Tetrahymena has a crossover pathway that combines features of both canonical pathways found in the majority of eukaryotes. The pathway uses a protein related to Zip3, which is commonly associated with the formation of a synaptonemal complex.
Abstract
When programmed meiotic DNA double-strand breaks (DSBs) undergo recombinational repair, genetic crossovers (COs) may be formed. A certain level of this is required for the faithful segregation of chromosomes, but the majority of DSBs are processed toward a safer alternative, namely noncrossovers (NCOs), via nonreciprocal DNA exchange. At the crossroads between these two DSB fates is the Msh4-Msh5 (MutSγ) complex, which stabilizes CO-destined recombination intermediates and members of the Zip3/RNF212 family of RING finger proteins, which in turn stabilize MutSγ. These proteins function in the context of the synaptonemal complex (SC) and mainly act on SC-dependent COs. Here we show that in the SC-less ciliate Tetrahymena, Zhp3 (a protein distantly related to Zip3/RNF212), together with MutSγ, is responsible for the majority of COs. This activity of Zhp3 suggests an evolutionarily conserved SC-independent strategy for balancing CO:NCO ratios. Moreover, we report a novel meiosis-specific protein, Sa15, as an interacting partner of Zhp3. Sa15 forms linear structures in meiotic prophase nuclei to which Zhp3 localizes. Sa15 is required for a wild-type level of CO formation. Its linear organization suggests the existence of an underlying chromosomal axis that serves as a scaffold for Zhp3 and other recombination proteins.
INTRODUCTION
During meiosis, homologous chromosomes must be paired to ensure their correct alignment and faithful segregation into haploid sets. Homologous chromosomes first recognize each other and align via mechanisms that are not yet fully elucidated. In most eukaryotes, the chromosomes then become connected by the synaptonemal complex (SC). During this process, known as synapsis, crossing over takes place. The chiasmata resulting from crossovers (COs) provide the physical bonds between chromosomes once the SC becomes resolved toward the end of meiotic prophase. Crossing over is initiated by programmed DNA double-strand breaks (DSBs) induced by Spo11 (Lam and Keeney, 2014). The ends of broken DNA are resected to produce single-stranded 3′-overhangs; one of these first invades a homologous double-strand DNA molecule to form a D-loop. This end is then extended by cDNA synthesis. At this point, it can be displaced and fused with the other end, thereby restoring an intact molecule by a noncrossover (NCO) process. In fact, the majority of DSBs are repaired via this synthesis-dependent strand-annealing (SDSA) pathway. In a subset of DSBs, the second end is also captured, resulting in the formation of a four-branched joint molecule (JM) that may be resolved by crossing over (Hunter, 2015). JMs can be converted into COs along two major pathways. Most eukaryotes use the class I or ZMM pathway (named after the Zip1/2/3/4, Msh4/5, and Mer3 proteins) as the major route to COs. Class I COs are formed with the involvement of SC proteins (although they do not require a fully mature tripartite SC; Voelkel-Meiman et al., 2016) and are subject to interference (i.e., mutual suppression). In the class I pathway, JMs are resolved by the Mlh1-Mlh3 endonuclease complex. A small proportion of SC-independent, noninterfering COs are generated by the class II pathway, also called the Mus81 pathway after the corresponding JM resolvase (de los Santos et al., 2003). The fission yeast, which does not form an SC, has only class II COs (Smith et al., 2003).
Meiosis in the ciliate Tetrahymena thermophila is similar to the fission yeast in that an SC is not formed. Instead, the nucleus elongates dramatically (Figures 1 and 2A), and chromosomes adopt an extreme bouquet arrangement in which all telomeres are clustered at one end of the nucleus and all centromeres are clustered at the opposite end. This arrangement juxtaposes homologous chromosome regions and facilitates homologous recombination (Loidl et al., 2012). The early steps of recombination, that is, DSB formation and processing, follow the canonical processes (Loidl and Lorenz, 2016); however, as in fission yeast, COs all depend on Mus81 and its cofactor Mms4/Eme1 (Lukaszewicz et al., 2013). Nevertheless, they are not considered typical class II COs because their formation involves the ZMM proteins Msh4 and Msh5 (Shodhan et al., 2014).
FIGURE 1:
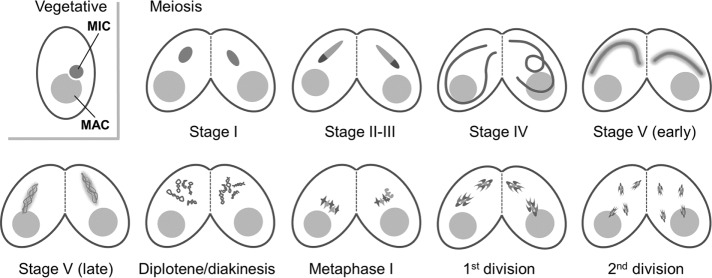
Schematic of meiotic progression in Tetrahymena. Meiosis takes place simultaneously in two conjugating cells. Generative micronuclei (MIC) undergo meiosis, whereas vegetative polyploid macronuclei (MAC) do not play a role in sexual reproduction. During prophase, MICs transiently elongate to about twice the length of the cell. At metaphase I, bivalents condense and assemble into a metaphase plate. Chromosomes segregate at anaphase I, and chromatids segregate at anaphase II. The classification of prophase stages is modified from Sugai and Hiwatashi (1974): stage I, egg-shaped nucleus; stage II-III, elongating nucleus shorter than the cell; stage IV, nucleus approximately twice the length of the cell; early stage V, shortening nucleus approximately the length of the cell and slightly bent; late stage V, nucleus shorter than the cell, straight, with inhomogeneously distributed chromatin.
FIGURE 2:
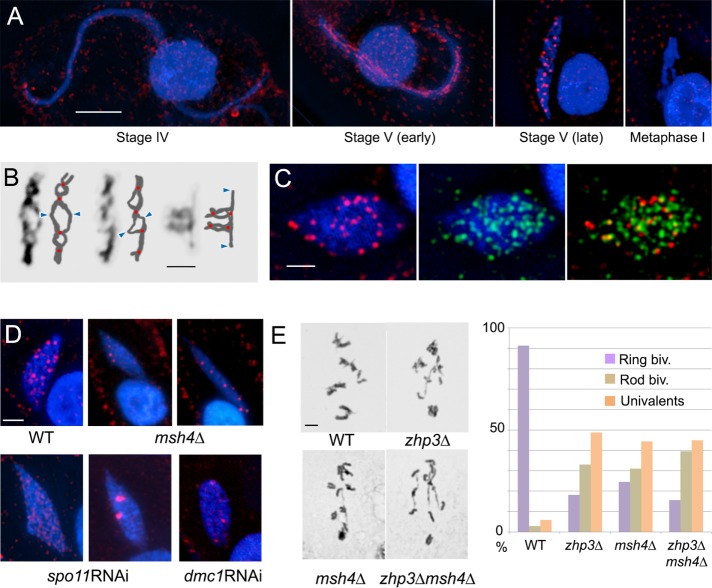
Meiotic phenotype of the ZHP3 deletion mutant. Meiotic progression in (A) wild-type (WT) and (B) zhp3Δ cells. At prophase, nuclear reorganization and chromosome dynamics are indistinguishable in WT and mutant cells. (C) Southern hybridization with a MIC-specific DNA probe to a PFGE gel shows transient DSB-generated DNA fragmentation. DSB formation and repair occur with similar dynamics in WT and zhp3Δ cells. Samples were run 0, 3, 4, 5, 6, and 7 h after induction of meiosis. DSB band intensity (a rough measure of the relative DSB abundance) was determined, normalized to the loading control (a 306-kb macronuclear chromosome), and plotted as the n-fold increase over background intensity. (D) Dmc1 foci in elongated meiotic prophase nuclei are similar in WT and zhp3Δ cells. (E) FISH was used to quantify homologous pairing in elongated meiotic prophase nuclei. Inset, examples of separate (top) and paired (bottom) homologous loci. Red bars indicate mean distances. (F) Schaudinn fixation and Giemsa staining were used to spread chromosomes and analyze bivalents in diakinesis or metaphase I. The WT has mainly ring bivalents, whereas zhp3Δ mostly has rod bivalents or univalents. Bars, 10 µm (A, B, D), 2 µm (F).
Here we assigned a meiotic function to the Tetrahymena protein Zhp3, which is similar to the Zip3/RNF212/HEI10 family of ZMM proteins that is involved in the CO versus NCO decision. Besides Msh4 and Msh5, Zhp3 represents another example of ZMM proteins coopted for use in SC-less meiosis. Moreover, we identified another protein, Sa15, which is an interaction partner of Zhp3 and is required for its proper localization and, possibly, its full activity.
RESULTS
Tetrahymena Zhp3 is a ring finger protein related to the Zip3/RNF212 protein family
We studied the gene TTHERM_00049220, previously known as COI20 (for Conjugation Induced; Woehrer et al., 2015), because it is specifically expressed in cells at the early conjugation stage, that is, when meiosis takes place (TetraFGD; http://tfgd.ihb.ac.cn; Miao et al., 2009; Xiong et al., 2012). The predicted protein, TTHERM 00049220p, consists of 415 amino acids and has a molecular weight of 49.6 kDa. Orthologous proteins can be identified in Tetrahymena ellioti, Tetrahymena malaccensis, Tetrahymena borealis, and Ichthyophthirius multifiliis as the only and reciprocal-best BLAST hits. The proteins share a common domain architecture, with an N-terminal RING-type zinc finger domain and a compositionally biased C-terminal region. Evidence for a RING finger domain in TTHERM_00049220p is found using PROSITE (PS50089 ZF RING 2 10-55aa) and CDD (zf-C3HC4 2, E = 0.02, 10-41aa) and confirmed in the orthologues (Supplemental Figure S1). Compositional bias analysis using CAST (Promponas et al., 2000) detects a Q-rich region from 157 to 390 (score 173) and an N-rich region from 137 to 376 (score 77) in TTHERM_00049220p.
To identify homologues in more distant species, we used the N-terminal protein sequences (positions 1–160) of TTHERM_00049220p (excluding the compositionally biased C-terminal segment) in profile-profile searches, which consistently pointed to a similarity to the conserved N-terminal segment of the Zip3/RNF212 protein family. HHsearch (Söding, 2005) against the PANTHER protein family database (Mi et al., 2016) reported the RING finger protein family PTHR22663 (which includes human ZHP3/RNF212 and RNF212B, budding yeast Cst9/Zip3, and Caenorhabditis elegans Zhp-3 and F55A12.10) as the only significant hit (E = 0.0014) and the only hit with E < 0.01. Using the HHpred server (Söding et al., 2005) to search against a database of nine eukaryotic proteomes resulted in three significant hits: Drosophila melanogaster CG31053, C. elegans F555A12.10, and human ZHP3, all of which belong to the PTHR22663 family. HMMsearch against the National Center for Biotechnology Information nonredundant protein database obtained a single significant hit in addition to TTHRM_00049220, namely protein XP_002996515 of the microsporidian Nosema ceranae, which is also a member of the PTHR22663 family. Iterated HMMsearch with this protein included other RNF212B-like proteins. However, whereas the RING finger domains of the nonciliate homologues mostly feature the conventional cysteine-histidine (C3HC4) amino acid motif, TTHERM_00049220p has a less common C8-type motif (Burroughs et al., 2011).
Despite its distant homology, we consider TTHERM_00049220p the most likely orthologue of Zip3/RNF212/HEI10 family members (Chelysheva et al., 2012) because of its similar nuclear localization and depletion phenotype (see later discussion). We therefore named it Zhp3 (for Zip3 homologous protein).
ZHP3 deletion reduces chiasma formation
ZHP3 deletion slightly reduced the viability of sexual progeny (46%, n = 100 mating pairs tested) compared with the wild type (75%, n = 100; for viability testing, see Karrer, 2000). Cytological analysis showed normal progression of meiotic stages (Figure 2, A and B, and Supplemental Figure S2). DSBs were detected as DSB-dependent DNA fragments under pulsed-field gel electrophoresis and appeared to be formed and repaired with normal dynamics (Figure 2C). In addition, as indicated by the number of foci of the DSB-associated recombination protein Dmc1 in stage IV nuclei (Figure 2D), the number of DSBs was similar in wild type and zhp3Δ mutant: 122 ± 16 foci (17 cells) in the wild type and 138 ± 14 foci (17 cells) in the mutant. Finally, homologous pairing in stage IV nuclei, as determined by fluorescence in situ hybridization (FISH) of homologous chromosomal loci, was similar in the wild type and mutant (Figure 2E). To investigate the cause of reduced fertility, we used a method (Schaudinn fixation plus Giemsa staining) in which nuclei are released from cells to enable the separation and inspection of individual chromosomal configurations from diplotene/diakinesis to metaphase I (Figure 2F). Evaluation of ∼80 diakinesis-metaphase I nuclei of the wild type and mutant showed that in zhp3Δ, 43% of chromosome pairs were present as univalents, 46% as rod bivalents, and only 11% as ring bivalents, whereas 90% of homologous pairs in the wild type were present as ring bivalents. Assuming an average of 20 COs (five bivalents with four COs each) per wild-type cell (see later discussion) and that ring and rod bivalents of the mutant most likely have only two and one COs, respectively, we estimate that CO frequency in the mutant is reduced to somewhat less than 20%. Given the normal appearance of DNA fragments upon electrophoresis and the moderate reduction in Dmc1 foci, this substantial reduction is unlikely to be due to decreased DSB formation. Therefore Zhp3 may play a role in the conversion of DSBs into COs.
Zhp3 protein localizes to the meiotic nucleus
To determine the localization of Zhp3, we produced strains expressing Zhp3-hemagglutinin (HA) and Zhp3–enhanced green fluorescent protein (EGFP) fusion proteins, with similar results. The Zhp3-HA strain showed wild-type bivalent formation, indicating that the tag is functional. Zhp3 first became visible as thread-like structures within early stage V nuclei (Figure 3A). Later, in late stage V nuclei, Zhp3 threads coalesced into 15–25 distinct foci (17.8 ± 4.0 SD, n = 53). The number of foci corresponds to the estimated number of 20 COs per nucleus. This estimate is based on the occasional observation of four presumptive chiasmata in diplotene bivalents (Figure 3B) and a chromosome number of 2n = 10. Zhp3 foci were formed only after the disappearance of Dmc1 foci, which mark the sites of DSBs during JM formation (Howard-Till et al., 2011), and, by diplotene, Zhp3 foci had completely disappeared (Figure 3A) The appearance of Zhp3 foci within this time window supports a possible role for Zhp3 at the CO–NCO decision step.
FIGURE 3:
Zhp3 localization and mutant phenotypes. (A) Zhp3 localization at different meiotic stages in the wild type (background staining outside nuclei comes from basal bodies of cilia). (B) Individual diplotene bivalents with schematic interpretations. Possible CO sites and centromere regions are indicated by red dots and blue arrowheads, respectively. (C) BrdU (incorporated during recombinational repair; green) and Zhp3 (red) coappear at late stage V. (D) Zhp3 foci in wild-type (WT), msh4Δ, spo11RNAi, and dmc1RNAi cells. (E) Quantification of ring bivalents, rod bivalents, and univalent pairs in WT, zhp3Δ and msh4Δ single mutants, and zhp3Δmsh4Δ double mutant. Bars, in 10 µm (A–D), 2 µm (E).
To investigate whether the appearance of Zhp3 foci correlates with the molecular recombination process, we stained the protein in late stage V nuclei, where DSB repair synthesis could be determined by bromodeoxyuridine (BrdU) incorporation (Loidl and Lorenz, 2016). In 93 of 100 nuclei, distinct Zhp3 foci appeared concomitantly with BrdU foci (Figure 3C). Thus most Zhp3 foci begin to form within nuclei after the start of repair synthesis. If Zhp3 foci mark a subset of recombination intermediates, then they would be expected to colocalize with a subset of BrdU foci. However, colocalization was limited (Figure 3C). It is possible that some Zhp3 may localize to dedicated CO sites before the incorporation of a detectable amount of BrdU. To investigate a possible link between Zhp3 localization and CO sites, we determined the appearance of Zhp3 foci in mutant backgrounds with reduced or missing COs. For this, we constructed ZHP3-HA spo11RNAi (RNA interference) and ZHP3-HA dmc1RNAi double mutants. In spo11RNAi nuclei, Zhp3 signals were dispersed: they did not form threads and coalesce into wild-type-looking foci, but either remained dispersed or formed a few large aggregates (Figure 3D). During meiosis in dmc1RNAi, a small number of Zhp3 foci appeared, which is consistent with the rare occurrence of COs (Figure 3D; Howard-Till et al., 2011). These observations indicate that Zhp3 foci depend on DSBs and suggest their association with COs.
Zhp3 genetically interacts with Msh4
The reduction in chiasma formation in zhp3Δ is similar to that found in msh4Δ (Figure 3E; Shodhan et al., 2014). Msh4 forms a heterodimeric complex (MutSγ) with Msh5, and together they form a sliding clamp on Holliday junctions (i.e., sites where single strands are exchanged on a DNA molecule) and stabilize these structures (Rakshambikai et al., 2013, and references therein). It has been proposed that in the meiosis of budding yeast and other organisms, these complexes promote CO over NCO formation from recombination intermediates (Nishant et al., 2010) and that they are supported by Zip3 family proteins (Agarwal and Roeder, 2000; Chelysheva et al., 2012; Reynolds et al., 2013; Qiao et al., 2014).
To investigate whether Zhp3 and Msh4 are involved in the same CO pathway, we created a double knockout. The reduction in chiasmata in the zhp3Δ msh4Δ double knockout was similar to that of the two single knockouts (Figure 3E), as expected if the two proteins functioned together. Next we tested the localization of Zhp3-HA in the msh4Δ background (Figure 3D). We found that early localization of Zhp3 (early stage V) was unaffected, whereas the number of distinct foci in late stage V nuclei was reduced to less than half, namely 6.9 ± 3.6 (53 nuclei evaluated). Thus, although we could not produce evidence to support a physical interaction or colocalization (see Discussion), Zhp3 and Msh4 are likely to cooperate.
Zhp3 interacts with a novel protein, Sa15
To identify potential Zhp3 interacting partners, we immunoprecipitated Zhp3-HA and analyzed coprecipitated proteins by mass spectrometry (MS; Supplemental Figure S3). Of the significant MS hits in two biological repeats (Supplemental Figure S3B), one protein (TTHERM_00460720p, also known as Sa15) shows a meiosis-specific expression pattern according to TFGD. Sa15 is predicted to have 562 amino acids and a molecular weight of 66.8 kDa. Previously this protein was shown to play a minor role in postmeiotic events (Hayashi and Mochizuki, 2015). No Sa15 homologues were identified outside the ciliates, and the protein does not contain any conserved domains.
Sa15 was mCherry-tagged to determine its localization. Sa15-mCherry first appeared as a dispersed signal in fully elongated (stage IV) meiotic nuclei. In nuclei at early stage V, the signal coalesced into thread-like structures (Figure 4A), and, of interest, Zhp3 localized to these threads (Figure 4B). This colocalization was lost at late stage V, when Zhp3 became concentrated in ∼20 distinct foci (see later discussion), whereas Sa15 assumed a granular distribution all over the nuclei (Figure 4A). To test whether the localization of Zhp3 and Sa15 was interdependent, we transformed zhp3Δ strains with Sa15-mCherry and sa15Δ strains with Zhp3-HA. The formation of Sa15 threads was unchanged in the absence of Zhp3 (unpublished data). To compare the strengths of the nuclear Zhp3 signals, we used the background staining of basal bodies of cilia as a reference. We found that in the wild type, the nuclear signals were stronger and in sa15Δ about the same compared with the basal bodies (Figure 4G). The slightly reduced intensity of Zhp3 foci in the sa15Δ background suggests that Sa15 may improve the association of Zhp3 with chromosomes.
FIGURE 4:
Sa15 localization and sa15Δ phenotype. (A) mCherry-tagged Sa15 protein first appears in fully elongated nuclei (stage IV), condenses into thread-like structures in shortening nuclei (early stage V), becomes dispersed at late stage V, and disappears before diakinesis. (B) Sa15 and Zhp3 colocalize in early stage V nuclei (Sa15, green; Zhp3, red; DAPI, blue). (C) Progression of meiosis is normal in sa15Δ (compare with Figures 1 and 2A). For nuclear stages, see Figure 1. (D) Dmc1 loading is normal. (E) Clumped bivalents and univalents in metaphase I. (F) Bivalents are abnormal in sa15Δ. Individual diakinesis-metaphase I bivalents with schematic interpretations. Arrowheads indicate centromere positions. (G) Left, 3D projections including Zhp3-stained nuclei and cell membranes with basal bodies of cilia. Right, 3D projections excluding cell membranes. Comparison of nuclear and basal body staining shows that nuclear Zhp3 signals are stronger in the wild type (top) than in the sa15Δ background (bottom). Bars, 10 µm (A–D), 2 µm (E–G).
We then studied the progression of sa15Δ meiosis. Viability of sa15Δ sexual progeny was 43% as compared with 71% of the wild type (110 mating pairs each). Similar to zhp3Δ, all stages of meiosis were present (Figure 4C). Normal appearance of Dmc1 foci (Figure 4D) and the close association of homologous FISH signals (Figure 2E) suggest that DSB formation and homologous paring are similar to the wild type. However, closer inspection by the Schaudinn-plus-Giemsa method revealed abnormal chromosome behavior at diakinesis-metaphase I: most often, chromosomes were clumped (Figure 4E), which precluded the quantification of chiasmata in intact nuclei. Therefore bivalent formation was determined among stray configurations. Of note, many atypical bivalents were found that had only one proximal chiasma per arm (Figure 4F). In contrast, wild-type bivalents have both arms closely associated (Figure 4F), which is consistent with two chiasmata per arm. The frequencies of ring bivalents, rod bivalents, and pairs of univalents in sa15Δ were 40, 43, and 17%, respectively (363 configurations scored). Thus, applying the same criteria as for zhp3Δ (see earlier discussion), we estimated chiasma frequency to be ∼33% of the wild type. Therefore Sa15 is another factor required for the full level of CO formation.
DISCUSSION
Eukaryotes have a conserved mechanism for the CO–NCO decision
In practically all eukaryotes, an excess of DSBs over COs is formed (Serrentino and Borde, 2012). In Tetrahymena, this excess is estimated to be ∼10-fold (Shodhan et al., 2014). It is generally believed that DSBs are required during the homology-searching process (Zickler, 2006) but that only a fraction of DSBs become homologous COs because too many might be harmful. Several pathways can direct recombination intermediates at various stages toward an NCO outcome: a subset of DSBs are repaired by recombination with the sister chromatid, and others end up as NCOs by dissolution of double Holliday junctions (Goldfarb and Lichten, 2010; Chapman et al., 2012). However, for most DSBs the decision between NCO formation by SDSA and CO formation is made at the D-loop stage (Figure 5).
FIGURE 5:
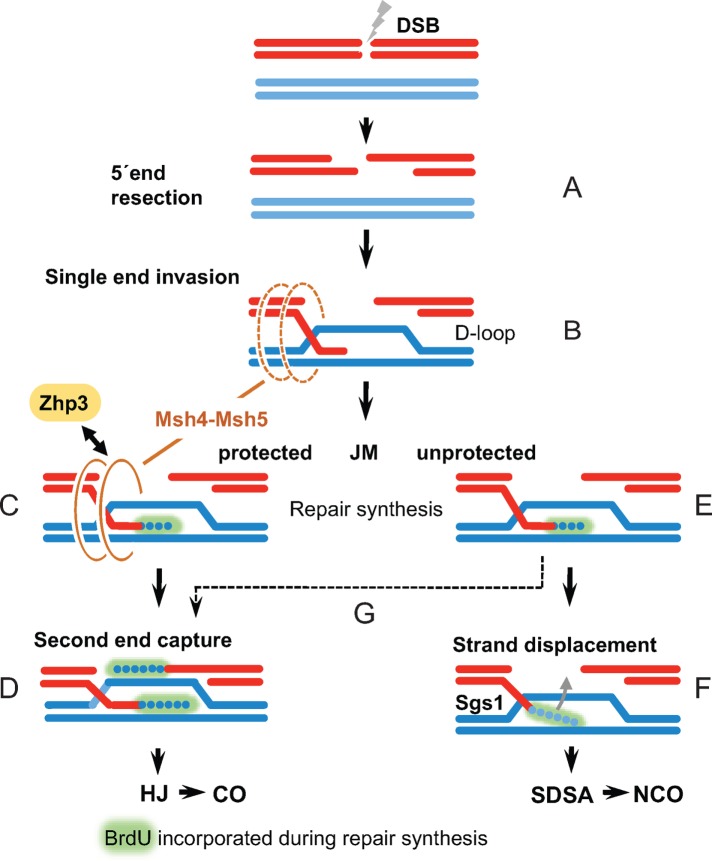
Schematic diagram of possible Zhp3 involvement in the CO–NCO decision in Tetrahymena. The processes shown are based on models proposed for the protection of meiotic joint molecules (JMs) by Msh-Msh5 (MutSγ) complexes (Snowden et al., 2004) and their stabilization by Zip3/RNF2012 family members (Reynolds et al., 2013). Extension of the heteroduplex by Mer3 (Mazina et al., 2004) is not shown because there is no evidence for this activity in Tetrahymena. Of the two single-stranded 3′ ends generated by strand resection around a DSB (A), one invades a DNA molecule for homology searching. Once a homologous region is found and a short heteroduplex has been formed, the Msh5-Msh5 (MutSγ) complex may associate with a JM (B). Once the invading end starts to extend by repair synthesis (which can be observed cytologically by BrdU incorporation), MutSγ is stabilized by Zhp3, allowing it to efficiently protect the JM (C). Zhp3 foci only rarely localize before BrdU incorporation. Protected JMs will then capture the second 3′ end and form COs (D). Most unprotected JMs are unwound and become NCOs via the SDSA pathway (E, F). However, because the absence of Zhp3 or MutSγ does not completely abolish CO formation, some unprotected JMs might persist for long enough to allow second-end capture to occur (G).
For progression toward a CO, a D-loop must be maintained until second-end capture occurs. Otherwise, heteroduplexes will be unwound (probably by Sgs1), the invading strand will be displaced, and the DSB will be repaired as a NCO via SDSA (Hunter, 2015). Two mechanisms have been proposed to help stabilize CO-proficient precursors. One is the unwinding of D-loops by the Mer3 helicase, allowing extension of the heteroduplex (in the 3′–5′ direction of the incoming strand; Mazina et al., 2004). In addition, a Msh4-Msh5 heterodimer (the MutSγ complex) stabilizes D-loops by embracing the involved duplexes (Snowden et al., 2004). Mutant and localization studies on these factors in numerous experimental systems have substantiated this model (Lynn et al., 2007). Findings in Tetrahymena are compatible with a role for MutSγ in protecting CO precursors (Shodhan et al., 2014), whereas a Mer3 orthologue has not been found (Loidl and Lorenz, 2016).
In the mouse, the putative SUMO E3 ligase RNF212 was identified as an additional pro-CO factor, and it was proposed that MutSγ is activated or stabilized at future CO sites by RNF212-dependent SUMOylation of a MutSγ component (Reynolds et al., 2013; Qiao et al., 2014). Budding yeast Zip3 and C. elegans ZHP-3 are RNF212 orthologues, have similar localization to the central region of the SC, colocalize or interact with Msh4 and Msh5, and promote COs (Agarwal and Roeder, 2000; Jantsch et al., 2004; Bhalla et al., 2008; Serrentino et al., 2013). In plants and in the ascomycete Sordaria macrospora, a similar function may be exerted by HEI10, which is a more distant member of the Zip3/RNF212 family (Chelysheva et al., 2012; Wang et al., 2012; De Muyt et al., 2014).
Protein sequence similarity, along with similar depletion phenotypes and localization, suggests that Tetrahymena Zhp3 is evolutionarily related to the Zip3/RNF212 family. Neither Msh4 nor Msh5 was detected as Zhp3 partners by coimmunoprecipitation followed by MS, and we could not investigate the potential colocalization of Zhp3 with Msh4 or Msh5 because we failed to produce antibodies against or functional tagged versions of the latter two proteins. However, the zhp3Δ msh4Δ double mutant did not have a more severe phenotype than either of the single mutants, and the localization of Zhp3 was affected by the absence of Msh4. Therefore it is possible that Tetrahymena Zhp3 and MutSγ cooperate in a similar manner in this evolutionarily distant protist as proposed for other model organisms (Figure 5).
Zip3/RNF212 proteins may function both within and outside the context of an SC
Two major CO pathways are proposed to operate in most eukaryotes: the first (class I) is related to the presence of an SC, produces interfering COs, and depends on ZMM proteins and the MutLγ complex; the second (class II) is independent of an SC, is largely independent of ZMM proteins, produces noninterfering COs, and requires the Mus81-Mms4 complex (Lynn et al., 2007).
The ZMM proteins Zip3, ZHP-3, and RNF212 exert their pro-CO function in the context of an SC. Moreover, albeit in different ways, they are involved in coordinating DSB processing and SC turnover: budding yeast Zip3 promotes SC polymerization via its SUMO ligase activity (Cahoon and Hawley, 2016, and references therein), and C. elegans ZHP-3 couples recombination to SC disassembly (Bhalla et al., 2008). Orthologues have not been detected in SC-less Schizosaccharomyces pombe, supporting a functional link between SC formation and ZMM proteins. Accordingly, deletion of Zip3 family proteins affects only class I CO formation in Saccharomyces cerevisiae, Sordaria, and Arabidopsis and abolishes CO formation in C. elegans (which only forms SC-dependent COs; Agarwal and Roeder, 2000; Jantsch et al., 2004; Chelysheva et al., 2012; De Muyt et al., 2014). However, in the mouse, COs are almost completely suppressed in the absence of RNF212 and MutSγ, indicating that these proteins promote both class I and class II COs (Holloway et al., 2014).
Tetrahymena presents another case of the involvement of MutSγ (Shodhan et al., 2014) and a Zip3-like protein (this study) in the formation of SC-independent Mus81-dependent COs. Conversely, MUS-81 was found to resolve class I COs in C. elegans (Agostinho et al., 2013; O'Neil et al., 2013; Saito et al., 2013) and Mms4 to facilitate proper second-end capture in class I COs in yeast (Oke et al., 2014). Together these studies show that the two CO pathways are not strictly separated.
Zhp3 and Sa15 interact and colocalize at an axial structure
In mammals and C. elegans, RNF212/ZHP-3 activity is governed by the metazoan-specific cyclin-like proteins CNTD and COSA-1, respectively (Yokoo et al., 2012; Holloway et al., 2014). In Tetrahymena, we found another factor, Sa15, to promote wild-type CO formation. Sa15 is unrelated to CNTD/COSA-1 and, although its coprecipitation with Zhp3 suggests that the two proteins may interact, its mode of action is unclear. The transient colocalization of these two proteins and the slight reduction in the intensity of nuclear Zhp3 signals in the absence of Sa15 suggest a role of Sa15 in the proper localization of Zhp3.
The thread-like structures formed by Zhp3 and even more extensively by Sa15 (Figures 3A and 4, C and D) resemble the linear elements of fission yeast, which are evolutionary remnants of SC lateral elements (Loidl, 2006). Similar structures were sporadically seen after immunostaining of Dmc1, tagged RPA, and phosphorylated S/T-Q motifs as part of an unidentified protein (J.L., unpublished data; Supplemental Figure S4). Because in Tetrahymena, microscopy and bioinformatics searches failed to identify SCs or SC proteins (Chi et al., 2014), we speculate that the meiotic chromosomes of Tetrahymena possess axes of a yet unknown composition to which different meiotic proteins become transiently attached. Sa15 is unlikely to be a core component of these axes because it appears as threads only during later stages than Dmc1 and RPA threads. The presence of such an axial structure on meiotic chromosomes of Tetrahymena would be consistent with the requirement for a loop-axis organization of meiotic chromatin to initiate DSBs in other organisms (Lam and Keeney, 2014).
MATERIALS AND METHODS
Cell culture, meiosis induction, and strain construction
Cells were vegetatively propagated in Neff's medium at 30°C (Orias et al., 2000). To make cells competent for meiosis, they were starved in 10 mM Tris, pH 7.5, at 30°C for 16–20 h. Meiosis was induced by mixing starved cultures of complementary mating types. Mutants were created in T. thermophila strains B2086 (mating type II) and Cu428 (mating type VII). All deletions were created in both mating types because to observe a phenotype in mating cells, both mating partners must be mutated. Otherwise, the protein produced by one partner rescues the defect of the other. In contrast, for most genes, efficient RNAi-induced protein depletion occurs in both mating partners if only one partner produces interfering RNA. Further, most tagged proteins expressed by one partner are transferred to the mating partner, where a (sometimes weaker) signal is produced.
For ZHP3 gene knockout, a plasmid comprising a pBluescript (Agilent Technologies, Santa Clara, CA) backbone and two ∼500–base pair fragments from either side of the ZHP3 open reading frame (ORF; TTHERM_00049220) surrounding the NEO4 resistance gene under the control of a Cd2+-inducible MTT1 metallothionein promoter was constructed using the Gibson DNA assembly system (New England Biolabs, Frankfurt, Germany). The linearized construct was introduced into strains of both mating types by biolistic transformation to replace the ZHP3 ORF in the macronuclear genome by homologous recombination between the flanking fragments. Clones carrying the deletion cassette were selected with increasing paromomycin concentrations (Cassidy-Hanley et al., 1997). Southern hybridization with a PCR-amplified probe from within the wild-type ZHP3 sequence was used to estimate the degree of replacement of the ∼50 copies of the wild-type gene by the truncated version (Supplemental Figure S5). A complete sa15 knockout was created similar to the zhp3Δ, but using the NEO5 instead of the NEO4 resistance marker (Supplemental Table S1).
For tagged Zhp3 protein expression, cassettes carrying a ZHP3-EGFP fusion gene and a ZHP3-HA fusion gene were constructed. Each cassette comprised a ∼500–base pair sequence that started just before the stop codon, followed by the EGFP or HA sequence, respectively, and the NEO4 gene under the control of a MTT1 promoter and a ∼500–base pair sequence from the gene's UTR. A similar expression plasmid was constructed for the SA15-mCherry fusion gene using the mCherry sequence and NEO5 resistance marker. All primers are listed in Supplemental Table S1. All constructs were transformed into Tetrahymena by particle bombardment. Replacement efficiency in the tagged strains was tested by qPCR using primers flanking the replaced regions (Supplemental Table S1).
Both spo11RNAi (Howard-Till et al., 2013) and dmc1RNAi (Howard-Till et al., 2011) constructs were transformed into cells expressing tagged Zhp3 to create double mutants. For both strains, full RNAi efficiency was confirmed by the absence of Dmc1 staining. For the msh4Δzhp3Δ double knockout, msh4Δ deletion constructs were created by Gibson assembly using a cycloheximide resistance cassette (for primers see Supplemental Table S1; Papazyan et al., 2014) and then transformed into the zhp3Δ strain. High-replacement clones were selected by sequentially increasing the cycloheximide concentration and tested for knockout efficiency by quantitative PCR (for primers see Supplemental Table S1).
DSB detection
Southern hybridization of DNA separated by pulsed-field gel electrophoresis (PFGE) was used to analyze DSB dynamics in mutants. For this, DNA was extracted from both wild-type and zhp3Δ cells at meiotic time points from 0 to 7 h postmixing. For each time point, cells were collected, mixed with low-melting agarose in CHEF-TE buffer (0.05 M Na2EDTA, 0.01 M Tris-Cl, pH 7.5), and allowed to solidify in plug molds. Agarose plugs were incubated with proteinase K in NDS buffer (0.5 M Na2EDTA, 0.01 M Tris-Cl, pH 7.5, 1% N-lauroylsarcosine) to lyse the cells and then treated with RNase and stored in CHEF-TE at 4°C (Lukaszewicz et al., 2010). DNA plugs were loaded into a 1% PFGE agarose gel and run on a Bio-Rad CHEF-DR III system in 0.5× TBE for 24 h at 14°C and 6 V/cm, with an inclination angle of 120° and a switch time of 60×120 s. DNA was then transferred onto a membrane, and DSB-dependent DNA fragments were visualized using a Southern probe for the MIC-specific TLR1 sequence (Lukaszewicz et al., 2010).
Cytological methods
Paraformaldehyde fixation and immunostaining.
To analyze meiotic progression, mating cells were fixed at various time points by adding formaldehyde (final concentration 4%) and Triton X-100 (final concentration 0.5%) to a 5-ml culture sample. After 30 min at room temperature, cells were pelleted and resuspended in a solution of 4% paraformaldehyde and 3.4% sucrose. The cell suspension was spread onto a slide and air dried. Slides were then washed twice with 1× phosphate-buffered saline (PBS) and once with 1× PBS containing 0.05% Triton X-100 (PBST). Cell preparations were stained with 4′,6′-diamidino-2-phenylindole (DAPI) in Vectashield (Vector Laboratories, Burlingame, CA). The same fixation method was used to visualize tagged proteins. After washing with PBST, primary antibody (anti-HA, H6908; Sigma-Aldrich, St. Louis, MO; anti-Rad51 MS-988; NeoMarkers, Fremont, CA; anti-GFP, mouse monoclonal; Takara Bio, Mountain View, CA; anti-mCherry, rabbit polyclonal; Takara Bio) was applied and incubated under a coverslip for 1–2 h. Slides were then washed, and a secondary fluorophore-coupled antibody was applied for 1 h, followed by washing and DAPI staining.
Immunodetection of chromatin-bound proteins.
An ice-cold mixture of 50 µl of 37% formaldehyde and 450 µl of 10% Triton X-100 was added to 5 ml of mating cells and mixed gently. The high detergent concentration was used to spread the nuclei and remove free proteins from the nuclei. The suspension was incubated on ice for 25 min, and then another 450 µl of 37% formaldehyde was added and incubated for an additional 5 min on ice. Fixed cells were then pelleted and resuspended in 4% paraformaldehyde containing 3.4% sucrose. The cell suspension was spread onto slides, air-dried, and stained as described.
Schaudinn fixation.
Cells (5 ml) were pelleted and resuspended in 500 µl of Schaudinn's fixative (2:1 saturated HgCl2: ethanol plus 5 µl of acetic acid). After 1 h of incubation at room temperature (with intermittent shaking), fixed cells were pelleted and resuspended in 1 ml of 70% ethanol. A 100-µl sample of the suspension was pelleted, resuspended in 300 µl of a methanol-acetic acid mixture (3:1), and dropped onto a slide to break open the cells. These cell preparations were used for Giemsa staining or FISH.
Giemsa staining.
Giemsa staining of Schaudinn-fixed cell preparations provides superior resolution of metaphase I bivalents. For this, cells were hydrated and incubated with 100 µl of 5 N HCl under a coverslip for 2 min, washed in distilled water, and air dried. Slides were then incubated in 4% Giemsa solution in 1× PBS for 10 min, washed under running water, and air dried before mounting with Euparal and visualization by light microscopy.
Fluorescence in situ hybridization.
For FISH, a Cy3-labeled, 22.1-kb probe against a MIC-specific intercalary chromosomal locus was used (Loidl and Mochizuki, 2009). Cell preparations on slides were soaked with water for a few minutes, and incubated with 90 µl of 1 M sodium thiocyanate under a coverslip at 90°C for 15 min, and then washed twice with 2× saline sodium citrate (SSC), followed by denaturation in 70% formamide in 2× SSC (pH 7.1) for 2 min at 68°C. Slides were then rinsed with ice-cold water and air dried. In the meantime, the DNA probe was dissolved in hybridization buffer (50% formamide and 10% dextran sulfate in 2× SSC), denatured at 95°C for 3 min, and put on ice. An 8-µl sample of the probe was dropped onto each slide and sealed under a coverslip; slides were then incubated in a moist chamber at 37°C for 48 h. After hybridization, coverslips were removed carefully, and slides were washed in hybridization buffer at 37°C for 5 min, followed by 5-min washes in 2× SSC, 1× SSC, and 1× PBS plus 0.05% Triton X-100. Finally, slides were mounted with Vectashield with DAPI for inspection by fluorescence microscopy.
BrdU incorporation and detection and signal evaluation.
BrdU (final concentration 0.2 mM) was added to mating cells at 3 h, 15 min after mixing, followed by incubation in the dark for 1 h, 15 min. Cells were then fixed on slides by the paraformaldehyde method. After air drying, cells were washed in 2× SSC and denatured in 70% formamide in 2× SSC for 2 min at 65°C to expose the labeled nucleotides for antibody binding. Slides were then transferred to ice-cold water and washed twice in ice-cold 1× PBS and once in ice-cold 1× PBST, followed by incubation with anti-BrdU, anti-HA, and secondary antibodies (Loidl et al., 2012).
Microscopy inspection and analysis.
Cells were evaluated using a fluorescence microscope fitted with appropriate filters. For documentation, three-dimensional (3D) stacks of images were recorded, deconvolved, and projected as previously described (Loidl and Mochizuki, 2009).
Immunoprecipitation
For immunoprecipitation of Zhp3-bound proteins, Zhp3-HA–expressing cells were collected 4.5–5 h after induction of meiosis, pelleted, resuspended in ice-cold Tris wash buffer (50 mM Tris, 100 mM NaCl, 100 mM EDTA), and repelleted. Cell pellets were resuspended in lysis buffer (100 mM Tris base, Tris-HCl, 1 M KCl, 1 M MgCl2, 1% Triton X-100, 0.01 M phenylmethylsulfonyl fluoride, pH 7.5) and ground in a Dounce homogenizer on ice. NaCl (final concentration 150 mM) was added to the lysate, and cell debris was removed by centrifugation and filtration of the supernatant. The HA fusion protein was captured from the lysate by incubation with anti-HA magnetic beads (Pierce Anti-HA Magnetic Beads; Thermo Fisher Scientific, Waltham, MA) for 2 h at 4°C. Beads were washed three times with wash buffer (100 mM Tris base, Tris-HCl, 1 M KCl, 1 M MgCl2, 150 mM NaCl, pH 7.5) containing 1% Triton X-100, once with wash buffer without Triton X-100, and then four times with 50 mM ammonium bicarbonate. All buffers except for ammonium bicarbonate contained cOmplete proteinase inhibitor (Roche, Indianapolis, IN). Beads were sent for mass spectrometry analysis, and Zhp3 was eluted from a small aliquot for Western blotting with an anti-HA antibody to confirm precipitation.
Note added in proof. Gene TTHERM_00460720 (SA15 ) is now named BIME1 in the Tetrahymena Genome Database (http://ciliate.org/).
Supplementary Material
Acknowledgments
This study was supported by Grants W1238-B20 and P27313-B20 from the Austrian Science Fund (F.W.F.).
Abbreviations used:
- CO
crossover
- DAPI
4′,6-diamidino-2-phenylindole
- DSB
double-strand break
- FISH
fluorescence in situ hybridization
- JM
joint molecule
- NCO
noncrossover
- PFGE
pulsed-field gel electrophoresis
- RNAi
RNA interference
- SC
synaptonemal complex.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0678) on January 18, 2017.
REFERENCES
- Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- Agostinho A, Meier B, Sonneville R, Jagut M, Woglar A, Blow J, Jantsch V, Gartner A. Combinatorial regulation of meiotic Holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 2013;9:e1003591. doi: 10.1371/journal.pgen.1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Iyer LM, Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol Biosyst. 2011;7:2261–2277. doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon CK, Hawley RS. Regulating the construction and demolition of the synaptonemal complex. Nat Struct Mol Biol. 2016;23:369–377. doi: 10.1038/nsmb.3208. [DOI] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MRG, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, Lemhemdi A, Vrielynck N, Le Guin S, Novatchkova M, Grelon M. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 2012;8:e1002799. doi: 10.1371/journal.pgen.1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J, Mahé F, Loidl J, Logsdon J, Dunthorn M. Meiosis gene inventory of four ciliates reveals the prevalence of a synaptonemal complex-independent crossover pathway. Mol Biol Evol. 2014;31:660–672. doi: 10.1093/molbev/mst258. [DOI] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Zhang LR, Piolot T, Kleckner N, Espagne E, Zickler D. E3 ligase Hei10: a multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev. 2014;28:1111–1123. doi: 10.1101/gad.240408.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, Lichten M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A, Mochizuki K. Targeted gene disruption by ectopic induction of DNA elimination in Tetrahymena. Genetics. 2015;201:55–64. doi: 10.1534/genetics.115.178525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Sun X, Yokoo R, Villeneuve AM, Cohen PE. Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J Cell Biol. 2014;205:633–641. doi: 10.1083/jcb.201401122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Till RA, Lukaszewicz A, Loidl J. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genet. 2011;7:e1001359. doi: 10.1371/journal.pgen.1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Till RA, Lukaszewicz A, Novatchkova M, Loidl J. A single cohesin complex performs mitotic and meiotic functions in the protist Tetrahymena. PLoS Genet. 2013;9:e1003418. doi: 10.1371/journal.pgen.1003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination: the essence of heredity. CSH Perspect Biol. 2015;7:a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J. Targeted gene knock out reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol. 2004;24:7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer KM. Tetrahymena genetics: two nuclei are better than one. In: Asai DJ, Forney JD, editors. Tetrahymena thermophila. San Diego, CA: Academic Press; 2000. pp. 127–186. [Google Scholar]
- Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. CSH Perspect Biol. 2014;7:a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 2006;115:260–271. doi: 10.1007/s00412-006-0047-7. [DOI] [PubMed] [Google Scholar]
- Loidl J, Lorenz A. DNA double-strand break formation and repair in Tetrahymena meiosis. Sem. Cell Dev Biol. 2016;54:126–134. doi: 10.1016/j.semcdb.2016.02.021. [DOI] [PubMed] [Google Scholar]
- Loidl J, Lukaszewicz A, Howard-Till RA, Koestler T. The Tetrahymena meiotic chromosome bouquet is organized by centromeres and promotes interhomolog recombination. J Cell Sci. 2012;125:5873–5880. doi: 10.1242/jcs.112664. [DOI] [PubMed] [Google Scholar]
- Loidl J, Mochizuki K. Tetrahymena meiotic nuclear reorganization is induced by a checkpoint kinase-dependent response to DNA damage. Mol Biol Cell. 2009;20:2428–2437. doi: 10.1091/mbc.E08-10-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Howard-Till RA, Loidl J. Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex. Nucleic Acids Res. 2013;41:9296–9309. doi: 10.1093/nar/gkt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Howard-Till RA, Novatchkova M, Mochizuki K, Loidl J. MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma. 2010;119:505–518. doi: 10.1007/s00412-010-0274-9. [DOI] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Börner V. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC. Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′heteroduplex extension by Rad51: implications for crossover control in meiotic recombination. Cell. 2004;117:47–56. doi: 10.1016/s0092-8674(04)00294-6. [DOI] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant KT, Chen C, Shinohara M, Shinohara A, Alani E. Genetic analysis of baker's yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability. PLoS Genet. 2010;6:e1001083. doi: 10.1371/journal.pgen.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil NJ, Martin JS, Youds JL, Ward JD, Petalcorin MIR, Rose AM, Boulton SJ. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet. 2013;9:e1003582. doi: 10.1371/journal.pgen.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oke A, Anderson CM, Yam P, Fung JC. Controlling meiotic recombinational repair - specifying the roles of ZMMs, Sgs1 and Mus81/Mms4 in crossover formation. PLoS Genet. 2014;10:e1004690. doi: 10.1371/journal.pgen.1004690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E, Hamilton EP, Orias JD. Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. In: Asai DJ, Forney JD, editors. Tetrahymena thermophila. San Diego, CA: Academic Press; 2000. pp. 189–211. [DOI] [PubMed] [Google Scholar]
- Papazyan R, Voronina E, Chapman JR, Luperchio TR, Gilbert TM, Meier E, Mackintosh SG, Shabanowitz J, Tackett AJ, Reddy KL, et al. Methylation of histone H3K23 blocks DNA damage in pericentric heterochromatin during meiosis. Elife. 2014;3:e02996. doi: 10.7554/eLife.02996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promponas VJ, Enright AJ, Tsoka S, Kreil DP, Leroy C, Hamodrakas S, Sander C, Ouzounis CA. CAST: an iterative algorithm for the complexity analysis of sequence tracts. Bioinformatics. 2000;16:915–922. doi: 10.1093/bioinformatics/16.10.915. [DOI] [PubMed] [Google Scholar]
- Qiao H, Rao HBDP, Yang Y, Fong JH, Cloutier JM, Deacon DC, Nagel KE, Swartz RK, Strong E, Holloway JK, et al. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet. 2014;46:194–199. doi: 10.1038/ng.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshambikai R, Srinivasan N, Nishant KT. Structural insights into Saccharomyces cerevisiae Msh4-Msh5 complex function using homology modeling. PLoS One. 2013;8:e78753. doi: 10.1371/journal.pone.0078753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet. 2013;45:269–278. doi: 10.1038/ng.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Lui DY, Kim H-M, Colaiácovo MP. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 2013;9:e1003586. doi: 10.1371/journal.pgen.1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrentino ME, Borde V. The spatial regulation of meiotic recombination hotspots: are all DSB hotspots crossover hotspots? Exp Cell Res. 2012;318:1347–1352. doi: 10.1016/j.yexcr.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Serrentino ME, Chaplais E, Sommermeyer V, Borde V. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 2013;9:e1003416. doi: 10.1371/journal.pgen.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shodhan A, Lukaszewicz A, Novatchkova M, Loidl J. Msh4 and Msh5 function in SC-independent chiasma formation during the streamlined meiosis of Tetrahymena. Genetics. 2014;198:983–993. doi: 10.1534/genetics.114.169698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai T, Hiwatashi K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J Protozool. 1974;21:542–548. doi: 10.1111/j.1550-7408.1974.tb03695.x. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Cheng SY, Morehouse SJ, MacQueen AJ. Synaptonemal complex proteins of budding yeast define reciprocal roles in MutSγ-mediated crossover formation. Genetics. 2016;203:1091–1103. doi: 10.1534/genetics.115.182923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KJ, Wang M, Tang D, Shen Y, Miao CB, Hu Q, Lu TG, Cheng ZK. The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 2012;8:e1002809. doi: 10.1371/journal.pgen.1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehrer SL, Aronica L, Suhren JH, Bush CJL, Noto T, Mochizuki K. A Tetrahymena Hsp90 co-chaperone promotes siRNA loading by ATP-dependent and ATP-independent mechanisms. EMBO J. 2015;34:559–577. doi: 10.15252/embj.201490062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Lu X, Zhou Z, Chang Y, Yuan D, Tian M, Zhou Z, Wang L, Fu C, Orias E, et al. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using deep RNA sequencing. PLoS One. 2012;7:e30630. doi: 10.1371/journal.pone.0030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve A. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.