Abstract
Heard Island in the Southern Ocean is a biological hotspot that is suffering the effects of climate change. Significant glacier retreat has generated proglacial lagoons, some of which are open to the ocean. We used pyrotag sequencing of SSU rRNA genes and environmental data to characterize microorganisms from two pools adjacent to animal breeding areas, two glacial lagoons and Atlas Cove (marine site). The more abundant taxa included Actinobacteria, Bacteroidetes and Proteobacteria, ciliates and picoflagellates (e.g. Micromonas), and relatively few Archaea. Seal Pool, which is rich in organic matter, was characterized by a heterotrophic degradative community, while the less eutrophic Atlas Pool had more eucaryotic primary producers. Brown Lagoon, with the lowest nutrient levels, had Eucarya and Bacteria predicted to be oligotrophs, possess small cell sizes, and have the ability to metabolize organic matter. The marine influence on Winston Lagoon was evident by its salinity and the abundance of marine-like Gammaproteobacteria, while also lacking typical marine eucaryotes indicating the system was still functioning as a distinct niche. This is the first microbiology study of Heard Island and revealed that communities are distinct at each location and heavily influenced by local environmental factors.
Heard Island (53°05′S, 73°30′E) is a 367 km2 volcanic island located ~4350 km southwest of Western Australia on the Kerguelen Plateau1. Surrounded by cold Antarctic waters and located south of the Antarctic Polar Front, it experiences strong and persistent westerly winds, low seasonal and daily temperature ranges, and high precipitation2. The vast bulk of the island is covered in glaciers, which radiate from the summit of the intermittently active volcanic peak Big Ben (2750 m above sea level) to the tidewater (those with northerly to southwesterly aspects) or terminate on land or in lagoons inland from the ocean (those with southerly to northeasterly aspects)3 (Fig. 1). Isolated vegetated headlands emerge between the glaciers, and together with the exposed plains and shoreline, are colonized by very high numbers of breeding and non-breeding mammals and seabirds4,5,6. Coastal eutrophication due to input from penguins, other seabirds and seals, combined with long daylight hours in summer, can lead to high primary production4,7. In contrast to the studies of the geology, climate, vegetation and wildlife (invertebrates, fish, marine mammals and birds) of Heard Island8, and despite the possible presence of microorganisms in lava tube caves9 and the potential of microorganisms to cause disease on the island10, no studies characterizing microorganisms from this World-Heritage-listed site have been reported.
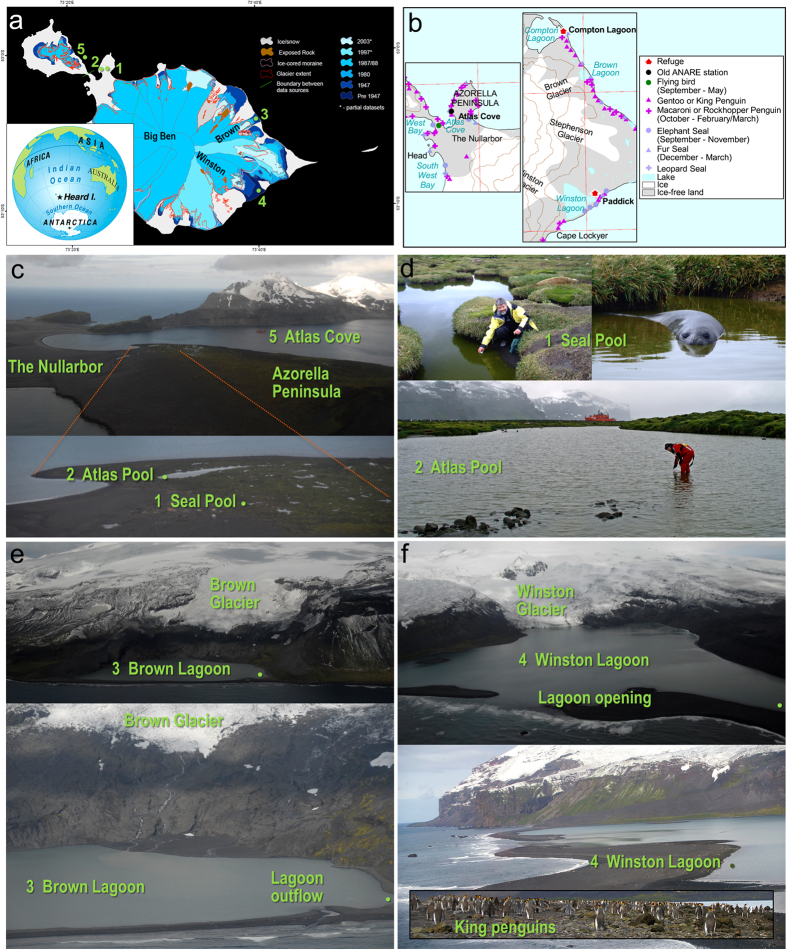
Figure 1. Heard Island sampling sites.
(a) Map showing change in glacier boundaries from pre 1947 to 2006–2009 (red outline), with sample collection sites numbered: 1, Seal Pool; 2, Atlas Pool; 3, Brown Lagoon; 4, Winston Lagoon; 5, Atlas Cove. Modified from a map courtesy of the Australian Antarctic Data Centre, CAASM metadata record ID Heard_Island_digitising_2009, map number 13691, © Commonwealth of Australia 2009. Inset: Depiction of Earth showing the isolation of Heard Island relative to surrounding continents with the closest being Antarctica (1,700 km to Davis Station), Australia (4,100 km to Perth) and Africa (4,800 to Johannesburg). Image courtesy of the Australian Geographic Cartographic Division. (b) Distribution of wildlife near the sample collection sites. Sections of a map courtesy of the Australian Antarctic Data Centre, map number 12583, © Commonwealth of Australia 2000. (c) Seal Pool (1) and Atlas Pool (2) on the Azorella Peninsula relative to the Atlas Cove (5) location where seawater was collected from the Aurora Australis (red ship). Photographer Gary Miller. (d) Sample collection at Seal Pool with elephant seal in Seal Pool prior to sampling, and Atlas Pool with Aurora Australis in the background. Photographers the Australian Antarctic Division and Rick Cavicchioli. (e) Aerial views of Brown Glacier above Brown Lagoon showing the rocky sill separating the lagoon from the ocean, and the lagoon outflow and approximate sampling sites. Photographer Gary Miller. (f) Aerial views of Winston Glacier and Winston Lagoon showing the glacier tongue in contact with the lagoon, and the opening of the lagoon permitting influx from the ocean, the approximate sampling site, and king penguins (inset) on the ocean-side shore of the lagoon at the time of sampling. Photographer Gary Miller and Stephen Brown.
Consistent with an almost 1 °C rise in the average annual air temperature of Heard Island from 1948–1954 to 1997–200111, total glacier coverage has decreased from 288 km2 in 1947 (79% coverage) to 257 km2 in 1988 (70%) and 231 km2 in 2008 (63%)12,13 (Fig. 1a). While all glaciers have retreated, individual glaciers to the east of Big Ben have been particularly affected and closely monitored, both during the infrequent research expeditions to the island and via satellite. In 1947 Winston Glacier had already retreated from the shoreline, and continued to retreat a further 1.6 km by 196314. It then advanced during the 1970s, only to retreat again in the 1980s12. The Brown and Stephenson glaciers began to retreat in the 1960s14, with proglacial lagoons developing by the mid-1980s. The terminus of Brown Glacier retreated at an average rate of 30 m per year between 1947 and 2004 to reach a total of 1.2 km from the coast, resulting in a loss of 29% of its original area, which is a remarkable change given its 25 m high terminal ice cliffs were calving into the ocean in 19473. Between January 2004 and January 2006, Brown Glacier retreated <0.1 km and Winston Glacier retreated by ~0.2 km13,15. Based on data from 1947 and 2008, ~55 km2 or 15% of the island’s terrain has been exposed by the retreating ice, revealing or expanding several large proglacial lagoons and making newly deglaciated land available for colonization. Together with higher ambient temperatures and increased availability of liquid water (more precipitation as rain rather than snow), animal, plant and microbial communities will be able to expand into new niches. The effect of these changes on terrestrial and lacustrine food webs will be important to establish and monitor.
Compared to other Sub-Antarctic islands which experience tourism or more regular research visits, Heard Island is largely pristine and remarkably free of invasive species16. However, climate change and warming conditions increase the potential for non-native species to become established. Moreover, the Kerguelen Plateau is the largest volcanic plateau in the Southern Ocean and provides a major source of iron that stimulates phytoplankton blooms around, and to its east1. Climate-related changes have implications for the marine environment surrounding Heard Island, as increased freshwater from melting glaciers will carry iron and nitrogen rich minerals, as well as microbial populations with as yet uncharacterized roles in C, N and P cycling, out into the sea and eastward along the flow of the Antarctic Circumpolar Current.
In order to begin the characterization of microbial communities present in pools and lagoons on Heard Island, pyrotag sequencing of SSU rRNA genes was performed on biomass from water samples collected in December 2008 from two proglacial lagoons (Brown Lagoon and Winston Lagoon), two pools formed from rainwater and snowmelt adjacent to seal and penguin breeding areas on the Azorella Peninsula, and seawater from Atlas Cove (Fig. 1). The biomass was captured by sequential fractionation through a 20 μm prefilter onto 3.0-, 0.8-, and 0.1-μm filters using an approach previously adopted for Southern Ocean17 and Antarctic lake systems18 that facilitates taxonomic analyses by reducing the overall complexity and enabling size partitioning of samples19,20. By combining environmental with pyrotag sequencing data, we were able to infer important functional processes and the environmental factors affecting communities and ecosystem function. This snapshot provides the first insight into microbial community diversity and richness in aquatic systems on Heard Island, and provides a baseline for ongoing monitoring of this unique environment that is subject to significant environmental change.
Results
Water composition of each location
Seal Pool and Atlas Pool were pools designated in this study (see Methods and Fig. 1). Chemical analysis revealed a great deal of variation among the five aquatic locations (Table 1). All waters were brackish, with Seal Pool the closest to freshwater, followed by Atlas Pool, and then Brown Lagoon with conductivity of 6580 μS cm−1. The conductivity of Winston Lagoon indicates it was ~72% as saline as typical seawater (taken as 55000 μS cm−1; ref. 21), with the conductivity of the Atlas Cove site being 88% of seawater salinity, which may reflect the input of glacial meltwater from Heard Island. Sulfur levels increased with increasing salinity. Brown Lagoon had the lowest phosphorus levels, with all samples containing ≥27 μg P L−1. Nitrogen compounds were most prevalent in Seal and Atlas pools, with highest levels of ammonia (24.9 mg N L−1) in Seal Pool and nitrate in Atlas Pool (4.32 mg N L−1). Brown Lagoon had the lowest levels of ammonia, nitrate, nitrite and total nitrogen, even though it had a moderate level of dissolved organic carbon (DOC) (27 mg L−1). Seal Pool had the highest DOC (170 mg L−1).
Table 1. Water composition analysis.
| Analyte | Units | Seal Pool | Atlas Pool | Brown Lagoon | Winston Lagoon | Atlas Cove |
|---|---|---|---|---|---|---|
| Conductivity | μS cm−1 | 715 | 2000 | 6580 | 39800 | 48200 |
| Ammonia | mg N L−1 | 24.9 | 7.18 | 0.01 | 0.011 | 0.036 |
| Nitrate | mg N L−1 | 1.48 | 4.32 | <0.002 | 0.713 | 0.391 |
| Nitrite | mg N L−1 | 0.022 | 0.091 | <0.002 | <0.002 | 0.005 |
| Nitrogen, total filtered1 | mg N L−1 | 27 | 13 | <0.04 | 0.89 | 0.63 |
| Nitrogen, total2 | mg N L−1 | nd3 | 18 | 0.19 | 0.88 | 0.63 |
| Phosphorus, dissolved reactive | mg P L−1 | 0.03 | 0.043 | 0.025 | 0.073 | 0.06 |
| Phosphorus, total filtered1 | mg P L−1 | 0.082 | 0.115 | 0.027 | 0.124 | 0.092 |
| Phosphorus, total2 | mg P L−1 | nd3 | 1.7 | 0.052 | 0.125 | 0.124 |
| S dissolved | μg S L−1 | 8870 | 20200 | 69000 | 671000 | 771000 |
| S total2 | μg S L−1 | nd3 | 16500 | 74900 | 609000 | 851000 |
| DOC | μg C L−1 | 170 | 12 | 27 | 14 | 34 |
Abbreviations: nd, not determined; DOC, dissolved organic carbon.
10.1 μm filtrate recovered after biomass filtration.
2Sample water filtrate through 20 μm pre-filter.
3Sample volume insufficient to permit analysis.
Microbial diversity
The biomass captured on each filter from each location (total of 15) is referred to as a sample. After quality control processing in QIIME22, a total of 198709 SSU rRNA gene pyrotag sequences (length 222–538 bp, mean 379 bp) were retained from the initial 209304 denoised, chimera checked and trimmed sequences obtained from the 15 samples. As pyrotags of length 250–400 bp have been shown to be adequate for determining accurate community diversity23,24 and taxonomy, even down to the genus level provided suitable reference species and trees are employed24,25, we proceeded with comparisons of community diversity and investigation of the taxa present. Note that all taxonomic identifications are considered putative – we report similarity of OTUs to closest matching characterized species in order to illustrate potential community members and function, rather than implying those species are actually present. Relative abundances of OTUs should be interpreted with caution due to the potential for organisms (particularly Eucarya) to possess multiple copies of SSU genes – see Methods for how this was addressed. The number of sequences per sample ranged from 5118 (Seal Pool 3.0 μm) to 36132 (Seal Pool 0.1 μm) (Table 2). A total of 2794 operational taxonomic units (OTUs) were formed when sequences were clustered at the level of 97% sequence identity. Seal Pool 0.1 μm filter had the greatest number of OTUs (859), followed by Atlas Pool 0.8 μm (650), while the Brown Lagoon 0.1 μm and 3.0 μm filters had the fewest (87 and 99 OTUs, respectively).
Table 2. Diversity metrics for SSU rRNA gene sequences obtained by 454 pyrotag sequencing.
| Sample Site | Seal Pool |
Atlas Pool |
Brown Lagoon |
Winston Lagoon |
Atlas Cove |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | −53.0183417°, 073.3922250° | −53.0195361°, 073.3895833° | −53.0690111°, 073.6648083° | −53.1503972°, 073.6760667° | −53.0125000°, 073.3706667° | ||||||||||
| Sample ID | 509 | 510 | 511 | 490 | 489 | 488 | 572 | 571 | 570 | 543 | 542 | 541 | 538 | 539 | 540 |
| Filter size (μm) | 0.1 | 0.8 | 3 | 0.1 | 0.8 | 3 | 0.1 | 0.8 | 3 | 0.1 | 0.8 | 3 | 0.1 | 0.8 | 3 |
| No. of sequences | 36132 | 5323 | 5118 | 13121 | 26968 | 18150 | 12472 | 20162 | 7678 | 8118 | 8647 | 6832 | 9518 | 11990 | 8480 |
| No. of OTUs at 97% similarity | 859 | 287 | 271 | 337 | 650 | 416 | 87 | 277 | 99 | 392 | 218 | 388 | 176 | 317 | 365 |
| Observed species* (Av. ± std. dev) | 391 ± 10 | 283 ± 1 | 271 ± 0 | 237 ± 10 | 367 ± 9 | 275 ± 5 | 63 ± 5 | 151 ± 8 | 90 ± 3 | 339 ± 4 | 173 ± 6 | 363 ± 5 | 148 ± 3 | 244 ± 5 | 321 ± 6 |
| Chao1* (Confidence interval) | 719 (615–872) | 337 (314–377) | 289 (280–309) | 360 (312–438) | 579 (507–689) | 406 (355–489) | 93 (74–145) | 281 (221–390) | 115 (100–154) | 441 (404–498) | 263 (223–335) | 431 (405–474) | 198 (173–248) | 360 (313–441) | 396 (365–447) |
| Faith’s Phylogenetic diversity* | 35.5 | 25.1 | 22.2 | 25.1 | 27.2 | 20.5 | 8.9 | 15.7 | 9.8 | 32.8 | 17.2 | 28.1 | 12.5 | 20.5 | 21.6 |
| Shannon H’ * | 4.46 | 5.46 | 5.61 | 4.10 | 5.75 | 5.13 | 2.51 | 2.79 | 3.50 | 4.52 | 3.38 | 5.34 | 4.11 | 5.10 | 6.49 |
| Effective species (e^H’)* | 87 | 235 | 272 | 60 | 314 | 168 | 12 | 16 | 33 | 92 | 29 | 210 | 61 | 164 | 660 |
| Simpson (1-D)* | 0.829 | 0.943 | 0.944 | 0.867 | 0.954 | 0.935 | 0.748 | 0.693 | 0.862 | 0.855 | 0.787 | 0.890 | 0.868 | 0.926 | 0.979 |
*Alpha diversity calculations are based on rarefaction to 5118 sequences.
In order to correct for variation in sequencing depth, samples were rarefied to 5118 sequences before alpha-diversity metrics were calculated. Faith’s Phylogenetic Diversity (Faith’s PD) provides a measure of how widely dispersed taxa are across a phylogenetic tree26, and the Chao1 metric provides an estimate of richness which prioritizes the number of rare species27. By both of these measures, the Seal Pool 0.1 μm filter had the greatest diversity (Table 2). Samples with the lowest number of OTUs also had the fewest observed species and the lowest Chao1 estimates and Faith’s PD values (Brown 0.1 μm, Brown 3.0 μm, Atlas Cove 0.1 μm and Winston Lagoon 0.8 μm).
Shannon index is a measure of the richness and evenness of a system, and was highest for the Atlas Cove 3.0 μm sample (6.49), followed by Altas Pool 0.8 μm (5.75), Seal Pool 3.0 μm (5.61) and Seal Pool 0.8 μm (5.46) samples. The non-linearity of the Shannon index curves can make direct comparison of samples difficult, so the effective number of species was also calculated (number of equally-common species required to give a particular Shannon index value28). This showed the Atlas Cove sample with 660 effective species to be more than twice as diverse as the next most diverse sample (Atlas Pool 0.8 μm, 314 effective species) and 55 times more diverse than the filter with the lowest Shannon index (Brown Lagoon 0.1 μm, 12 effective species). Full alpha-diversity metrics (diversity of each site) are provided in Fig. S1.
Overall microbial composition
Based on assignments of OTUs (see Methods), the distribution of taxa in each location and sample was examined. A total of six OTUs were present in all five sampling locations, with one eucaryotic and five bacterial representatives (Fig. 2 and Table S1). Their relative abundance varied between locations, with for example, the Comamonadaceae-related OTU denovo1428 comprising a maximum of 1.9%, 0.75%, 16.5%, 8.2% and 0.008% reads from Seal Pool, Atlas Pool, Brown Lagoon, Winston Lagoon, and Atlas Cove, respectively (Table S1). Brown Lagoon had 26% of OTUs unique to its location while the other four had ≥55% unique sequences (Fig. 2). In pair-wise comparisons, Seal and Atlas pools shared the greatest number of OTUs (366), followed by Brown and Winston lagoons (107), and Winston Lagoon and Atlas Cove (84).
Figure 2. Venn diagram showing OTUs shared between the Heard Island sampling locations.
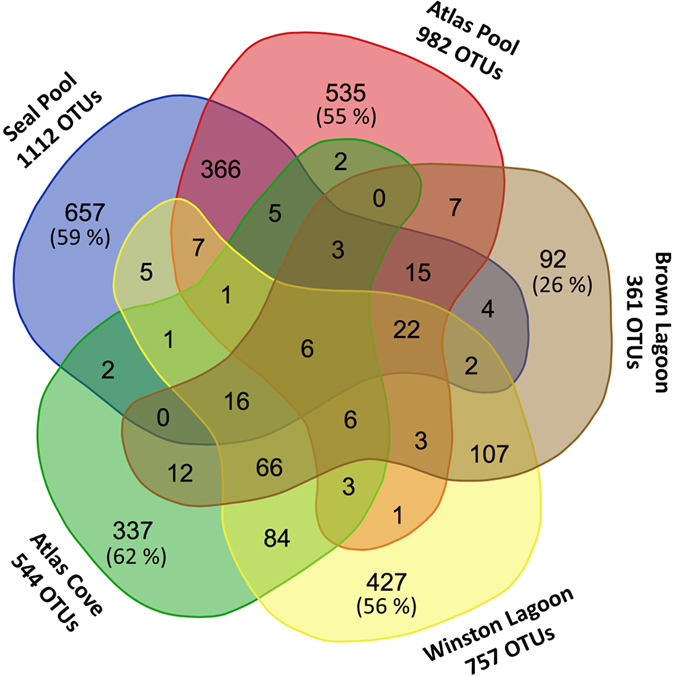
Total number of OTUs per site, percentage of non-shared OTUs at each location (outer lobe), and shared OTUs are shown. The six OTUs present in all locations were denovo1904, denovo179, denovo1428, denovo2631, denovo1094 and denovo1322. Image created using the Venn diagram tool at http://bioinformatics.psb.ugent.be/cgi-bin/liste/Venn/calculate_venn.htpl.
A total of 47% of the Heard Island OTUs had matches ≥95% to sequences in the nt database, including 90 with 100% match. OTUs representing novel species, genera or families, included those with no matches (78 sequences), <80% identity (139), 80–90% identity (526) or 90–95% identity (710) to available sequences in the nt database. Most of the novel taxa were not very abundant, with 78 of the top 100 most abundant OTUs (totaling 82% of sequences) having a best BLAST match ≥97%. The proportion of sequences in samples that could not be taxonomically assigned was 0.4–3.1%. We acknowledge that the differential susceptibilities of particular species to lysis by the DNA extraction method used, along with possible primer bias, may mean that additional species are present on Heard Island but were not detected in this study.
Bacteria
The major bacterial phyla obtained from the Heard Island samples, namely Actinobacteria, Bacteroidetes and Proteobacteria, showed a varied distribution across the five locations and three filter sizes (Fig. 3), with the total proportion of sequences on each filter belonging to bacterial phyla ranging from 33% (Brown 3.0 μm) to 99.5% (Brown 0.1 μm). Bacteria belonging to the Firmicutes, Candidate Division OP11, Candidate Division OD1, Fusobacteria, Candidate Division TM7, Deferribacteres, Verrucomicrobia, Cyanobacteria and Planctomycetes were present at very low levels, with each representing 0.3% or less of total sequences.
Figure 3. Relative abundance of the major microbial groups detected in Heard Island samples.
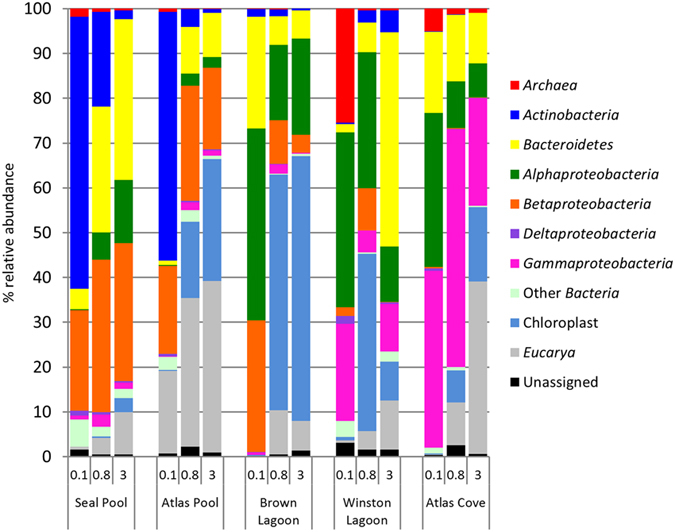
Samples are ordered by filter size (0.1 μm, 0.8 μm and 3.0 μm) and by increasing salinity from Seal Pool to Atlas Cove.
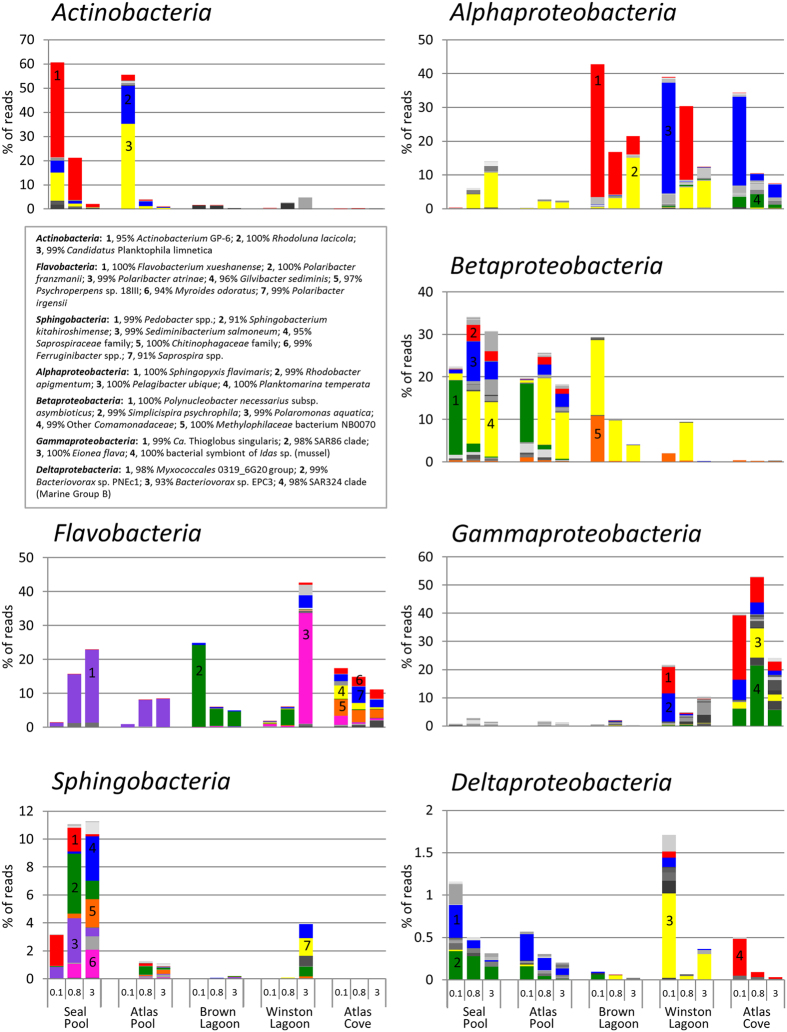
OTUs for Actinobacteria comprised greater than 55% of both the Seal Pool and Atlas Pool 0.1 μm filters (Fig. 4). The predominant OTU from Seal Pool had 95% identity to Actinobacterium GP-6 and likely represents a novel species. In contrast, the predominant OTUs from Atlas Pool had 100% sequence identity to the well characterized Rhodoluna lacicola strain MWH-Ta829 and 98–99% to Planktophila limnetica MWH-EgelM2-3.acl30.
Figure 4. Relative abundance of OTUs for individual phyla of Bacteria.
Colours represent different OTUs at the level of 97% sequence identity. For selected dominant OTUs an indicative best BLAST match for the taxa is shown in the key, and percent identity has been rounded to a whole number. If the taxa could not be assigned to a genus, the next most specific grouping was reported. Full OTU data is provided in Table S4. The scales for relative abundance (% of reads) vary between panels. Samples are ordered by filter size and increasing salinity as for Fig. 3.
Flavobacteria were detected in every location and filter size, but specific species dominated particular locations: OTUs with highest identity to Flavobacterium xueshanense from Seal Pool and Atlas Pool; Polaribacter franzmannii from Brown Lagoon; Polaribacter atrinae from Winston Lagoon. Atlas Cove was the only location to have relatively even proportions of OTUs related to multiple species of Flavobacteria: Polaribacter irgensii, Psychropserpens sp., Gilvibacter sediminis and a distant relative of Myroides odoratus.
Sphingobacterial OTUs from Seal Pool included those with high similarity to characterized organisms (Pedobacter spp., Sediminibacterium salmoneum and Ferruginibacter spp.), and low similarity to known taxa (91% to Sphingobacterium kitahiroshimense, and Saprospria spp., or best matches to Saprospiraceae family or Chitinophagaceae family); these OTUs were absent from 9 of the other 12 filters from the other four locations.
In terms of Proteobacteria, OTUs with 100% identity to Pelagibacter ubique and Sphingopyxis flavimaris were the dominant Alphaproteobacteria. Size fractionation of these OTUs was apparent in Winston Lagoon, but Sphingopyxis OTUs were present in all filter fractions of Brown Lagoon, and Pelagibacter dominated the Atlas Cove samples. Two OTUs closely related to the non-photosynthetic alphaproteobacterium Rhodobacter apigmentum were found in all locations except Atlas Cove. Betaproteobacteria included an OTU with 100% identity to Polynucleobacter necessarius subsp. asymbioticus in the 0.1 μm filters of Seal Pool and Altas Pool, and species which were classified as “Other Comamonadaeae” were detected in all locations except Atlas Cove. OTUs for Gammaproteobacteria were present at very low levels (<3% of total) in Seal Pool, Atlas Pool and Brown Lagoon, whereas the Winston Lagoon 0.1 μm filter contained members of the SAR86 clade and close relatives of the facultatively aerobic sulfur-oxidizing carbon-fixing mixotroph, Candidatus Thioglobus singularis PS1 (denovo611, 99.4% identity)31. In addition to SAR86 and Ca. T. singularis, the Atlas Cove samples contained high levels of sequences related to a mussel symbiont32 (denovo1277, 99.7% identity, 21% of reads on 0.8 μm filter) and to Eionea flava IMCC196233 (10.4% of 0.8 μm filter). Deltaproteobacteria were present in low numbers with OTUs for the SAR324 clade members present from all three Atlas Cove filter fractions, an OTU with 93% similarity to a Bacteriovorax species dominating the Winston Lagoon cohort, and a mixture of Myxococcales-related OTUs and Bacteriovorax sp. PNec1-related (99.3%) OTUs present in both Seal and Altas pools.
Eucarya
The relative abundance of eucaryal sequences, detected as chloroplast 16S rRNA gene sequences and 18S rRNA gene sequences, ranged from 0.23–65.6%. For each location, the 0.1 μm filter had the fewest signatures of eucaryotes and the 3.0 μm filter had the greatest proportion, except for Winston Lagoon where the 0.8 μm filter had a higher level (44%) than the 3.0 μm filter (20%) (Fig. 3).
Over 52% of the reads from the Brown Lagoon 0.8 μm filter belonged to a single OTU with 100% sequence identity to Micromonas pusilla chloroplast (10485 seq) (Fig. 5). This OTU was also abundant on the Winston Lagoon 0.8 μm filter, comprising 39% of total sequences. The presence of Micromonas in the two glacial lagoons was corroborated by the detection of 18S rRNA gene sequences for this organism from the 0.8 μm filters. In addition to the dominant OTUs matching Micromonas pusilla in Brown Lagoon, other OTUs were predominantly affiliated with the Cryptophyceae - Guillardia theta and the Antarctic/Sub-Antarctic species Geminigera cryophila34.
Figure 5. Relative abundance of OTUs for Eucarya.
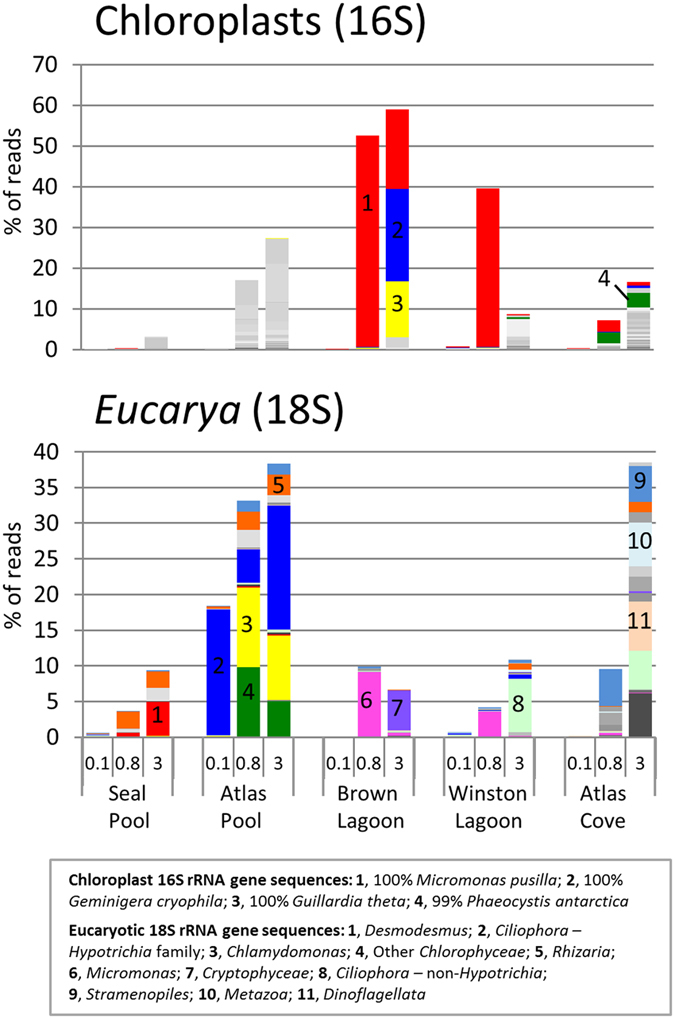
For chloroplast sequences, selected dominant OTUs and their best BLAST match are shown in the key. For Eucarya detected as 18S rRNA gene sequences, OTUs are grouped at Order/Family level. Note comments about the colours, sequence identity, abundance scale, sample order and OTU table as for Fig. 4.
The characteristics of the other 18S rRNA gene sequences indicated the presence of location-specific eucaryotic consortia (Fig. 5). Atlas Pool had OTUs for members of the Hypotrichia family, Chlamydomonas and other Chlorophyceae, while Atlas Cove included Dinoflagellata, non-Hypotrichia Ciliophora, and Metazoa. OTUs for Stramenopiles and Rhizaria were detected in all five locations at low to moderate levels, while the main OTU detected in Seal Pool had 100% identity to Desmodesmus communis, a common freshwater green alga.
Archaea
Archaea were present at only low levels (0 to 5%) in all samples except for the Winston Lagoon 0.1 μm filter where they comprised 25.4%. OTUs belonged to four main groups (Fig. 6), with 153 OTUs from the Euryarchaeal Deep Sea Hydrothermal Vent Group 6 family (DHVEG-6) which includes Candidatus Parvarchaeum. The best BLAST matches related to this group of OTUs were 91.6% to the genome of Archaeon GW2011_AR1535, and 99.7% to 90.2% to environmental sequence data. These OTUs represent a diverse population of archaea present in Winston Lagoon (0.1 μm filter, 109 OTUs; 0.8 μm filter, 4 OTUs; 3.0 μm filter, 6 OTUs), Seal Pool (0.1 μm filter, 32 OTUs) and Atlas Pool (0.1 μm filter, 18 OTUs). The other major archaeal group of OTUs in Winston Lagoon was closely related to ammonia-oxidizing Nitrosopumilis maritimus. Low levels of N. maritimus-related OTUs were also detected in Atlas Cove and Brown Lagoon, while a small number of Methanoregula-related sequences were recovered from Seal Pool and Atlas Pool. Atlas Cove was the only sample in which Marine Group II (MGII) Euryarchaeota were detected (all three filters). Although initial classification based on the Silva database could not identify them beyond family-level, BLAST searches and construction of a 16S rRNA gene phylogenetic tree (Fig. S2) confirmed that two OTUs (denovo2505 and denovo420) were closely related to Candidatus Thalassoarchaea mediterranei from Marine group IIb36 and five OTUs (denovo486, denovo2226, denovo1995, denovo1041 and denovo1042) were closely related to the MGIIa uncultured strain MG2-GG3 for which a whole genome sequence has been reconstructed37.
Figure 6. Relative abundance of OTUs for Archaea.
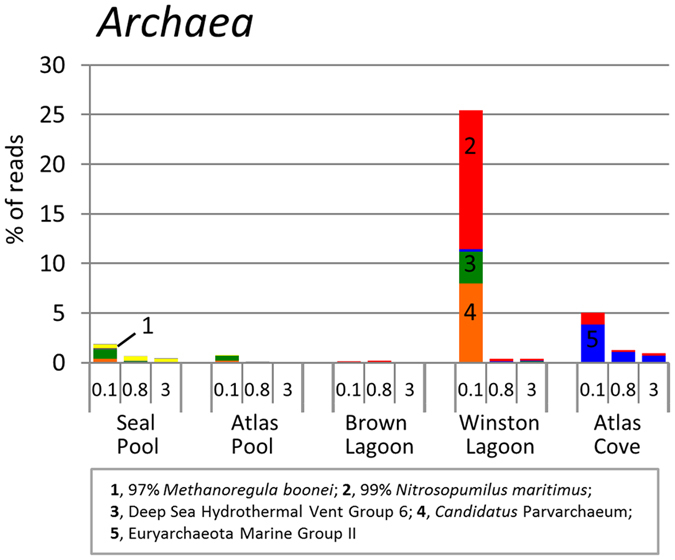
Note comments about the colours, sequence identity, BLAST matches, abundance scale, sample order and OTU table as for Fig. 4.
Community comparisons and correlation with environmental parameters
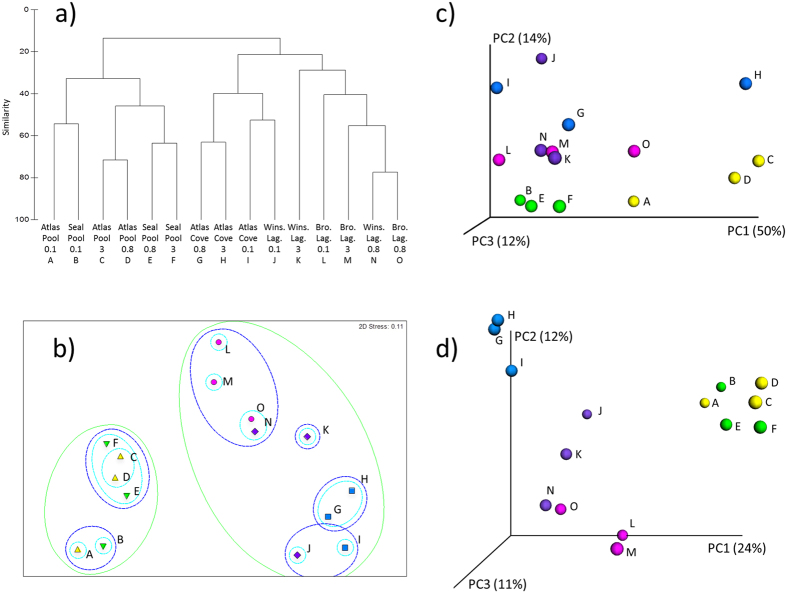
Similarities and differences between the samples were explored with Hierarchical Cluster Analysis (HCA) and non-metric multidimensional scaling (nMDS) at genus-level (Fig. 7) or kingdom/phylum-level (Fig. S3). In the clustering dendrogram, the pools clustered together on the left, while the glacial lagoons and the Atlas Cove sample grouped together on the right (Fig. 7a). Similarity Profile (SIMPROF) analysis yielded a significant Pi statistic (Pi = 9.151, p < 0.1%) rejecting the null hypothesis that no multivariate structure exists within the data, and 2-way crossed Analysis of Similarity (ANOSIM) also confirmed these differences (by location: R = 0.786, p < 0.1%; by filter size: R = 1, p < 0.3%). Pairwise testing (ANOSIM, Table S2) confirmed that the profile of Atlas Pool and Seal Pool overlapped and could not be distinguished (R = 0.111), as was the case for Winston Lagoon and Brown Lagoon (R = 0.222). The remainder of the pairwise comparisons gave values of R greater than 0.5 (all except one were close to 1) indicating the pairs of samples were well separated. These differences between locations were also reflected in the SIMPER average dissimilarity values which ranged from 60 to 92%. A full listing of which taxa most contributed to the dissimilarities between locations is provided in Table S3.
Figure 7. Clustering of Heard Island samples based on genus-level taxon abundance.
Taxon abundance data calculated as Bray-Curtis dissimilarity, normalised and square-root transformed. Samples which clustered together with 20%, 40% and 60% similarity in the dendrogram (a) are circled by green, blue and aqua ovals, respectively, in the nMDS plot (b). Clustering based on beta-diversity measured as weighted (c) and unweighted Unifrac principal coordinates analysis (d). For beta-diversity calculations, samples were rarefied to 5118 sequences. Samples are labelled A – O as per the dendrogram (a), with colour coding to indicate sampling location: Seal Pool (green), Atlas Pool (yellow), Brown Lagoon (pink), Winston Lagoon (purple) and Atlas Cove (blue).
Placement of samples in 2-dimensional space by nMDS showed that the Altas Pool and Seal Pool 0.1 μm fractions grouped together (40% similarity), as did the Altas Pool 0.8 μm and 3.0 μm filters (60% similarity) and the Seal Pool 0.8 μm and 3.0 μm fractions (60% similarity) (Fig. 7b). The placement of the Brown Lagoon, Winston Lagoon and Atlas Cove samples followed a rough trajectory from least to most saline, with the 0.8 μm filters of Brown and Winston lagoons grouped together (60% similarity) and the Atlas Cove 0.8 and 3.0 μm filters grouped together (60% similarity).
The quantitative measure of beta-diversity (weighted unifrac) first dimension (PC1) explains a full 50% of the variation among samples (Fig. 7c). This measure highlights community differences among sites including changes in relative taxon abundance. The qualitative measure (unweighted unifrac), which utilizes presence/absence data but not abundance information, shows the samples from each location grouping with themselves, with those from both Atlas Pool and Seal Pool particularly near to each other (Fig. 7d). The first three dimensions of PCoA in the unweighted plot do not explain as great a total proportion of the observed variation among samples as the first dimension of the weighted plot.
In order to assess to what degree the abiotic factors (Table 1) explained the biotic patterns observed, the BEST analysis routine in PRIMER-E was employed. The null hypothesis of no correlation between biotic and abiotic factors was strongly rejected (R = 0.824, p < 0.001), with the most important single factor being either conductivity (proxy for salinity) or dissolved sulfur (equal best correlation of 0.740). The best total correlation of 0.824 was obtained when the abiotic variables conductivity, nitrite, total nitrogen and dissolved sulfur were considered.
Discussion
This study presents the first assessment of microbial diversity in Heard Island’s aquatic systems. The distribution patterns of abundance of individual OTUs support the existence of distinct microbial communities at each location that are influenced by the environmental factors characteristic of each site (Figs 4–6). Only six OTUs were present across all five systems (Table S1), and many OTUs were only present in a single location indicating each community was likely to perform specific microbial processes. A large number of OTUs possessed little similarity to characterized strains (53%, <95% 16S/18S rRNA gene identity), underscoring the undersampling and/or novelty of microorganisms in Antarctic and Sub-Antarctic environments.
There are limits to the extent to which inferences about functional processes can be made from purely taxonomic data, with the literature supporting active discussion about methodological approaches and the validity of rationalizing function38,39,40,41,42. While pyrotag data are not a substitute for shotgun metagenome data20, this method has been successfully applied to study changes in community composition and infer microbial processes in Antarctic lakes43,44, and to assess the biogeography and effects of advection on Southern Ocean microbial communities17,45,46. Here we demonstrate that in combination with analyses of measured environmental variables, pyrotag sequencing can provide a relatively inexpensive approach for gaining valuable insight into distinctive ecological properties of previously unstudied sites. Naturally, functional inferences are putative and provide a broad approximation of ecosystem processes based upon the OTUs detected.
Seal Pool and Atlas Pool
The high levels of nutrients evident in Seal Pool (Table 1) are provided by seals in and around the pool, and the wet mixed herbfield vegetation nearby (Fig. 1). Eucaryotic OTUs, including those related to the photosynthetic green alga Desmodesmus, and OTUs for Cyanobacteria were detected at low relative abundance, possibly indicating that limited primary production was occurring (Figs 5 and 6). OTUs related to bacteria that are capable of organic matter turnover, including Actinobacteria, Sphingobacteria and Flavobacteria, were abundant (Fig. 5). The Actinobacteria have been linked to Antarctic lake eutrophication and Flavobacteria to degradation of Antarctic aquatic detritus19,47,48, and Sphingobacteria more broadly to a degradative capacity49. Myxococcales were also present and likely contribute to degradation of organic matter49.
The bacterial OTUs for Seal Pool are also characterized by a relatively high degree of novelty compared to characterized strains. For example, the most abundant Actinobacteria OTU has 95% identity to Actinobacterium GP-6, and Sphingobacteria have <91% identity. It is possible that the novel taxa in Seal Pool may possess novel physiology or particular adaptations allowing them to dominate in this environment.
The presence of OTUs related to Bacteriovorax suggests bacterial predation as these organisms feed on Gram-negative prey50. This activity would also contribute to the release and recycling of organic matter. Denitrifying Betaproteobacteria also appear to be present, as OTUs with 99–100% identity to Comamonas spp. and Simplicispira psychrophila were identified; bacteria that are known to perform chemoorganotrophic growth51,52,53. OTUs matching the ubiquitous and cosmopolitan heterotrophic freshwater bacterium, P. necessarius subsp. asymbioticus54 were also abundant in Seal Pool. All of these findings are consistent with the rich organic matter of Seal Pool supporting a microbial ecosystem driven by heterotrophic degradation.
The moderate relative abundance of OTUs (Table S4) for chloroplasts and multiple types of green algal 18S rRNA genes (Chlamydomonas and Other Chlorophyceae, Fig. 5.) suggest primary production plays a much greater role in Atlas Pool than Seal Pool. The eucaryal OTUs included Halteria sp. bLaN2 (98.5% identity) and the abundance of such ciliates has been shown to correspond to summertime runoff of nutrients, with this marine species involved in the incorporation of particulate and dissolved organic carbon into the food web55. Alphaproteobacteria and Gammaproteobacteria are almost entirely absent from Atlas Pool. In contrast, the communities of Flavobacteria, Betaproteobacteria and Deltaproteobacteria largely mirror that of Seal Pool suggesting they play a similar role in turnover of organic matter. OTUs for Sphingobacteria are greatly reduced in Atlas Pool compared to Seal Pool. This may indicate that the nutrients metabolized by Sphingobacteria in Seal Pool have been degraded (oxidized) by the time any runoff reaches Atlas Pool (see pool locations Fig. 1; DOC of 170 μg C L−1 in Seal pool c.f. 12 μg C L−1 in Atlas Pool, Table 1).
The abundance of Actinobacteria appears similar to that of Seal Pool, but the proportions of specific types are swapped with OTUs closely related to Rhodoluna lacicola and Candidatus Planktophila limnetica dominant over those for Actinobacterium GP-6. Cand. P. limnetica has only been isolated in stable co-culture with P. necessarius subsp. asymbioticus, with the latter likely providing some essential nutrients for growth30. R. lacicola and Cand. P. limnetica both possess actinorhodopsins for photoheterotrophy30,56,57 and have been reported as dominant members of some freshwater lakes29,58,59. The detection of OTUs matching these bacteria in Atlas and Seal pools suggests that interspecies interactions may have important roles in these less saline food webs.
Brown Lagoon
Brown Lagoon had the lowest levels of nitrogen (Table 1) and the lowest community diversity (Table 2) and was dominated by OTUs matching the unicellular photosynthetic picoflagellate Micromonas pusilla (both 18S rRNA and chloroplast 16S rRNA gene sequences) (Fig. 5). This picoflagellate can be numerically dominant in tropical, temperate and polar waters, is very small in cell size (<2 μm) with a small genome (15 Mb), and is able to effectively scavenge ammonium under nitrogen limiting conditions60,61. The draft genome of an Antarctic Micromonas sp. is reported to encode many enoyl-CoA hydratases which may enhance fatty acid metabolism, and an antifreeze protein62. Such properties would likely contribute to the ability of Antarctic Micromonas spp., including those in Brown Lagoon, to grow in the cold.
OTUs for Sphingopyxis (99.6% similarity to S. flavimaris) were abundant, particularly on the 0.1 μm filter (39%) compared to 0.8 μm (12%) and 3.0 μm (5%) filters. The increase in OTU abundance with decreasing filter size may indicate the Brown Lagoon species have properties in common with Sphingopyxis alaskensis, which was isolated from cold Alaskan waters, the North Sea and the North Pacific, has a small cell size and can thrive under oligotrophic conditions63,64,65. The abundance (16.5%, 0.1 μm filter) of an OTU 99% similar to the recently isolated oligotrophic Comamonadaceae bacterium LSUCC012366 is consistent with Brown Lagoon supporting the growth of oligotrophic bacteria. Oligotrophic bacteria may be innately suited to Antarctic environments due to their ability to weather periods of energy limitation67.
OTUs were also abundant for close relatives of Methylophilaceae bacterium strain NB0070 (10.7%, 0.1 μm filter) indicating a capacity to metabolize methanol and high molecular weight organic matter68, and the psychrophilic heterotrophic Polaribacter franzmanii (24%, 0.1 μm filter) indicating an ability to degrade algal polysaccharides and proteins69,70. Overall, the OTU data suggest Brown Lagoon is characterized by Eucarya and Bacteria with relatively small cell sizes, oligotrophic strategies, and capacities to metabolize organic matter (including of high molecular weight).
Atlas Cove and Winston Lagoon
The major taxa of the marine Atlas Cove community consisted of OTUs for Gammaproteobacteria (Eionea, Thioglobus), Alphaproteobacteria (SAR11, Roseobacter), and Flavobacteria, with primary producers being Eucarya (e.g. Phaeocystis antarctica) rather than cyanobacteria; findings typical of Southern Ocean surface waters48,71. A notable difference was for Archaea, with a higher proportion of OTUs for MGII Euryarchaeota than for Marine Group I nitrifying chemolithoautotrophs (e.g. Nitrosopumilus maritimus); the latter being reported to dominate Southern Ocean surface waters19,71. Genome reconstruction of MGII strains indicates they are facultative photoheterotrophs that can degrade organic carbon36,37,72. The OTUs from Atlas Cove had a wide-range of identity (84–99%) to known MGII strains indicating they may possess novel physiological traits.
In Winston Lagoon, the low relative abundance of OTUs for Eucarya (18S rRNA and chloroplast genes), but high representation (31% total reads) of Flavobacteria (99.7% identity to Polaribacter atrinae), is likely to indicate sampling occurred towards the end of an algal bloom cycle. Consistent with this, an unusually high level of brown material (e.g. algal detritus) was present on the 20 μm prefilter that was used to filter samples. P. atrinae possesses a wide array of amidase, DNase, esterase, lipase, phosphatase and sugar degradative enzymes73, and Polaribacter spp. are known to degrade algal polysaccharides and proteins and can represent a high proportion of the population during Antarctic algal blooms19,48,70,74, including in the iron-rich waters on the Kerguelen Plateau75. In coastal Southern Ocean waters, Alphaproteobacteria utilize labile substrates liberated from algal detritus by Flavobacteria48. The abundance (~30–40%) of OTUs for Alphaproteobacteria (e.g. Pelagibacter ubique and Sphingopyxis) on 0.1 and 0.8 μm filters, and Flavobacteria (~40%) on 3.0 μm filters, suggests these bacterial taxa in Winston Lagoon associate with detritus from algal blooms in a similar way to their oceanic counterparts. Sphingobacteria (26 OTUs on the 3 μm filter) were also likely to have been involved in algal organic matter turnover49, whereas OTUs with 99% identity to Nitrosopumilus maritimus indicate possible chemolithoautotrophic or mixotrophic ammonia-oxidiation being performed by Archaea.
Being open to the ocean, the salinity and nutrient composition of Winston Lagoon was much more similar to Atlas Cove than Brown Lagoon (Table 1), and the presence of Gammaproteobacteria in Winston Lagoon was an indicator of oceanic influx as this class of Bacteria was abundant in Atlas Cove but essentially absent in Brown Lagoon, Atlas Pool and Seal Pool (Fig. 4). Interestingly, Winston Lagoon lacked the signatures of the types of Eucarya present in Atlas Cove, suggesting the environmental conditions were sufficiently distinct that oceanic Stramenopiles, Metazoa and dinoflagellates currently struggle to colonize Winston Lagoon (Table S4). As a proxy for Antarctic freshwater glacial lagoons experiencing marine influx, Winston Lagoon will be valuable to monitor to assess whether salinity levels and community composition remain stable. By determining if total diversity increases or if endemic species decline, we will be able to learn about the interplay between species competing for changing niches.
Factors influencing each ecosystem
Location specific factors can greatly influence microbial communities76,77,78. In wetland streams, dissolved organic matter (DOM) was identified as the main factor controlling community composition77, whereas microbial populations ultimately persisted in a tundra lake despite seasonal inputs of DOM and allochthonous species76. In a lake on Ardley Island (north of the Antarctic Peninsula), the bacterial sediment community correlated with the historical presence of colonizing penguins, and elemental composition of penguin guano influenced the relative abundance of many taxa78. Here we show for the Heard Island systems that salinity, dissolved sulfur, total nitrogen and nitrite are important for explaining the present patterns of community composition. Detailed investigation of S- and N- cycling in the Heard Island pools and lagoons will be of interest for further study – for example, examining the role of dissolved sulfur in Winston Lagoon in supporting sulfur oxidizers such as Cand. Thioglobus singularis31, which in turn provide organic reduced sulfur for species like SAR86 that lack the capacity to reductively assimilate sulfate79.
In addition to water chemistry, we can comment on the possible influences of dispersal, limnology and biological factors. Strong winds across the island would undoubtedly cause Aeolian transport of microorganisms. However, our data shows that communities are largely specific to each location indicating that environmental selection provided by each system overcomes the effects of dispersal. The ground around Brown and Winston lagoons includes recently deglaciated rock and moraine which does not yet support complex vegetation, and glacial meltwater travels a greater distance across deglaciated ground to reach Brown Lagoon compared to Winston Lagoon where the glacier still calves into the lagoon. In addition to water source differences between the five locations (rain, snow, glacier, marine), the microbial communities in the pools on the Azorella Peninsula will be influenced by the high nutrient and microbial inputs from elephant and fur seal excreta and runoff from herbfield vegetation and soil. The lagoons are much larger bodies of water so penguin and seal input will have less influence, although the different seal populations breeding near each lagoon (Brown Lagoon, fur; Winston Lagoon, elephant) may seed each with animal specific microorganisms (Fig. 1). The effects of climate change, particularly glacial retreat and formation of new lagoons, are expected to cause the redistribution of plants, sea birds and seal populations80, thereby influencing nutrient and microbial inputs into lagoons and pools across the island. Cooperation between microbial community members may also have an important influence, as this has been observed for some Antarctic sea ice, lake and oceanic communities20,81, and our data suggests as-yet unstudied factors may influence the syntrophic interactions between Cand. P. limnetica and P. nucleobacter subsp. asymbioticus30, allowing Cand. P limnetica to become the dominant Actinobacteria in Atlas Pool but not Seal Pool. In future work, it will be important to augment understanding of the food web relationships by also including the potentially important roles of viruses20.
Perspective
The range of environmental parameters, community structures, and putative functions observed across the five study sites suggests a significant reservoir of microbial biodiversity is found on the island. Are these snapshots of a stable community structure; communities which periodically oscillate between several stable states; a temporary response to perturbation; or, environments in genuine and ongoing flux due to climate change? To answer these questions, temporal studies covering days, seasons and years need to be performed20,82. Obtaining sufficient replicates will be necessary for distinguishing random fluctuations from genuine trends. It will be no small challenge to achieve all this while recognizing Heard Island’s World-Heritage status and preserving and protecting its unique and valuable natural resources. However, it is important to perform this research in view of the rapid changes occurring on the island, as the findings will heighten awareness amongst policy makers of how urgently we need international action to mitigate the effects of anthropocentric climate change.
Methods
Study sites and sampling
Five study sites were selected around Heard Island (Fig. 1 and Table 2). Seal Pool (name designated in this study) is a small shallow pool formed by elephant seals within wet mixed herbfield vegetation (Tussock grass, Cushion plants and Kerguelen Cabbage). Its waters were visibly eutrophic (green) at the time of sampling. Nearby Atlas Pool (name designated in this study) is a moderately sized shallow pool located to the west and shorewards of Seal Pool. Both of these pools lie on the Azorella Peninsula and receive freshwater from snow melting rather than from glacial sources, as well as rain, with Seal Pool likely draining towards Atlas Pool. Large populations of elephant and fur seals, macaroni, rockhopper, gentoo and king penguins are prevalent on the peninsula and nearby Nullarbor Plain.
Brown Lagoon was first observed in 1963 when Brown Glacier retreated 100 m inland leaving a small lagoon separated from the ocean by a boulder beach14. By 1971 the glacier had retreated a further 350 m creating a proglacial lagoon, and the current size is ~900 m long along the coastal edge and ~600 m wide at the widest point (calculated from maps3,83). A gravel bar separates the lagoon from the ocean, and the wildlife present are fur seals and macaroni, rockhopper, gentoo and king penguins. Winston Lagoon formed prior to the commencement of glacial monitoring in 1947, is one of the largest glacial lagoons on the island, and is currently ~2.3 km long along the coastal edge with a maximum width of ~2.3 km (calculated from maps83). Winston Glacier still terminates at and calves ice into the lagoon, and the wildlife in this region are mostly elephant seals and gentoo or king penguins, with the closest fur seals recorded at Paddick Valley to the east. Coastal prograding has occurred and although the opening from Winston Lagoon to the sea first observed in 1947 appeared to be superficially blocked in the 1980s2, it was open to the ocean at the time of sampling.
The marine sample from the site designated Atlas Cove was collected while the research vessel Aurora Australis was moored near Atlas Cove between Lauren’s Peninsula and the Azorella Peninsula (Fig. 1), and shotgun metagenome data from this site was previously described (Sample ID GS39484). Here, DNA was used for pyrotag sequencing with the SSU rRNA gene data being representative of the marine environment adjacent to Heard Island and serving as a comparison to the pool and lagoon study sites.
Water samples were collected on the 16th December (Atlas Lagoon, 2 L; Seal Pool, 1.75 L) or 17th December 2008 (Brown Lagoon, 17.4 L; Winston Lagoon, 13.4 L; Atlas Cove, 400 L). Biomass was harvested concurrently with- or within a few hours of water collection by passing the water from each location through a 20 μm pre-filter, and serially size fractionating through 293 mm diameter polyethersulfone membrane filters (Pall, Port Washington, USA) with 3.0, 0.8 and 0.1 μm pore sizes, as described previously17,18,85. Water sample collection and processing at Atlas Lagoon and Seal Pool were terminated at relatively small volumes (1.75–2 L) due to biomass saturation of the filters. All filters were placed in storage buffer, frozen in liquid nitrogen and cryogenically maintained at −80 °C until processing could be performed at the University of New South Wales (Sydney, Australia), as described previously17,18.
Water analysis
Analysis of water composition (DOC, nitrate, nitrite, ammonia, total nitrogen, total dissolved nitrogen, dissolved reactive phosphorous, total phosphorus, total dissolved phosphorus, total sulfur and total dissolved sulfur) was performed by Analytical Services, Tasmania according to American Public Health Association’s Standard Methods, as previously described86. Nutrients were measured from water collected after filtration through the on-site 20 μm pore size pre-filter (250 ml), with dissolved nutrients measured after filtration through a 0.1 μm pore size membrane filter (250 ml).
DNA extraction
DNA extraction was performed on each entire filter using a phenol-chloroform method85 with modifications87. Briefly, filters were cut into fine strips (~3 mm × 10 mm) and incubated shaking in sucrose lysis buffer (final concentration 25 mM EDTA, 25 mM EGTA, 0.75 M sucrose, 5 mM Tris-HCl [pH 8.0]) with 2.5 mg mL−1 lysozyme for 30–60 min at 37 °C. Proteinase K was added to a final concentration of 200 μg mL−1 and three freeze-thaw cycles (−80 °C for 20–30 min, 55 °C for 20–30 min) were performed. An additional 200 μg mL−1 of Proteinase K, and SDS to a final concentration of 1% were added, and samples incubated in a shaking water bath at 55 °C for 2 h. This was followed by two extractions with phenol and one extraction with phenol/chloroform/iso-amyl alcohol (25:24:1). DNA was precipitated with sodium acetate and 1-propanol, and the pellet washed with 70% ethanol and dried before re-suspension in TRIS-EDTA buffer. All reagents were from Sigma-Aldrich (St. Louis, USA).
454 pyrotag sequencing and data processing
Amplification of SSU rRNA gene sequences and pyrosequencing was carried out at the Research and Testing Laboratory (Lubbock, TX, USA) using Roche 454 sequencing technologies and methodology. Samples were amplified for pyrosequencing using a forward and reverse fusion primer. The forward primer was constructed with (5′-3′) the Roche A linker (CCATCTCATCCCTGCGTGTCTCCGACTCAG), an 8 bp barcode to multiplex samples, and the 926wF primer (AAACTYAAAKGAATTGRCGG88). The reverse fusion primer was constructed with (5′-3′) a biotin molecule, the Roche B linker (CCTATCCCCTGTGTGCCTTGGCAGTCTCAG), and the 1392R primer (ACGGGCGGTGTGTRC89). These primers amplify the V6-V8 region of the SSU rRNA gene and were selected as they cover about 84%, 71% and 91% of 16S rRNA gene sequences from Bacteria, Archaea and Eucarya, respectively, based on interrogation of sequences spanning the V6-V8 region in the curated SILVA Ref NR SSU rRNA gene dataset. This coverage was determined using TestPrime90 (https://www.arb-silva.de/search/testprime/) with a stringency setting of 1 bp mismatch permitted but 0 bp mismatches allowed in the 5 bases at 3′ end, and phylogenetic coverage details for 926wF - 1392R are provided in Table S5 and S6.
Amplifications were performed in 25 μl reactions with Qiagen HotStar Taq master mix (Qiagen Inc, Valencia, California), 1 μl of each 5 μM primer, and 1 μl of template. Reactions were performed on ABI Veriti thermocyclers (Applied Biosytems, Carlsbad, California) under the following thermal profile: 95 °C for 5 min, then 35 cycles of 94 °C for 30 sec, 54 °C for 40 sec, 72 °C for 1 min, followed by one cycle of 72 °C for 10 min and 4 °C hold. Amplification products were visualized with eGels (Life Technologies, Grand Island, New York). Products were pooled with equimolar quantities and each pool was cleaned and size selected using Agencourt AMPure XP (BeckmanCoulter, Indianapolis, Indiana) following Roche 454 protocols (454 Life Sciences, Branford, Connecticut). Size selected pools were quantified and diluted to be used in emulsion-based clonal amplifications (emPCR), which were performed and subsequently enriched. Sequencing followed established manufacturer protocols (454 Life Sciences) using Roche reagents and the GS FLX platform (Roche, Branford, USA). Denoising (using USEARCH91), chimera checking (using the de novo method built into UCHIIME92), and removal of poor quality reads (criteria: failed sequence reads; sequences with low quality tags, primers or ends; sequences less than 250 bp in length) were performed by the sequencing facility. The raw data have been deposited in the Sequence Read Archive with BioProject number: PRJNA335685.
Data were processed using the Quantitative Insight Into Microbial Ecology (QIIME) pipeline v1.8.0 with default parameters22. Briefly, samples were de-multiplexed and filtered for quality (length range 200–1000 bp; number of ambiguous bases must not exceed 6; mean quality score must be 25 or greater; maximum homopolymer run of 6 bp; zero mismatches in primer), then de novo OTUs were picked and assigned taxonomy93 by reference with the Silva 111 dataset94 using UCLUST91 with clustering at 97% identity as this threshold has been shown to substantially reduce artificial inflation of diversity estimates due to pyrosequencing errors95. A full OTU table with relative abundances, Silva-based classification and best BLAST match of all OTUs is provided in Table S4. The number of 16S rRNA genes may vary between different species of Bacteria and Archaea, and Eucarya may possess large numbers of 18S rRNA genes and/or chloroplasts (including multiple copies of 16S rRNA genes within each chloroplast)96, thereby precluding accurate estimates of the true abundance of species from the relative abundances of SSU rRNA gene sequences. Nevertheless, no ‘correction’ was applied to adjust relative abundances as the likelihood of performing this accurately is low given the novelty of the systems studied and demonstration that a high proportion of OTUs had low identity to characterized bacterial, archaeal and algal species. Sequences were also aligned using PyNAST97 and used to construct a phylogenetic tree using FastTree98. Alpha-diversity (shannon, simpson, goods_coverage, PD_whole_tree, chao1, and observed_species) and Beta-diversity (weighted and unweighted unifrac99, principal coordinate analysis) metrics were calculated in QIIME and visualized using Emporer100. The relatedness of Heard Island taxa to their closest characterized relatives was assessed by BLAST against the NCBI nt database with environmental and metagenome sequences removed101. Relatedness to all known taxa was assessed by BLAST against the full NCBI nt database.
Abundance counts of taxa summarized at Level 2 (Kingdom – Phyla) and Level 6 (Kingdom – Phyla – Class – Order – Family – Genus) were converted into relative percentages for each of the 15 samples and a Bray-Curtis dissimilarity matrix102 was generated in PRIMER-E v6103 from square-root transformed data in preparation for statistical analysis. Water chemistry data were also transformed and normalized, and Principal Component Analysis (PCA) was performed. Taxonomic composition-based similarities between samples were investigated using Cluster (HCA), and the non-parametric permutation procedure ANOSIM was used to test the differences in composition of the communities detected in each location and at each filter size. Evidence of structure in an a priori unstructured set of samples was assessed using global SIMPROF analysis. nMDS was used to plot the position of each sample based on its distance from all other samples, and then used for BEST analysis. BEST analysis compares the 2-dimensional distribution of samples obtained based on biotic (taxonomic) factors and abiotic factors (water chemistry data) to identify which factors best explained the observed patterns. The contribution of particular genera to the observed dissimilarity between groups in the nMDS plots was calculated using SIMPER (similarity percentages procedure). The ANOSIM, BEST, Cluster, nMDS, PCA, SIMPER and SIMPROF statistical tests were all performed using the software PRIMER-E v6103 with recommended settings.
Additional Information
How to cite this article: Allen, M. A. and Cavicchioli, R. Microbial communities of aquatic environments on Heard Island characterized by pyrotag sequencing and environmental data. Sci. Rep. 7, 44480; doi: 10.1038/srep44480 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Australian Research Council (DP1095504) and the Australian Antarctic Science program (project 2899). We thank Mark Brown, John Rich, Federico Lauro and the expeditioners and crew of Aurora Australis for assistance with collecting, documenting (including photographs) and processing of water samples, Suhaila Mohd Omar for extracting DNA from sample biomass, and Timothy Williams for critical comments on the manuscript. We acknowledge Gary Miller, Stephen Brown and the Australian Antarctic Division for provision of photographs, and the Australian Antarctic Data Centre and the Australian Geographic Cartographic Division for provision of maps and images used to construct Fig. 1, and David Smith from the Australian Antarctic Division for valuable assistance with resourcing the content for Fig. 1.
Footnotes
The authors declare no competing financial interests.
Author Contributions R.C. conceived the research. M.A. analyzed the data and interpreted the findings. M.A. and R.C. wrote the manuscript.
References
- Park Y.-H. et al. Polar front around the Kerguelen Islands: An up-to-date determination and associated circulation of surface/subsurface waters. J. Geophys. Res. Oceans 119, 6575–6592 (2014). [Google Scholar]
- Allison I. F. & Keage P. L. Recent changes in the glaciers of Heard Island. Polar Rec. 23, 255–271 (1986). [Google Scholar]
- Thost D. E. & Truffer M. Glacier recession on Heard Island, Southern Indian Ocean. Arct. Antarct. Alp. Res. 40, 199–214 (2008). [Google Scholar]
- Bergstrom D. M. & Chown S. L. Life at the front: history, ecology and change on southern ocean islands. Trends Ecol. Evol. 14, 472–477 (1999). [DOI] [PubMed] [Google Scholar]
- Woehler E. Status and conservation of the seabirds of Heard Island. In Green K. & Woehler E. (eds) Heard Island: Southern Ocean Sentinel. Surrey Beatty & Sons, Chipping Norton, pp 128–165 (2006). [Google Scholar]
- Green K. The marine mammals of Heard Island. In Green K. & Woehler E. (eds) Heard Island: Southern Ocean Sentinel. Surrey Beatty & Sons, Chipping Norton, pp 166–183 (2006). [Google Scholar]
- Laybourn-Parry J. & Marchant H. J. Daphniopsis studeri (Crustacea: Cladocera) in lakes of the Vestfold Hills, Antarctica. Polar Biol. 11, 631–635 (1992). [Google Scholar]
- Green K. & Woehler E. (eds) Heard Island: Southern Ocean Sentinel. 270 pp. Surrey Beatty & Sons, Chipping Norton (2006). [Google Scholar]
- Kiernan K. & McConnell A. Management considerations for the Heard Island lava tube caves. Papers and Proceedings of the Royal Society of Tasmania 133, 13–22 (2000). [Google Scholar]
- Jones H. I. Notes on parasites in penguins (Spheniscidae) and petrels (Procellariidae) in the Antarctic and Sub-antarctic. J. Wildlife Dis. 24, 166–167 (1988). [DOI] [PubMed] [Google Scholar]
- Thost D. & Allison I. The climate of Heard Island. In Green K. & Woehler E. (eds) Heard Island: Southern Ocean Sentinel. Surrey Beatty & Sons, Chipping Norton, pp 52–68 (2006). [Google Scholar]
- Ruddell A. An inventory of present glaciers on Heard Island and their historical variation. In Green K. & Woehler E. (eds) Heard Island: Southern Ocean Sentinel. Surrey Beatty & Sons, Chipping Norton, pp 28–51 (2006). [Google Scholar]
- Cogley J. G., Berthier E. & Donoghue S. Chapter 32: Remote sensing of glaciers of the Subantarctic islands. In Kargel J. S. et al. (eds) Global Land Ice Measurements from Space. Springer-Verlag Berlin Heidelberg, pp 205–228 (2014). [Google Scholar]
- Budd G. M. & Stephenson P. J. Recent glacier retreat on Heard Island (IAHS No. 86) International Association of Hydrological Sciences, Rennes, France, pp 449–458 (1970). [Google Scholar]
- Donoghue S. Changes in the morphology, mass balance and dynamics of Brown Glacier, Heard Island, with comparisons to the surrounding sub-Antarctic islands. PhD. Thesis, University of Tasmania, Hobart, Australia (2009). [Google Scholar]
- Frenot Y. et al. Biological invasions in the Antarctic: extent, impacts and implications. Biol. Rev. Camb. Philos. Soc. 80, 45–72 (2005). [DOI] [PubMed] [Google Scholar]
- Wilkins D., van Sebille E., Rintoul S. R., Lauro F. M. & Cavicchioli R. Advection shapes Southern Ocean microbial assemblages independent of distance and environment effects. Nat. Commun. 4, 2457 (2013). [DOI] [PubMed] [Google Scholar]
- Lauro F. M. et al. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J. 5, 879–895 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins D. et al. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol. Rev. 37, 303–335 (2013). [DOI] [PubMed] [Google Scholar]
- Cavicchioli R. Microbial ecology of Antarctic aquatic systems. Nat. Rev. Microbiol. 13, 691–706 (2015). [DOI] [PubMed] [Google Scholar]
- Conductivity, salinity and total dissolved solids. In, Fundamentals of Environmental Measurements. Fondriest Environmental, Inc. Web. http://www.fondriest.com/environmental-measurements/parameters/water-quality/conductivity-salinity-tds/ (2014).
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Lozupone C., Hamady M., Bushman F. D. & Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35, e120 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse S. M. et al. Exploring Microbial Diversity and Taxonomy Using SSU rRNA Hypervariable Tag Sequencing. PLoS Genet. 4, e1000255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., DeSantis T. Z., Andersen G. L. & Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 36, e120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith D. P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10 (1992). [Google Scholar]
- Chao A. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270 (1984). [Google Scholar]
- Jost L. Partitioning diversity into dependent alpha and beta components. Ecology 88, 2427–2439 (2007). [DOI] [PubMed] [Google Scholar]
- Hahn Schmidt J., Taipale S. J., Doolittle W. F. & Koll U. Rhodoluna lacicola gen. nov., sp. nov., a planktonic freshwater bacterium with stream-lined genome. Int. J. Syst. Evol. Microbiol. 64, 3254–3263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezbera J., Sharma A. K., Brandt U., Doolittle W. F. & Hahn M. W. ‘Candidatus Planktophila limnetica’. An actinobacterium representing one of the most numerically important taxa in freshwater bacterioplankton. Int. J. Syst. Evol. Microbiol. 59, 2864–2869 (2009). [DOI] [PubMed] [Google Scholar]
- Marshall K. T. & Morris R. M. Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J. 7, 452–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues C. F., Cunha M. R., Génio L. & Duperron S. A complex picture of associations between two host mussels and symbiotic bacteria in the Northeast Atlantic. Naturwissenschaften 100, 21–31 (2013). [DOI] [PubMed] [Google Scholar]
- Baek K., Choi A. & Cho J. C. Eionea flava sp. nov., isolated from coastal seawater, and emended description of the genus Eionea. Int. J. Syst. Evol. Microbiol. 65, 2975–2979 (2015). [DOI] [PubMed] [Google Scholar]
- Guiry M. D. & Guiry G. M. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org searched on 07 June 2016.
- Castelle C. J. et al. Genomic expansion of domain Archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr. Biol. 25, 690–701 (2015). [DOI] [PubMed] [Google Scholar]
- Martin-Cuadrad A.-B. et al. A new class of marine Euryarchaeota group II from the mediterranean deep chlorophyll maximum. ISME J. 9, 1619–1634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson V. et al. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335, 587–590 (2012). [DOI] [PubMed] [Google Scholar]
- Shah N., Tang H., Doak T. G. & Ye Y. Comparing bacterial communities inferred from 16S rRNA gene sequencing and shotgun metagenomics. Pac. Symp. Biocomput. 2011, 165–176 (2010). [DOI] [PubMed] [Google Scholar]
- Langille M. G. et al. (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Malmer D., Langille M. G., Way S. F. & Knight R. Which is more important for classifying microbial communities: who’s there or what they can do? ISME J. 8, 2357–2359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aßhauer K. P., Wemheuer B., Daniel R. & Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S. R., Robeson M. S., Hauser L. J., Schadt C. W. & Gorin A. A. PanFP: pangenome-based functional profiles for microbial communities. BMC Res. Notes 8, 479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. E. et al. Microbial life at −13 °C in the brine of an ice-sealed Antarctic lake. Proc. Natl Acad. Sci. USA 109, 20626–20631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick-Majors T. J., Priscu J. C. & Amaral-Zettler L. A. Modular community structure suggests metabolic plasticity during the transition to polar night in ice-covered Antarctic lakes. ISME J. 8, 778–789 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione J. -. F. et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl Acad. Sci. USA 109, 17633–17638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sul W. J., Oliver T. A., Ducklow H. W., Amaral-Zettler L. A. & Sogin M. L. Marine bacteria exhibit a bipolar distribution. Proc. Natl Acad. Sci. USA 110, 2342–2347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D. A., van der Gast C. J., Woodward K. & Newsham K. K. Significant changes in the bacterioplankton community structure of a maritime Antarctic freshwater lake following nutrient enrichment. Microbiology (SGM) 151, 3237–3248 (2005). [DOI] [PubMed] [Google Scholar]
- Williams T. J. et al. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 15, 1302–1317 (2013). [DOI] [PubMed] [Google Scholar]
- Krieg N. R. et al. (eds) Bergey’s Manual of Systematic Bacteriology: Volume 4: The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae and Planctomycetes (2nd Edition). Springer, New York (2010). [Google Scholar]
- Davidov Y. & Jurkevitch E. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax–Peredibacter clade as Bacteriovoracaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54, 1439–1452 (2004). [DOI] [PubMed] [Google Scholar]
- Terasaki Y. Transfer of five species and two subspecies of Spirillum to other genera (Aquaspirillum and Oceanospirillμm), with emended descriptions of the species and subspecies. Int. J. Syst. Evol. Microbiol. 29, 130–144 (1979). [Google Scholar]
- Grabovich M. et al. Proposal of Giesbergeria voronezhensis gen. nov., sp. nov. and G. kuznetsovii sp. nov. and reclassification of [Aquaspirillum] anulus, [A.] sinuosum and [A.] giesbergeri as Giesbergeria anulus comb. nov., G. sinuosa comb. nov. and G. giesbergeri comb. nov., and [Aquaspirillum] metamorphum and [A.] psychrophilum as Simplicispira metamorpha gen. nov., comb. nov. and S. psychrophila comb. nov. Int. J. Syst. Evol. Microbiol. 56, 569–576 (2006). [DOI] [PubMed] [Google Scholar]
- Ma Y. F. et al. The complete genome of Comamonas testosteroni reveals its genetic adaptations to changing environments. Appl. Environ. Microbiol. 75, 6812–6819 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezbera J., Jezberova J., Brandt U. & Hahn M. W. Ubiquity of Polynucleobacter necessarius subspecies asymbioticus results from ecological diversification. Environ. Microbiol. 13, 922–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K. P., Lin C. Y., Lee C. H., Shiah F. K. & Chang J. (2003) The coupling of oligotrich ciliate populations and hydrography in the East China Sea: spatial and temporal variations. Deep Sea Res. Pt II 50, 1279–1293 (2003). [Google Scholar]
- Sharma A. K. et al. Actinorhodopsin genes discovered in diverse freshwater habitats and among cultivated freshwater Actinobacteria. ISME J. 3, 726–737 (2009). [DOI] [PubMed] [Google Scholar]
- Keffer J. L., Hahn M. W. & Maresca J. A. Characterization of an unconventional rhodopsin from the freshwater Actinobacterium Rhodoluna lacicola. J. Bacteriol. 197, 2704–2712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner F. O. et al. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl. Env. Microbiol. 66, 5053–5065 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlov D. S., Marie D., Chuvochina M. S., Alekhina I. A. & Bulat S. A. Microbial communities of water column of Lake Radok, East Antarctica, dominated by abundant actinobacterium “Candidatus Planktophila limnetica”. Microbiology. 80, 576–579 (2011). [PubMed] [Google Scholar]
- Corhlan W. P. & Harrison P. J. Uptake of nitrate, ammonium, and urea by nitrogen-starved cultures of Micromonas pusilla (Prasinophyceae): transient responses. J. Phycol. 27, 673–679 (1991). [Google Scholar]
- Slapeta J., López-García P. & Moreira D. Global dispersal and ancient cryptic species in the smallest marine eukaryotes. Mol. Biol. Evol. 23, 23–29 (2006). [DOI] [PubMed] [Google Scholar]
- Delmont T. O., Eren A. M., Vineis J. H. & Post A. F. Genome reconstructions indicate the partitioning of ecological functions inside a phytoplankton bloom in the Amundsen Sea, Antarctica. Front. Microbiol. 6, 1090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavicchioli R., Ostrowski M., Fegatella F., Goodchild A. & Guixa-Boixereu N. Life under nutrient limitation in oligotrophic marine environments: an eco/physiological perspective of Sphingopyxis alaskensis (formerly Sphingomonas alaskensis). Microb. Ecol. 45, 203–217 (2003). [DOI] [PubMed] [Google Scholar]
- Lauro F. M. et al. The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. USA 106, 15527–15533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. J., Ertan H., Ting L. & Cavicchioli R. Carbon and nitrogen substrate utilization in the marine bacterium Sphingopyxis alaskensis strain RB2256. ISME J. 3, 1036–1052 (2009). [DOI] [PubMed] [Google Scholar]
- Henson M. W. et al. Artificial seawater media facilitates cultivating members of the microbial majority from the Gulf of Mexico. mSphere 1, e00028–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M. A., et al. Life under extreme energy limitation: a synthesis of laboratory- and field-based investigations. FEMS Microbiol. Rev. 39, 688–728 (2015). [DOI] [PubMed] [Google Scholar]
- Sosa O. A., Gifford S. M., Repeta D. J. & DeLong E. F. High molecular weight dissolved organic matter enrichment selects for methylotrophs in dilution to extinction cultures. ISME J. 9, 2725–2739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink J. J., Woese C. R. & Staley J. T. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus’ as Polaribacter glomeratus comb. nov. Int. J. Syst. Bacteriol. 48, 223–235 (1998). [DOI] [PubMed] [Google Scholar]
- Xing P. et al. Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J. 9, 1410–1422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins D. et al. Biogeographic partitioning of Southern Ocean microorganisms revealed by metagenomics. Environ. Microbiol. 15, 1318–1333 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang C. L., Xie W., Martin-Cuadrado A. B. & Rodriguez-Valera F. Marine Group II Archaea, potentially important players in the global ocean carbon cycle. Front. Microbiology. 6, 1108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun D. W. et al. Polaribacter atrinae sp. nov., isolated from the intestine of a comb pen shell, Atrina pectinata. Int. J. Syst. Evol. Microbiol. 64, 1654–1661 (2014). [DOI] [PubMed] [Google Scholar]
- Delmont T. O., Hammar K. M., Ducklow H. W., Yager P. L. & Post A. F. Phaeocystis antarctica blooms strongly influence bacterial community structures in the Amundsen Sea polynya. Front. Microbiol. 5, 646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- West N. J., Obernosterer I., Zemb O. & Lebaron P. Major differences of bacterial diversity and activity inside and outside of a natural iron-fertilized phytoplankton bloom in the Southern Ocean. Environ. Microbiol. 10, 738–756 (2008). [DOI] [PubMed] [Google Scholar]
- Crump B. C., Kling G. W., Bahr M. & Hobbie J. E. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69, 2253–2268 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty K. M., Young K. C., Maurice P. A. & Bridgham S. C. Dissolved organic matter concentration and quality influences upon structure and function of freshwater microbial communities. Microb. Ecol. 52, 378–388 (2006). [DOI] [PubMed] [Google Scholar]
- Zhu R. et al. Bacterial diversity is strongly associated with historical penguin activity in an Antarctic lake sediment profile. Sci. Rep. 5, 17231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C. L. et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 6, 1186–1199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Climate Change Impacts in the sub-Antarctic Islands Technical Report N°2 of ONERC February. Observatoire National sur les Effets du Réchauffement Climatique. Republic of France (2009). [Google Scholar]
- Bertrand E. M. et al. Phytoplankton-bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc. Natl. Acad. Sci. USA 112, 9938–9943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K., Lahti L., Gonze D., de Vos W. M. & Raes J. Metagenomics meets time series analysis: unraveling microbial community dynamics. Curr. Opin. Microbiol. 25, 56–66 (2015). [DOI] [PubMed] [Google Scholar]
- Australian Antarctic Data Center Map 12583 - Heard Island – main wildlife concentrations (Helicopter Operations) Accessed on June 18th 2016: https://data.aad.gov.au/database/mapcat/heard/heli-heard-wildlife_aug00.pdf (2000).
- Brown M. V. et al. Global biogeography of SAR11 marine bacteria. Mol. Syst. Biol. 8, 595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch D. B. et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5, e77 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau S. et al. Metagenomic insights into strategies of carbon conservation and unusual sulfur biogeochemistry in a hypersaline Antarctic lake. ISME J. 7, 1944–1961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins D. F. Microbial ecology and biogeography of the Southern Ocean. PhD Thesis, The University of New South Wales, Sydney, Australia (2013).
- DeMaere M. Z. et al. High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc. Natl. Acad. Sci. USA 110, 16939–16944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J. 16S/23S rRNA sequencing. In Stackebrandt E. & Goodfellow M. (eds) Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom, pp 115–175 (1991). [Google Scholar]
- Klindworth A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucl. Acids Res. 41, e1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST, Bioinformatics. 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C. & Knight R. UCHIIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V., Engelbrektson A., Ochman H. & Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates, Env. Microbiol. 12, 118–123 (2010). [DOI] [PubMed] [Google Scholar]
- Zhu F., Massana R., Not F., Marie D. & Vaulot D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52, 79–92 (2005). [DOI] [PubMed] [Google Scholar]
- Caporaso J. G. et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 26, 266–267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. & Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Baeza Y., Pirrung M., Gonzalez A. & Knight R. Emperor: A tool for visualizing high-throughput microbial community data. Gigascience 2, 16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Bray J. R. & Curtis J. T. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27, 325–349 (1957). [Google Scholar]
- Clarke K. R. & Gorley R. N. PRIMER v6: User Manual/Tutorial. 1–190, PRIMER-E Ltd, Plymouth, UK (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.