The circadian clock regulates IGF-1 production and signaling through proteins called cryptochromes, which regulate the activity of transcriptional factor STAT5B and control mouse body and organ size.
Abstract
Insulin-like growth factor (IGF) signaling plays an important role in cell growth and proliferation and is implicated in regulation of cancer, metabolism, and aging. Here we report that IGF-1 level in blood and IGF-1 signaling demonstrates circadian rhythms. Circadian control occurs through cryptochromes (CRYs)—transcriptional repressors and components of the circadian clock. IGF-1 rhythms are disrupted in Cry-deficient mice, and IGF-1 level is reduced by 80% in these mice, which leads to reduced IGF signaling. In agreement, Cry-deficient mice have reduced body (∼30% reduction) and organ size. Down-regulation of IGF-1 upon Cry deficiency correlates with reduced Igf-1 mRNA expression in the liver and skeletal muscles. Igf-1 transcription is regulated through growth hormone–induced, JAK2 kinase–mediated phosphorylation of transcriptional factor STAT5B. The phosphorylation of STAT5B on the JAK2-dependent Y699 site is significantly reduced in the liver and skeletal muscles of Cry-deficient mice. At the same time, phosphorylation of JAK2 kinase was not reduced upon Cry deficiency, which places CRY activity downstream from JAK2. Thus CRYs link the circadian clock and JAK-STAT signaling through control of STAT5B phosphorylation, which provides the mechanism for circadian rhythms in IGF signaling in vivo.
INTRODUCTION
Insulin-like growth factor-1 (IGF-1) is an essential paracrine and autocrine factor for tissue homeostasis, growth, and development (Rother and Accili, 2000; Dupont and Holzenberger, 2003; Rosenfeld and Hwa, 2009; Clemmons, 2012). The production of IGF-1 and IGF signaling needs to be precisely regulated because both low and high IGF-1 production may have negative effect on physiology: a low level of IGF-1 might compromise development and tissue regeneration (Rosenfeld and Hwa, 2009; Vottero et al., 2013), whereas a high level of IGF-1 is implicated in the development of cancer, metabolic diseases, and aging (Anisimov and Bartke, 2013; Junnila et al., 2013). The majority of the blood-circulating (plasma) IGF-1 is produced by the liver, with a small contribution from other tissues (Liu et al., 1998). Production of IGF-1 in the liver and in other tissues is regulated by growth hormone (GH)–dependent control of transcription (Herrington et al., 2000; Herrington and Carter-Su, 2001; Rotwein, 2012; Chia, 2014). GH binds its receptor (GHR) and induces activation of the receptor-associated tyrosine kinase Janus kinase2 (JAK2). JAK2 phosphorylates the intracellular part of GHR, which results in engagement of the JAK-signal transducer and activator of transcription (STAT) signaling pathway (Lanning and Carter-Su, 2006). Transcriptional factors from the STAT family are recruited to the phosphorylated receptor and get phosphorylated on the Tyr residue by JAK2 (Herrington et al., 2000; Rotwein and Chia, 2010; Chia, 2014). After dissociation from the receptor, phosphorylated STATs form dimers, translocate to the nucleus, and bind to specific sites and regulate transcription. STAT1, STAT3, STAT5A, and STAT5B are phosphorylated upon GH/GHR interaction, and STAT5B is recognized as a critical intermediate of GH action in rodents and humans (Hwa et al., 2011; Rotwein, 2012; Varco-Merth and Rotwein, 2014).
The plasma IGF-1 level changes during the day, suggesting some circadian control, but molecular mechanisms are not known. The circadian clock generates rhythms in physiology and behavior known as the circadian rhythms (Schibler and Sassone-Corsi, 2002; Dardente and Cermakian, 2007; Dibner et al., 2010) and synchronizes the processes in organisms with the environment. The mammalian central circadian clock is located in the suprachiasmatic nucleus of the hypothalamus and controls the activities of peripheral clocks that are found in almost every organ system (Mohawk et al., 2012). Disruption of the circadian clock causes the desynchronization between the environment and central and peripheral clocks, which leads to metabolic defects in animal models and may increase the risk of a variety of diseases, such as diabetes and cancer in humans (Schernhammer et al., 2003; Levi et al., 2007; Green et al., 2008; Maury et al., 2010; Kondratova and Kondratov, 2012). The circadian clock has been implicated in regulation of aging and development of cancer (Fu et al., 2002; Froy and Miskin, 2010; Khapre et al., 2014a) but the mechanisms are mostly unknown. On the cellular level, the circadian clock is formed by interlinked negative and positive feedback loops (Shearman et al., 2000; Schibler and Sassone-Corsi, 2002). Transcriptional factor BMAL1 in complex with another circadian clock transcriptional factor CLOCK or NPAS2 positively regulates expression of genes for cryptochromes 1 and 2 (Cry1 and Cry2) and periods 1 and 2 (Per1 and Per2; Vitaterna et al., 1999). In turn, the cryptochrome 1 and 2 proteins (CRY1, 2) interact with PER1, 2 proteins and act as negative regulators of the BMAL1/CLOCK transcriptional activity and their own expression. This negative feedback loop formed by BMAL1, CLOCK, CRYs and PERs is essential for circadian clock activities. The BMAL1:CLOCK (NPAS2) complex also regulates the expression of Rev-Erbs and RORs genes, whose products in turn negatively and positively regulate the expression of the Bmal1 gene and therefore form another feedback loop (Schibler and Sassone-Corsi, 2002). There are two mammalian Cry genes, Cry1 and Cry2, with redundant functions; double knockout of these genes results in disruption of circadian rhythms in behavior and gene expression (Van der Horst et al., 1999; Carter-Su et al., 2000). Here we report that CRYs are involved in regulation of IGF-1 production and signaling.
RESULTS
Cry1,2−/− mice develop dwarfism
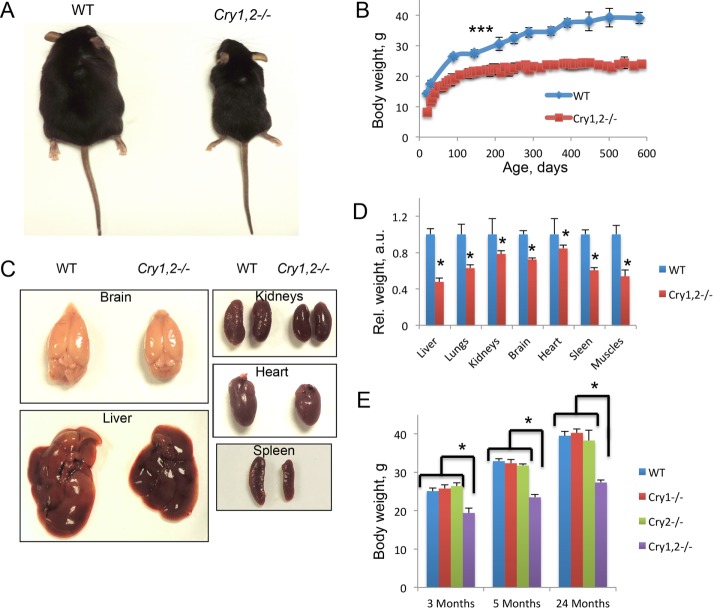
We noticed that Cry1,2−/− mice are smaller than wild-type (WT) mice of the same age. The sizes of 5-mo-old WT and Cry1,2−/− males are compared in Figure 1A; a similar size difference was observed for female mice. We decided to investigate the difference in size in more detail. Figure 1B presents growth curves of male mice of both genotypes (see Supplemental Figure S1A for females). The difference between genotypes is seen already at weaning (3 wk of age); the observed difference was statistically significant for all ages. The weight of male Cry1,2−/− mice was ∼70–75% of the weight of WT males; the weight of female Cry1,2−/− mice was ∼65–70% of the weight of WT females (the difference changed with age). We checked whether the size and weight reduction are proportional or the reduced weight is due to the reduction of the weights of some particular organs and tissues. We measured the weights of all major organs in 5-mo-old animals, when both WT and Cry1,2−/− mice reached their adult size. As shown in Figure 1, C and D, for males and Supplemental Figure S1B for females, all major organs, including brain, liver, heart, and kidneys, were proportionally reduced in their size and weight; therefore we conclude that Cry1,2−/− mice are proportionally smaller than WT mice. Cry1 and Cry2 genes are partially redundant; therefore we checked whether a deficiency of either of them would be sufficient to induce dwarfism. Body weights of Cry1−/− and Cry2−/− mice at three different ages (Figure 1E for males and Supplemental Figure S1C for females) were similar to the body weight of WT mice. Thus deficiency of both Cry1 and Cry2 genes is necessary for body size reduction.
FIGURE 1:
Cry1, 2−/− mice have reduced body weight and size. (A) Cry1, 2−/− males are smaller than WT males of the same age. (B) Cry1, 2−/− males (red squares) have reduced body weight compared with WT (blue diamonds) males (N = 16 for each genotype). The difference between genotypes is statistically significant (p < 0.001) at all ages, starting at 10 d of age. (C) Size of visceral organs are reduced in Cry1, 2−/− males. (D) Weight of visceral organs is reduced in Cry1, 2−/− males. Organ weights were assayed for WT (blue bars) and Cry1, 2−/− (red bars) for 5-mo-old males (N = 16 for each genotype); the difference between genotypes is statistically significant (p < 0.001). (E) Weight of WT (blue bars), Cry1−/− (red bars), Cry2−/− (green bars), and Cry1, 2−/− (purple bars) males at three different ages (N = 6 for every age group for every genotype). *Statistically significant difference between genotypes (p < 0.05).
Cry1,2−/− mice have a reduced production of IGF-1
Reduced activity of the somatotropic (GH/IGF-1) axis is one of the major causes of dwarfism in rodent and humans (Bartke and Brown-Borg, 2004; Chia, 2014). We assayed the levels of IGF-1 in the plasma of WT and Cry1,2−/− mice. We observed that in WT mice, the levels of the circulating IGF-1 demonstrated a statistically significant daily oscillation, with the highest value at zeitgeber time 2 h (ZT2)–ZT6 and the lowest value at ZT14–ZT22 (Figure 2A). We observed a significant reduction in the levels of the circulating IGF-1 in Cry1,2−/− mice at all six time points tested (Figure 2A). The circulating IGF-1 level was reduced by ∼60–80% for Cry1,2−/− (depending on the time of the day). Thus, similar to other models of dwarfism, a reduced level of circulating IGF-1 correlated with the reduced body size of the Cry1, 2−/− mice (Bartke and Brown-Borg, 2004).
FIGURE 2:
CRY deficiency affects IGF-1 level in plasma and tissues. (A) Daily rhythms of plasma IGF-1 in WT AL (blue diamonds) and Cry1, 2−/− AL (red squares) males (N = 3 per time point). (B) Plasma IGF-1 levels for WT (blue bars), Cry1−/− (red bars), Cry2−/− (green bars), and Cry1, 2−/− (purple bars) males at ZT6 and ZT18 (N = 6 per time point). (C) Representative Western blotting (pooled extracts from three mice per time point) of IGF-1 expression in the liver (left) and skeletal muscle (right) of WT and Cry1, 2−/− male mice. (D) Quantification of IGF-1 in the liver (left) and skeletal muscle (right) of WT AL (blue diamonds) and Cry1, 2−/− (red squares) male mice (N = 3 per time point). The quantification data for the liver and skeletal muscles are presented as relative arbitrary units; note that these units are different for the liver and skeletal muscle graphs because the levels of IGF1 protein and Igf1 mRNA in the liver are orders of magnitude higher than in skeletal muscle. Different exposure time was used for Western blots in the liver and skeletal muscles. *Statistically significant difference between genotypes (p < 0.05). Light was on at ZT0 and off at ZT12.
Cry1−/− and Cry2−/− mice did not demonstrate any reduction in body size (Figure 1E); therefore we decided to assay whether deficiency of both Cry1 and Cry2 is necessary for the effects on the circulating IGF-1. We compared plasma IGF-1 levels for WT and single or double Cry knockouts at two times of the day, ZT6 and ZT18, which corresponded to the high and low levels of plasma IGF-1 in WT mice. We found a 30% reduction of the IGF-1 level in Cry1−/− mice at ZT6 (compare with 80% reduction in Cry1,2−/− mice; Figure 2B). There was no difference between WT, Cry1−/−, and Cry2−/− at ZT18 and no difference between WT and Cry2−/− at ZT6. Thus deficiency of both Cry genes is necessary for down-regulation in the circulating IGF-1 level, which correlates well with the effects of CRY deficiency on the body size.
Thus we observed significant down-regulation of the circulating IGF-1 levels in Cry1,2−/− mice. The major source of the circulating IGF-1 is the liver (Yakar et al., 1999); therefore we compared the levels of the IGF-1 protein in the livers of WT and Cry1, 2−/− mice (Figure 2C). In WT mice, the liver IGF-1 level displayed oscillations (with the highest level of IGF-1 expression at ZT18–ZT2), and Cry1,2 deficiency resulted in reduction of liver IGF-1 levels (the difference between the genotypes was significant at three of six time points). Many other tissues also produce IGF-1, and, although IGF-1 from these sources does not significantly affect the levels of the circulating hormone, it still plays an important role in paracrine signaling (Liu et al., 1998). Therefore we also determined the IGF-1 level in skeletal muscle (Figure 2C). In WT, the level of skeletal muscle IGF-1 was significantly lower at ZT10 than at other time points. Similar to the effect in the liver, we observed significant down-regulation of skeletal muscle IGF-1 in Cry1,2−/− mice (the difference between the genotypes was significant at all time points except for ZT10). Thus Cry1,2 deficiencies resulted in down-regulation of IGF-1 production in different tissues.
IGF-1 signaling is reduced in Cry1,2−/− mice
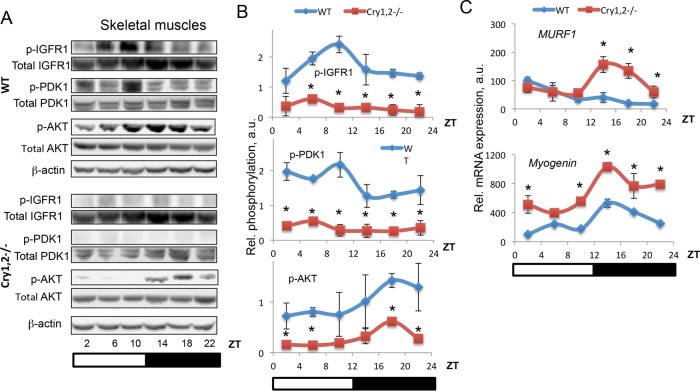
IGF-1 promotes growth of muscle, bone, and adipose tissues in mammals (Anisimov and Bartke, 2013; Chia, 2014); skeletal muscles are one of the main targets of IGF-1. We decided to see whether IGF-1 signaling is affected in skeletal muscles of Cry1,2−/− mice. The receptor for IGF-1 is the tyrosine kinase IGF-1R (Kavran et al., 2014). Binding of IGF-1 to IGF-1R leads to receptor activation and autophosphorylation on Y1137/1138 in mouse (Y1135/1136 in human IGF-1R; Ullrich et al., 1986). We compared IGF-1R phosphorylation in skeletal muscles of WT and Cry1,2−/− mice (Figure 3, A and B). In skeletal muscles of WT mice, IGF-1R phosphorylation significantly oscillated during the day, with the highest level of phosphorylation at ZT6–ZT10, in agreement with the highest level of the circulating IGF-1. The level of Y1137/Y1138 phosphorylation was significantly reduced in Cry1, 2−/− muscles, again in agreement with significantly reduced IGF-1 in plasma. PDK1 and AKT are protein kinases that act downstream of IGF-1R (Schiaffino and Mammucari, 2011). They are not direct targets of IGF-1R but are phosphorylated and activated upon IGF-1 binding to IGF-1R. We assayed the phosphorylation of PDK1 on S241 and AKT on S473 (Figure 3, A and B). In WT mice, PDK1 S241 phosphorylation oscillated during the day, with the highest level at ZT10–ZT14, which correlated with IGF-1R Y1137/1138 phosphorylation. There was an additional peak of high PDK1 S241 phosphorylation at ZT2. AKT-S473 phosphorylation was high at the dark phase of the day; thus AKT phosphorylation was delayed compared with IGF-1R phosphorylation or PDK1 phosphorylation. In skeletal muscles of Cry1, 2−/− mice, phosphorylation of both PDK1 and AKT was significantly reduced (reduction by 30–70% for PDK1 and by 60–80% for AKT, depending on the time of the day). Of interest, AKT phosphorylation was still rhythmic in skeletal muscles of Cry1, 2−/− mice, with a high level of phosphorylation at night.
FIGURE 3:
IGF signaling is reduced in skeletal muscle of Cry1, 2−/− mice. (A) Representative Western blotting (pooled extracts from three mice per time point) of phospho–IGF-1R on Y1137/1138, PDK1 on S241, and AKT on S473 in skeletal muscle of WT (blue diamonds) and Cry1, 2−/− (red squares) male mice. (B) Quantification of phospho–IGF-1R on Y1137/1138, PDK1 on S241, and AKT on S473 in skeletal muscle of WT (blue diamonds) and Cry1, 2−/− (red squares) male mice (N = 3 per time point). (C) mRNA expression of FOXO transcriptional targets in skeletal muscle of WT (blue diamonds) and Cry1, 2−/− (red squares) male mice (N = 3 per time point). *Statistically significant difference between genotypes (p < 0.05). Light was on at ZT0 and off at ZT12.
IGF-1 signaling is evolutionarily conserved; in many organisms, reduction of IGF-1 signaling leads to activation of the transcriptional factor FOXO. We assayed the expression of FOXO targets MURF1 and Myogenin in skeletal muscles of WT and Cry1, 2−/− mice (Figure 3C). We found that expression was up-regulated at ZT14–ZT22 for MURF1 and at ZT2 and ZT10–ZT22 for Myogenin. These results are in good agreement with reduced IGF-1 signaling. Of interest, we also found that expression of the FOXO target genes significantly oscillated across the day in both WT and the circadian mutant, with different patterns, suggesting circadian clock–dependent and –independent effects of time of the day. Data in Figure 3 argue that IGF-1 signaling is significantly reduced in skeletal muscles of Cry1, 2−/− mice. Thus deficiency of CRYs resulted in reduced plasma IGF-1 levels and reduced tissue IGF-1 levels and IGF-1 signaling in skeletal muscles, which may be a reason for the observed reduced body size of Cry1, 2−/− mice.
STAT5-dependent regulation of IGF-1 expression is disrupted in the liver and skeletal muscles of Cry1, 2−/− mice
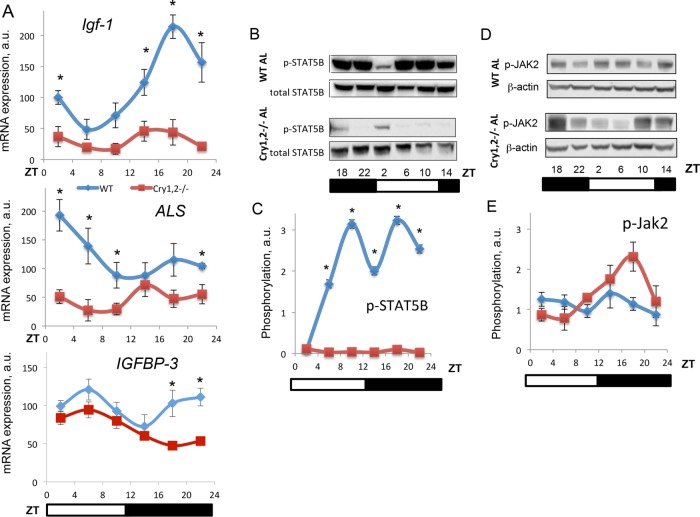
Reduced levels of IGF-1 in the plasma and tissues of Cry1, 2−/− mice indicate that production of IGF-1 is compromised in these mice. We assayed the expression of Igf1 on the mRNA level across the circadian cycle in the liver of Cry1, 2−/− mice and compared it with the expression in the liver of WT mice. As shown in Figure 4A, Igf1 mRNA expression in the liver of WT mice significantly changed during the day, with the highest expression between ZT14 and ZT22 and the lowest expression at ZT2–ZT6. Cry deficiency resulted in a significant reduction of Igf-1 expression in the liver (the difference was significant at all six time points). In skeletal muscle of WT mice (Supplemental Figure S2A), the Igf-1 mRNA level did not show any significant rhythms across the day, whereas in skeletal muscle of Cry1, 2−/− mice, expression was reduced at all six time points, which correlates with the reduced IGF1 protein level in skeletal muscles. Thus reduced Igf-1 mRNA expression may contribute to the observed reduced IGF1 protein level in the tissues of Cry1, 2−/− mice.
FIGURE 4:
Igf-1 mRNA expression and STAT5 phosphorylation but not JAK2 phosphorylation is down-regulated in the liver of Cry1, 2−/− mice. (A) Daily rhythms of Igf-1 (top) and ALS (bottom) mRNA expression in the liver of WT (blue diamonds) and Cry1, 2−/− (red squares) males. (B) Representative Western blotting (pooled extracts from three mice per time point) of phosphorylation of STAT5B on Y699 in the liver of WT and Cry1, 2−/− male mice. (C) Quantification of phosphorylation of STAT5B on Y699 in the liver of WT (blue diamonds) and Cry1, 2−/− (red squares) male mice. (D) Representative Western blotting (pooled extracts from three mice per time point) of phosphorylation of JAK2 on Y1007 in the liver of WT and Cry1, 2−/− male mice. (E) quantification of phosphorylation of JAK2 on Y1007 in the liver of WT (blue diamonds) and Cry1, 2−/− (red squares) male mice (N = 3 per time point). *Statistically significant difference between genotypes (p < 0.05). Light was on at ZT0 and off at ZT12.
The transcriptional factor STAT5B is the major regulator of Igf-1 transcription (Herrington et al., 2000). STAT5B activity is regulated by phosphorylation: phosphorylated STAT5B is transported from the cytoplasm to the nucleus and drives the expression of its target genes (Herrington et al., 2000). We assayed the levels of STAT5B phosphorylation on Y699 (used as a marker of STAT5B activation). We found that in the liver of WT mice (Figure 4B), STAT5B phosphorylation significantly changed across the day, with the lowest phosphorylation level at ZT2, which correlates with the lowest level of Igf-1 mRNA transcription, and the highest phosphorylation level at ZT10 and ZT18, which correlates with the highest Igf-1 mRNA level. Of interest, the level of total STAT5B protein did not change significantly across the day, which suggests that regulation occurs predominantly on the level of phosphorylation. In the liver of Cry1, 2−/− mice, STAT5B phosphorylation was significantly reduced. We also observed some small reduction in the level of total STAT5B in the liver of Cry1, 2−/− mice, but this reduction was not significant compared with the effect on STAT5B phosphorylation. The difference in phosphorylation was dramatic even after normalization on total protein level (the quantification of relative STAT5B phosphorylation presented in Figure 4C). We also assayed the expression of other known transcriptional targets of STAT5B—acid-labile subunit (ALS) and IGF-binding protein 3 (IGFBP-3; Woelfle and Rotwein, 2004). As shown in Figure 4A (middle and bottom), ALS and IGFBP-3 expression was significantly reduced in the liver of Cry1, 2−/− mice at several times of the day, in agreement with reduced STAT5B transcriptional activity. In skeletal muscles, we observed similar effects of Cry deficiency on STAT5B phosphorylation (Supplemental Figure 2, B and C). In WT, STAT5B phosphorylation oscillated across the day, with highest level at ZT10-ZT14. Phosphorylation was statistically significantly reduced in Cry1, 2−/− at ZT10 and ZT14 (Supplemental Figure S2B).
STAT5B is phosphorylated by the protein kinase JAK2. JAK2 kinase is activated by GHR upon binding to GH. Activated JAK2 can be monitored by assaying JAK2 phosphorylation on Y1007. Data on JAK2 phosphorylation on T1007 are presented in Figure 4D. In the liver of WT mice, the level of phosphorylation changed across the day with low amplitude. In the liver of Cry1, 2−/− mice, we also observed daily changes in JAK2 phosphorylation. The pattern of JAK2 phosphorylation was different compared with WT, but statistical analysis did not reveal a significant difference. In skeletal muscles of WT mice, JAK2 phosphorylation showed some moderate changes (Supplemental Figure 2, D and E). Of importance, we did not detect any reduction in JAK2 phosphorylation in the tissues of Cry1, 2−/− mice: on the contrary, we observed a significant increase in the phosphorylation at several time points; therefore the reduced phosphorylation of STAT5B was not a consequence of reduced JAK2 phosphorylation.
Expression of CISH is up-regulated in the liver of Cry1, 2−/− mice
Proteins from the suppressors of the cytokine signaling (SOCS) family are negative regulators of JAK2 signaling. SOCS1, SOCS2, SOCS3, and cytokine-inducible SH2-containing protein (CISH or CIS) have been implicated in control of the GHR/JAK2/STAT5B pathway. We analyzed the expression of these proteins in the liver of WT and Cry1, 2−/− mice (Figure 5). We did not detect any significant changes in SOCS2 expression. Expression of SOCS1 and SOCS3 was reduced at some times of the day (at ZT2 for SOCS1 and ZT10 for SOCS3). Expression of CISH was significantly up-regulated at all time points in the CRY-deficient liver, and this change in expression correlated with reduced STAT5B phosphorylation and could be one of the contributing factors. To further understand the mechanism of regulation, we also assayed the expression of mRNA for these genes (Figure 6). We found that, in agreement with protein data, expression of SOCS2 mRNA was not significantly affected by CRY deficiency. Expression of SOCS1 and SOCS3 genes was reduced at some time points. This down-regulation of expression was not surprising; indeed, the transcription of these genes is positively regulated by STAT5B, and therefore their expression is expected to be reduced under conditions of low STAT5B activity. The expression of CISH was significantly increased across the day, which suggests that CRYs—known transcriptional repressors—might regulate CISH on the level of mRNA expression.
FIGURE 5:
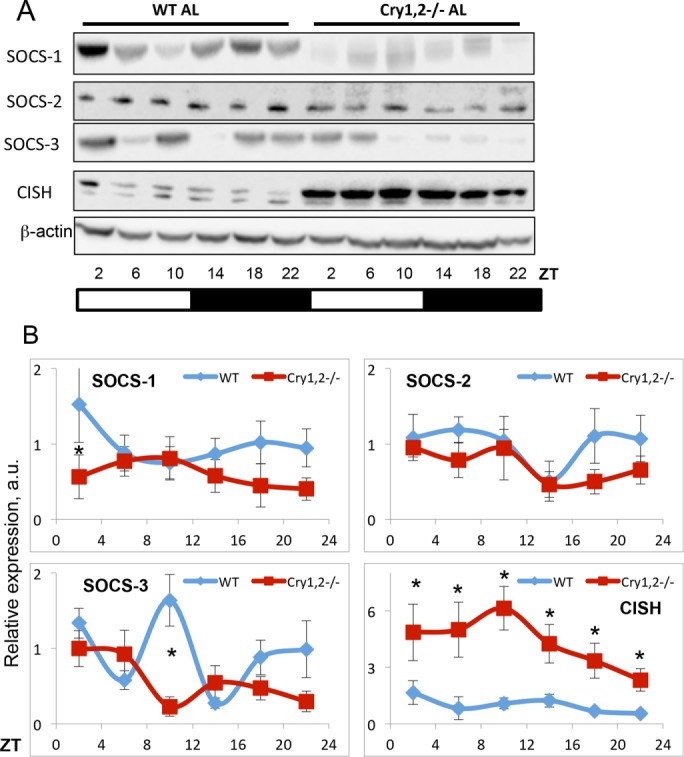
The expression of CISH is up-regulated in the liver of Cry1, 2−/− mice. (A) Representative Western blotting (pooled extracts from three mice per time point) of expression of SOCS1, SOCS2, SOCS3, and CISH in the liver of WT and Cry1, 2−/− male mice. (B) Quantification of expression of SOCS1, SOCS2, SOCS3, and CISH in the liver of WT (blue diamonds) and Cry1, 2−/− (red squares) male mice (N = 3 per time point). *Statistically significant difference between genotypes (p < 0.05). Light was on at ZT0 and off at ZT12.
FIGURE 6:
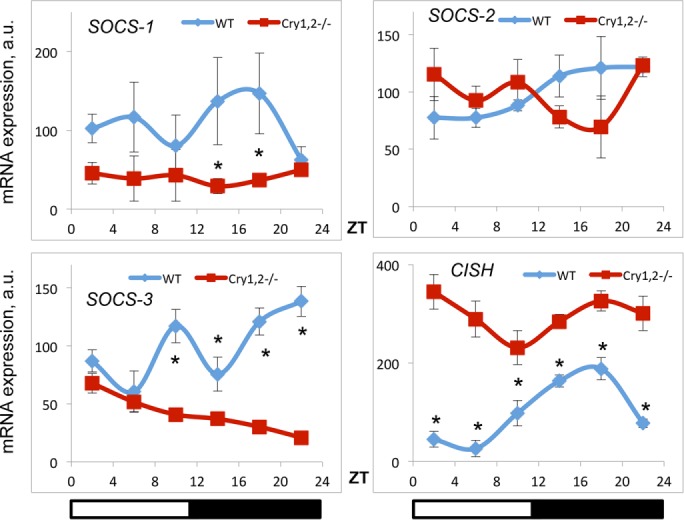
effect of CRY deficiency on the expression of SOCS and CISH mRNA in the liver. Daily profiles of mRNA expression of indicated SOCS family member genes in the liver of WT (blue diamonds) and Cry1, 2−/− (red squares) male mice (N = 3 per time point). *Statistically significant difference between genotypes (p < 0.05). Light was on at ZT0 and off at ZT12.
DISCUSSION
Deciphering the mechanisms of regulation of IGF-1 signaling will contribute to understanding mechanisms of cancer, metabolic diseases, and aging. IGF-1 production is regulated through the GH/GHR/JAK2/STAT5 signaling cascade (Herrington and Carter-Su, 2001). Interaction of GH with GHR leads to activation and autophosphorylation of receptor-associated protein kinase JAK2 and phosphorylation of the receptor. Receptor phosphorylation creates binding sites for STAT5B transcriptional factors that are also phosphorylated by JAK2, and as a result of this phosphorylation, STAT5B factors form a dimer and translocate to the nucleus, where STAT5B drives the expression of target genes, including Igf-1 (Herrington et al., 2000).
We found that CRYs are essential players in the regulation of IGF-1 production and IGF-1 signaling. CRYs are photolyase-like proteins, but in mammals, they do not have any detectable photolyase activity (Sancar, 2000). CRYs are essential for circadian clock functions: indeed, Cry1, 2−/− mice do not demonstrate circadian rhythms in physiology and behavior in constant darkness (van der Horst et al., 1999; Vitaterna et al., 1999). CRYs act as transcriptional repressors and suppress the expression of genes controlled by the BMAL1:CLOCK transcriptional complex (Kume et al., 1999). CRYs also modulate the transcriptional activity of glucocorticoid receptor (Lamia et al., 2011) and CREB (Zhang et al., 2010). We observed that Cry1, 2−/− mice developed dwarfism, manifested by reduced weight and size of all major organs and the body as a whole. The reduced size of Cry1, 2−/− mice is associated with reduced production of IGF-1 by the liver and skeletal muscles and reduced IGF-1 signaling in skeletal muscles. We analyzed plasma IGF-1 levels across the day and observed the circadian nature of rhythms of plasma IGF-1 in WT mice. These rhythms were impaired in Cry1, 2−/− mice. In agreement with the oscillation of plasma IGF-1, we detected daily oscillations of IGF-1 receptor phosphorylation and phosphorylation of its downstream targets PDK1 and AKT. IGF-1R and PDK1 phosphorylation did not show circadian rhythms, whereas AKT phosphorylation was rhythmic. In Cry1, 2−/− mice, phosphorylation of IGF-1R, PDK1, and AKT was significantly reduced; however, phosphorylation of AKT was still rhythmic in the circadian manner. Thus rhythms in the IGF-1 and IGF-1 signaling can be regulated through circadian clock–dependent and –independent mechanisms. For example, there are other biological clocks, such as the food-entrainable oscillator, able to generate 24-h rhythms (Mistlberger, 2011). Bioavailability of IGF-1 is also regulated by IGF-BPs (Leroith, 1996), whose plasma levels might also oscillate during the day; in addition, some internal feedback mechanisms might be involved.
There are several models of dwarfism in mice, which are associated with reduced IGF-1 secretion and reduced IGF-1 signaling (Bartke and Brown-Borg, 2004; Quarrie and Riabowol, 2004). Cry1, 2−/− mice represent a novel model of dwarfism. Indeed, Ames and Snell mice lack cells producing GH in the pituitary; both mice do not produce detectable GH and also do not produce prolactin and thyroid-stimulating hormone. Mice with a mutation in GH-releasing hormone receptor, called “Little” mice, have significantly reduced GH levels. Thus, for all of the aforementioned models, reduced IGF-1 production is secondary to reduced GH level. GHR- deficient mice (Laron mice) have increased circulating GH level but are resistant to the action of GH due to absence of the receptor. These mice were developed as an animal model of human Laron dwarfism syndrome. Thus, in contrast to Ames, Snell, or Little mice, both Laron and Cry1, 2−/− mice have reduced IGF-1 but normal GH levels. The difference between Cry1, 2−/− mice and Laron mice is that Laron mice do not have GHR and downstream signaling; in addition, they lack the GH-binding protein, which is a product of GHR processing, whereas Cry1, 2−/− mice have GHR activation, as judged by JAK2 phosphorylation. In Cry1, 2−/− mice, the daily pattern of JAK2 phosphorylation was affected but there was no reduction in its level, which, to the contrary, was rather increased. Thus activation of JAK2 is not affected by CRY deficiency, in agreement with a report stating that the GH level is not significantly affected in Cry1, 2−/− mice (Bur et al., 2009). Therefore Cry1, 2−/− mice represent the model of reduced GH sensitivity down from the GHR receptor. The body size of Cry1, 2−/− mice was similar to that of mice overexpressing fibroblast growth factor 21 (FGF21; Inagaki et al., 2008). Also similar to Cry1, 2−/−, FGF21 transgenic mice have reduced STAT5B phosphorylation but unaffected JAK2 phosphorylation (Inagaki et al., 2008). It would be interesting to study whether CRYs and FGF21 are part of the same mechanism or regulate IGF-1 production independently. Whereas regulation of IGF-1 production is considered a major outcome of GHR signaling, further study is necessary to find whether all signaling downstream of GHR (Herrington and Carter-Su, 2001) is not affected in these mice or is specific for STAT5 phosphorylation and IGF-1 production.
Circadian disruption is associated with reduction of lifespan in several animal models (Yu and Weaver, 2010). It is unclear whether the rhythms per se or the physiological activity of a particular clock protein are important for lifespan regulation. All of the aforementioned dwarf mice have increased lifespan. Lifespan of Cry1, 2−/− mice is unknown; deficiency of either Cry1 or Cry2 does not significantly affect lifespan in mice (Destici et al., 2013), probably due to partial redundancy of CRY proteins, in agreement with our not detecting any significant effect of single-gene deficiency in IGF-1 plasma level. It will be of interest to determine the lifespan of Cry1, 2−/− mice.
Reduced IGF-1 signaling results in reduced cell proliferation and contributes to the known anticancer activity of CR in rodents (Anisimov and Bartke, 2013). For example, CR reduces the tumor rate of p53-deficient tumors in mice through an IGF-1 dependent mechanism (Dunn et al., 1997). Of interest, mice deficient for p53 and Cry1, 2 genes have significantly delayed tumor formation and increased survival compared with p53−/− mice (Ozturk et al., 2009). This observation was to some extent paradoxical, because disruption of the circadian clocks and rhythms is considered as a risk factor for development of cancer (Schernhammer et al., 2003). The mechanism of Cry1, 2–dependent tumor suppression is not known. On the basis of our data, we propose that reduced IGF-1 signaling upon Cry1, 2 deficiency contributes to the reduction of tumorigenesis in p53/Cry1, 2 triple-knockout mice. However, Cry1, 2 deficiency does not improve survival of mice in the model of chemically induced hepatocellular carcinoma (Lee et al., 2010). Therefore the effect of CRYs on tumorigenesis might be tumor type dependent and requires more study.
We observed that in the liver and in skeletal muscle of Cry1, 2−/− mice, the expression of Igf-1 was reduced on both mRNA and protein levels, which correlated with the reduced level of the circulating IGF-1 in the blood of these mice. We cannot exclude increased turnover of the circulating IGF-1 due to changes in the production of IGF-1-binding proteins that regulate IGF-1 bioavailability and half-life. Here we provide evidence that CRYs are involved in the control of Igf-1 expression. Figure 7 presents the molecular model of IGF-1 production through a CRY-dependent mechanism.
FIGURE 7:

Model of regulation of Igf-1 expression by CRYs in response to GH stimulation. Solid lines represent previously reported activation of STAT5B phosphorylation by JAK2 and suppression of this phosphorylation by members of the SOC family and/or protein tyrosine phosphatases (PTPs). CRYs might directly interact with JAK2-containing complexes and stimulate JAK2 activity toward STAT5B; CRYs might recruit STAT5B to the JAK2/GHR complex; and CRYs might also suppress SOC or PTP expression/activity. Thus CRYs stimulate STAT5B phosphorylation and activation, which in turn drives Igf-1 expression in the liver and other tissues.
Phosphorylation of JAK2 was not reduced; therefore CRYs are acting between JAK2 and STAT5. The level of STAT5 phosphorylation is controlled by several known negative regulators of JAK signaling. Members of the SOCS family of proteins play major roles in down-regulation of GH/JAK2 signaling and act through several different mechanisms by blocking interaction between JAK2 and its targets, suppressing JAK2 kinase activity or regulating the turnover rate for JAK2 (Flores-Morales et al., 2006). We found that CRYs regulate the expression of CISH on the mRNA and protein levels and that increased expression of CISH can be a contributing factor to the observed reduced STAT5B phosphorylation. We also cannot exclude the other possible mechanisms illustrated in Figure 7. One possibility is that CRYs directly interact with JAK2, STAT5, or GHR and either suppress JAK2 kinase activity or prevent formation of the GHR/JAK2/STAT5 complex, which is necessary for STAT5 phosphorylation (Herrington et al., 2000). It is known that CRYs directly interact with several proteins: PER1 and PER2 (Ye et al., 2014), the BMAL1/CLOCK complex (Kume et al., 1999), and glucocorticoid receptor (Lamia et al., 2011). Interaction with CRYs affects the activity and posttranslational modifications of these proteins. Protein tyrosine phosphatases such as PTP1B and PTP-1H dephosphorylate JAK2 targets (Gu et al., 2003). CRYs, known transcriptional suppressors, might control their mRNA expression or regulate their activities through direct or indirect interaction. Further study will determine the molecular mechanisms of CRY-dependent regulation.
GH/IGF-1 signaling plays an important role in tissue homeostasis. Too high or too low a level of IGF-1 will affect cell metabolism and proliferation, compromising tissue-regenerative ability or contributing to cancer progression (Anisimov and Bartke, 2013; Chia, 2014). IGF-1 production is tightly regulated, and understanding mechanisms of Igf-1 expression under physiological or stress conditions is important to develop strategies for somatotropic axis manipulations. We reported here that integral components of the circadian clock CRY proteins regulate IGF-1 production through STAT5-dependent mechanism.
MATERIALS AND METHODS
Experimental animals
Wild-type and Cry1, 2−/− mice were previously described (Vitaterna et al., 1999). Mice of all genotypes were of C57B6J background. Mice were maintained on the 12:12 light:dark cycle with lights on at 7:00 am. All groups had unrestricted access to the 18% protein rodent diet (Harlan) and water. All tissue collection experiments were performed on 5-mo-old mice. For all experiments, at least three animals of each genotype and gender were used, with at least three animals per time point. All animal studies were performed with approval from and within the guidelines of the Institutional Animal Care and Use Committee of Cleveland State University.
RNA isolation and analysis of mRNA expression
For gene expression studies, tissues were collected every 4 h throughout the day and stored at −80°C. Total RNA was isolated from the liver using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. RNA quantification was performed using real-time reverse transcription PCR with Universal SYBR Green Mix (Bio-Rad, Hercules, CA) as described in Khapre et al. (2014b). Primers used for the analysis of mRNA expression are listed in Supplemental Table S1.
Analysis of protein expression and phosphorylation
For the analysis of protein expression, tissues were collected every 4 h throughout the day and stored at −80°C. For representative Western blotting, three liver samples from three different mice were pooled at each time point. For quantitative data, liver samples were run individually to estimate variability among biological replicates. For lysate preparation, frozen liver pieces were lysed in Cell Signaling lysis buffer with Protease/Phosphatase Inhibitor Cocktail (Cell Signaling Technology, Beverly, MA) using a sonicator. Protein concentration was determined with the aid of the Bradford protein assay kit according to the manufacturer's protocol, after which lysates were stored at −20°C. Protein (45 μg) was loaded into 3–8% Tris-acetate and 4–12% Bis-Tris gels (Invitrogen). After SDS–PAGE, protein was transferred on the polyvinylidene fluoride membrane at 110 mA. Equal loading of proteins was checked by Ponceau staining. The membranes were probed with the primary antibodies listed in the Supplemental Materials. Probing the same membranes with anti–β-actin or anti–glyceraldehyde-3-phosphate dehydrogenase antibodies was used for normalization of the signal. Images were obtained with Odyssey FC imaging system (LI-COR, Lincoln, NE), and quantification was performed with the Odyssey FC imaging system, version 3.0, software.
Measurement of the plasma IGF-1 level
Plasma samples were collected every 4 h throughout the day and stored at −80°C. Plasma IGF-1 levels were determined using a RayBio Mouse IGF-1 96-well plate enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's protocol. Plasma samples were diluted 50-fold with the solution provided by the manufacturer. Sandwich ELISA was done using two different antibodies; the second antibody was conjugated with biotin. For detection, the manufacturer-provided streptavidin-conjugated horseradish peroxidase and TMB substrate were used for colorimetric reaction, and optical density was detected at 450 nm. Manufacturer-provided mouse IGF-1 was used to generate the standard concentration curve; starting concentration was 2 ng/ml, with serial dilutions. All experimental samples and standards were run in duplicate on the same plate; experiments were repeated three times with different plates. The intraassay coefficient of variation was between 4 and 9% for different plates; interassay coefficient of variation was 12%. The detection limit for IGF-1 measurement was 4 pg/ml.
Statistical analysis
For all experiments, we used at least three animals for every time point for each feeding type and each genotype. Data are shown as average ± SD. IBM SPSS Statistics 20 and GraphPad Prism Version 5.04 software packages were used for statistical analysis. To assay the effects of genotypes and the time of the day, the analysis was performed using two-way or one-way analysis of variance, followed by pairwise comparison of genotypes over time. p < 0.05 was considered as a statistically significant difference.
Supplementary Material
Acknowledgments
This work was supported by Grant R01AG039547 from the National Institutes of Health and funds from the Center for Gene Regulation and Health and Disease at Cleveland State University to R.V.K. and a Dissertation Research Award from Cleveland State University to S.P.
Abbreviations used:
- ALS
acid-labile subunit
- CISH
cytokine-inducible SH2-containing protein
- CRY
cryptochrome
- FOXO
forkhead box O transcription factor
- GH
growth hormone
- IGF1
insulin-like growth factor 1
- IGF1R
insulin-like growth factor 1 receptor
- IGFBP3
IGF-binding protein 3
- JAK2
Janus kinase 2
- Murf1
muscle RING-finger protein-1
- SOCS
suppressor of cytokine signaling
- STAT
signal transducers and activators of transcription
- WT
wild type
- ZT
zeitgeber.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-08-0624) on January 18, 2017.
REFERENCES
- Anisimov VN, Bartke A. The key role of growth hormone-insulin-IGF-1 signaling in aging and cancer. Crit Rev Oncol Hematol. 2013;87:201–223. doi: 10.1016/j.critrevonc.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A, Brown-Borg H. Life extension in the dwarf mouse. Curr Top Dev Biol. 2004;63:189–225. doi: 10.1016/S0070-2153(04)63006-7. [DOI] [PubMed] [Google Scholar]
- Bur IM, Cohen-Solal AM, Carmignac D, Abecassis PY, Chauvet N, Martin AO, van der Horst GTJ, Robinson IC, Maurel P, Mollard P, Bonnefont X. The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J Biol Chem. 2009;284:9066–9073. doi: 10.1074/jbc.M808360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Su C, Rui L, Herrington J. Role of the tyrosine kinase JAK2 in signal transduction by growth hormone. Pediatr Nephrol. 2000;14:550–557. doi: 10.1007/s004670000366. [DOI] [PubMed] [Google Scholar]
- Chia DJ. Minireview: mechanisms of growth hormone-mediated gene regulation. Mol Endocrinol. 2014;28:me20141099. doi: 10.1210/me.2014-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmons DR. Metabolic actions of insulin-like growth factor-I in normal physiology and diabetes. Endocrinol Metab Clin North Am. 2012;41:425–443. doi: 10.1016/j.ecl.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardente H, Cermakian N. Molecular circadian rhythms in central and peripheral clocks in mammals. Chronobiol Int. 2007;24:195–213. doi: 10.1080/07420520701283693. [DOI] [PubMed] [Google Scholar]
- Destici E, Jacobs EH, Tamanini F, Loos M, Van der Horst GTJ, Oklejewicz M. Altered phase-relationship between peripheral oscillators and environmental time in Cry1 or Cry2 deficient mouse models for early and late chronotypes. PLoS One. 2013;8:e83602. doi: 10.1371/journal.pone.0083602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dunn SE, Kari FW, French J, Leininger JR, Travios G, Wilson R, Barrett JC. Dietary restriction reduces insulin-like growth factor I levels, which modulates apoptosis, cell proliferation, and tumor progression in p53-deficient mice. Cancer Res. 1997;57:4667–4672. [PubMed] [Google Scholar]
- Dupont J, Holzenberger M. Biology of insulin-like growth factors in development. Birth Defects Res C Embryo Today. 2003;69:257–271. doi: 10.1002/bdrc.10022. [DOI] [PubMed] [Google Scholar]
- Flores-Morales A, Greenhalgh CJ, Norstedt G, Rico-Bautista E. Negative regulation of growth hormone receptor signaling. Mol Endocrinol. 2006;20:241–253. doi: 10.1210/me.2005-0170. [DOI] [PubMed] [Google Scholar]
- Froy O, Miskin R. Effect of feeding regimens on circadian rhythms: implications for aging and longevity. Aging (Albany NY) 2010;2:7–27. doi: 10.18632/aging.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Dubé N, Kim JW, Cheng A, Maria de J Ibarra-Sanchez, Tremblay ML, Boisclair YR. Protein tyrosine phosphatase 1B attenuates growth hormone-mediated JAK2-STAT signaling. Mol Cell Biol. 2003;23:3753–3762. doi: 10.1128/MCB.23.11.3753-3762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J, Carter-Su C. Signaling pathways activated by the growth hormone receptor. Trends Endocrinol Metab. 2001;12:252–257. doi: 10.1016/s1043-2760(01)00423-4. [DOI] [PubMed] [Google Scholar]
- Herrington J, Smit LS, Schwartz J, Carter-Su C. The role of STAT proteins in growth hormone signaling. Oncogene. 2000;19:2585–2597. doi: 10.1038/sj.onc.1203526. [DOI] [PubMed] [Google Scholar]
- Hwa V, Nadeau K, Wit JM, Rosenfeld RG. STAT5b deficiency: lessons from STAT5b gene mutations. Best Pract Res Clin Endocrinol Metab. 2011;25:61–75. doi: 10.1016/j.beem.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Lin VY, Goetz R, Mohammadi M, Mangelsdorf DJ, Kliewer S. Inhibition of growth hormone signaling by the fasting-induced hormone FGF21. Cell Metab. 2008;8:77–83. doi: 10.1016/j.cmet.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junnila RK, List EO, Berryman DE, Murrey JW, Kopchick JJ. The GH/IGF-1 axis in ageing and longevity. Nat Rev Endocrinol. 2013;9:366–376. doi: 10.1038/nrendo.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, McCabe JM, Byrne PO, Connacher MK, Wang Z, Ramek A, Sarabipour S, Shan Y, Shaw DE, Hristova K, et al. How IGF-1 activates its receptor. Elife. 2014;3:7554/eLife.03772. doi: 10.7554/eLife.03772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, Kondratov RV. BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging (Albany NY) 2014a;6:48–57. doi: 10.18632/aging.100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khapre RV, Patel S, Kondratova AA, Chauhary A, Vilingkaar N, Antoch M, Kondratov RV. Metabolic clock generates nutrient anticipation rhythms in mTOR signaling. Aging (Albany NY) 2014b;6:675–689. doi: 10.18632/aging.100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13:325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanning NJ, Carter-Su C. Recent advances in growth hormone signaling. Rev Endocr Metab Disord. 2006;7:225–235. doi: 10.1007/s11154-007-9025-5. [DOI] [PubMed] [Google Scholar]
- Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One. 2010;5:e10995. doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroith D. Insulin-like growth factor receptors and binding proteins. Baillieres Clin Endocrinol Metab. 1996;10:49–73. doi: 10.1016/s0950-351x(96)80298-9. [DOI] [PubMed] [Google Scholar]
- Levi F, Filipski E, Iurisci I, Li XM, Innominato P. Cross-talks between circadian timing system and cell division cycle determine cancer biology and therapeutics. Cold Spring Harb Symp Quant Biol. 2007;72:465–475. doi: 10.1101/sqb.2007.72.030. [DOI] [PubMed] [Google Scholar]
- Liu JL, Grinberg A, Westphal H, Sauer B, Accili D, Karas M, LeRoith D. Insulin-like growth factor-I affects perinatal lethality and postnatal development in a gene dosage-dependent manner: manipulation using the Cre/loxP system in transgenic mice. Mol Endocrinol. 1998;12:1452–1462. doi: 10.1210/mend.12.9.0162. [DOI] [PubMed] [Google Scholar]
- Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106:447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE. Neurobiology of food anticipatory circadian rhythms. Physiol Behav. 2011;104:535–545. doi: 10.1016/j.physbeh.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci USA. 2009;106:2841–2846. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie JK, Riabowol KT. Murine models of life span extension. Sci Aging Knowledge Environ. 2004;2004:re5. doi: 10.1126/sageke.2004.31.re5. [DOI] [PubMed] [Google Scholar]
- Rosenfeld RG, Hwa V. The growth hormone cascade and its role in mammalian growth. Horm Res. 2009;71(Suppl 2):36–40. doi: 10.1159/000192434. [DOI] [PubMed] [Google Scholar]
- Rother KI, Accili D. Role of insulin receptors and IGF receptors in growth and development. Pediatr Nephrol. 2000;14:558–561. doi: 10.1007/s004670000351. [DOI] [PubMed] [Google Scholar]
- Rotwein P. Mapping the growth hormone-Stat5b-IGF-I transcriptional circuit. Trends Endocrinol Metab. 2012;23:186–192. doi: 10.1016/j.tem.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotwein P, Chia DJ. Gene regulation by growth hormone. Pediatr Nephrol. 2010;25:651–658. doi: 10.1007/s00467-009-1258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu Rev Biochem. 2000;69:31–67. doi: 10.1146/annurev.biochem.69.1.31. [DOI] [PubMed] [Google Scholar]
- Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, Fuchs CS, Colditz GA. Night-shift work and risk of colorectal cancer in the nurses' health study. J Natl Cancer Inst. 2003;95:825–828. doi: 10.1093/jnci/95.11.825. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet Muscle. 2011;1:4. doi: 10.1186/2044-5040-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, Van der Horst GTJ, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Ullrich A, Gray A, Tam AW, Yang-Feng T, Tsubokawa M, Collins C, Henzel W, Le Bon T, Kathuria S, Chen E. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 1986;5:2503–2512. doi: 10.1002/j.1460-2075.1986.tb04528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Varco-Merth B, Rotwein P. Differential effects of STAT proteins on growth hormone-mediated IGF-I gene expression. Am J Physiol Endocrinol Metab. 2014;307:E847–E855. doi: 10.1152/ajpendo.00324.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vottero A, Guzzetti C, Loche S. New aspects of the physiology of the GH-IGF-1 axis. Endocrine Dev. 2013;24:96–105. doi: 10.1159/000342573. [DOI] [PubMed] [Google Scholar]
- Woelfle J, Rotwein P. In vivo regulation of growth hormone-stimulated gene transcription by STAT5b. Am J Physiol Endocrinol Metab. 2004;286:E393–E401. doi: 10.1152/ajpendo.00389.2003. [DOI] [PubMed] [Google Scholar]
- Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Selby CP, Chiou Y-Y, Ozkan-Dagliyan I, Gaddameedhi S, Sancar A. Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev. 2014;28:1989–1998. doi: 10.1101/gad.249417.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EA, Weaver DR. Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging (Albany NY) 2010;3:479–493. doi: 10.18632/aging.100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.