leaves of each transgenic plant. Unlike transgenic rice plants carrying pUbi:AtFD and pUbi:AtFDP, transgenic rice plants containing pHd3a:AtFD or pHd3a:AtFDP did not show any significant phenotypic alteration in plant height and grain size (Figs 2 and 3). If AtFD and/or AtFDP interact with Hd3a (or RFT1) in rice phloem companion cells, alteration of heading date can be expected by the up-regulation of downstream genes such as OsMADS15 (early heading) or retardation of Hd3a movement to the shoot apex (delayed heading). However, no significant changes in either heading date or expression of OsMADS14 and OsMADS15 were detected in the transgenic plants (Figs 3 and 5). To verify the activity of Hd3a promoter we used, pHd3a:OsFD1 transgenic rice plants were generated. Interestingly, they showed an early heading time with slightly reduced plant height but had no significant reduction in spikelet/grain size demonstrating a molecular functional discrepancy between AtFD/AtFDP and OsFD1 in rice (Supplementary Fig. S7).
Identification of candidate genes responsible for the reduction in stem elongation and spikelet size
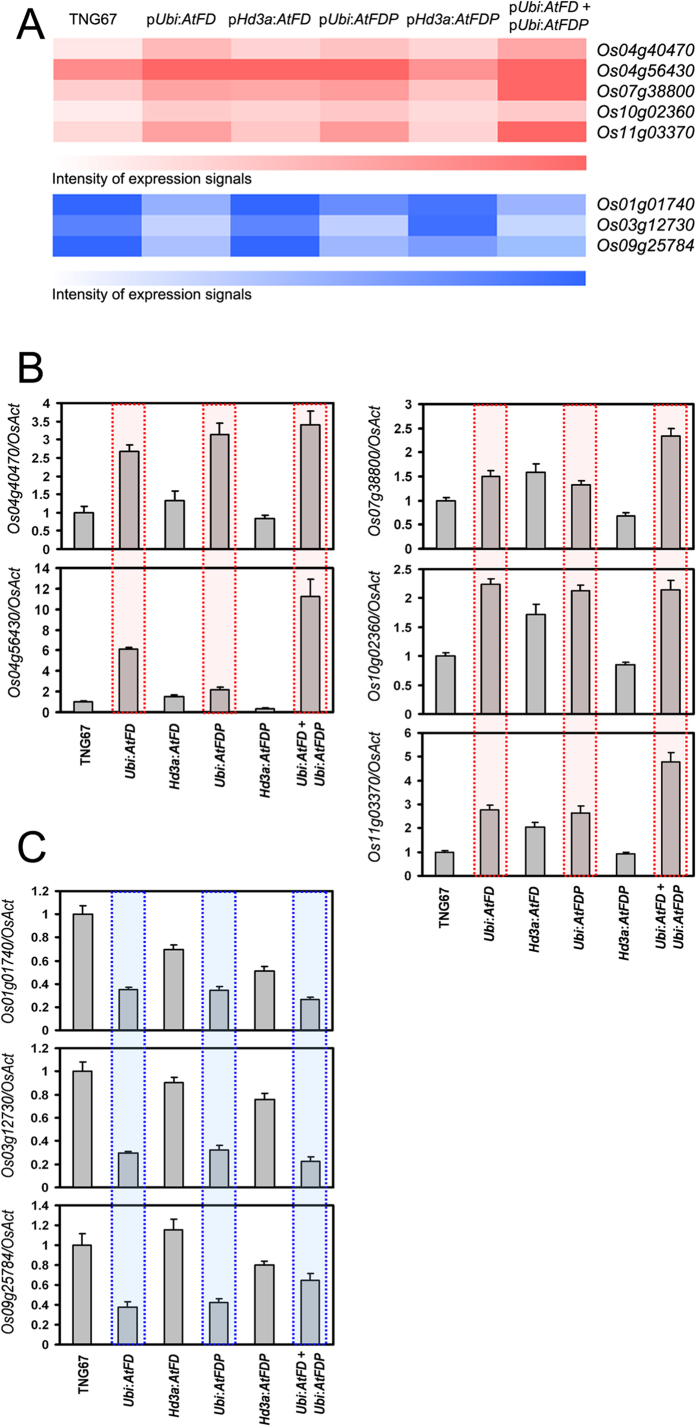
Since transgenic rice plants of pUbi:AtFD, pUbi:AtFDP and pUbi:AtFD-pUbi:AtFDP showed a significant growth defect especially in stem elongation and spikelet development compared to the wild type, TNG67 or transgenic rice plants expressing AtFD or AtFDP in a phloem-specific manner, candidate rice genes responsible for the defective phenotypes of AtFD, AtFDP and AtFD-AtFDP dual overexpressors were identified by analyses of microarray hybridization experiments. Total RNAs from the above-ground parts of 50-day-old transgenic plants for pUbi:AtFD, pUbi:AtFDP, pHd3a:AtFD, pHd3a:AtFDP and pUbi:AtFD-pUbi:AtFDP together with control TNG67 were extracted for microarray hybridization. By comparison of each microarray hybridization result, we identified eight genes displaying significant changes of expression in plants that showed phenotypes: Os04g40470 (encoding a cytochrome p450 family protein), Os04g56430 (encoding a cysteine-rich receptor-like protein kinase), Os07g38800 (encoding a lectin-like receptor kinase), Os10g02360 (encoding a wall-associated kinase), Os11g03370 (encoding a NAC-domain containing transcription factor), Os01g01740 (encoding a protein kinase-domain protein), Os03g12730 (encoding a leucine rich repeat-receptor like kinase) and Os09g25784 (encoding a auxin-induced protein 5NG4). First, we selected up-regulated genes in pUbi:AtFD, pUbi:AtFDP and AtFD-AtFDP dual overexpressors without significant changes of expression in pHd3a:AtFD and pHd3a:AtFDP compared with WT. Next, among the candidates, we narrowed down genes showing higher levels of expression in AtFD-AtFDP dual overexpressors, which showed more severe phenotypes than either of the single gene overexpressors. Down-regulated genes were also selected by the same logic. Thus, the first five genes, Os04g40470, Os04g56430, Os07g38800, Os10g02360 and Os11g03370 were picked up as up-regulated genes and the other three genes, Os01g01740, Os03g12730 and Os09g25784 were selected as genes down-regulated by AtFD and AtFDP overexpression with phenotypes. Quantitative RT-PCR analyses showed that some genes are similar but others such as Os07g38800, Os10g02360 and Os09g25784 have different expression patterns compared to the results of microarray hybridization (Fig. 6).
Figure 6. Confirmation of the expression patterns of candidate genes identified by microarray-based expression changes in the transgenic plants that showed phenotypes.
(A) A heat map showing expression level of candidate genes in each genotype through microarray hybridization. Red color and blue color represent up- and down-regulated candidate genes, respectively. Expression of candidates identified by microarray analyses as up-regulated genes (B) and down-regulated genes (C) in 50-day old plants of pUbi:AtFD, pUbi:AtFDP and pUbi:AtFD-pUbi:AtFDP double plants. Values are means ± SD of three PCR replicates.
Expression changes of candidate genes responsible for short stems
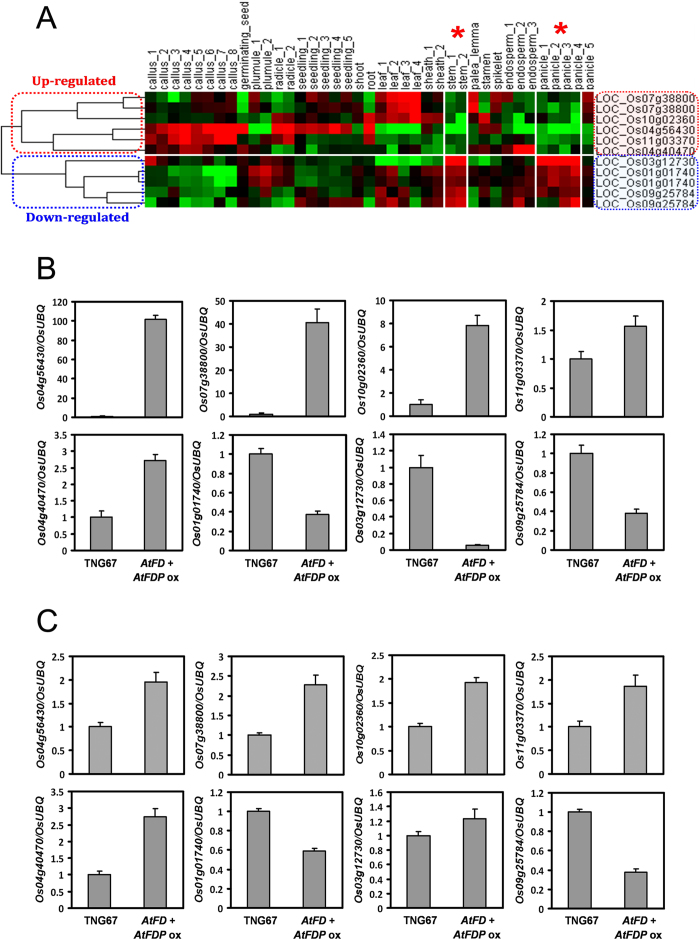
We investigated the expression patterns of each gene identified using different rice organs at different developmental stages (Supplementary Fig. S8). The spatiotemporal expression patterns of the candidate genes are similar to the microarray results available in the public database (http://www.ricearray.org). Notably, candidate genes up-regulated by AtFD and AtFDP overexpression exhibited lower expression levels, whereas genes down-regulated by AtFD and AtFDP overexpression showed a higher expression level in stems and panicles of wild-type rice, in common, based on the public microarray database (Fig. 7). To test whether this expressional relationship is correct, we investigated expression of candidate genes in panicles and stems of AtFD-AtFDP dual overexpressors compared with WT. Indeed, all candidate genes exhibited similar changes in expression between WT and AtFD-AtFDP dual overexpressing stems and panicles (except for Os03g12730 expression in panicles) compared with those in the above-ground parts of 50-day-old WT and AtFD-AtFDP dual overexpressors (Figs 6 and 7).
Figure 7. Generation of a heat map showing spatiotemporal expression and expression analyses of candidate genes in the stems and panicles.
(A) Up-regulated and down-regulated genes are differentially grouped into two clades. Expression of up-regulated genes in stems and panicles is low and that of down-regulated genes is high, in common. Red asterisks indicate stems and panicles. In the heat map, the red color represents an increase and the green color represents a decrease in gene expression, respectively. Verification of expression level of candidate genes in stems (B) and young panicles (C) between WT and AtFD-AtFDP dual overexpressors. Stem and panicle samples we used for expression analyses are matched with the stem_2 stage (heading stage) and panicle_4 stage (developing panicle with length between 40 and 50 mm), respectively in the heat map in (A). Values are means ± SD of three PCR replicates.
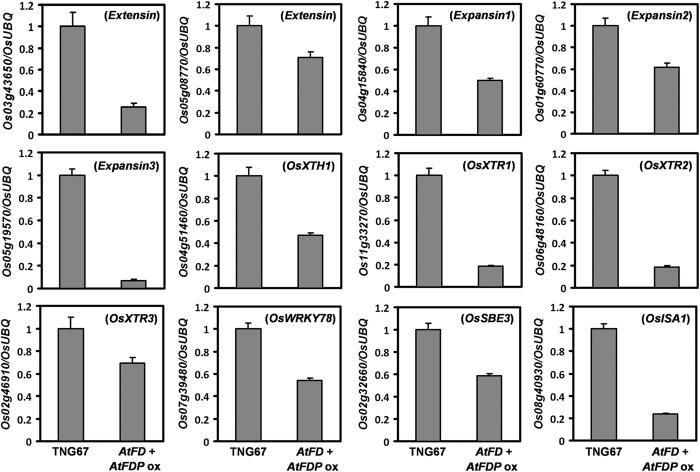
Since we observed short stems composed of smaller epidermal cells in the transgenic rice plants (Supplementary Fig. S5), we compared expression levels of several genes involved in cell elongation in the stems of AtFD-AtFDP dual overexpressors and WT rice plants. Reduced level of expression from all the genes examined was detected in the transgenic stems indicating retarded cell elongation (Fig. 8).
Figure 8. Expression analyses of genes involved in cell elongation in the stems between 100-day-old WT and AtFD-AtFDP dual expressing plants.
Values are means ± SD of three PCR replicates.
Discussion
As florigen-interacting proteins, bZIP transcription factors AtFD and AtFDP are regarded as important players in the flowering of Arabidopsis21. Rice OsFD1 as a component of FAC of rice has also been shown to be responsible for the activation of downstream genes such as OsMADS15, a floral meristem identity gene1. However, it seems that transgenic rice plants with overexpression or knockdown of OsFD1 do not show significant alterations in heading date despite overexpression of OsFD1 for the phosphomimetic form (OsFD1 S192E) causes early heading1. Based on the results of yeast two-hybrid system and BiFC assays in rice protoplasts, AtFD and AtFDP are able to interact not only with AtFT but also Hd3a and RFT1. In rice, 14-3-3 proteins are intracellular Hd3a receptors and the Hd3a-OsFD1 interaction is dependent on the existence of 14-3-3 proteins, which are eukaryote-specific in glutathione S-transferase (GST) pull-down assays1,22. It has also been suggested that the interaction of AtFT with AtFD is indirect, mediated by the 14-3-3 adaptors, with yeast 14-3-3s being able to substitute for plant 14-3-3s in yeast two-hybrid assays for the formation of FAC in the nucleus1 although no direct evidence is available4,17. Interestingly, however, we observed distinct interaction profiles between Hd3a and AtFT with different bZIP proteins in the MBP pull-down assays. We observed positive interactions between AtFT and bZIP proteins under the same conditions as for the negative interaction between Hd3a and OsFD1, which mirrors published results1 implying molecular discrepancy between the two proteins, AtFT and Hd3a. Moreover, transgenic rice plants overexpressing Hd3a always produce spikelets/panicles from the callus during transformation, a trait which was not observed in transgenic rice with AtFT overexpression.
Transgenic rice overexpressing AtFD and AtFDP simultaneously exhibited reduced plant height and size of organs including spikelets, and these phenotypes were more severe than either of the single gene overexpressors. However, phloem-specific expression of AtFD or AtFDP did not cause these phenotypic alterations suggesting that the two bZIP proteins had roles in mesophyll cells. Indeed, transgenic Arabidopsis expressing AtFD under the control of mesophyll cell-specific CHLOROPHYLL A/B BINDING PROTEIN 3 (CAB3) promoter exhibit reduced plant body size with dwarfism23 (Supplementary Fig. S9). Remarkably, overexpressors of OsbZIP23 and OsbZIP72 encoding proteins interacting with Hd3a, RFT1 and GF14c were indistinguishable from wild-type rice implying that the phenotypes driven by AtFD and AtFDP overexpression are gene-specific (Fig. 4, Supplementary Fig. S10). A recent report showing an early heading of transgenic rice plants (TNG67 cultivar) containing pUbi:PaFD, an AtFD homologue of Phalaenopsis orchid also support this notion24.
Of note, the reduced spikelet size of pUbi:OsFD1 or pUbi:OsFD1(S192E) plants indicates that smaller spikelet size is not linked to heading date because there is a significant difference in heading date between pUbi:OsFD1 and pUbi:OsFD1(S192E) transgenic rice plants although the effect of OsFD1 on rice height is unknown1. If the action of AtFD and/or AtFDP introduced into rice is through the formation of FAC-like complexes, it would be worthwhile checking whether GF14s are available in phloem companion cells since no phenotypic alterations were observed in transgenic plants expressing AtFD or AtFDP in the phloem companion cells. Even though there were no significant changes in heading date, notably, the expression of OsMADS14 and OsMADS15 was increased in the leaves but not in the stems of pUbi:AtFD, pUbi:AtFDP and pUbi:AtFD-pUbi:AtFDP double transgenic rice plants indicating that downstream genes affected by AtFD and/or AtFDP are likely to be distinct based on cell types. Also, retarded stem elongation may influence the heading date of the transgenic plants. In that case, the transition of the shoot apex to the reproductive phase may occur early based on increased expression of two rice AP1 genes but retarded elongation of stem may compromise the early flowering phenotype since days for panicle emergence are counted as heading date in rice. Moreover, we cannot exclude the possibility that formation of complexes may occur between introduced AtFD/AtFDP and endogenous RCN proteins whose transcripts are preferentially abundant in the vegetative and reproductive meristems. Actually, RCN1/RCN2-overexpressing transgenic rice plants under the control of 35S promoter resulted in delayed flowering25.
AtFD has been reported to be a flowering activator encoding a bZIP transcription factor that interacts with AtFT, with fd mutants exhibiting delayed flowering17,26. However, there has been no report about its possible effect on vegetative tissue or organ development. In Arabidopsis, Abe et al.17 demonstrated that p35S:AtFD rescued the late flowering of fd-1. Although they did not describe the reduced plant size, the complemented mutant was very tiny. We observed such a small sized plants caused by p35S:AtFD even in wild-type Arabidopsis with a mild early flowering phenotype although phloem-specific AtFD expression in fd-3 mutant did not exhibit such phenotypes showing similar plant size and flowering time to control plants (Supplementary Fig. S9). These results gained from transgenic Arabidopsis approaches are similar to the phenotypes we observed in our transgenic rice plants suggesting common effects of AtFD or AtFDP in mesophyll cells of the two model plant species. In addition, expression of OsFD1 under the control of shoot apex-specific AtFD promoter was able to rescue the late flowering phenotype of fd-3 without affecting plant size indicating that OsFD1 can functionally replace the AtFD gene in Arabidopsis (Supplementary Fig. S9).
Fukazawa et al.27,28 reported that tobacco bZIP-domain protein, REPRESSION OF SHOOT GROWTH (RSG) is also implicated in cell elongation linked to active GA biosynthesis. In addition, RSG interacts with 14-3-3 proteins29, which is similar to several bZIP proteins including wheat EmBP130, Arabidopsis FD, FDP, rice OsFDs5, OsbZIP23 and OsbZIP72 (Supplementary Fig. S10). BZI-1, another tobacco bZIP protein has been implicated in the fine-tuning of auxin-mediated transcription in plants31. Although several rice bZIP proteins interact with Hd3a or RFT1, it is difficult to anticipate whether all of them play similar roles in plant development (Supplementary Fig. S10). For example, overexpression of OsbZIP72, which is regarded as a positive regulator of ABA response and drought tolerance32, did not cause any dwarfism or reduction in organ size in rice despite its binding pattern being similar to those seen in AtFD, AtFDP and OsFD1 with florigenic proteins and GF14c in yeast systems (Supplementary Fig. S10, Fig. 4). Since bZIP proteins are also known to form homo and/or heterodimers33, ectopic expression of bZIP proteins may induce novel phenotypes by interrupting original interactions and/or formation of de novo interactions resulting in transcriptional changes in the cells. Recruitment of transcriptional co-activators and co-repressors can also not be excluded in the formation of FT-FD complexes for the downstream gene regulation2. Moreover, distinct FAC-like complexes depending on transcription factors and/or PEBPs may occur to regulate various developmental and physiological processes in plants. OsFD2, a homologue of OsFD1 can be an example of the latter since OsFD2 forms a FAC with Hd3a and 14-3-3 for controlling leaf development5.
Overexpression of AtFD and AtFDP in rice caused dwarfism with reduced size of the plant body, which is likely due to retarded cell elongation. Indeed, expression of a series of genes involved in cell elongation such as extensins34 and expansins35 is reduced in the stems of AtFD-AtFDP dual overexpressors. In addition, genes that encode cell-wall loosening enzymes necessary for cell elongation such as xyloglucan endotransglycosylase-related (XTR) proteins36,37, xyloglucan endotransglycosylase/hydrolase (XTH) proteins38 as well as OsWRKY78 that encode a transcription factor regulating stem elongation and its putative targets, OsSBE3 and ISA1 show reduced expression in the short stems of AtFD-AtFDP overexpressors compared to WT indicating defective cell elongation in the dual gene overexpressors39. Reduced expression level of genes encoding expansins, extensins and XTHs is also observed in p35S:AtFD transgenic Arabidopsis compared to WT (Supplementary Fig. S9) demonstrating a common molecular effect of AtFD in rice and Arabidopsis. Since the transgenic dual overexpressors still respond to exogenous GA3 treatment, phenotypes of dwarfism and small organs are likely linked to impaired GA biosynthesis in plants.
Candidate rice genes responsible for the phenotypes were identified based on transcriptomic analyses using microarrays. Interestingly, the majority of the candidates were found to encode various types of protein kinases whose expression is influenced by overexpression of AtFD and AtFDP in rice. It is also known that bZIP proteins such as AtFD and RSG are phosphorylated by calcium-dependent protein kinases in Arabidopsis4 and tobacco40.
The dynamic nature of the plant cell wall allows growing cells to expand. Although multiple receptor-like kinases (RLKs) that likely play a role in regulating cell wall function have been identified, many questions remain unanswered about the downstream signaling targets and effects on plant growth and development41. Recently, it was reported that rice leucine-rich repeat receptor-like kinase 1 (LRK1) overexpression increased rice growth by promoting cellular proliferation. One of our candidate genes, Os03g12730 encoding a similar type kinase to LRK1 showed reduced expression in transgenic dwarf plants42. Moreover, higher expression of Os11g03370 that codes for a NAC transcription factor is observed in transgenic rice containing pUbi:AtFD/AtFDP and, recently, it was reported that transgenic rice plants constitutively expressing OsNAC2 (Os04g38720) caused shorter plant height with shorter spikelets, which may support our results since both NAC transcription factors, Os11g03370 and OsNAC2 belong to the same NAM/CUC3 subgroup of NAC transcription factor family in rice43,44. Functional characterization of the candidate genes identified in this study as being likely responsible for the phenotypes of transgenic rice may provide valuable information for potential application in the manipulation of plant architecture in cereals.
Methods
Plant materials and growth conditions
Japonica rice (Oryza sativa L.) variety Tainung67 (TNG67) and Arabidopsis Columbia-0 (Col) were used as wild type. TNG67, a photoperiod insensitive flowering rice cultivar containing defective Hd1 and Ehd1 sequences the same as those of Taichung65 (T65)45,46 was used to produce transgenic rice plants and the transgenic plants were grown in a growth chamber (14 h L, 28 °C/10 h D, 26 °C) for 2 weeks after germination and moved to the outdoor GMO greenhouse of the Academia Sinica Biotechnology Center in Southern Taiwan (23°04′N 120°17′E). Phenotypes of transgenic rice were examined from the second crop period (July to November) of 2011 to the first crop period (February to June) 2016. Transgenic plants used for analyses in this work are all T3 or T4 independent homozygous lines. Control rice plants with an empty vector were also generated and it was verified that they are almost identical to TNG67 in heading date, plant height and grain size. Generally, Arabidopsis plants were grown in the growth chamber under LD conditions (16/8-h photoperiod at 100 μmol m−2 s−1) at 22 °C. fd-3 in the Columbia background has been previously described17. For Arabidopsis flowering time measurement, 8 to 12 plants per line were counted for total leaf numbers when their first flowers were at anthesis. Days of heading of 10 to 12 rice plants per each line were measured when panicles were emerged.
GA3 treatment
Rice seedlings were germinated and incubated on MS media for 10 days and transferred to test tubes containing water (mock) or GA3 (5 μM) solution. The plants were grown for 2 days under 12 h-light photoperiod at constant temperature of 28 °C, and then taken out for cryo-scanning electron microscopy47.
Yeast two-hybrid experiments and BiFC assays
AtFT, Hd3a, RCN1 and RCN2 full-length ORFs were cloned in-frame in the pBD-GAL4 Cam vector (Stratagene) to generate bait constructs. The pAD-GAL4 vector (Stratagene) was used for generation of preys including pAD:AtFD, pAD:AtFDP and pAD:OsFD1 constructs. X-gal filter assays were performed as described previously24,47. For cellular localization of Hd3a, RFT1, AtFD and AtFDP in rice, either yellow florescence protein (YFP):gateway (GW) or cyan fluorescent protein (CFP):GW vector was used for the florescence fusion as described previously24. For bimolecular florescence complementation (BiFC) assays in rice, each cDNA of Hd3a and RFT1 was cloned into pVYCE vector for Hd3a:cYFP and RFT1:cYFP fusions. AtFD and AtFDP cDNAs were cloned into pVYNE vector for nYFP:AtFD and nYFP:AtFDP fusions, respectively48,49. Isolation and transfection of rice protoplasts were followed as described by Zhang et al.50 and images of cells with fluorescence were taken by confocal microscopy (LSM 510 META NLO DuoScan, Carl Zeiss).
MBP pull-down assay
pMAL-4cx vector (NEB) and pET201 vector51 were used for production of maltose binding protein (MBP) fusion proteins and 6× histidine (His)-tag fusion proteins, respectively. AtFT and Hd3a cDNA fragments for each coding region with XbaI/SalI and EcoRI/SalI ends, respectively, were ligated into SpeI/SalI or EcoRI/SalI-treated pMAL-4cx vector for MBP:AtFT and MBP:Hd3a proteins. AtFD, AtFDP and OsFD1 cDNA fragments digested by SpeI/XhoI were introduced between NheI and XhoI sites of pET201 for His-tag fusion. Transformed cells [E. coli strain Rosetta (DE3), Novagen] were grown and the tagged proteins were induced with 1 mM IPTG when the culture reached an OD600 value of 0.6. After 20 hours of induction at 16 °C, the cells were harvested and homogenized by sonication in either MBP buffer (20 mM Tris pH 8.0, 1 mM EDTA, 200 mM NaCl), for MBP-tagged proteins, or 1XPBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), for HIS-tagged proteins. MBP-tagged proteins were immobilized onto Amylose Resin (NEB) and incubated with His-tagged proteins-containing crude extract in the binding buffer (20 mM Tris pH 8.0, 1 mM EDTA, 100 mM NaCl, 0.1% NP40, 1 mM PMSF) for 3 hours at 4 °C with constant rotation. The unbound residues were removed by washing with binding buffer 6 times. Proteins with affinity to the resins were separated on 12% SDS polyacrylamide gels and detected by western blot.
Expression analysis
Total RNAs from plant materials were extracted using an RNeasy Plant Mini Kit (Qiagen) and treated with RNase-free DNase (Invitrogen) following the manufacturer’s protocol to remove any residual genomic DNA. DNase-treated RNA was subjected to reverse transcriptase reactions using oligo (dT) primer and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. Subsequent PCR was performed with the first-strand cDNA mixture and EX-Taq polymerase (Takara Bio). qPCR was performed on a CFX96TM real-time system (Bio-Rad) using Maxima SYBR Green qPCR Master Mix (Thermo). The primers used for quantification are listed in Supplementary Table S1. For PCR, each sample was analyzed in triplicate. The run protocol was: denaturation at 95 °C for 10 min and annealing/extension repeated 45 times (95 °C for 15 s and 60 °C for 30 s, data acquisition was performed). Housekeeping genes such as OsUBQ11, OsAct52, AtPEX453 and AtACT54 were included in the reactions as internal controls for normalizing the variations in the amount of cDNA used55. The threshold cycle (CT) was automatically determined for each reaction by the system set with default parameters. The specificity of the quantitative reverse transcription-PCR (qRT-PCR) was determined by curve analysis of the amplified products using the standard method installed in the system. Information on primers used is presented in Supplementary Table S1.
Plant transformation and analyses of transgenic plants
For pSUC2:AtFD, pSUC2:AtFDP and pAtFD:OsFD1 constructs, AtFD, AtFDP and OsFD1 entry clones were inserted into pSUC2:GW and/or pAtFD:GW destination vectors, respectively24,56. For pCAB3:AtFD, 1,537 bp CAB3 promoter was fused to AtFD as described previously57. All plasmids for plant transformation were introduced into Agrobacterium strain GV3101 (pMP90RK) and transformed into Col WT or fd-3 plants by the floral-dip method58. For overexpression of target genes in rice, the binary vector pGA3426 and its derivative pGA3777 were used, and each transgene construct was introduced into the rice genome by Agrobacterium-mediated transformation59. For pHd3a:AtFD, pHd3a:AtFDP and pHd3a:OsFD1, ubiquitin promoter was exchanged for 2.2 Kb-promoter of Hd3a; 2,153 bp Hd3a promoter fragment amplified by primers 5′CGCGGATCCAGGCATCAGATTAAGACAGCAACGCAG3′ and 5′GCGACTAGTCGATCTTGCAAAAAACCCTGAAGGTTTATAG3′ were used. Restriction enzyme sites for BamHI and SpeI are underlined, respectively, which is longer than the one Hayama et al.60 used previously and then AtFD, AtFDP and OsFD1 were introduced in the plasmid, respectively. More than 15 independent primary transgenic plants were obtained and analyzed, and at least two independent homozygous lines were selected for phenotypic description.
Microarray analyses
The whole above ground parts of two independent homozygous plants per each genotype (50-day old plants) were harvested for RNA extraction for microarray. We used the Rice Whole Genome OneArray v1.1 (Phalanx Biotech Group, Taiwan), which contains 22,003 DNA oligonucleotide probes, and each probe is a 60-mer designed in the sense direction. Among the probes, 21,179 probes corresponded to the annotated genes in the RGAP v.6.1 and BGI database. In addition, we included 824 control probes. The detailed descriptions of the gene array list are available from http://www.phalanx.com.tw/products/RiOA_Probe.php. Fluorescent antisense RNA (aRNA) targets were prepared from 1 μg total RNA samples with the OneArray Amino Allyl aRNA Amplification Kit (Phalanx Biotech Group, Taiwan) and Cy5 dyes (Amersham Pharmacia, Piscataway, NJ, USA). Fluorescent targets were hybridized to the Rice OneArray with Phalanx hybridization buffer using Phalanx Hybridization System. After hybridization for 16 h at 50 °C, non-specific binding targets were washed away by three different washing steps (Wash I, 42 °C 5 min; Wash II, 42 °C, 5 min; 25 °C, 5 min; Wash III, rinse 20 times), and the slides were dried by centrifugation and scanned by an Agilent G2505C scanner (Agilent Technologies, Santa Clara, CA, USA). The Cy5 fluorescent intensities of each spot were analyzed by GenePix 4.1 software (Molecular Devices). The signal intensity of each spot was loaded into Rosetta Resolver System (Rosetta Bio-software) to process data analysis. The error model of the Rosetta Resolver System could remove both systematic and random errors from the data. We filtered out the spots whose flag was less than 0. Spots that passed the criteria were normalized by 50% media scaling normalization method. The technical repeat data was tested by Pearson correlation coefficient calculation to check the reproducibility (R value > 0.975). Normalized spot intensities were transformed to gene expression log2 ratios between the control and treatment groups. The interest spots which show significant differences were selected by log2 ratio ≥1 or log2 ratio ≤−1 and P < 0.05. Two independent biological replicates of hybridizations were performed.
Hierarchical cluster analysis
The raw data of expression values were downloaded from the microarray experiment, and the developmental transcriptomes of rice were dissected using the Internet website “Rice Oligonucleotide Array Database” (http://www.ricearray.org). Then, the raw data were inputted into Gene Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/). The graphic of clustering heat map was created using with Alok Saldanha’s Java TreeView v1.1.6r2.
Additional Information
Accession codes: AtFD (At4g35900), AtFDP (At2g17770, AtbZIP27), Hd3a (Os06g06320), OsFD1 (Os09g36910), RCN1 (Os11g05470), RCN2 (Os02g32950), RFT1 (Os06g06300), GEO accession number for microarray results (GSE93822)
How to cite this article: Jang, S. et al. Ectopic expression of Arabidopsis FD and FD PARALOGUE in rice results in dwarfism with size reduction of spikelets. Sci. Rep. 7, 44477; doi: 10.1038/srep44477 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank members of core facility laboratories of Academia Sinica for microscopy and DNA sequencing. We also thank Dr. Julien Kuraba and Dr. Iju Chen for set up of the MBP pull-down assays, Dr. Gynheung An for pGA vectors for rice transformation, Ms. Pei-Chun Liao for technical assistance in plant transformation and Ms. Miranda Loney for help with English editing. This work was supported in part by grants from NSC, Taiwan (101-2313-B-001-005- to SJ) and BCST of ABRC, Academia Sinica.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.J. designed the experiments. S.J., H.-Y.L. and M.-L.K. performed experiments, analyzed the results and prepared the figures. S.J. wrote the paper. All authors reviewed the manuscript.
References
- Taoka K.-i. et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335, doi: http://www.nature.com/nature/journal/v476/n7360/abs/nature10272.html#supplementary-information (2011). [DOI] [PubMed] [Google Scholar]
- Ho W. W. H. & Weigel D. The Plant Cell 26, 552–564, doi: 10.1105/tpc.113.115220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M. et al. The Plant Cell 25, 1228–1242, doi: 10.1105/tpc.112.109090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto N., Sasabe M., Endo M., Machida Y. & Araki T. Scientific Reports 5, 8341, doi: 10.1038/srep08341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Nakamura H., Taoka K.-i. & Shimamoto K. Plant and Cell Physiology 54, 385–397, doi: 10.1093/pcp/pct005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Taoka K.-i. & Shimamoto K. Current Opinion in Plant Biology 14, 45–52, doi: 10.1016/j.pbi.2010.08.016 (2011). [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H. L., Yokoi S. & Shimamoto K. Science 316, 1033–1036, doi: 10.1126/science.1141753 (2007). [DOI] [PubMed] [Google Scholar]
- Faure S., Higgins J., Turner A. & Laurie D. A. Genetics 176, 599–609, doi: 10.1534/genetics.106.069500 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T. et al. Genes & Development 16, 2006–2020, doi: 10.1101/gad.999202 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S. et al. Plant and Cell Physiology 43, 1096–1105, doi: 10.1093/pcp/pcf156 (2002). [DOI] [PubMed] [Google Scholar]
- Komiya R., Ikegami A., Tamaki S., Yokoi S. & Shimamoto K. Development 135, 767–774 (2008). [DOI] [PubMed] [Google Scholar]
- Navarro C. et al. Nature 478, 119–122, doi: http://www.nature.com/nature/journal/v478/n7367/abs/nature10431.html#supplementary-information (2011). [DOI] [PubMed] [Google Scholar]
- Hsu C.-Y. et al. Proceedings of the National Academy of Sciences of the United States of America 108, 10756–10761, doi: 10.1073/pnas.1104713108 (2011). [DOI] [PMC free article] [PubMed]
- Krieger U., Lippman Z. B. & Zamir D. Nat Genet 42, 459–463, doi: http://www.nature.com/ng/journal/v42/n5/suppinfo/ng.550_S1.html (2010). [DOI] [PubMed] [Google Scholar]
- Kinoshita T. et al. Current Biology 21, 1232–1238, doi: 10.1016/j.cub.2011.06.025 (2011). [DOI] [PubMed] [Google Scholar]
- Lee R., Baldwin S., Kenel F., McCallum J. & Macknight R. Nat Commun 4, doi: 10.1038/ncomms3884 (2013). [DOI] [PubMed] [Google Scholar]
- Abe M. et al. Science 309, 1052–1056, doi: 10.1126/science.1115983 (2005). [DOI] [PubMed] [Google Scholar]
- Denison F. C., Paul A.-L., Zupanska A. K. & Ferl R. J. Seminars in Cell & Developmental Biology 22, 720–727, doi: 10.1016/j.semcdb.2011.08.006 (2011). [DOI] [PubMed] [Google Scholar]
- Purwestri Y. A., Ogaki Y., Tamaki S., Tsuji H. & Shimamoto K. Plant and Cell Physiology 50, 429–438, doi: 10.1093/pcp/pcp012 (2009). [DOI] [PubMed] [Google Scholar]
- Maiti R., Satya P., Rajkumar D. & Ramaswamy A. Crop plant anatomy. (Wallingford, UK, CAB International, 2012) [Google Scholar]
- Jaeger K. E., Pullen N., Lamzin S., Morris R. J. & Wigge P. A. The Plant Cell 25, 820–833, doi: 10.1105/tpc.113.109355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R. & Robatzek S. Molecular Plant-Microbe Interactions 28, 511–518, doi: 10.1094/MPMI-10-14-0322-CR (2015). [DOI] [PubMed] [Google Scholar]
- Susek R. E., Ausubel F. M. & Chory J. Cell 74,787–799. [DOI] [PubMed] [Google Scholar]
- Jang S., Choi S.-C., Li H.-Y., An G. & Schmelzer E. PLoS ONE 10, e0134987, doi: 10.1371/journal.pone.0134987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Shimamoto K. & Kyozuka J. The Plant Journal 29, 743–750, doi: 10.1046/j.1365-313X.2002.01255.x (2002). [DOI] [PubMed] [Google Scholar]
- Wigge P. A. et al. Science 309, 1056–1059 (2005). [DOI] [PubMed] [Google Scholar]
- Fukazawa J. et al. Plant Signaling & Behavior 6, 26–28, doi: 10.4161/psb.6.1.14114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J. et al. The Plant Cell 12, 901–916 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S., Fukazawa J., Yuasa T. & Takahashi Y. The Plant Cell 16, 2641–2651, doi: 10.1105/tpc.104.024604 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz T. F., Medina J., Hill A. & Quatrano R. S. The Plant Cell 10, 837–847 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinekamp T. et al. The Plant Journal 38, 298–309, doi: 10.1111/j.1365-313X.2004.02043.x (2004). [DOI] [PubMed] [Google Scholar]
- Lu G., Gao C., Zheng X. & Han B. Planta 229, 605–615, doi: 10.1007/s00425-008-0857-3 (2009). [DOI] [PubMed] [Google Scholar]
- Iven T. et al. The Plant Journal 63, 155–166, doi: 10.1111/j.1365-313X.2010.04230.x (2010). [DOI] [PubMed] [Google Scholar]
- Lamport D. T. A., Kieliszewski M. J., Chen Y. & Cannon M. C. Plant Physiology 156, 11–19, doi: 10.1104/pp.110.169011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H. T. & Kende H. The Plant Cell 9, 1661–1671, doi: 10.1105/tpc.9.9.1661 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K. et al. The Plant Journal 47, 519–531, doi: 10.1111/j.1365-313X.2006.02804.x (2006). [DOI] [PubMed] [Google Scholar]
- Uozu S., Tanaka-Ueguchi M., Kitano H., Hattori K. & Matsuoka M. Plant Physiology 122, 853–860 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R., Rose J. K. C. & Nishitani K. Classification and Expression Analysis. Plant Physiology 134, 1088–1099, doi: 10.1104/pp.103.035261 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-Q. et al. Planta 234, 541–554, doi: 10.1007/s00425-011-1423-y (2011). [DOI] [PubMed] [Google Scholar]
- Ishida S., Yuasa T., Nakata M. & Takahashi Y. The Plant Cell 20, 3273–3288, doi: 10.1105/tpc.107.057489 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinwand B. J. & Kieber J. J. Plant Physiology 153, 479–484, doi: 10.1104/pp.110.155887 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha X. et al. Plant Biotechnology Journal 7, 611–620, doi: 10.1111/j.1467-7652.2009.00428.x (2009). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. The Plant Journal 82, 302–314, doi: 10.1111/tpj.12819 (2015). [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M. et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 465, 30–44, doi: 10.1016/j.gene.2010.06.008 (2010). [DOI] [PubMed] [Google Scholar]
- Doi K. et al. Genes Dev. 18, 926–936 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. D. et al. Botanical Studies 54, 12, doi: 10.1186/1999-3110-54-12 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S. Plant and Cell Physiology 56, 2234–2247, doi: 10.1093/pcp/pcv130 (2015). [DOI] [PubMed] [Google Scholar]
- Citovsky V. et al. Journal of Molecular Biology 362, 1120–1131, doi: 10.1016/j.jmb.2006.08.017 (2006). [DOI] [PubMed] [Google Scholar]
- Tzfira T. et al. Plant Molecular Biology 57, 503–516, doi: 10.1007/s11103-005-0340-5 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Plant Methods 7, 30–30, doi: 10.1186/1746-4811-7-30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M., Farrona S., Huettel B., Koncz Z. & Koncz C. CDKF;1 and CDKD Protein Kinases Regulate Phosphorylation of Serine Residues in the C-Terminal Domain of Arabidopsis RNA Polymerase II. The Plant Cell 24, 1626–1642, doi: 10.1105/tpc.112.096834 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W. et al. Annals of Botany, doi: 10.1093/aob/mct212 (2013). [DOI] [Google Scholar]
- Gregis V. et al. Genome Biology 14, 1–26, doi: 10.1186/gb-2013-14-6-r56 (2013). [DOI] [Google Scholar]
- Jang S., Torti S. & Coupland G. Plant J 60 (2009). [DOI] [PubMed] [Google Scholar]
- Guénin S. et al. Journal of Experimental Botany 60, 487–493, doi: 10.1093/jxb/ern305 (2009). [DOI] [PubMed] [Google Scholar]
- An H. et al. Development 131, 3615–3626, doi: 10.1242/dev.01231 (2004). [DOI] [PubMed] [Google Scholar]
- Ranjan A., Fiene G., Fackendahl P. & Hoecker U. Development 138, 1851–1862, doi: 10.1242/dev.061036 (2011). [DOI] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. The Plant Journal 16, 735–743, doi: 10.1046/j.1365-313x.1998.00343.x (1998). [DOI] [PubMed] [Google Scholar]
- Kim S.-R., Lee D.-Y., Yang J.-I., Moon S. & An G. Journal of Plant Biology 52, 73–78, doi: 10.1007/s12374-008-9008-4 (2009). [DOI] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M. & Shimamoto K. Nature 422, 719–722, doi: http://www.nature.com/nature/journal/v422/n6933/suppinfo/nature01549_S1.html (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.