Abstract
Chelerythrine (CHE), a natural benzo[c]phenanthridine alkaloid, shows anti-cancer effect through a number of mechanisms. Herein, the effect and mechanism of the CHE-induced autophagy, a type II programmed cell death, in non-small cell lung cancer (NSCLC) cells were studied for the first time. CHE induced cell viability decrease, colony formation inhibition, and apoptosis in a concentration-dependent manner in NSCLC A549 and NCI-H1299 cells. In addition, CHE triggered the expression of phosphatidylethanolamine-modified microtubule-associated protein light-chain 3 (LC3-II). The CHE-induced expression of LC3-II was further increased in the combination treatment with chloroquine (CQ), an autophagy inhibitor, and large amounts of red-puncta were observed in the CHE-treated A549 cells with stable expression of mRFP-EGFP-LC3, indicating that CHE induces autophagy flux. Silence of beclin 1 reversed the CHE-induced expression of LC3-II. Inhibition of autophagy remarkably reversed the CHE-induced cell viability decrease and apoptosis in NCI-H1299 cells but not in A549 cells. Furthermore, CHE triggered reactive oxygen species (ROS) generation in both cell lines. A decreased level of ROS through pretreatment with N-acetyl-L-cysteine reversed the CHE-induced cell viability decrease, apoptosis, and autophagy. Taken together, CHE induced distinctive autophagy in A549 (accompanied autophagy) and NCI-H1299 (pro-death autophagy) cells and a decreased level of ROS reversed the effect of CHE in NSCLC cells in terms of cell viability, apoptosis, and autophagy.
Keywords: Chelerythrine, Autophagy, Apoptosis, ROS, NSCLC
Highlights
-
•
Chelerythrine induces autophagic flux in non-small cell lung cancer (NSCLC) A549 and NCI-H1299 cells.
-
•
Chelerythrine induces an accompanied autophagy in A549 cells, while a pro-death autophagy in NCI-H1299 cells.
-
•
A decreased level of ROS reverses the chelerythrine-induced apoptosis and autophagy in NSCLC cells.
1. Introduction
Chelerythrine (CHE), a natural benzo[c]phenanthridine alkaloid, extracts from a number of plant species, such as Chelidonium majus, Macleaya cordata, and Sanguinaria canadensis etc. [1], [2]. It exerts a wide spectrum of biological activities, including anti-cancer [3], anti-diabetes [4], anti-fungus [5], as well as protective effect against the ethanol-induced gastric ulcer [6] and the lipopolysaccharide-induced endotoxic shock [7] etc. The anti-cancer effect of CHE has been studied both in vitro and in vivo. Activation of the RAF/mitogen-activated protein/extracellular signal-regulated kinase kinase pathway resulted in the CHE-induced apoptosis in osteosarcoma cells [8]. Inhibition of Bcl-2 expression and activation of a mitochondrial pathway contributed to the CHE-induced apoptosis in hepatocellular carcinoma cells [9]. CHE also induced apoptosis and the G1-phase cell cycle arrest in human promyelocytic leukemia HL-60 cells [10]. Moreover, CHE inhibited tumor growth in the mice bearing uterine leiomyoma cells [11], head and neck squamous cell carcinoma cells [12], as well as lymphoma cells [13]. Most of the current anti-cancer study of CHE focused on its ability for induction of apoptosis, a type I programmed cell death [14]. However, whether autophagy, a type II programmed cell death [15], contributes to the anti-cancer effect of CHE is unclear.
Autophagy is an evolutionary conserved degradation system that induces the degradation of cytoplasmic contents in a lysosome-dependent manner [16]. It connects to numerous of human diseases and physiologies, including cancer, neurodegeneration, microbial infection, and ageing [17], [18] etc. Autophagy can be stimulated through reactive oxygen species (ROS) generation, hypoxia, nutrient starvation, and virus infection [19], [20], [21], [22]. So far, some autophagy regulators have been obtained [23], [24]. Our previous studies have identified a series of autophagy inducers from natural products, such as glycyrrhetinic acid, platycodin D, baicalein, licochalcone A, cryptotanshinone, isocryptotanshinone [25], [26], [27], [28], [29], [30], [31] etc. The role of the compound-induced autophagy (pro-survival, pro-death, or accompanied effect) in cancer therapy is complex [25], [26], [27], [28], [29], [30], [31].
Therefore, the effect and potential mechanism of the CHE-induced autophagy in non-small cell lung cancer (NSCLC) A549 and NCI-H1299 cells were performed, and the role of autophagy in the CHE-induced cell viability and apoptosis was studied.
2. Materials and methods
2.1. Reagents
CHE, with the purity of 99.34% that determined by using high-performance liquid chromatography assay, was purchased from the National Institutes for Food and Drug Control (Beijing, China) and dissolved in dimethyl sulfoxide (DMSO) at a concentration of 40 mM, stored at −20 °C. 3-(4,5-dimethylthiazol-2-yl)-2, 5-Diphenyltetrazolium bromide (MTT), chloroquine (CQ), puromycin, paraformaldehyde (PFA), and DMSO were obtained from the Sigma (St. Louis, MO, USA). Dulbecco's modified Eagle's medium (DMEM) medium, RPMI 1640 medium, fetal bovine serum (FBS), penicillin, streptomycinwere, phosphate-buffered saline (PBS), and hanks' balanced salt solution (HBSS) were obtained from the Gibco Life Technologies (Grand Island, NY, USA). 2′,7′-dichlorofluorescin-diacetate (DCFH2-DA) and crystal violet staining solution were purchased from the Beyotime Biotechnology Corporation (Shanghai, China). The primary antibodies, i.e. poly (ADP-ribose) polymerase (PARP) (#9532), cleaved caspase 3 (#9664), LC3 (#12741), beclin 1 (#3495), GAPDH (#2118), and anti-rabbit IgG, HRP-linked (#7074) were obtained from Cell Signaling Technology (Beverly, MA, USA).
2.2. Cell line and culture
The NSCLC A549 and NCI-H1299 cells were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA), and cultured in a RPMI 1640 medium supplemented with 10% (v/v) FBS and antibiotics (100 units/mL penicillin and 100 μg/mL streptomycin). A549 cells with mRFP-EGFP-LC3 stable expression were cultured in a DMEM medium supplemented with 10% (v/v) FBS, 1% (v/v) antibiotics (100 units/mL penicillin and 100 μg/mL streptomycin), and 2 μg/mL puromycin. All cells were cultured in a 5% CO2 incubator at 37 °C.
2.3. MTT assay
The effect of CHE on cell viability was detected by using MTT assay as described in the previous report [32]. Exponentially growing cells were seeded into 96-well plates and treated as indicated. The cell viability was examined through incubation of the cells with 1 mg/mL MTT for 4 h. DMSO was then added into solubilize the formazan and shaking in the dark. The absorbance at 570 nm was recorded with a microplate reader (Perkin Elmer, 1420 Multilabel Counter Victor3, Wellesley, MA, USA).
2.4. Colony formation assay
Cells were seeded into 6-well plates. After attachment, cells were incubated with the different concentrations of CHE for 24 h. The medium was placed with fresh medium and cells were cultured for another 14 d until the visible colonies were observed. The colonies were fixed with 4% PFA and stained with crystal violet staining solution. The images of cell colony were captured by using an ordinary NIKON camera.
2.5. Annexin V-FITC/PI staining assay
After incubation with the indicated concentrations of CHE in the presence and absence of CQ (10 μΜ, 1 h), NAC (5 mM, 1 h), or silence of beclin 1, cells were trypsinized, washed, and collected. The dead cells (apoptotic and necrotic cells) were detected by using annexin V-FITC/PI dual labeling assay kit (BioVision, CA, USA) in accordance with the protocol provided by the manufacturer. At least 10,000 cells were collected and analyzed by using a flow cytometer (Becton Dickinson FACS Canto, Franklin Lakes, NJ).
2.6. Western blot assay
The total protein was obtained by using a radioimmunoprecipitation lysis buffer containing 1% phenylmethanesulfonyl fluoride and 1% protease inhibitor cocktail. Then, the protein concentrations were calculated with the BCA™ protein assay kit (Pierce, Rockford, IL, USA). Equal amounts of proteins were separated by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to a polyvinylidene difluoride membrane followed by blocking in 5% non-fat dried milk in PBST at room temperature for 1 h. The membrane was incubated with primary antibodies overnight at 4 °C. After washing with PBST, the membranes were incubated with corresponding secondary antibodies at room temperature for 1 h. The specific protein bands were visualized with an ECL advanced Western blot analysis detection kit (BD Biosciences, Bedford, MA, USA). Equal protein loading was verified by probing with anti-GAPDH antibodies. The quantification of Western blot images was calculated by the following steps: 1) the grey level of each indicated protein was obtained through the ChemiDocTM MP imaging system. 2) The ratio of indicated protein/GAPDH was obtained. 3) The fold of control value was obtained by calculating “treatment group value”/“control group value”. 4) Three independent experiments were performed and mean±standard deviation (SD) was calculated.
2.7. Immunofluorescence staining assay
A549 cells with mRFR-EGFR-LC3 constitutive expression were treated with 15 μΜ CHE, HBSS, and 10 μM CQ for 24 h, respectively. Cells were then fixed with 4% PFA for 30 min and washed with PBS for 3 times. The immunofluorescent images were obtained by using a confocal laser scanning microscope (Leica TCS SP8, Solms, Germany) and typical images were presented.
For quantitative assay, the red-puncta and green-puncta numbers were counted as described previously [33], [34]. Briefly, at least 20 cells (per experiment) were randomly selected for counting the number of mRFP-LC3 or EGFR-LC3 puncta in each group, and three independent experiments were performed.
2.8. Small interfering RNA (siRNA) transfection assay
The specific target sequences of beclin1 (sense 5′-GGAGCCAUUUAUUGAAACUTT-3′, antisense 5′-AGUUUCAAUAAAUGGCUCCTT-3′) and the scrambled siRNA (sense 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense 5′-ACGUGACACGUUCGGAGAATT-3′) were purchased from the GenePharma (Shanghai, China). Cells were seeded into 6-well plates for overnight and then transfected with siRNA of beclin 1 and the scrambled siRNA by using the Lipofectamine™ 2000 transfection reagent (Invitrogen Corp., Carlsbad, CA, USA) in strict accordance with the manufacturer's instructions. Cells were then treated with 15 μΜ CHE for 24 h. The protein expression levels of beclin 1, LC3, and GAPDH were detected by using Western blot assay.
2.9. Reactive oxygen species (ROS) generation assay
The generation of intracellular ROS was detected with DCFH2-DA probe. Cells were pretreated with DCFH2-DA probe for 30 min, and followed by incubation with 15 μM CHE for 1.5 h in the presence or absence of NAC (5 mM, 1 h). Cells were harvested and washed with PBS. The generation of ROS was studied by using a flow cytometry (Becton Dickinson FACS Canto, Franklin Lakes, NJ). At least 10,000 cells were recorded for each sample and typical images were presented.
2.10. Statistical analysis
The mean±SD was determined for each group. Statistical analysis was performed with one-way analysis of variance and Tukey's test. Differences were considered statistically significant for *P<0.05 and **P<0.01. The “ns” means “no statistical difference”. At least three independent experiments were performed for each assay.
3. Results
3.1. CHE induced cell viability decrease, cell death, and apoptosis in A549 and NCI-H1299 cells
First, the effect of CHE on the cell viability of NSCLC cells was detected by using MTT assay. The cell viability of A549 and NCI-H1299 cells were remarkably reduced in a concentration-dependent manner after treatment with CHE for 24 h (Fig. 1A), with cell viabilities remaining at 70.63%, 47.86%, and 14.42% in A549 cells and 70.59%, 38.04%, and 7.82% in NCI-H1299 cells after incubation with 10, 15, and 20 μM CHE, respectively. The inhibitive effect of CHE on NSCLC cells was further confirmed through colony formation assay. As shown in Fig. 1B, almost no colonies were observed in A549 and NCI-H1299 cells after treatment with 10 and 15 μM CHE, respectively. To investigate the potential mechanisms of the CHE-induced cell viability decrease, the dead cells were detected by using annexin V-FITC/PI dual labeling assay. As shown in Fig. 1C, the numbers of annexin V-FITC positive cells were increased after treatment with CHE in A549 (9.50%, 29. 60%, and 59.93% for 10, 15, and 20 μM CHE treatment, respectively) and NCI-H1299 (9.3%, 37.97%, and 67.43% for 10, 15, and 20 μM CHE treatment, respectively) cells. In addition, Western blot assay suggested that the exposure of cells to CHE increased the protein expressions of cleaved PARP and cleaved caspase 3 (Fig. 1D), which are protein biomarkers of apoptosis [35]. These results suggested that CHE induces cell viability decrease, cell death, and apoptosis in NSCLC A549 and NCI-H1299 cells.
Fig. 1.
CHE induced cell viability decrease and apoptosis in A549 and NCI-H1299 cells. (A) A549 and NCI-H1299 cells were treated with different concentrations of CHE for 24 h. Cell viability was detected by using MTT assay. *P<0.05, **P<0.01, compared with 0 μM CHE treatment. (B) Cells were treated with indicated concentrations of CHE for 24 h and then cultured in fresh medium for about 14 d until the visible colonies were observed. The colonies were stained with crystal violet and photographed. Representative images were presented. (C) After treatment with indicated concentrations of CHE for 24 h, the dead cells were evaluated through annexin V-FITC/PI dual staining assay. *P<0.05 and ** P<0.01. (D) A549 and NCI-H1299 cells were treated with different concentrations of CHE for 24 h, the protein expressions of PARP, cleaved caspase 3, and GAPDH were detected by using Western blot assay.
3.2. CHE induced beclin 1-dependent autophagy in A549 and NCI-H1299 cells
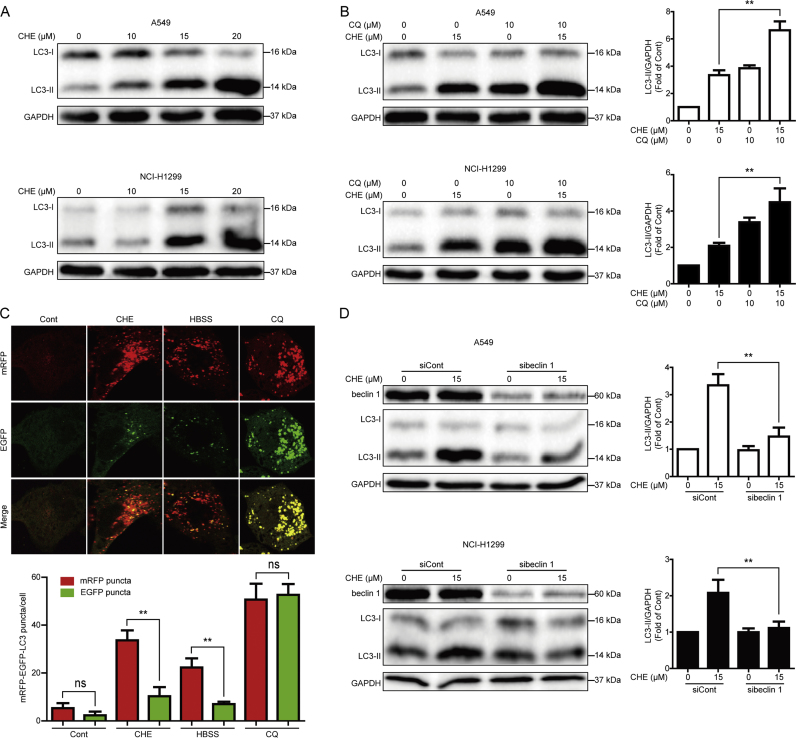
We then studied whether or not CHE induces autophagy in NSCLC cells. The protein expression of LC3-II, a protein marker of autophagy [36], was detected by using Western blot assay. As shown in Fig. 2A, CHE induced the expression of LC3-II protein in a concentration-dependent manner in A549 and NCI-H1299 cells. Combinative treatment with CQ, an autophagy inhibitor that disrupts the function of lysosome to inhibit autophagy [37], further enhanced the CHE-induced expression of LC3-II (Fig. 2B). The A549 cells with mRFP-EGFP-LC3 stable expression were used to confirm the CHE-induced autophagy. As shown in Fig. 2C, more red-puncta than green-puncta was observed after exposing of CHE in A549 cells with mRFP-EGFP-LC3 stable expression, this result was similar to HBSS, an autophagy inducer [37], while opposite to CQ. Moreover, silence of beclin 1, a part of a lipid kinase complex that induces the initial stages of autophagosome formation [38], by using the specific siRNA remarkably reversed the CHE-induced expression of LC3-II (Fig. 2D), suggesting that CHE induces beclin 1-dependent autophagy in A549 and NCI-H1299 cells.
Fig. 2.
CHE induced autophagy in A549 and NCI-H1299 cells. (A) A549 and NCI-H1299 cells were treated with different concentrations of CHE for 24 h, and cell extracts were analyzed to detect the changes of protein expression by Western blot assay. (B) A549 and NCI-H1299 cells were treated with 15 μM CHE for 24 h with or without pretreatment with CQ (10 μM, 1 h). Cell extracts were analyzed for protein expression by using Western blot assay. *P<0.05 and **P<0.01. (C) A549 cells with mRFP-EGFP-LC3 stable expression were treated with CHE, HBSS, and CQ for 24 h, respectively. The formation of puncta was imaged by using a confocal microscope and typical images were presented. For quantification, at least 20 cells (per experiment) were randomly selected for counting the number of mRFP-LC3 or EGFR-LC3 puncta in each group. Data were presented from one representative experiment of three independent experiments. *P<0.05, **P<0.01. The “ns” means “no statistical difference”. (D) A549 and NCI-H1299 cells were transiently transfected with siRNA of beclin 1 for 24 h. 15 μM CHE was added into the cells and cultured for another 24 h. The expression changes of indicated proteins were examined through Western blot assay. *P<0.05 and **P<0.01.
3.3. Inhibition of autophagy reversed the CHE-induced cell viability decrease in NCI-H1299 cells while not in A549 cells
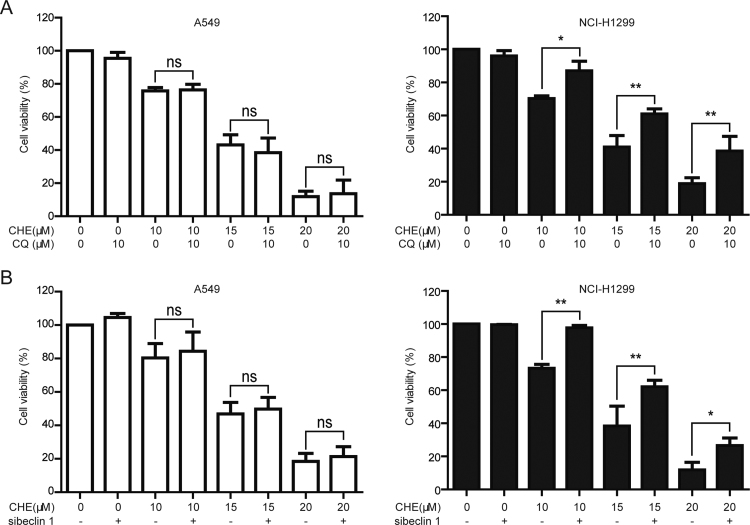
The role of autophagy in cancer therapy is complex and controversial [39]. Herein, the CHE-induced cell viability decrease was examined after pharmacological and genetic inhibition of autophagy. As shown in Fig. 3A, the CHE-induced cell viability decrease was not remarkably changed when pretreatment with CQ (10 μM, 1 h) in A549 cells (the cell viabilities of 10, 15, and 20 μM CHE-treated cells were changed from 75.79%, 43.20%, and 11.86 to 76.33%, 38.46%, and 13.42% after pretreatment with CQ, respectively). However, pretreatment with CQ statistically reversed the CHE-induced cell viability decrease in NCI-H1299 cells (the cell viabilities of 10, 15, and 20 μM CHE-treated cells were changed from 70.22%, 40.99%, and 18.87 to 87.05%, 60.96%, and 38.55% after pretreatment with CQ, respectively). The distinctive effect of the CHE-induced autophagy was further studied by using siRNA of beclin 1. As shown in Fig. 3B, the cell viabilities in A549 cells were changed from 80.38% (10 μM CHE), 48.63% (15 μM CHE), and 18.46% (20 μM CHE) to 84.37% (10 μM CHE + siRNA of beclin 1), 49.79% (15 μM CHE + siRNA of beclin 1) and 21.34% (20 μM CHE + siRNA of beclin 1), respectively, while the cell viabilities in NCI-H1299 cells were changed from 73.29% (10 μM CHE), 38.39% (15 μM CHE), and 11.84% (20 μM CHE) to 97.60% (10 μM CHE + siRNA of beclin 1), 62.02% (15 μM CHE + siRNA of beclin 1) and 26.58% (20 μM CHE + siRNA of beclin 1), respectively. These results suggested that the CHE-induced autophagy contributes to the cell viability decrease in NCI-H1299 cells, meanwhile inhibition of autophagy could not remarkably change the CHE-induced cell viability decrease in A549 cells.
Fig. 3.
Inhibition of autophagy reversed the CHE-induced cell viability decrease in NCI-H1299 cells while not in A549 cells. (A) A549 and NCI-H1299 cells were treated with various concentrations of CHE for 24 h with or without pretreatment with CQ (10 μM, 1 h). Cell viability was studied by using MTT assay. *P<0.05 and **P<0.01. The “ns” means “no statistical difference”. (B) After inhibition of autophagy through silence of beclin 1, A549 and NCI-H1299 cells were treated with 10, 15, and 20 μM CHE for 24 h. Cell viability was examined through MTT assay. *P<0.05 and **P<0.01. The “ns” means “no statistical difference”.
3.4. Inhibition of autophagy reversed the CHE-induced cell death and apoptosis in NCI-H1299 cells while not in A549 cells
The distinctive effect of the CHE-induced autophagy in NSCLC cells was further confirmed by evaluating the CHE-induced cell death and apoptosis after inhibition of autophagy. First, the annexin V-FITC/PI dual labeling assay was detected. The annexin V-FITC positive cell numbers of the CHE-treated A549 and NCI-H1299 cells were changed from 28.33% to 28.93% and from 36.77% to 22.83% when pretreatment with CQ (10 μM, 1 h), respectively (Fig. 4A). Inhibition of autophagy through siRNA of beclin 1 could not remarkably reverse the CHE-induced cell death in A549 cells (the annexin V-FITC positive cell numbers were changed from 29.57% to 27.33% under siRNA of beclin 1). In contrast, the CHE-induced annexin V-FITC positive cell numbers in NCI-H1299 cells was obviously revered under siRNA of beclin 1 (the annexin V-FITC positive cell numbers were 36.90% in the CHE-treated group and 22.97% in the CHE + siRNA of beclin 1-treated group) (Fig. 4B). Moreover, silence of beclin 1 remarkably decreased the CHE-induced expression of cleaved PARP in NCI-H1299 cells while not in A549 cells (Fig. 4C). The aforementioned results suggested that CHE induces a pro-death autophagy in NCI-H1299 cells while an accompanied autophagy in A549 cells.
Fig. 4.
Inhibition of autophagy reversed the CHE-induced cell death and apoptosis in NCI-H1299 cells while not in A549 cells. (A) A549 and NCI-H1299 cells were treated with 15 μM CHE for 24 h with or without pretreatment with CQ (10 μM, 1 h). The dead cells were evaluated through annexin V-FITC/PI dual staining assay. *P<0.05 and **P<0.01. The “ns” means “no statistical difference”. (B–C) After silence of belcin 1, cells were treated with 15 μM CHE for 24 h. The dead cells were evaluated through annexin V-FITC/PI dual staining assay. The protein expressions of PARP and GAPDH were evaluated by using Western blot assay. *P<0.05 and **P<0.01. The “ns” means “no statistical difference”.
3.5. A decreased level of ROS reversed the CHE-induced cell viability decrease in A549 and NCI-H1299 cells
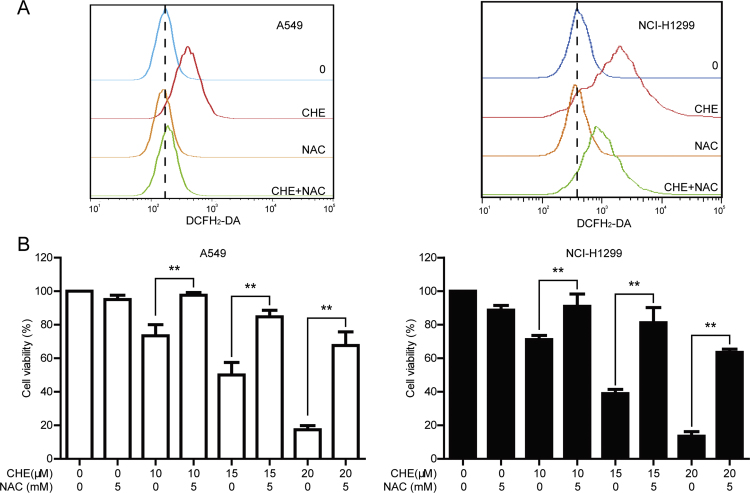
Reactive oxygen species (ROS), a series of oxygen-containing and active species, plays a critical effect on regulation of cellular programs and signal transduction [40], [41]. Whether CHE increases the generation of ROS in A549 and NCI-H1299 cells was detected by using DCFH2-DA probe. As shown in Fig. 5A, CHE increased the generation of ROS in both cell lines after treatment for 1.5 h, and pretreatment with NAC (5 mM, 1 h) decreased the CHE-induced ROS generation. The effect of ROS in the CHE-induced cell viability decrease was then determined. Pretreatment with NAC (5 mM, 1 h) reversed the CHE-induced cell viability decrease in both cell lines (Fig. 5B). The cell viabilities of 10, 15, and 20 μM CHE-treated A549 cells were changed from 73.41%, 50.06%, and 17.30 to 97.62%, 84.71%, and 67.61% under pretreatment with NAC, respectively, and the cell viabilities of 10, 15, and 20 μM CHE-treated NCI-H1299 cells were changed from 71.16%, 38.95%, and 13.51 to 91.00%, 81.29%, and 63.58% under pretreatment with NAC, respectively (Fig. 5B).
Fig. 5.
A decreased level of ROS reversed the CHE-induced cell viability decrease in A549 and NCI-H1299 cells. (A) A549 and NCI-H1299 cells were incubated with 15 μM CHE for 1.5 h with or without pretreatment with NAC (5 mM, 1 h). The level of ROS was monitored with DCFH2-DA probe. (B) The different concentrations of CHE were added into A549 and NCI-H1299 cells for 24 h with or without pretreatment with NAC (5 mM, 1 h). Cell viability was detected through MTT assay. *P<0.05, and **P<0.01.
3.6. A decreased level of ROS reversed the CHE-induced cell death, apoptosis, and autophagy in A549 and NCI-H1299 cells
Whether the CHE-induced cell death, apoptosis, and autophagy depend on the generation of ROS was detected by using annexin V-FITC/PI staining and Western blot assays. As shown in Fig. 6A, the CHE-induced annexin V-FITC positive cell numbers were reversed when pretreatment with NAC (the annexin V-FITC positive cell numbers were changed from 30.07% (15 μM CHE) to 5.33% (15 μM CHE +5 mM NAC) in A549 cells and from 37.40% (15 μM CHE) to 6.67% (15 μM CHE +5 mM NAC) in NCI-H1299 cells). In addition, the CHE-induced expressions of cleaved PARP and LC3-II were reversed after pretreatment with NAC (5 mM, 1 h) (Fig. 6B–C). The overall data suggested that ROS contributes to the CHE-induced cell viability decrease, cell death, apoptosis, and autophagy in A549 and NCI-H1299 cells.
Fig. 6.
A decreased level of ROS reversed the CHE-induced cell death, apoptosis, and autophagy in A549 and NCI-H1299 cells. (A–C) A549 and NCI-H1299 cells were treated with indicated concentration of CHE for 24 h with or without pretreatment with NAC (5 mM, 1 h). The dead cells were evaluated through annexin V-FITC/PI dual staining assay. The protein expressions of PARP, LC3-II, and GAPDH were evaluated by using Western blot assay. *P<0.05 and **P<0.01.
4. Discussion
As far as we know, this is the first time to report that CHE induced autophagic flux in cancer cells, as evidence by 1) further enhanced the CHE-induced expression of LC3-II when combinative treatment with CQ. Combinative treatment with CQ further increases the autophagy inducer-induced level of LC3-II, while not increases the autophagy inhibitor-induced expression of LC3-II [37]. 2) More red-fluorescent punta than green-fluorescent punta were observed in the CHE-treated group. If an agent induces autophagic flux, more red puncta are observed in the cells because of the easy-to-quench GFP and the relatively stable mRFP in acidic environment, while inhibition of autophagic flux leads to most puncta exhibiting both green and red fluorescence, and these puncta present yellow in the merged images [37], [42]. 3) Silence of beclin 1, a critical molecule in the process of autophagic flux [43], reversed the CHE-induced expression of LC3-II.
The effect of the compound-induced autophagy in cancer therapy is controversial [39], [44]. Our previous studies indicated that glycyrrhetinic acid and platycodin D induced a pro-surviving autophagy, cryptotanshinone and isocryptotanshinone induced a pro-death autophagy, and licochalcone A induced an accompanied autophagy in cancer cells [25], [26], [27], [28], [29]. Interesting, we here showed that even one compound induced distinctive effect of autophagy in the same type of cancer cells (i.e. EGFR wide type NSCLC cells). CHE induced an accompanied autophagy in NSCLC A549 cells, while a pro-death autophagy in NSCLC NCI-H1299 cells. This inconsistent effect of autophagy might be due to the different background of cancer cells. The antioxidant ability of A549 cells was higher than NCI-H1299 cells [45]. Herein, it was much easier to generate excessive ROS in NCI-H1299 than A549 cells, and the generation of excessive ROS often resulted in autophagic cell death [46]. In addition, the different autophagy-degraded cargo might contribute to the distinctive effect of autophagy [47]. Therefore, some proteins that critical for cell survival might be degraded in NCI-H1299 while not in A549 cells when treatment with CHE.
Previous studies indicated that ROS triggers the pro-death or pro-surviving autophagy in cancer cells [48], [49]. In this study, the ROS-induced autophagy contributed to the CHE-induced cell death in NCI-H1299 cells. Meanwhile, the CHE-induced autophagy through ROS generation in A549 cells was an accompanied effect, suggested that the ROS-induced autophagy in cancer therapy could be pro-surviving, pro-death, or accompanied effect. Induction of ROS production from nicotinamide adenine dinucleotide phosphate oxidase, mitochondria, and cyclooxygenase etc. or reduction of scavenging capacity of ROS contributed to the intercellular accumulation of ROS [50], [51], [52]. The generation of intercellular ROS by selectively degrading the protein expression of catalase, an enzyme that decomposes the hydrogen peroxide in cells, resulted in cell death [53]. Meanwhile, induction of ROS generation from mitochondria contributed to pro-surviving autophagy [54], indicating that the effect of the ROS-induced autophagy in cancer therapy might depend on the origin of ROS. Therefore, the source of the CHE-induced ROS generation in A549 and NCI-H1299 cells might be different, and more experiments are required to clarify this issue in the future.
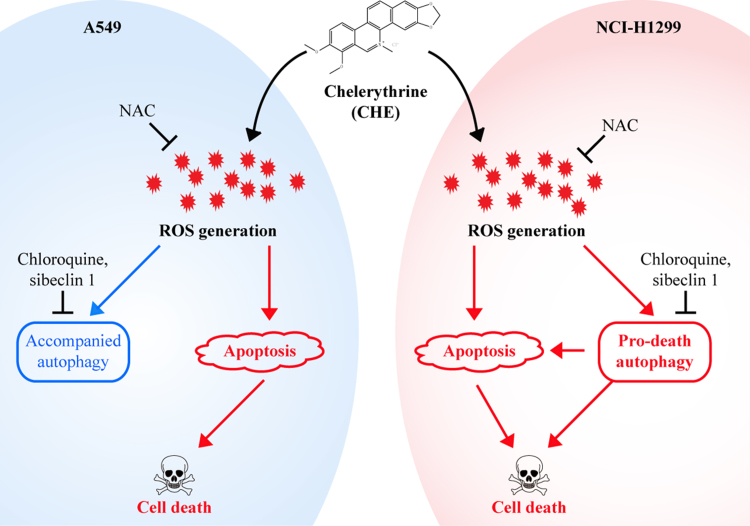
In conclusion, we demonstrated for the first time that CHE induced a distinctive effect of ROS-dependent autophagy in A549 (an accompanied autophagy) and NCI-H1299 (a pro-death autophagy) cells, and a decreased level of ROS reversed the CHE-induced cell viability decrease, cell death, apoptosis, and autophagy in NSCLC cells (Fig. 7).
Fig. 7.
Schematic of the CHE-induced apoptosis and autophagy in A549 and NCI-H1299 cells. CHE induced the ROS-dependent autophagy and apoptosis in NSCLC A549 and NCI-H1299 cells. Inhibition of autophagy reversed the CHE-induced cell viability decrease and apoptosis in NCI-H1299 cells while not in A549 cells. A decreased level of ROS reversed the CHE-induced apoptosis, and autophagy in A549 and NCI-H1299 cells.
Author contributions
Z.H.T., W.X.C., and Z.Y.W. performed the research; Z.H.T. and J.J.L. designed the research study; X.P.C. and J.J.L. contributed essential reagents or tools; Z.H.T., W.X.C., B.L., J.H.L, X.P.C., and J.J.L. analyzed the data; Z.H.T. and J.J.L. wrote the paper. All authors reviewed the manuscript.
Competing financial interests
The authors declare no conflict of interest.
Acknowledgements
This work was supported by Science and Technology Development Fund, Macao S.A.R (FDCT) (038/2014/A1), the Research Fund of University of Macau (MYRG2015-00091-ICMS-QRCM and MYRG2015-00101-ICMS-QRCM) and Science and Technology Program of Guangzhou, China (201607010338).
References
- 1.Zhang F., Chen B., Xiao S., Yao S. Optimization and comparison of different extraction techniques for sanguinarine and chelerythrine in fruits of Macleaya cordata (Willd) R. Br. Sep Purif. Technol. 2005;42(3):283–290. [Google Scholar]
- 2.Malikova J., Zdarilova A., Hlobilkova A. Effects of sanguinarine and chelerythrine on the cell cycle and apoptosis. Biomed. Pap. 2006;150(1):5–12. doi: 10.5507/bp.2006.001. [DOI] [PubMed] [Google Scholar]
- 3.Jana J., Mondal S., Bhattacharjee P., Sengupta P., Roychowdhury T., Saha P., Kundu P., Chatterjee S. Chelerythrine down regulates expression of VEGFA, BCL2 and KRAS by arresting G-quadruplex structures at their promoter regions. Sci. Rep. 2017;7:40706. doi: 10.1038/srep40706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng W., Qiu L., Wang R., Feng X., Han Y., Zhu Y., Chen D., Liu Y., Jin L., Li Y. Selective targeting of PPARγ by the natural product chelerythrine with a unique binding mode and improved antidiabetic potency. Sci. Rep. 2015;5:12222. doi: 10.1038/srep12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X., Miao F., Yao Y., Cao F., Yang R., Ma Y., Qin B., Zhou L. In vitro antifungal activity of sanguinarine and chelerythrine derivatives against phytopathogenic fungi. Molecules. 2012;17(11):13026–13035. doi: 10.3390/molecules171113026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W.-F., Hao D.-J., Fan T., Huang H.-M., Yao H., Niu X.-F. Protective effect of chelerythrine against ethanol-induced gastric ulcer in mice. Chem.-Biol. Inter. 2014;208:18–27. doi: 10.1016/j.cbi.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Niu X., Mu Q., Li W., Huang H., Yao H., Li H. Protective effects of chelerythrine against lipopolysaccharide-induced endotoxic shock in mice. Inflammation. 2014;37(6):1968–1975. doi: 10.1007/s10753-014-9929-7. [DOI] [PubMed] [Google Scholar]
- 8.Yang R., Piperdi S., Gorlick R. Activation of the RAF/mitogen-activated protein/extracellular signal-regulated kinase kinase/extracellular signal-regulated kinase pathway mediates apoptosis induced by chelerythrine in osteosarcoma. Clin. Cancer Res. 2008;14(20):6396–6404. doi: 10.1158/1078-0432.CCR-07-5113. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z., Guo Y., Zhang J., Wei X. Induction of apoptosis by chelerythrine chloride through mitochondrial pathway and Bcl-2 family proteins in human hepatoma SMMC-7721 Cell. Arch. Pharm. Res. 2011;34(5):791–800. doi: 10.1007/s12272-011-0513-5. [DOI] [PubMed] [Google Scholar]
- 10.Vrba J., Doležel P., Vičar J., Modrianský M., Ulrichová J. Chelerythrine and dihydrochelerythrine induce G1 phase arrest and bimodal cell death in human leukemia HL-60 cells. Toxicol. Vitr. 2008;22(4):1008–1017. doi: 10.1016/j.tiv.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Medvetz D., Sun Y., Li C., Khabibullin D., Balan M., Parkhitko A., Priolo C., Asara J.-M., Pal S., Yu J. High-throughput drug screen identifies chelerythrine as a selective inducer of death in a TSC2-null setting. Mol. Cancer Res. 2015;13(1):50–62. doi: 10.1158/1541-7786.MCR-14-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chmura S.-J., Dolan M.-E., Cha A., Mauceri H.-J., Kufe D.-W., Weichselbaum R.-R. In vitro and in vivo activity of protein kinase C inhibitor chelerythrine chloride induces tumor cell toxicity and growth delay in vivo. Clin. Cancer Res. 2000;6(2):737–742. [PubMed] [Google Scholar]
- 13.Kumar S., Tomar M.-S., Acharya A. Chelerythrine delayed tumor growth and increased survival duration of Dalton's lymphoma bearing BALB/c H 2d mice by activation of NK cells in vivo. J. Cancer Res. Ther. 2015;11(4):904–910. doi: 10.4103/0973-1482.143342. [DOI] [PubMed] [Google Scholar]
- 14.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsujimoto Y., Shimizu S. Another way to die: autophagic programmed cell death. Cell Death Differ. 2005;2:1528–1534. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- 16.Klionsky D.-J., Emr S.-D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N., Levine B., Cuervo A.M., Klionsky D.-J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan G., Xie W., Liu Z., Xu W., Lao Y., Huang N., Cui K., Liao M., He J., Jiang Y. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014;10(1):70–79. doi: 10.4161/auto.26534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tallóczy Z., Jiang W., Virgin H.-W., Leib D.-A., Scheuner D., Kaufman R.-J., Eskelinen E.-L., Levine B. Regulation of starvation-and virus-induced autophagy by the eIF2α kinase signaling pathway. P Natl. Acad. Sci. USA. 2002;99(1):190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azad M.-B., Chen Y., Gibson S.-B. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxid. Redox Sign. 2009;11(4):777–790. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- 23.Ding Q., Bao J., Zhao W., Hu Y., Lu J., Chen X. Natural autophagy regulators in cancer therapy: a review. Phytochem Rev. 2015;14(1):137–154. [Google Scholar]
- 24.VakifahmetogluNorberg H., Xia H., Yuan J. Pharmacologic agents targeting autophagy. J. Clin. Invest. 2015;125(1):5–13. doi: 10.1172/JCI73937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo S., Luo W., Liu L., Pang X., Zhu H., Liu A., Lu J., Ma D., Leung C., Wang Y. Isocryptotanshinone, a STAT3 inhibitor, induces apoptosis and pro-death autophagy in A549 lung cancer cells. J. Drug Target. 2016;24(10):1–9. doi: 10.3109/1061186X.2016.1157882. [DOI] [PubMed] [Google Scholar]
- 26.Hao W., Zhang X., Zhao W., Zhu H., Liu Z., Lu J., Chen X. Cryptotanshinone induces pro-death autophagy through JNK signaling mediated by reactive oxygen species generation in lung cancer cells. Anti-Cancer Agent M. 2016;16(5):593–600. doi: 10.2174/1871520615666150907093036. [DOI] [PubMed] [Google Scholar]
- 27.Tang Z.-H., Zhang L.-L., Li T., Lu J.-H., Ma D.-L., Leung C.-H., Chen X.-P., Jiang H.-L., Wang Y.-T., Lu J.-J. Glycyrrhetinic acid induces cytoprotective autophagy via the inositol-requiring enzyme 1α-c-Jun N-terminal kinase cascade in non-small cell lung cancer cells. Oncotarget. 2015;6(41):43911–43926. doi: 10.18632/oncotarget.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T., Xu X.-h., Tang Z.-h., Wang Y.-f., Leung C.-h., Ma D.-l., Chen X.-p., Wang Y.-t., Chen Y., Lu J.-j. Platycodin D induces apoptosis and triggers ERK-and JNK-mediated autophagy in human hepatocellular carcinoma BEL-7402 cells. Acta Pharm. Sin. 2015;36(12):1503–1513. doi: 10.1038/aps.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Z.-H., Chen X., Wang Z.-Y., Chai K., Wang Y.-F., Xu X.-H., Wang X.-W., Lu J.-H., Wang Y.-T., Chen X.-P. Induction of C/EBP homologous protein-mediated apoptosis and autophagy by licochalcone A in non-small cell lung cancer cells. Sci. Rep. 2016;6:26241. doi: 10.1038/srep26241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y.-F., Xu Y.-L., Tang Z.-H., Li T., Zhang L.-L., Chen X., Lu J.-H., Leung C.-H., Ma D.-L., Qiang W.-A., Wang Y., Lu J.-J. Baicalein induces beclin 1-and extracellular signal-regulated kinase-dependent autophagy in ovarian cancer cells. Am. J. Chin. M. 2017;45(11):1–14. doi: 10.1142/S0192415X17500094. [DOI] [PubMed] [Google Scholar]
- 31.Tang Z.-H., Li T., Chang L.-L., Zhu H., Tong Y.-G., Chen X.-P., Wang Y.-T., Lu J.-J. Glycyrrhetinic Acid triggers a protective autophagy by activation of extracellular regulated protein kinases in hepatocellular carcinoma cells. J. Agr. Food Chem. 2014;62(49):11910–11916. doi: 10.1021/jf503968k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Z.-H., Li T., Gao H.-W., Sun W., Chen X.-P., Wang Y.-T., Lu J.-J. Platycodin D from Platycodonis Radix enhances the anti-proliferative effects of doxorubicin on breast cancer MCF-7 and MDA-MB-231 cells. Chin. Med. 2014;9(1):2014. doi: 10.1186/1749-8546-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J.-H., Tan J.-Q., Durairajan S.-S., Liu L.-F., Zhang Z.-H., Ma L., Shen H.-M., Chan H.-Y., Li M. Isorhynchophylline, a natural alkaloid, promotes the degradation of alpha-synuclein in neuronal cells via inducing autophagy. Autophagy. 2012;8(1):98–108. doi: 10.4161/auto.8.1.18313. [DOI] [PubMed] [Google Scholar]
- 34.Ganley I.-G., Wong P.-M., Gammoh N., Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol. Cell. 2011;42(6):731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen G.-M. Caspases: the executioners of apoptosis. Biochem. J. 1997;326(1):1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3(6):542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang R., Zeh H., Lotze M., Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathew R., Karantza-Wadsworth V., White E. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7(12):961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apel K., Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 41.Khan A.-U., Wilson T. Reactive oxygen species as cellular messengers. Chem. Biol. 1995;2(7):437–445. doi: 10.1016/1074-5521(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 42.Mizushima N. Methods for monitoring autophagy. Int J. Biochem Cell B. 2004;36(12):2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Cao Y., Klionsky D.-J. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17(10):839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z.-J., Chee C.-E., Huang S., Sinicrope F.-A. The role of autophagy in cancer: therapeutic implications. Mol. Cancer Ther. 2011;10(9):1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun X., Wang Q., Wang Y., Du L., Xu C., Liu Q. Brusatol enhances the radiosensitivity of A549 cells by promoting ROS production and enhancing DNA damage. Int J. Mol. Sci. 2016;17(7):997. doi: 10.3390/ijms17070997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin K., Barrett J. Reactive oxygen species as double-edged swords in cellular processes: low-dose cell signaling versus high-dose toxicity. Hum. Exp. Toxicol. 2002;21(2):71–75. doi: 10.1191/0960327102ht213oa. [DOI] [PubMed] [Google Scholar]
- 47.Gump J.-M., Staskiewicz L., Morgan M.-J., Bamberg A., Riches D.-W., Thorburn A. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat. Cell Biol. 2014;16(1):47–54. doi: 10.1038/ncb2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y., McMillan-Ward E., Kong J., Israels S., Gibson S. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ. 2008;15(1):171–182. doi: 10.1038/sj.cdd.4402233. [DOI] [PubMed] [Google Scholar]
- 49.Nair R.-R., Emmons M.-F., Cress A.-E., Argilagos R.-F., Lam K., Kerr W.-T., Wang H.-G., Dalton W.-S., Hazlehurst L.-A. HYD1-induced increase in reactive oxygen species leads to autophagy and necrotic cell death in multiple myeloma cells. Mol. Cancer Ther. 2009;8(8):2441–2451. doi: 10.1158/1535-7163.MCT-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holmström K.-M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Bio. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 51.Kalyanaraman B., Darley-Usmar V., Davies K.-J., Dennery P.-A., Forman H.-J., Grisham M.-B., Mann G.-E., Moore K., Roberts L.-J., Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52(1):1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalyanaraman B. Teaching the basics of redox biology to medical and graduate students: oxidants, antioxidants and disease mechanisms. Redox Biol. 2013;1(1):244–257. doi: 10.1016/j.redox.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu L., Wan F., Dutta S., Welsh S., Liu Z., Freundt E., Baehrecke E.H., Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc. Natl. Acad. Sci. USA. 2006;103(13):4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karna P., Zughaier S., Pannu V., Simmons R., Narayan S., Aneja R. Induction of reactive oxygen species-mediated autophagy by a novel microtubule-modulating agent. J. Biol. Chem. 2010;285(24):18737–18748. doi: 10.1074/jbc.M109.091694. [DOI] [PMC free article] [PubMed] [Google Scholar]