Upon mitogenic induction of immediate-early genes, phosphorylation of histone H3 at S10 or S28 occurs on different alleles. S28ph depends on CBP/p300-mediated K27ac, whereas H3 acetylated on K9 by PCAF is phosphorylated on S10. The redundant roles of S10ph and S28ph and their random targeting on distinct alleles may enable a fast response.
Abstract
Stimulation of the MAPK pathway results in mitogen- and stress-activated protein kinase 1/2 (MSK1/2)-catalyzed phosphorylation of histone H3 at serine 10 or 28 and expression of immediate-early (IE) genes. In 10T1/2 mouse fibroblasts, phosphorylation of H3S10 and H3S28 occurs on different H3 molecules and in different nuclear regions. Similarly, we show that mitogen-induced H3S10 and H3S28 phosphorylation occurs in separate pools in human primary fibroblasts. High-resolution imaging studies on both cell types reveal that H3S10 and H3S28 phosphorylation events can be induced in a single cell but on different alleles, giving rise to H3S10ph and H3S28ph epialleles. Coimmunoprecipitation and inhibition studies demonstrate that CBP/p300-mediated H3K27 acetylation is required for MSK1/2 to phosphorylate S28. Although the K9ac and S10ph marks coexist on H3, S10 phosphorylation is not dependent on K9 acetylation by PCAF. We propose that random targeting of H3S10 or H3S28 results from the stochastic acetylation of H3 by CBP/p300 or PCAF, a process comparable to transcriptional bursting causing temporary allelic imbalance. In 10T1/2 cells expressing Jun, at least two of three alleles per cell were induced, a sign of high expression level. The redundant roles of H3S10ph and H3S28ph might enable rapid and efficient IE gene induction.
INTRODUCTION
In eukaryotes, DNA is wrapped around a histone octamer composed of two H2A/H2B dimers, two H3, and two H4 to form a nucleosome, the first level of chromatin organization. The nucleosome is not only a packaging unit; it is also a signaling module that enables a quick transcriptional response to external and internal stimuli. Each of the core histones is subject to an array of dynamic posttranslational modifications (PTMs; Huang et al., 2014), including acetylation or methylation of lysine and phosphorylation of serine. PTMs or marks are recognized by reader proteins via specific domains. In turn, readers recruit chromatin modifiers and remodelers, altering nucleosome structure and function (Bannister and Kouzarides, 2011; Turner, 2014).
On mitogenic or stress stimuli, the mitogen- and stress-activated protein kinases (MSKs) 1 and 2 (MSK1 and MSK2), activated by the ERK or p38 mitogen-activated protein kinase (MAPK) pathways, mediate the rapid and transient H3 phosphorylation at serine 10 (H3S10ph) and serine 28 (H3S28ph) in the regulatory regions of responsive genes (Soloaga et al., 2003). The 14-3-3 proteins bind to H3S10ph and H3S28ph (Macdonald et al., 2005; Winter et al., 2008b) and recruit BRG1, the ATPase subunit of the SWI/SNF remodeler (Drobic et al., 2010). After remodeling, transcription factors such as JUN are able to bind to regulatory regions of immediate-early (IE) genes, ensuring the onset of transcription (Drobic et al., 2010). Chromatin immunoprecipitation (ChIP) assays provide direct evidence that both H3S10ph and H3S28ph are associated with transcriptional induction (Cheung et al., 2000; Clayton et al., 2000; Thomson et al., 2001; Drobic et al., 2010; Josefowicz et al., 2016).
However, phosphorylation events at S10 and S28 are independent and act separately to promote gene expression. First, sequential immunoprecipitation studies and high-resolution fluorescence microscopy analyses show that H3S10ph and H3S28ph do not coexist on the same histone tail or adjacent nucleosomes (Dunn and Davie, 2005; Dyson et al., 2005). Concordantly, sequential ChIP assays have demonstrated that IE gene regulatory regions are associated with either H3S10ph or H3S28ph (Drobic et al., 2010). It has also been shown that H3S28ph, but not H3S10ph, is associated with the activation of Polycomb-silenced genes (Gehani et al., 2010; Lau and Cheung, 2011). Second, although both H3S10ph and H3S28ph are dynamically acetylated, H3S28ph has a higher steady-state of acetylation than H3S10ph (Dunn and Davie, 2005). Moreover, cell treatment with a histone deacetylase (HDAC) inhibitor elevates the level of phosphorylation on S28 but not S10 and also results in a higher steady state of acetylation for H3S28ph than for H3S10ph (Soloaga et al., 2003; Dunn and Davie, 2005; Dyson et al., 2005). This increase in S28 phosphorylation associated with HDAC inhibition suggests a dependence of S28 phosphorylation on acetylation. The cross-talk between acetylation and S10 or S28 phosphorylation marks has not been elucidated in the context of IE gene induction. Regarding the cotargeting of S10 phosphorylation and K9/K14 acetylation, one proposed model suggests the coupled and synergistic recruitment of kinases and K-acetyltransferases (KATs) to an H3 tail (Cheung et al., 2000), while another theory suggests that acetylation and phosphorylation of the H3 tail are independently regulated dynamic events (Thomson et al., 2001). According to more recent results of MSK1/2 inhibition studies, it is likely that H3K9 acetylation at the regulatory regions of IE genes is partly dependent on MSK1 activity (Drobic et al., 2010). In the case of activation of Polycomb-silenced genes, S28ph has been found associated with either K27me3 or K27ac resulting from a methyl-acetylation switch on K27.
To study the nucleosomal response at a single-cell level, we used high-resolution fluorescence microscopy combined with fluorescence in situ hybridization (FISH). We addressed the question of whether the distinct alleles associated with H3S10ph or H3S28ph, which we called epialleles, could coexist in a single cell or if only one type of epiallele could occur in a cell. We also asked whether both epialleles were transcribed. Finally, we performed coimmunoprecipitation (coIP) and HDAC/KAT inhibition studies to discover the mechanism targeting MSK1/2 to phosphorylate H3 at either S10 or S28.
RESULTS
Mitogen-induced H3S10ph and H3S28ph exist in separate pools in human primary fibroblasts
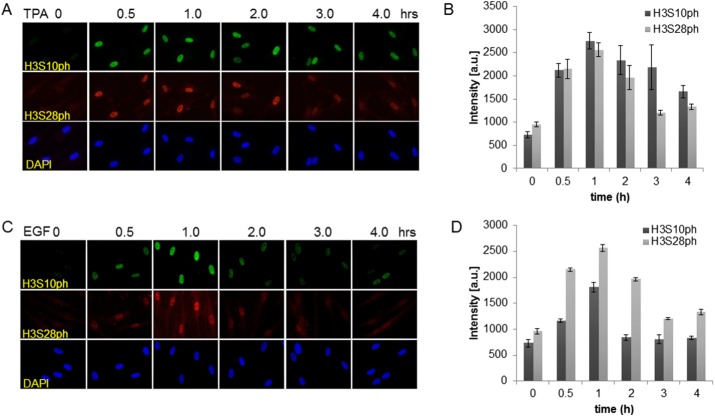
In mouse 10T1/2 fibroblast cells, mitogen-induced S10 and S28 phosphorylation events happen on distinct populations of H3 (Dunn and Davie, 2005; Dyson et al., 2005). To verify that this occurrence is physiologically relevant, we analyzed the distribution of these two modifications in human primary fibroblast CCD-1070Sk cells. First, we analyzed the temporal induction pattern of H3S10 and H3S28 phosphorylation upon treatment of serum-starved cells with 100 nM 12-O-tetradecanoyl-phorbol-13-acetate (TPA) or 30 ng/ml epidermal growth factor (EGF). This was done by fluorescence microscopy after indirect immunofluorescence labeling of cells grown and fixed on coverslips. Figure 1 shows that phosphorylation of both S10 and S28 was induced in response to mitogenic stimulation of CCD-1070Sk cells. It is also noticeable that the TPA-induced phosphorylation of S10 and S28 was sustained for a longer period of time than the EGF-induced phosphorylation.
FIGURE 1:
Mitogen-induced H3 phosphorylation in human primary fibroblasts. Serum-starved CCD-1070Sk cells exposed to (A) 100 nM TPA or (C) 30 ng/ml EGF for the indicated times were fixed and subjected to immunofluorescence labeling with anti-H3S10ph and anti-H3S28ph antibodies. Cells were counterstained with DAPI. The distribution of H3S10ph and H3S28ph was visualized by fluorescence microscopy. (B, D) Overall signal intensities within the nucleus were determined using FIJI. The data were then exported as Excel files for statistical analysis and graphical representation.
We quantified the overall changes of this stimulation by measuring the overall intensities within the nucleus using the open-source platform FIJI (Schindelin et al., 2012). Phosphorylation of H3S10ph and H3S28ph peaked 1 h after stimulation with both EGF and TPA. Stimulation with EGF resulted in a 2.5-fold increase of H3S10 phosphorylation and a 2.7-fold increase of H3S28ph phosphorylation. However, we observed a stronger response of TPA stimulation of serum-starved CCD-1070Sk cells, with a 3.8-fold increase in H3S10ph modification. Of interest, the TPA-induced phosphorylation of H3S28ph did not differ from that of EGF stimulated cells (2.7-fold increase).
Besides the changes in the phosphorylation of H3S10 and H3S28, we noticed an effect on the nuclear area, depending on the stimulant used (Supplemental Figure S1). Whereas the EGF-stimulation of serum-starved CCD-1070Sk cells did not result in a change of the nuclear area, TPA stimulation led to reduction by >20%.
Next we visualized the relative spatial nuclear distributions of H3S10ph and H3S28ph after 60 min of TPA stimulation of serum-starved CCD-1070Sk cells by fluorescence microscopy and image deconvolution. The merged images show that H3S10ph and H3S28ph had distinct subnuclear sites (Figure 2). The measurement of the H3S10ph and H3S28ph in a three-dimensional space showed that only a small percentile of signals was colocalizing. Of 267 H3S10ph and 426 H3S28ph signals, only 31 signals showed a perfect match, and 83 were in close vicinity. Similarly, H3S10ph and H3S28ph were found in separate pools when serum-starved CCD-1070Sk cells were stimulated with EGF (Supplemental Figure S2). Thus, in human primary fibroblasts, as in mouse established 10T1/2 cells, mitogen-induced S10 and S28 phosphorylation occurs on nucleosomes physically separated into distinct foci in the interphase nuclei.
FIGURE 2:
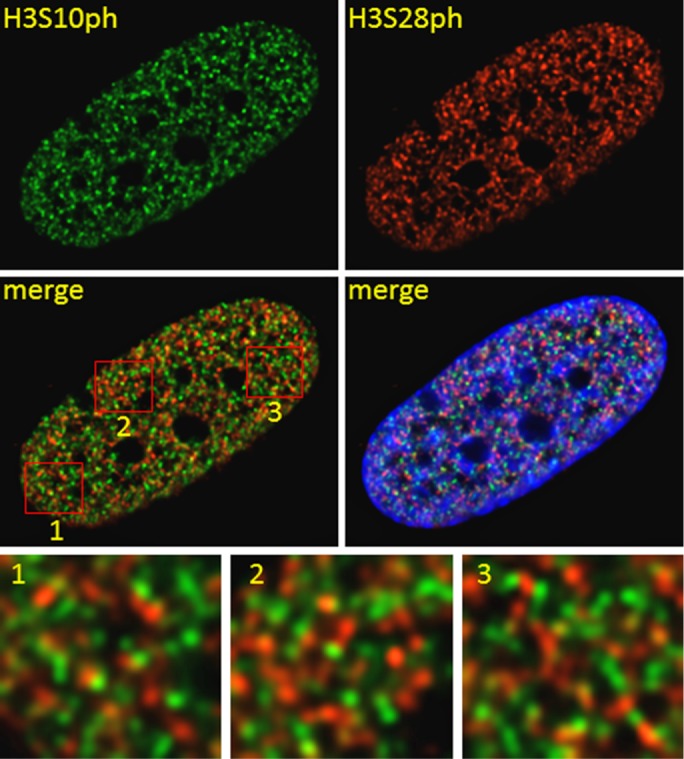
Relative intranuclear localization of H3S10ph and H3S28ph in TPA-treated human primary fibroblasts. Serum-starved CCD-1070Sk cells treated with 100 nM TPA for 60 min were subjected to immunofluorescence staining using anti-H3S10ph and anti-H3S28ph antibodies. Cells were digitally imaged. Boxed areas of merged image are shown enlarged in insets.
Transcribed H3S10ph and H3S28ph epialleles coexist in a single TPA-treated fibroblast cell
The observed absence of H3S10ph and H3S28ph colocalization in 10T1/2 and CCD-1070Sk nuclei can be explained in two different ways. MSK1/2 would phosphorylate H3S10 on one allele and H3S28 on another allele within a cell, or MSK1/2 would phosphorylate exclusively H3S10 or H3S28 on both alleles in a single cell. To distinguish between the two scenarios, we used DNA immuno-FISH. We first explored the nucleosomal response in serum-starved mouse 10T1/2 cells exposed to a 30 min of TPA stimulation. We used antibodies against H3S10ph and H3S28ph and a probe for detection of the Jun alleles. Figure 3 and Supplemental Figures S3 and S4 present representative images, showing the cooccurrence of distinct epialleles in a single 10T1/2 cell. An average of three Jun alleles per cell was detected, in agreement with the known aneuploidy makeup of the 10T1/2 cell line (Reznikoff et al., 1973; Dunn et al., 2009). To determine whether Jun colocalizes with H3S10ph and/or H3S28ph, we examined 30 nuclei using the analysis software TeloView (Vermolen et al., 2005). Of the H3S10ph/28ph signals colocalizing with Jun-signals, the majority was with H3S10ph (42%), whereas only a few Jun-signals colocalized with H3S28 (3%). A minority of H3S10ph and H3S28ph colocalized with Jun (2%). Thirteen percent of the cells showed colocalization of Jun signals with either H3S10ph or H3S28ph within the same nucleus, and 60% of cells showed Jun signals colocalizing with H3S10ph. However, ∼27% of the examined nuclei did not show any colocalization of Jun with H3S10ph or H3S28ph. To conclude, the TPA-induced nucleosome response generated heterozygous Jun epialleles in 10T1/2 cells.
FIGURE 3:
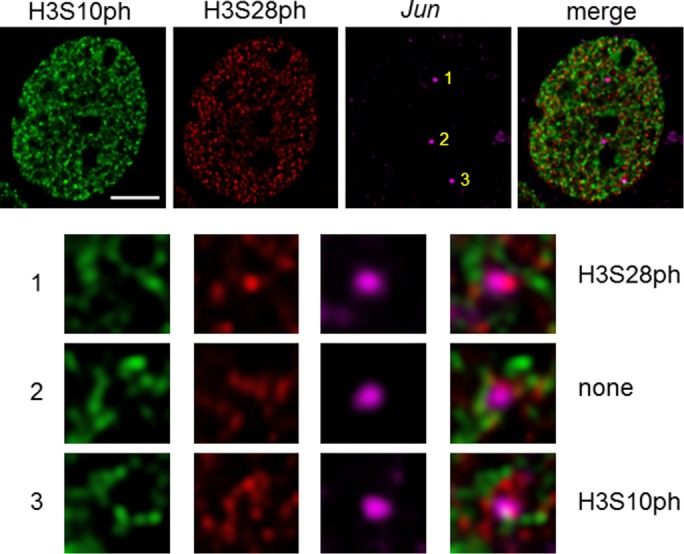
H3S10ph and H3S28ph epialleles coexist in a single mouse fibroblast cell downstream of MAPK signaling. DNA immuno-FISH to detect Jun loci with H3S10ph/H3S28ph staining in 10T1/2 cells, which were serum starved and TPA treated for 30 min. Bar, 5 µm. Enlargements of each area with a Jun signal are shown.
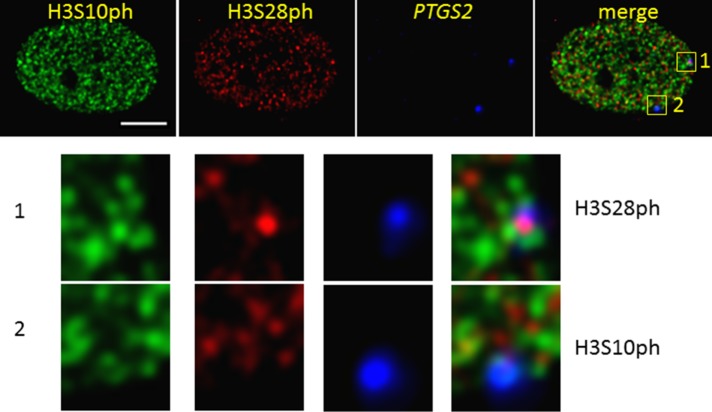
We then visualized the nucleosomal response for the prostaglandin-endoperoxide synthase 2 (PTGS2) gene upon TPA induction of the CCD-1070Sk primary fibroblasts (Figure 4 and Supplemental Figures S5 and S6). The cell shown in Figure 4 had one PTGS2 allele with H3S10ph and one with H3S28ph. Other epiallele combinations (i.e., H3S10ph/H3S10ph or H3S10ph/none) were observed in other cells (Supplemental Figures S5 and S6).
FIGURE 4:
H3S10ph and H3S28ph epialleles coexist in a single human primary fibroblast cell downstream of MAPK signaling. DNA immuno-FISH to detect PTGS2 loci with H3S10ph/H3S28ph staining in a CCD-1070Sk cell, which was serum-starved and TPA-treated for 60 min. Bar, 5 µm. Enlargements of each area with a PTGS2 signal are shown.
To determine whether PTGS2 colocalizes with H3S10ph and/or H3S28ph, we examined 30 nuclei using TeloView. Similar to the colocalization study involving Jun, H3S10ph, and H3S28ph, we measured colocalization of PTGS2 with predominately H3S10ph (27%). Seven percent of the PTGS2 signals colocalized with H3S28ph, and 8% of the FISH signals showed simultaneous colocalization with the immunostained H3S10ph and H3S28ph. Analyzing the distribution of colocalization on nuclear level, we found that ∼47% of all examined nuclei showed PTGS2/H3S10ph colocalization. Ten percent of the nuclei showed colocalization of PTGS2/H3S28ph. Approximately 30% of the nuclei showed simultaneous colocalization of H3S10ph with PTGS2 or H3S28ph with PTGS2, and 13% of the examined nuclei did not show any colocalization of PTGS2 and H3S10ph/28ph.
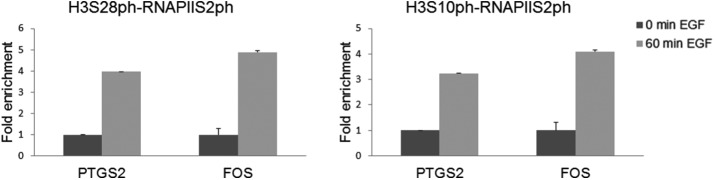
To address the question of whether H3S10ph and H3S28ph epialleles were both transcribed, we performed sequential ChIP assays to determine whether each modification would occur simultaneously with the occupancy at the 5′ end of the gene by the elongation-competent RNA polymerase II, which has its CTD phosphorylated at serine 2 (RNAPIIS2ph). To be able to coimmunoprecipitate the elongating RNAPII with the promoter upstream regions marked by H3S10ph or H3S28ph, we sonicated the cells to a lesser extent than in standard ChIP assays, generating an average fragment size of 800 base pairs. Figure 5 shows that RNAPIIS2ph was associated with H3S10ph or H3S28ph at the 5′ end of the FOS and PTGS2 genes upon EGF treatment of the CCD-1070Sk cells.
FIGURE 5:
IE genes are actively transcribed from H3S10ph or H3S28ph epialleles in human primary fibroblasts. Re-ChIP experiments were performed on formaldehyde–cross-linked chromatin fragments prepared from serum-starved CCD-1070Sk cells treated with 30 ng/ml EGF. Equal amounts of input and immunoprecipitated DNA were quantified by real-time quantitative PCR. The enrichment values of the 5′ end of the FOS and PTGS2 genes are the mean of three independent experiments, and the error bars represent the SD.
In conclusion, mitogen-induced H3S10ph and H3S28ph epialleles were both actively transcribed in primary fibroblast cells.
Phosphorylation of S28 cooccurs with K27 acetylation on H3, whereas phosphorylation of S10 cooccurs with K9 acetylation
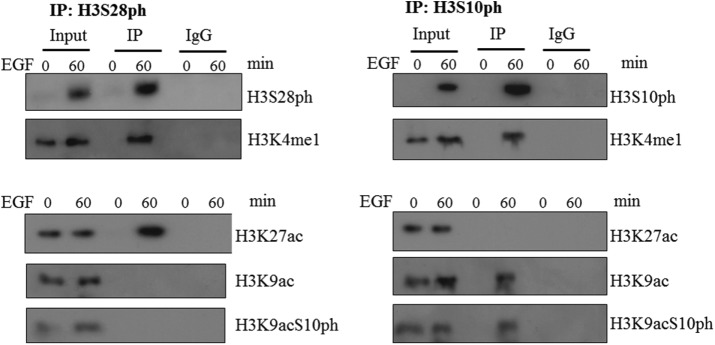
It was previously shown that CREB-binding protein (CBP)/p300 (also known as KAT3A/3B) and GCN5/p300/CBP-associated factor (PCAF; also known as KAT2A/2B) were associated with MSK1 (Janknecht, 2003; Drobic et al., 2010; Sundar et al., 2012). Moreover, MSK1 and PCAF were corecruited to the regulatory regions of IE genes upon their induction by TPA (Drobic et al., 2010). PCAF mediates H3K9 acetylation, whereas CBP/p300 acetylate H3K27 (Jin et al., 2011). H3 acetylated at lysine 27 (H3K27ac) is the signature of active enhancers and promoters (Heintzman et al., 2009; Creyghton et al., 2010; Rada-Iglesias et al., 2011), whereas H3K4me1 is implemented by MLL3/4 at poised or active enhancers (Heintzman et al., 2007; Herz et al., 2012; Hu et al., 2013). To explore the cross-talk between these marks deposited at regulatory regions by chromatin-modifier complexes, we performed coIP assays on SDS-treated nuclear lysates from serum-starved CCD-1070Sk cells treated with EGF. Figure 6 shows that K27ac coexisted with S28ph on H3 but not with S10ph, whereas K9ac was found with S10ph but not with S28ph. K4me1 was found on the tail of both H3S10ph and H3S28ph. The cooccurrence on H3 of S28ph and K27ac, but not S10ph and K27ac, was confirmed in nuclear extracts from serum-starved 10T1/2 cells stimulated with TPA (Supplemental Figure S7).
FIGURE 6:
Cooccurrence of S28 phosphorylation with K27 acetylation or S10 phosphorylation with K9 acetylation on H3, but not vice versa. Nuclear extracts from serum-starved CCD-1070Sk cells stimulated with 30 ng/ml EGF were incubated with anti-H3S28ph, anti-H3S10ph, or control nonspecific IgG antibodies. Input and immunoprecipitated fractions (IP and IgG) were resolved by SDS–15%-PAGE and immunoblotted with the indicated antibodies.
Mitogen-induced H3S28 phosphorylation depends on K27 acetylation
Analysis of DNA immuno-FISH images (as shown in Figure 3) from serum-starved and TPA-induced 10T1/2 cells revealed a preferential phosphorylation of H3S10 over H3S28 at Jun alleles. To determine whether histone hyperacetylation could shift this distribution, we incubated serum-starved 10T1/2 cells with the HDAC inhibitor trichostatin A (TSA) for 30 min before TPA stimulation. Histone was acid-extracted and analyzed by immunoblotting (Figure 7A). After HDAC inhibition, a clear increase in H3S28ph, but not H3S10ph, was observed simultaneously with H3K9 and H3K27 hyperacetylation. Moreover, even when acetylation levels were increased, S10ph and S28ph did not cooccur on H3 tails, as H3S28ph did not coimmunoprecipitate with H3S10ph (Supplemental Figure S8).
FIGURE 7:
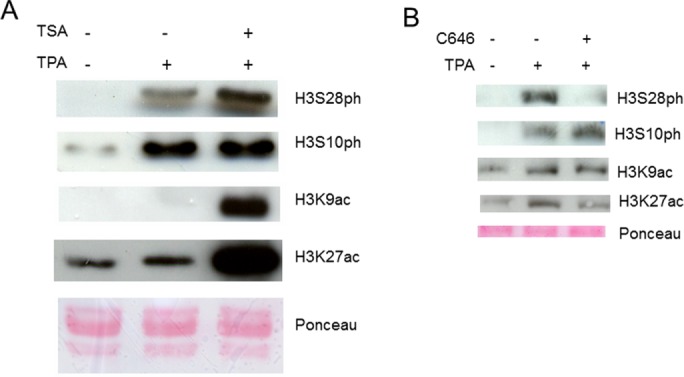
TPA-induced H3S28 phosphorylation depends on K27 acetylation. Histones acid extracted from serum-starved, TPA-stimulated 10T1/2 cells were resolved by 15% SDS–PAGE and immunoblotted with the indicated antibodies. Before blotting, membranes were stained with Ponceau-S. Before 30-min TPA stimulation, cells were treated or not with (A) TSA or (B) C646.
Results in Figure 7A suggested that H3K27ac might direct S28 phosphorylation by MSK1/2. To determine whether this was the case, we treated serum-starved 10T1/2 cells with C646 before TPA stimulation. C646 is a potent and selective inhibitor of p300/CBP (Bowers et al., 2010; Josefowicz et al., 2016), the KATs acetylating K27 (Jin et al., 2011). Figure 7B shows that K27ac level, and, even more, so S28ph level were decreased when cells were treated with C646, whereas S10ph and K9ac levels were not affected. Thus, K27ac directs MSK1/2 to phosphorylate H3 on S28.
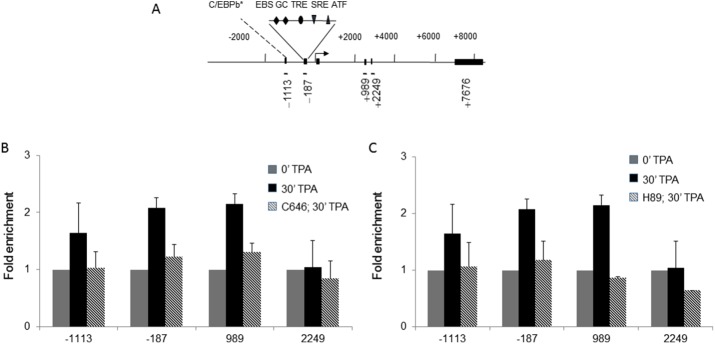
We next determined whether H3K27ac was increased at the regulatory region of the IE Fosl1 gene when serum-starved 10T1/2 cells were stimulated with TPA for 30 min. ChIP assays showed that H3K27ac levels were increased at the upstream promoter (−1113, −187) and internal enhancer (+989) regions but not further downstream in the coding region (+2249), paralleling the increase in H3S10ph or H3S28ph (Figure 8; Drobic et al., 2010). Inhibition of p300/CBP with C646 before addition of TPA to the 10T1/2 serum-starved cells prevented the increase in H3K27ac at the regulatory regions of the Fosl1 gene (Figure 8B).
FIGURE 8:
TPA-induced HK27 acetylation at the Fosl1 regulatory regions. (A) Schematic representation of the Fosl1 gene showing regions amplified in the ChIP assays. Each region is labeled according to the 5′ position of the forward primer relative to the transcription start site. Exons are represented by boxes, and binding sites of relevant transcription factors located in the amplified regions are displayed. C/EBP, CCAAT-enhancer binding protein; EBS, Ets binding site; GC, GC box, which is a binding site for the Sp family transcription factors; SRE, serum-responsive element; TRE, TPA-responsive element. AP-1 constitutes a combination of dimers formed of members of the JUN, FOS, and ATF families of transcription factors. Asterisk indicates a putative binding site. (B, C) ChIP experiments were performed using antibodies against H3K27ac on formaldehyde–cross-linked mononucleosomes prepared from serum-starved 10T1/2 cells treated or not with (B) C646 or (C) H89 before induction with TPA for 0 or 30 min. Equal amounts of input and immunoprecipitated DNA were quantified by real-time quantitative PCR. Enrichment values are the mean of three independent experiments, and the error bars represent the SD.
We showed previously that inhibition of MSK1/2 activity with H89 prevented the recruitment of the MSK1/2 complex to the regulatory regions of IE genes (Drobic et al., 2010). Because MSK, which is associated with CBP/p300, stimulates the transactivation activity of CBP (Janknecht, 2003), we examined whether MSK1/2 inhibition prevented the TPA-responsive increases in H3K27ac at the regulatory regions of Fosl1. Figure 8C shows that MSK1/2 inhibition prevented the TPA-induced increase in H3K27ac. Together, these results provide evidence that MSK-CBP/p300 acetylates H3K27ac and H3S28ph on the H3 tail of nucleosomes located in the regulatory regions of TPA-induced IE genes.
DISCUSSION
Analysis of the nucleosomal response at a single-cell level showed that mitogen induction of IE genes happened randomly on H3S10ph and/or H3S28ph epialleles in individual fibroblast cells from immortalized or finite cell lines. Our results showed that CBP/p300-mediated K27 acetylation was a signal for MSK1/2 to phosphorylate S28. On the other hand, H3 acetylated on K9 by PCAF would be phosphorylated on S10 because K9ac and S28ph did not coexist on a H3 tail. However, S10 phosphorylation did not appear to depend on K9 acetylation because the increase in K9ac after HDAC inhibition was not paralleled by an increase in S10ph. In fact, a previous study using the MSK1/2 inhibitor H89 showed that H3K9 acetylation at the regulatory regions of mitogen-induced IE genes in part resulted from MSK1/2-mediated phosphorylation (Drobic et al., 2010). Note that the antibody used in that study was specific for H3 acetylated at lysine 9 (H3K9ac), although it was raised against H3K9acK14ac (Edmondson et al., 2002).
A recent study on the stimulation of mouse macrophages with bacterial lipopolysaccharide demonstrated a role of MSK-mediated H3S28ph and CBP-stimulated transactivation in the induction of inflammatory genes (Josefowicz et al., 2016). However, in contrast to our study, that work found that inhibition of CBP/p300 with C646 did not prevent the induced phosphorylation of H3S28.
MSK1/2 are in multiprotein complexes containing 14-3-3 proteins, KATs, BRG1, and transcription factors such as p65 of NF-κB and JUN (Janknecht, 2003; Drobic et al., 2010; Sundar et al., 2012). Because H3K9acS10ph and H3K27acS28ph are typically mutually exclusive on a given allele, it is likely that MSK1/2 complexes with PCAF or CBP/p300 are recruited separately to different alleles. Specific transcription factors (e.g., NF-κB, ELK-1, and AP-1) recruit MSK1/2 complexes to the regulatory regions of IE genes (Espino et al., 2006; Joo and Jetten, 2008; Zhang et al., 2008). Once recruited, depending on the acetylation state of K27, MSK phosphorylates H3 at S10 or S28 of nucleosomes located at these regulatory regions. The fact that both S28ph and S10ph cooccur with K4me1 suggests that the nucleosomal response takes place at enhancers, as well as at upstream promoter regions. Indeed, TPA-induced, MSK1/2-mediated phosphorylation of H3S10 was previously evidenced at the trefoil factor 1 enhancer in MCF7 cells (Khan et al., 2013).
According to our proposed model, scaffolding phosphobinding proteins 14-3-3 protect the phosphorylated H3 from protein phosphatases and stabilize the interaction of the remodeling SWI/SNF complex with H3S10ph or S28ph, resulting in IE gene transcription (Drobic et al., 2010). The homodimer or heterodimer of 14-3-3ε and/or ξ binds to H3S10ph or H3S28ph, with the affinity of binding being S28ph > S10phK14Ac or S10phK9ac > S10ph (Macdonald et al., 2005; Winter et al., 2008a; Sharma et al., 2013). However, 14-3-3ε and 14-3-3ζ are recruited to the enhancer and upstream promoter regions of IE genes in different mammalian cell types with a similar time course as the induction of H3S10ph or H3S28ph (Drobic et al., 2010; Khan et al., 2013). An obvious question is why there are two sets of PTMs acting separately to achieve the mitogen or stress induction of IE genes.
The random targeting of H3S10 or H3S28 on distinct alleles is reminiscent of random and dynamic monoallelic expression, that is, unsynchronized transcriptional bursting of the two alleles. A recent RNA-sequencing study on individual cells of mouse preimplantation embryos discovered that 12–24% of autosomal genes were expressed from a single allele at any given time (Deng et al., 2014). Over time, independent transcription occurs in bursts from both alleles, pointing to a stochastic pattern of monoallelic expression quite different from stable allelic regulation processes like genomic imprinting, in which different alleles have active or inactive marks (Eckersley-Maslin and Spector, 2014; Reinius and Sandberg, 2015). This random monoallelic expression was also observed in human primary fibroblast single cells for the majority of genes (Borel et al., 2015). Moreover, the authors observed that rare highly expressed genes were transcribed from both alleles at any given time in the majority of single cells, whereas less expressed genes had a monoallelic expression (Borel et al., 2015). In our analyses of the mouse fibroblast cells showing a Jun epiallele associated with H3S10ph or H3S28ph, 70% of the cells had two or more Jun epialleles associated with an H3 phosphorylation mark, which is consistent with a high level of expression.
To conclude, it is likely that the distribution of H3K9acS10ph and H3K27acS28ph epialleles in a single cell results from the stochastic acetylation of H3 by CBP/p300 or PCAF. Moreover, it is conceivable that the independent implementation of these two sets of marks having duplicate roles would facilitate a rapid and sharp increase in IE gene induction in response to mitogen or stress.
MATERIALS AND METHODS
Cell culture
Mouse fibroblast 10T1/2 cells (American Type Culture Collection [ATCC] CCL-226) and human primary fibroblast CCD-1070Sk cells (ATCC CRL-2091) were grown at 37°C in a humidified atmosphere containing 5% CO2 in α-MEM and MEM (Life Technologies, Grand Island, NY), respectively, supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 1% antibiotic-antimycotic (Life Technologies). Subconfluent cells (80–90%) were serum starved for 48 h in their respective media supplemented with 0.1% FBS. Flow cytometry analysis of cells stained with ethidium bromide showed 89% of the serum-starved cells were in the G1 phase of the cell cycle. To induce MAPK signaling, serum-starved cells were treated with 30 ng/ml EGF (Invitrogen, Carlsbad, CA) or 100 nM TPA (Sigma-Aldrich, St. Louis, MO) for the indicated time periods. When required, 10T1/2 cells were pretreated with 250 nM TSA (Sigma-Aldrich), 10 µM C646 (EMD Millipore, Billerica, MA), or 10 µM H89 (Calbiochem) for 30 min before TPA treatment.
Indirect immunolocalization
Indirect immunolocalization was performed as described previously (He and Davie, 2006), using rabbit polyclonal antibodies against H3S10ph (1:250 dilution; Santa Cruz Biotechnology, Dallas, TX) and rat monoclonal antibodies against H3S28ph (1:250 dilution; Sigma-Aldrich). Peptide Competition assay was done by Standard Novus protocol (www.novusbio.com/support/support-by-application/peptide-competition/protocol.html) using H3 peptides from Abcam (human histone H3 (phospho S10) peptide [ab11477]; human histone H3 (phospho S28) peptide [ab14793; Supplemental Figure S9). Alexa Fluor 488 donkey anti-rabbit immunoglobulin G (IgG; Molecular Probes, Eugene, OR) and Texas red– or AMCA-conjugated donkey anti-rat IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) were used as secondary antibodies. DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Control experiments including epitope peptide blocking or primary antibody omission demonstrated the specificity of the antibodies used.
Fluorescence microscopy
Digital images were captured and quantitative image analysis was performed as described previously (He et al., 2013).
Quantification of fluorescence signals
For the analysis of the overall signal intensities within the nucleus, we used FIJI, an open source platform (Schindelin et al., 2012). For the filter, we used Gaussian blur at 10 for the counter stain (DAPI) throughout the experiments. The thresholds were adjusted to match the counterstained area (DAPI). The overall intensities, as well as the area, were measured for each channel. The data were then exported as Excel files for statistical analysis.
DNA immuno-FISH
DNA FISH assays were performed as described (Lomvardas et al., 2006). BAC clones RP24-282B18 and RP11-1122M4, carrying the mouse Jun and human PTGS2 loci, respectively (BACPAC Resources, CHORI, Oakland, CA), were labeled with Digoxigenin (DIG) by nick translation (Roche Diagnostics, Indianapolis, IN) to generate probes for FISH. 10T1/2 cells were grown on coverslips, serum starved for 24 h, and treated with 100 nM TPA for 30 min; CCD-1070Sk cells were serum starved for 48 h and treated with 100 nM TPA for 60 min. After indirect immunolocalization of H3S10ph and H3S28ph, the antigen–antibody complexes were cross-linked with 1 mM dithiobis(succinimidyl propionate) (Thermo Fisher Scientific, Waltham, MA) for 30 min at room temperature. The genomic DNA was denatured using 0.1 N NaOH for 2 min and hybridized to the denatured probes (72°C for 10 min) overnight at 37°C. After extensive washing steps, the probe was detected using primary anti-DIG antibody (Roche Diagnostics) and DyLight 594-conjugated secondary antibody (Jackson ImmunoResearch Laboratories).
Measurement of colocalizing signals
For the colocalization studies, we used TeloView (Vermolen et al., 2005). This program allows the three-dimensional measurement of the signal coordinates (x, y, z) from their center of gravity. The measurements were performed for H3S10ph and H3S28ph, as well as for H3S10ph, H3S28ph, and the FISH-signals Jun and PTGS2. Thirty nuclei were analyzed for each immuno-FISH experiment. Matching coordinates were classified as colocalizing signals.
ChIP and sequential ChIP assays
High-resolution ChIP using micrococcal nuclease digestion and sequential ChIP assays were performed as previously described (Drobic et al., 2010), except that cross-linked chromatin was sheared to ∼800–base pair fragments by sonication. Antibodies against H3S10ph (Santa Cruz Biotechnology), H3S28ph (Sigma-Aldrich), RNAPIIS2ph (Abcam, Cambridge, MA), or an isotype-matched nonrelated IgG as negative control (EMD Millipore) were used. Primers used to amplify UPRs were as follows: FOS forward, 5′-CCCCTTACACAGGATGTCCA-3′, and reverse, 5′-CCCCCAAGATGAGGGGTTTC-3′; and PTGS2 forward, 5′-AGGAGAGGGAGGGATCAGAC-3′, and reverse, 5′-AAAATCGGAAACCCAGGAAG-3′. Mouse Fosl1 primers are described in Drobic et al. (2010). Enrichment values are the mean of three independent experiments. Error bars indicate SD.
Histone extraction and immunoblot analysis
Histones were acid extracted from 10T1/2 and CCD-1070Sk cells, resolved by 15% SDS–PAGE, and stained immunochemically with anti-H3 (Abcam), anti-H3S10ph (Santa Cruz Biotechnology,), anti-H3S28ph (Sigma-Aldrich), anti-H3K27ac (Millipore), anti-H3K9ac (Abcam), anti-H3K14ac (Abcam), or H3K4me1 (Abcam) antibodies (Chadee et al., 1999). The specificity of the antibodies was verified by immunoblot using 2–3 µg of synthetic peptides derived from human histone H3.1 and carrying the following PTMs: H3S10ph (Abcam), H3S28ph (Abcam), H3K27ac (Abcam), H3K9ac (Abcam), H3K9ac/S10ph (Abcam), or H3K27ac/S28ph (EpiCypher, Research Triangle Park, NC). Three biological repeats were performed.
Histone coimmunoprecipitation
Cells were resuspended in cell lysis buffer (5 mM 1,4-piperazinediethanesulfonic acid, pH 8.0, 85 mM KCl, 0.5% NP-40) containing phosphatase and protease inhibitor cocktails (PhosSTOP and cOmplete, Mini, EDTA-free; Roche Diagnostics), incubated with shaking at 4°C for 10 min, and centrifuged at 2000 × g for 10 min. The nuclear pellet was resuspended in 1–2 ml of micrococcal nuclease (MNase) digestion buffer (10 mM Tris-HCl, pH 7.5, 0.25 M sucrose, 75 mM NaCl) supplemented with the phosphatase/protease inhibitor cocktails, and A260 was measured. To fragment chromatin, 2.5 U of MNase (Worthington Biochemical Corporation, Lakewood, NJ) per A260 of nuclear suspension was added in the presence of 3 mM CaCl2 and incubated at 37°C for 25 min. The reaction was stopped with the addition of EDTA, pH 8.0, to a final concentration of 5 mM. The nuclei were lysed by adding SDS to a final concentration of 0.5% and rotating at room temperature for 60 min, resulting in the dissociation of the histone from DNA. Insoluble material was removed by centrifugation at 2000 × g for 10 min, and the soluble material was diluted to 0.1% SDS with RIPA buffer (10 mM Tris-HCl, pH 8.0, 1% Triton-X-100, 0.1% SDS, 0.1% sodium deoxycholate) containing the phosphatase/protease inhibitors. The extract was precleared with 50% slurry of protein A/G Plus agarose (Santa Cruz Biotechnology). The beads were removed by centrifugation at 2000 × g for 10 min, and the A260 was measured. A 1-µg amount of antibody per A260 of precleared extract was added and incubated overnight at 4°C. Anti-H3S10ph (Abcam or Santa Cruz Biotechnology), anti-H3S28ph (Abcam), or isotype-specific nonrelated IgG as negative control (EMD Millipore) were used. From 40 to 80 µl of Dynabeads Protein G (Invitrogen), resuspended in RIPA buffer, was added and incubated for 3 h at 4°C. The beads were washed five times with RIPA buffer, resuspended in 2× loading buffer (4% SDS, 20% glycerol, 0.12 M Tris, pH 6.8, 10% β-mercaptoethanol, 0.2% bromophenol blue), and boiled at 95°C for 5 min. Samples were resolved by 15% SDS–PAGE and subjected to immunoblot analysis. Two (CCD-1070Sk cells) or three (10T1/2 cells) biological repeats were performed.
Supplementary Material
Acknowledgments
This work was supported by funding from Research Manitoba, the Children's Hospital Research Institute of Manitoba, the CancerCare Manitoba Foundation, a Canada Research Chair (to J.R.D.), a Cancer Research Society grant (76-313-6829a) to S.M., a Manitoba Health Research Council/CancerCare Manitoba studentship (to D.H.K.), and a Manitoba Health Research Council postdoctoral fellowship (to S.H.). We acknowledge the excellent support of the Genomic Center for Cancer Research and Diagnosis, which is housed in the Research Institute of Oncology and Hematology, CancerCare Manitoba. We thank 3D Signatures for permission to use the software TeloView for the analysis of the fluorescence signals included in this project.
Abbreviations used:
- CBP
CREB-binding protein
- ChIP
chromatin immunoprecipitation
- EGF
epidermal growth factor
- FISH
fluorescence in situ hybridization
- H3K9ac
H3 acetylated at lysine 9
- H3K27ac
H3 acetylated at lysine 27
- H3S10ph
H3 phosphorylated at serine 10
- H3S28ph
H3 phosphorylated at serine 28
- HDAC
histone deacetylase
- IE
immediate-early
- KAT
K-acetyltransferase
- MSK
mitogen- and stress-activated protein kinase
- PCAF
p300/CBP-associated factor
- PTGS2
prostaglandin-endoperoxide synthase 2
- PTM
posttranslational modification
- RNAPIIS2ph
RNA polymerase II phosphorylated at serine 2
- TPA
12-O-tetradecanoyl-phorbol-13-acetate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-08-0618) on January 11, 2017.
REFERENCES
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel C, Ferreira PG, Santoni F, Delaneau O, Fort A, Popadin KY, Garieri M, Falconnet E, Ribaux P, Guipponi M, et al. Biased allelic expression in human primary fibroblast single cells. Am J Hum Genet. 2015;96:70–80. doi: 10.1016/j.ajhg.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadee DN, Hendzel MJ, Tylipski CP, Allis CD, Bazett-Jones DP, Wright JA, Davie JR. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J Biol Chem. 1999;274:24914–24920. doi: 10.1074/jbc.274.35.24914. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Clayton AL, Rose S, Barratt MJ, Mahadevan LC. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 2000;19:3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- Drobic B, Perez-Cadahia B, Yu J, Kung SK, Davie JR. Promoter chromatin remodeling of immediate-early genes is mediated through H3 phosphorylation at either serine 28 or 10 by the MSK1 multi-protein complex. Nucleic Acids Res. 2010;38:3196–3208. doi: 10.1093/nar/gkq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KL, Davie JR. Stimulation of the Ras-MAPK pathway leads to independent phosphorylation of histone H3 on serine 10 and 28. Oncogene. 2005;24:3492–3502. doi: 10.1038/sj.onc.1208521. [DOI] [PubMed] [Google Scholar]
- Dunn KL, He S, Wark L, Delcuve GP, Sun JM, Yu Chen H, Mai S, Davie JR. Increased genomic instability and altered chromosomal protein phosphorylation timing in HRAS-transformed mouse fibroblasts. Genes Chromosomes Cancer. 2009;48:397–409. doi: 10.1002/gcc.20649. [DOI] [PubMed] [Google Scholar]
- Dyson MH, Thomson S, Inagaki M, Goto H, Arthur SJ, Nightingale K, Iborra FJ, Mahadevan LC. MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J Cell Sci. 2005;118:2247–2259. doi: 10.1242/jcs.02373. [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Spector DL. Random monoallelic expression: regulating gene expression one allele at a time. Trends Genet. 2014;30:237–244. doi: 10.1016/j.tig.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Davie JK, Zhou J, Mirnikjoo B, Tatchell K, Dent SY. Site-specific loss of acetylation upon phosphorylation of histone H3. J Biol Chem. 2002;277:29496–29502. doi: 10.1074/jbc.M200651200. [DOI] [PubMed] [Google Scholar]
- Espino PS, Li L, He S, Yu J, Davie JR. Chromatin modification of the trefoil factor 1 gene in human breast cancer cells by the Ras/mitogen-activated protein kinase pathway. Cancer Res. 2006;66:4610–4616. doi: 10.1158/0008-5472.CAN-05-4251. [DOI] [PubMed] [Google Scholar]
- Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- He S, Davie JR. Sp1 and Sp3 foci distribution throughout mitosis. J Cell Sci. 2006;119:1063–1070. doi: 10.1242/jcs.02829. [DOI] [PubMed] [Google Scholar]
- He S, Khan DH, Winter S, Seiser C, Davie JR. Dynamic distribution of HDAC1 and HDAC2 during mitosis: association with F-actin. J Cell Physiol. 2013;228:1525–1535. doi: 10.1002/jcp.24311. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33:4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458–458. doi: 10.1016/j.cell.2014.09.037. e451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R. Regulation of the ER81 transcription factor and its coactivators by mitogen- and stress-activated protein kinase 1 (MSK1) Oncogene. 2003;22:746–755. doi: 10.1038/sj.onc.1206185. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Jetten AM. NF-kappaB-dependent transcriptional activation in lung carcinoma cells by farnesol involves p65/RelA(Ser276) phosphorylation via the MEK-MSK1 signaling pathway. J Biol Chem. 2008;283:16391–16399. doi: 10.1074/jbc.M800945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Shimada M, Armache A, Li CH, Miller RM, Lin S, Yang A, Dill BD, Molina H, Park HS, et al. Chromatin kinases act on transcription factors and histone tails in regulation of inducible transcription. Mol Cell. 2016;64:347–361. doi: 10.1016/j.molcel.2016.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan P, Drobic B, Perez-Cadahia B, Healy S, He S, Davie JR. Mitogen- and stress-activated protein kinases 1 and 2 are required for maximal trefoil factor 1 induction. PLoS One. 2013;8:e63189. doi: 10.1371/journal.pone.0063189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PN, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci USA. 2011;108:2801–2806. doi: 10.1073/pnas.1012798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, Moggs JG, Orphanides G, Thomson S, Edmunds JW, et al. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius B, Sandberg R. Random monoallelic expression of autosomal genes: stochastic transcription and allele-level regulation. Nat Rev Genet. 2015;16:653–664. doi: 10.1038/nrg3888. [DOI] [PubMed] [Google Scholar]
- Reznikoff CA, Brankow DW, Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973;33:3231–3238. [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Mansukh A, Varma A, Gadewal N, Gupta S. Molecular modeling of differentially phosphorylated serine 10 and acetylated lysine 9/14 of histone H3 regulates their interactions with 14-3-3zeta, MSK1, and MKP1. Bioinform Biol Insights. 2013;7:271–288. doi: 10.4137/BBI.S12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar IK, Chung S, Hwang JW, Lapek JD, Jr, Bulger M, Friedman AE, Yao H, Davie JR, Rahman I. Mitogen- and stress-activated kinase 1 (MSK1) regulates cigarette smoke-induced histone modifications on NF-kappaB-dependent genes. PLoS One. 2012;7:e31378. doi: 10.1371/journal.pone.0031378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Mahadevan LC. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol Cell. 2001;8:1231–1241. doi: 10.1016/s1097-2765(01)00404-x. [DOI] [PubMed] [Google Scholar]
- Turner BM. Nucleosome signalling; an evolving concept. Biochim Biophys Acta. 2014;1839:623–626. doi: 10.1016/j.bbagrm.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Vermolen BJ, Garini Y, Mai S, Mougey V, Fest T, Chuang TC, Chuang AY, Wark L, Young IT. Characterizing the three-dimensional organization of telomeres. Cytometry A. 2005;67:144–150. doi: 10.1002/cyto.a.20159. [DOI] [PubMed] [Google Scholar]
- Winter S, Fischle W, Seiser C. Modulation of 14-3-3 interaction with phosphorylated histone H3 by combinatorial modification patterns. Cell Cycle. 2008a;7:1336–1342. doi: 10.4161/cc.7.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S, Simboeck E, Fischle W, Zupkovitz G, Dohnal I, Mechtler K, Ammerer G, Seiser C. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 2008b;27:88–99. doi: 10.1038/sj.emboj.7601954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Li L, Papadopoulou N, Hodgson G, Evans E, Galbraith M, Dear M, Vougier S, Saxton J, Shaw PE. Mitogen-induced recruitment of ERK and MSK to SRE promoter complexes by ternary complex factor Elk-1. Nucleic Acids Res. 2008;36:2594–2607. doi: 10.1093/nar/gkn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.