Abstract
Progranulin (PGRN) restrains inflammation and is therapeutic against inflammatory arthritis; however, the underlying immunological mechanism remains unknown. In this study, we demonstrated that anti-inflammatory cytokine IL-10 was a critical mediator for PGRN-mediated anti-inflammation in collagen-induced arthritis by using PGRN and IL-10 genetically modified mouse models. IL-10 green fluorescent protein reporter mice revealed that regulatory T (Treg) cells were the predominant source of IL-10 in response to PGRN. In addition, PGRN-mediated expansion and activation of Treg cells, as well as IL-10 production, depends on JNK signaling, but not on known PGRN-activated ERK and PI3K pathways. Furthermore, microarray and chromatin immunoprecipitation sequencing screens led to the discovery of forkhead box protein O4 and signal transducer and activator of transcription 3 as the transcription factors required for PGRN induction of IL-10 in Treg cells. These findings define a previously unrecognized signaling pathway that underlies IL-10 production by PGRN in Treg cells and present new insights into the mechanisms by which PGRN resolves inflammation in inflammatory conditions and autoimmune diseases, particularly inflammatory arthritis.—Fu, W., Hu, W., Shi, L., Mundra, J. J. Xiao, G., Dustin, M. L., Liu, C. Foxo4- and Stat3-dependent IL-10 production by progranulin in regulatory T cells restrains inflammatory arthritis.
Keywords: TNFR2, JNK, Treg
Progranulin (PGRN), a 593-aa secretory anti-inflammatory growth factor, is an extracellular glycoprotein that contains 7 and one-half cysteine-rich motif repeats. PGRN is abundantly expressed in cycling epithelial cells, immune cells, chondrocytes, and neurons (1, 2). Although PGRN is most known for its neuroprotective and proliferative properties implicated in neurodegenerative disease (3–5), PGRN is involved in diverse biological processes, including early embryogenesis (6), wounding healing (7), inflammation (8–15), cartilage development and degradation (2, 16–20), and tumorigenesis (1).
Our previous studies demonstrated that PGRN plays a critical role in the regulation of inflammatory arthritis, as evidenced by the discovery that in vivo systematic treatment with PGRN reverses the severe inflammatory arthritis seen in PGRN-deficient mice and significantly delays the onset of the arthritic phenotype that is characteristic of TNF transgenic mice (11). Moreover, PGRN-mediated immunosuppression during the course of collagen-induced arthritis (CIA) may be attributable to the up-regulation of regulatory T (Treg) cells and IL-10 production, as suggested by observations that PGRN selectively up-regulates forkhead box protein P3 (Foxp3) and promotes Treg differentiation in vitro, and that elevated IL-10 serum levels were observed in PGRN-treated CIA mice (11). However, the immunological mechanism that underlies PGRN-mediated therapeutic effects in inflammatory arthritis and, in particular, the origin of IL-10–producing cells and the molecular regulation of PGRN-mediated IL-10 production, remains unknown. In this study, we identify IL-10 as a necessary mediator of PGRN-induced anti-inflammatory effect. We also establish a mechanistic link between PGRN and TNF receptor 2 (TNFR2) signaling in the regulation of Treg cells and IL-10 via promoting phosphorylation of JNK. Furthermore, forkhead box protein O4 (Foxo4), which has not been implicated in IL-10 transcription previously, together with signal transducer and activator of transcription 3 (Stat3), known to regulate IL-10 transcription in other cell types, cooperate to govern IL-10 production in response to PGRN.
MATERIALS AND METHODS
Mice
DBA1J, TNFR2−/−, IL-10−/−, IL-10 green fluorescent protein (GFP), and Foxo4F/F and Stat3F/F mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). PGRN-deficient mice were maintained in the laboratory (11). All animals were maintained in a specific pathogen–free environment on a B6 background and were sex- and age-matched for experiments, typically between 8 and 10 wk of age. All animal studies were performed in accordance with institutional guidelines and approved by the Institutional Animal Care and Use Committee of New York University.
Preparation of rhPGRN
Generation of our recombinant PGRN stable cell line and purification of recombinant PGRN have been described in our previous publications (2). In brief, stable cells were cultured in DMEM that contained 1 mg/ml G418. PGRN was affinity-purified from the medium of starved cells by using nickel-nitrilotriacetic-agarose. The purity of recombinant PGRN was determined by SDS-PAGE.
CIA model
Eight-week-old mice were immunized via 0.1-ml intradermal injection of 100 μg chicken type II collagen (Chondrex, Seattle, WA, USA) emulsified with an equal volume of complete Freund's adjuvant (CFA) that contained 4 mg/ml heat-denatured Mycobacterium (Chondrex) at the base of the tail (d 0). In CIA mouse model, clinical signs of arthritis in the paws were evaluated and scored individually by using a 0–4 point scoring system. Scores from each individual paw were summed to yield an overall score for each mouse, with a maximum score of 16 (21). Scores were attributed as follows: a paw score of 0, no signs; 1, mild swelling confined to the tarsal bones or ankle joint; 2, mild swelling extending from ankle to the tarsal bones; 3, moderate swelling extending from ankle to the metatarsal joints; and 4, severe swelling encompassing the ankle, foot, and digits and/or ankylosis of the limb. To determine therapeutic effects, recombinant PGRN (5 mg/kg body weight) was intraperitoneally injected into mice with established mild arthritis (clinical score 1–2) on alternating days until euthanasia.
Histopathological examination of joints
Mouse joint tissues were fixed in 4% paraformaldehyde, decalcified in EDTA, and embedded in paraffin. Tissue sections were then prepared and stained with hematoxylin and eosin (H&E) or Safranin O staining to detect proteoglycans. H&E-stained sections were scored for inflammation and bone erosion. Inflammation was scored according to the following criteria: 0, no inflammation; 1, slight thickening of the lining layer or some infiltrating cells in the underlying layer; 2, slight thickening of the lining layer plus some infiltrating cells in the underlying layer; 3, thickening of the lining layer, an influx of cells in the underlying layer, and presence of cells in the synovial space; and 4, synovium highly infiltrated with many inflammatory cells. Cartilage damage was determined by using Safranin O staining, and the extent of cartilage damage was scored according to the following criteria: 0, no destruction; 1, minimal erosion limited to single spots; 2, slight-to-moderate erosion in a limited area; 3, more extensive erosion; and 4, general destruction (22).
Flow cytometry analysis
Single-cell suspensions from draining lymph nodes or spleen were subjected to flow cytometry using the following Abs: FITC-conjugated anti-CD4, PE-conjugated anti-CD25, eFluor 450–conjugated anti–IL-17, Alexa Fluor 700–conjugated anti–IL-10, and PE-conjugated anti-CD120b (BD Biosciences, Brea, CA, USA); biotin-–conjugated anti-Foxp3, APC-conjugated anti-CD25, eFluor 450–conjugated anti-CD11c, Alexa Fluor 700–conjugated anti-CD 19, APC-eFluor 780–conjugated anti-F4/80, PE-cyanine5–conjugated anti-CD4, and PE-cyanine7–conjugated anti-CD8 (eBioscience, San Diego, CA, USA); and streptavidin-conjugated Qdot 605 (Thermo Fisher Scientific, Waltham, MA, USA). Cells were acquired by using an LSRII (BD Biosciences) and data were analyzed by using FlowJo (Tree Star, Ashland, OR, USA).
T-cell purification and activation
Total CD4+ cells were separated from the spleens of 8-wk-old mice by using a CD4+ T-cell isolation kit (Miltenyi Biotec, San Diego, CA, USA). Isolated cells were activated with plate-bound anti-CD3 Ab (2 µg/ml) and soluble anti-CD28 Ab (2 µg/ml), then induced to differentiate into Treg cells with TGF-β (5 ng/ml) and recombinant mouse IL-2 (50 U/ml) with or without PGRN (200 ng/ml).
Quantification of cytokines
ELISA kits were obtained from R&D Systems (Madison, WI, USA). Cytokines were quantified in culture supernatant or in sera. For analysis of cytokine production by splenocytes, supernatants were collected from CD4+ cell culture after treatment with TGF-β with or without PGRN.
Gene expression profiling and analysis
CD4+ T cells were isolated from spleens of wild-type (WT) mice as previously described. RNA was isolated after 24-h activation using CD3 and CD28 Abs with or without PGRN. RNA integrity was assessed by standard denaturing agarose gel electrophoresis. Mouse 4 × 44 Gene expression Array, v2 (Agilent Technology, Santa Clara, CA, USA) with approximately ≥39,000 mouse genes and transcripts represented, all with public domain annotations, was employed for genomic profiling. Agilent Feature Extraction software (ver. 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data analysis were performed by using the bioconductor/limma package (23).
Real-time PCR
RNA was extracted with RNeasy mini kit (Qiagen, Valencia, CA, USA) and cDNA was transcribed by using the Reverse Transcription kit (Improm-II Reverse Transcription System; Promega, Madison, WI, USA) according to manufacturer instructions. Real-time PCR reactions were performed in multiple replicates and run on StepOnePlus Real-time PCR systems (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR mastermix (Applied Biosystems). Results are reported as δ Ct, the difference between the threshold cycle (Ct) for a sequence of interest and of a house-keeping gene.
Luciferase assays
Foxo4 or Stat3 expression plasmid was obtained from Origene (Rockville, MD, USA) or Thermo Fisher Scientific, respectively. Full-length and sequential truncated Il10 promoter fragments were PCR-amplified and cloned into luciferase vector pGL3-Basic (Promega). Jurkat T cells were transfected with Il10 promoter luciferase reporter constructs and a control dual luciferase Renilla construct, together with Foxo4 and/or Stat3 expression plasmids for 24 h. Cells were then collected and lysed to measure luciferase activity.
Identification of conserved noncoding regions
The University of California, Santa Cruz (UCSC) Genome Browser (Santa Cruz, CA, USA) was used to identify the conserved noncoding regions that surrounding Il10 locus among 100 vertebrates.
Chromatin immunoprecipitation and chromatin immunoprecipitation sequencing
Anti-Foxo4, normal goat IgG, and normal rabbit IgG were acquired from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-Stat3 Ab was obtained from EMD Millipore (Billerica, MA, USA). CD4+ T cells isolated from spleen of WT mice were treated with PGRN for 24 h followed by fixation with 1% formaldehyde for 10 min at room temperature and incubation for 10 min in 0.125 M glycine. Chromatin immunoprecipitation (ChIP) was carried out according to manufacturer instructions (Thermo Fisher Scientific). In ChIP-quantitative PCR analyses, values from immunoprecipitated samples were normalized to that from input DNA. ChIP sequencing (ChIP-seq) libraries preparation and sequencing was done at the New York University Genome Technology Center. Sequence reads were mapped to the mouse genome (mm10) with Bowtie2 program (24). Duplicate sequences were removed by using the Picard package (http://picard.sourceforge.net/) and peaks were called using a model-based analysis of MACS2 (25). Peaks were then annotated by Homer peak annotation program (26).
Statistical analyses
Statistical analyses were performed by using 2-tailed Student’s t tests for independent samples. A value of P < 0.05 was considered statistically significant. All data are presented as means ± sd.
RESULTS
IL-10 is a critical mediator of PGRN-mediated anti-inflammation
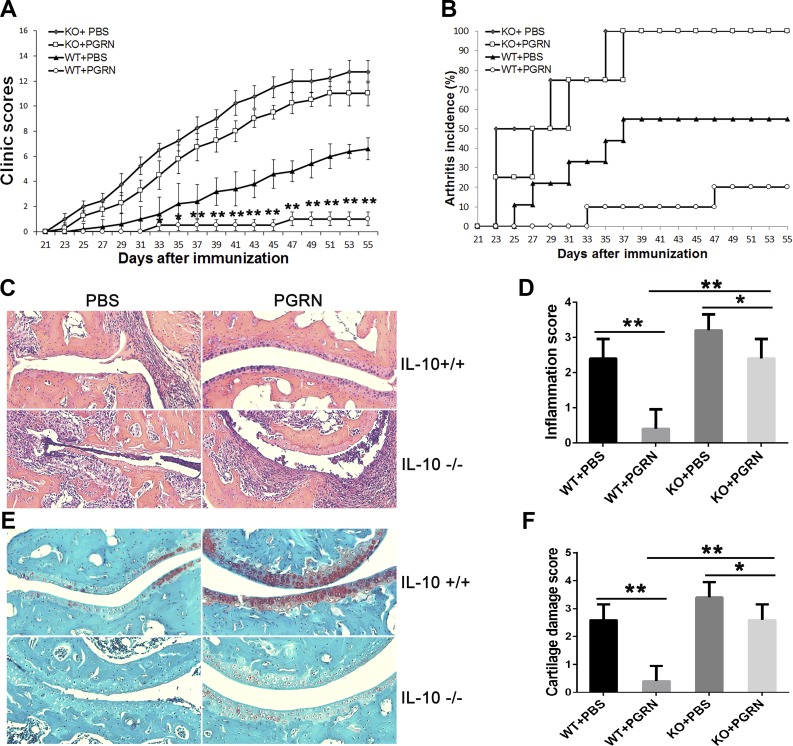
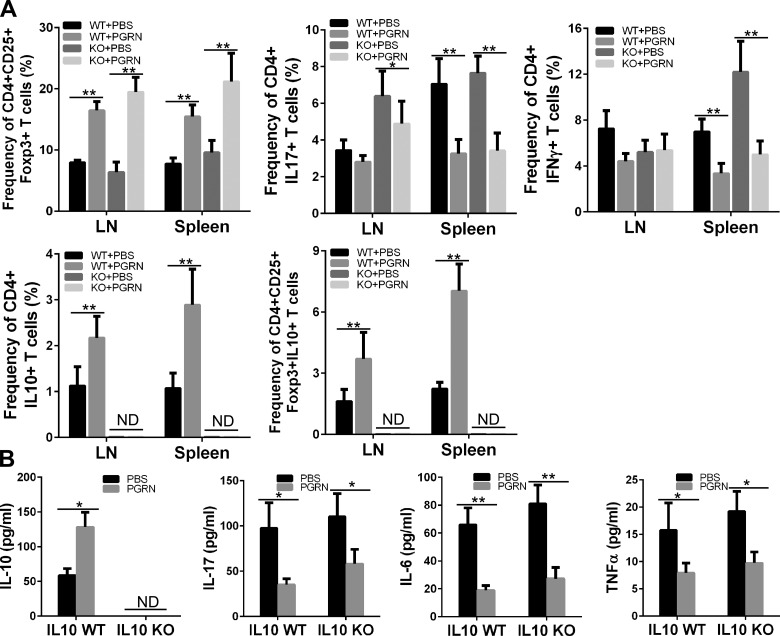
We previously reported that PGRN protects WT mice from CIA and that serum levels of IL-10 were selectively elevated in PGRN-treated mice among cytokine tested (11). We sought to determine whether induced IL-10 by PGRN is important for the protective action of PGRN in CIA. We first confirmed that intraperitoneal injection of PGRN effectively prevented the development of inflammatory arthritis after CIA induction by injection of chicken type II collagen in CFA in DBA1J mice, as evidenced by a decreased arthritis clinical score and lower incidence of disease (Supplemental Fig. 1). Consistent with these results, histological findings in the joint tissue after staining with H&E and Safranin O showed less inflammatory infiltration and attenuated cartilage damage in collagen/CFA-injected mice that were treated with PGRN compared with PBS-treated control group (Supplemental Fig. 2). To evaluate the consequences of interfering with IL-10 in joint inflammation and to determine whether the therapeutic effect of PGRN on CIA is mediated by IL-10, we then employed the CIA model in IL-10−/− mice or IL-10+/+ mice on C57BL/6J genetic background. IL-10−/− mice displayed increased severity of CIA compared with IL-10+/+ mice, evidenced by severe swelling, erythema, and joint rigidity in the hind paws upon immunization, which is consistent with previous reports that CIA is exacerbated in IL-10−/− mice (27, 28). Of note, IL-10+/+ C57BL/6J mice developed arthritis 4 d later and eventually reached severity at a lower level compared with DBA1J mice, which is consistent with previous reports (29). However, IL-10−/− C57BL/6J mice developed arthritis 2 d later compared with DBA1J mice, and eventually reached severity at a level comparable to arthritis in DBA1J mice. Of note, clinical arthritis score and incidence of arthritis was significantly lower in PGRN-treated IL-10+/+ mice compared with PBS-treated control IL-10+/+ group (Fig. 1A, B). Although PGRN suppressed CIA between d 31 and 47 in IL-10−/− mice in which CIA was induced, the effect size was significantly smaller than in IL-10+/+ mice in which we attempted to induce CIA. In addition, PGRN could only slow the onset of CIA but could not reduce the incidence of arthritis in IL-10−/− mice (Fig. 1A, B). Consistent with these results, histological analysis of joint tissue after H&E and Safranin O staining revealed less inflammatory infiltration and attenuated cartilage damage in PGRN-treated IL-10+/+ and IL-10−/− mice, although the effect in IL-10−/− mice was to a much lesser degree (Fig. 1C–F). Of note, PGRN-treated IL-10−/− mice displayed an increased frequency of CD4+CD25+Foxp3+ Treg cells and a decreased frequency of CD4+IL17+ T cells and CD4+IFN-γ+ T cells, concomitant with the secretion of lower levels of proinflammatory cytokines IL-17 and IFN-γ, similar to those observed in IL-10+/+ mice (Fig. 2). However, as expected, the serum level and Treg-cell–associated IL-10 were undetectable in IL-10−/− mice. In contrast, both the serum level and Treg-cell–associated IL-10 were significantly up-regulated in PGRN-treated IL-10+/+ mice (Fig. 2). Given that Treg cells can function by mechanisms other than IL-10 production (30, 31), it is not surprising that increased Treg-cell numbers in PGRN-treated mice in which CIA was induced have some anti-inflammatory effects. Nevertheless, the fact that the effect of PGRN is largely blocked in IL-10−/− mice highlights IL-10 as a crucial mediator of PGRN’s effect on CIA. Of more importance, these findings imply that the therapeutic effect of PGRN is attributable to increased IL-10–dependent Treg-cell function rather than simply increasing Treg-cell number.
Figure 1.
Therapeutic effects of PGRN on CIA are IL-10 dependent. A) WT and IL−/− mice (n = 12 per group) were immunized with type II collagen to establish the CIA model, then mice were treated with PBS or PGRN for a total of 5 wk starting from the onset of disease. Clinical arthritis scores were recorded. B) Incidence of arthritis in each treatment group with genetic background indicated. C, D) Ankle joint tissues were obtained from each group on wk 8 after immunization and stained with H&E (C) to assess the severity of inflammation (D). E, F) Ankle joint tissues were stained with Safranin O staining (E) to evaluate the cartilage damage (F). KO, knockout. Data are presented as means ± sd. *P < 0.05; **P < 0.01 vs. PBS-treated control group.
Figure 2.
PGRN increased Treg cells in CIA independent of IL-10. A) FACS analysis of Treg cells, CD4+IL17+ and CD4+IFN-γ+ T cells, and IL-10–producing CD4+ T cells and Treg cells in the spleens and lymph nodes (LNs) of CIA mice treated with or without PGRN (n = 12 per group). Bar graphs represent means ± sd of total mice per group. B) ELISA analysis of sera levels of IL-10, IL-17, IL-6, and TNF-α in CIA mice treated with or without with PGRN for 5 wk. KO, knockout; ND, nondetectable. Values are means ± sd. *P < 0.05; **P < 0.01 vs. PBS-treated control group.
Treg cells are the predominant new source of IL-10 in response to PGRN
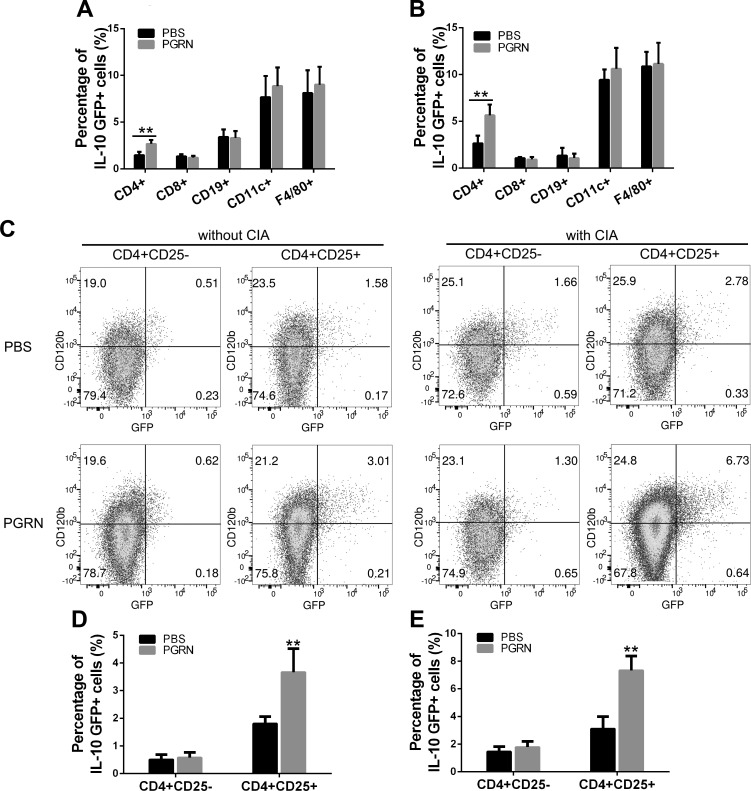
Although IL-10 has been implicated in the regulation of inflammation and autoimmune diseases, the source and stimuli that drive its production have not yet been defined. To determine the cellular source of IL-10 in response to PGRN, we used VERT-X IL-10eGFP reporter mice that encode enhanced GFP in the 3′-untranslated region of Il10 (32). Mice were treated with PGRN (100 µg, i.p. per mouse) every other day for a total of 2 wk, and GFP expression was determined in subpopulations of T cells, B cells, macrophages, and dendritic cells. Of interest, IL-10eGFP was only increased in CD4+ T cells. Other cell populations displayed no shift in GFP intensity (Fig. 3A and Table 1). Of note, this observation was consistent under inflammatory and physiological conditions (Fig. 3B and Table 1). In addition, among CD4+ cells, PGRN treatment stimulated a significant proportion of CD4+CD25+ T cells to coexpress IL-10, whereas no increase was observed in CD4+CD25− T cells, which indicated that Treg cells were the predominant new source of IL-10 source upon PGRN treatment in vivo under physiological conditions without any inflammatory challenge (Fig. 3C, D and Table 1). Significantly, the majority of Treg cells coexpressing IL-10 (>90%) also coexpressed TNFR2, a marker of activated potent suppressor CD4+CD25+FoxP3+ Treg cells (33) (Fig. 3C). Accordingly, we found that in PGRN-treated WT mice in which CIA induction was attempted, Treg cells coexpressing TNFR2 were also the predominant new source of IL-10 (Fig. 3C, E and Table 1). Of interest, although the proportion of TNFR2-positive Treg coexpression of IL-10 was significantly increased under inflammatory conditions compared with under physiological conditions, not all TNFR2-positive Treg cells are responsive to PGRN, which suggests the possibility that other inflammatory molecules that also signal through TNFR2, such as TNF-α, could antagonize the effect of PGRN on IL-10 production during inflammation.
Figure 3.
Treg cells are a predominant new source of PGRN-induced IL-10. IL-10eGFP reporter mice were challenged with or without CIA, followed by treatment with PBS or PGRN (n = 12 per group). A, B) Splenocytes were stained with specific Abs to CD4, CD8, CD19, VD11c, or F4/80, then subjected to FACS analysis. Statistical analysis of the percentage of each cell type in response to PGRN under physiological (A) or inflammatory (B) conditions. Bar graphs represent means ± sd of total mice per group. C) Expression of IL-10eGFP and TNFR2 (also known as CD120b) on CD4+ T-cell subtypes. Dot plots are of individual mice, numbers in quadrants indicate percent cells in each. D, E) Statistical analysis of percentage of subtype of CD4+ T cells in response to PGRN under physiological (D) or inflammatory (E) conditions. Bar graphs represent means ± sd of total mice per group. **P < 0.01 vs. control group.
TABLE 1.
Percentages of IL-10eGFP+ cell in VERT-X mice in response to PGRN
| Cell type | Physiological conditions |
Inflammatory conditions |
||
|---|---|---|---|---|
| PBS | PGRN | PBS | PGRN | |
| CD4+IL-10eGFP+ | 1.47 ± 0.35 | 2.68 ± 0.42* | 2.66 ± 0.81 | 5.64 ± 1.15* |
| CD8+IL-10eGFP+ | 1.34 ± 0.24 | 1.21 ± 0.20 | 1.08 ± 0.13 | 0.93 ± 0.26 |
| CD19+IL-10eGFP+ | 3.42 ± 0.81 | 3.31 ± 0.74 | 1.34 ± 0.83 | 1.08 ± 0.46 |
| CD11c+IL-10eGFP+ | 7.68 ± 2.27 | 8.87 ± 1.98 | 9.44 ± 1.11 | 10.62 ± 2.24 |
| F4/80+IL-10eGFP+ | 8.12 ± 2.44 | 9.02 ± 1.91 | 10.87 ± 1.55 | 11.14 ± 2.27 |
| CD4+CD25−IL-10eGFP+ | 0.51 ± 0.18 | 0.58 ± 0.19 | 1.46 ± 0.36 | 1.79 ± 0.42 |
| CD4+CD25+IL-10eGFP+ | 1.81 ± 0.25 | 3.67 ± 0.85* | 3.12 ± 0.87 | 7.34 ± 1.03* |
Data are presented as percentage ± sd for VERT-X mice (n = 12 per each group) in the presence or absence of PGRN under physiological or inflammatory conditions. *P < 0.01 vs. control group.
To determine whether Treg cells are also the solo cell type responsible for production of IL-10 in PGRN-deficient mice under both physiological and inflammatory conditions, we crossed PGRN−/− mice with IL-10 reporter mice. Results indicated PGRN deficiency did not significantly alter IL-10 production in healthy mice without inflammatory challenge (Supplemental Fig. 3A and Table 2). However, when these mice were challenged with collagen/CFA to induce CIA, PGRN deficiency was associated with decreased frequency of IL-10 expression by CD4+CD25+ T cells compared with WT mice (Supplemental Fig. 3B–E). PGRN deficiency also led to a small but statistically significantly decreased frequency of F4/80+ macrophage expression during the course of CIA compared with WT mice (Supplemental Fig. 3B and Table 2). Similar to IL-10–expressing Treg cells in WT mice under physiological and inflammatory conditions in response to PGRN, the majority of IL-10–expressing Treg cells also coexpressed TNFR2 in PGRN-deficient mice under CIA challenge (Supplementary Fig. 3C). Collectively, these data demonstrate that Treg cells are the predominant new source of IL-10 in response to PGRN.
TABLE 2.
Percentages of IL-10eGFP+ cell in VERT-X, PGRN+/+ or VERT-X, and PGRN−/− mice
| Cell type | Physiological conditions |
Inflammatory conditions |
||
|---|---|---|---|---|
| WT | KO | WT | KO | |
| CD4+IL-10eGFP+ | 1.24 ± 0.33 | 0.93 ± 0.17 | 2.22 ± 0.71 | 1.21 ± 0.42** |
| CD8+IL-10eGFP+ | 1.17 ± 0.23 | 1.02 ± 0.31 | 0.95 ± 0.25 | 1.07 ± 0.41 |
| CD19+IL-10eGFP+ | 2.86 ± 0.67 | 2.73 ± 0.49 | 1.06 ± 0.32 | 0.96 ± 0.26 |
| CD11c+ IL-10eGFP+ | 6.45 ± 1.67 | 5.40 ± 1.38 | 9.45 ± 2.42 | 8.37 ± 2.79 |
| F4/80+IL-10eGFP+ | 7.47 ± 1.56 | 7.18 ± 1.12 | 11.27 ± 1.77 | 8.69 ± 1.32** |
| CD4+CD25−IL-10eGFP+ | 0.64 ± 0.17 | 0.53 ± 0.12 | 1.73 ± 0.42 | 1.51 ± 0.39 |
| CD4+CD25+IL-10eGFP+ | 1.79 ± 0.38 | 1.87 ± 0.43 | 3.47 ± 0.56 | 1.42 ± 0.28* |
Data are presented as percentage ± sd for VERT-X, PGRN+/+ or VERT-X, or PGRN−/− mice (n = 12 per each group under physiological or inflammatory conditions. KO, knockout. *P < 0.01; **P < 0.05 vs. control group.
PGRN induces Treg cells and IL-10 in vitro and in vivo
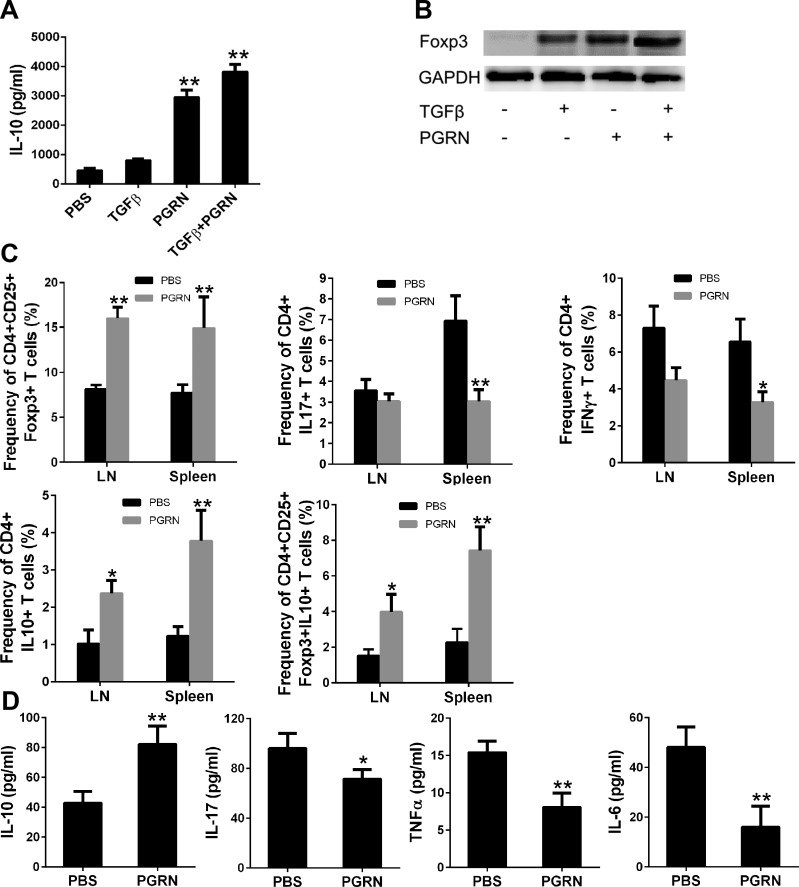
To establish whether PGRN induces Foxp3+ Treg cells and subsequent IL-10 production in vitro, T cells that were isolated from WT spleen were activated in vitro with anti-CD3e and anti-CD28 in the presence of PGRN, and the expression of Foxp3 and production of IL-10 were examined by Western blot and ELISA, respectively. TGF-β was used as a positive control. Treatment with PGRN resulted in an increase in IL-10 and Foxp3 expression. Remarkably, up-regulation of Foxp3 in PGRN-treated cells was comparable to that detected in TGF-β–treated cells; however, TGF-β alone resulted in only a modest increase in IL-10 production compared with PGRN. The effects of PGRN and TGF-β were additive for both IL-10 production and Foxp3 expression (Fig. 4A, B). Taken together, these data suggest that PGRN induces IL-10–secreting CD4+Foxp3+ cells from naive CD4+ T cells.
Figure 4.
PGRN induces Treg cells and IL-10 in vitro and in vivo. A, B) PGRN stimulates Treg cell conversion and IL-10 production in vitro. CD4+ T cells isolated from WT mice were incubated with PGRN in the presence or absence of TGFβ for 4 d, and expression of IL-10 production and Foxp3 in the culture were determined by ELISA (A) and immunoblotting (B), respectively. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is shown as a loading control. C, D) PGRN regulates IL-10 production by Treg cells in vivo. Lymph nodes (LNs), spleens, and serum were obtained from PBS-treated (n = 10) or PGRN-treated (n = 9) DBA1J mice with CIA on wk 8 after immunization. Statistical analysis of percentage of each cell type of CIA mice treated with PBS or PGRN (C). ELISA analysis of sera levels of IL-10, IL-17, TNF-α, and IL-6 (D). Values are means ± sd of total mice per group. *P < 0.05; **P < 0.01 vs. PBS-treated control group.
Stimulation of Treg cells and IL-10 by PGRN was further examined in vivo. Inhibition of CIA induction in DBA1J mice by PGRN was associated with a significant increase in the frequency of CD4+CD25+FoxP3+ T cells and a decrease in the frequency of CD4+IL17+ T cells and CD4+IFN-γ+ T cells in the spleen and draining lymph node (Fig. 4C). Moreover, PGRN treatment led to an increase in the frequency of CD4+IL-10+ T cells in the draining lymph node and spleen (Fig. 4C). Of more interest, the frequency of CD4+CD25+Foxp3+IL-10+ T cells showed a marked increase in PGRN-treated mice compared with control mice (Fig. 4C). Accordingly, when compared with the serum level of inflammatory cytokines from control animals, sera from PGRN-treated mice contained higher amounts of IL-10 and lower amounts of IL-17, IL-6, and TNF-α (Fig. 4D). Collectively, these data suggested that PGRN treatment resulted in the generation of CD4+CD25+Foxp3+IL-10+ Treg cells, which led to the effective prevention of inflammatory arthritis.
PGRN induces Treg cells and IL-10 expression via the TNFR2/JNK pathway
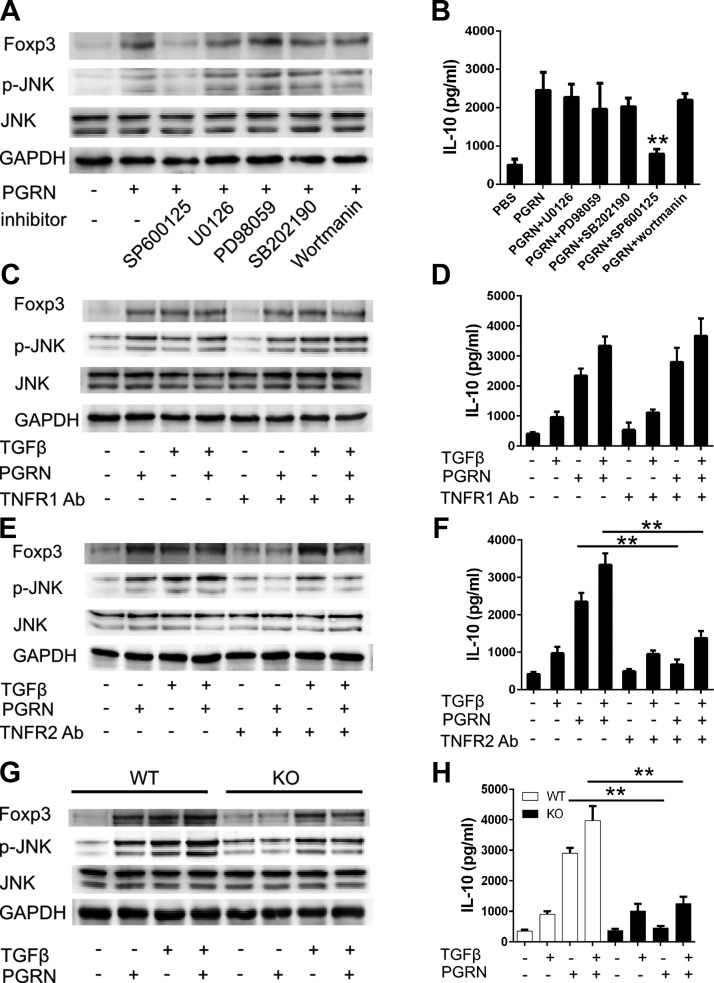
Given that PGRN can stimulate multiple intracellular pathways to promote cell proliferation, migration, and transformation, including the ERK signaling and PI3K pathways (34, 35), and that PGRN can directly bind to TNFR, with preferential binding to TNFR2, which impedes inflammatory response via inhibition of TNF-α–induced intracellular signaling pathways, including MAPK signaling pathways (11, 36), we sought to determine which signaling pathway was involved in the induction of Treg cells in response to PGRN. Of interest, Foxp3 up-regulation in response to PGRN seemed to be a result of the activation of the JNK signaling pathway, as the addition of JNK inhibitor (SP600125) completely abolished the enhancement of Foxp3 expression and IL-10 elevation observed in PGRN-treated T cells; addition of neither ERK (PD98059 or U0126), p38 (SB203580), nor PI3K inhibitor (Wortmannin) resulted in any noticeable effect on Foxp3 expression or IL-10 secretion (Fig. 5A, B). Collectively, these data indicate PGRN induces Foxp3 expression in a JNK-dependent manner.
Figure 5.
PGRN-stimulated expression of Foxp3 and IL-10 depend on TNFR2/JNK pathway. A, B) CD4+ T cells that were isolated from WT mice were treated with indicated inhibitors for 1 h before incubation with PGRN. Cells were isolated after 4 d to determine expression of FoxP3 or 4 h to determine the phosphorylation of JNK by immuoblotting (A), and IL-10 in 4-d culture supernatant was determined by ELISA (B). C, D) Isolated CD4+ T cells were pretreated with or without TNFR1 Abs for 1 h. Cells were then cultured with PGRN in the presence or absence of TGF-β. Cells were isolated after 4 d to determine expression of Foxp3 or 4 h to determine the phosphorylation of JNK by immuoblotting (C), and IL-10 in 4-d culture supernatant was determined by ELISA (D). E, F) Isolated CD4+ T cells were pretreated with or without TNFR2 Abs for 1 h. Cells were then cultured with PGRN in the presence or absence of TGF-β. Cells were isolated after 4 d to determine expression of Foxp3 or 4 h to determine the phosphorylation of JNK by immuoblotting (E), and IL-10 in 4-d culture supernatant was determined by ELISA (F). G, H) CD4+ T cells were isolated from WT or TNFR2−/− mice. Isolated cells were incubated with PGRN in the presence or absence of TGF-β. Cells were isolated after 4 d to determine expression of FoxP3 or 4 h to determine the phosphorylation of JNK by immuoblotting (G), and IL-10 in 4-d culture supernatant was determined by ELISA (H). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KO, knockout. Results are means ± sd from 3 independent experiments. **P < 0.01 vs. control group.
PGRN binds to TNFRs, and TNFRs have been linked to PGRN function in vitro and in vivo (11, 14). To evaluate the involvement of TNFRs in the effect of PGRN on Treg cells, we activated naive T cells in the presence of blocking Abs to TNFR1 or TNFR2, with or without PGRN. Blocking Abs to TNFR2, but not to TNFR1, abolished Foxp3 activation and IL-10 secretion in response to PGRN (Fig. 5C–F). To further reveal the involvement of TNFR1 or TNFR2 in controlling expression of Foxp3 and IL-10 production, we isolated spleenocytes from TNFR1−/− or TNFR2−/− mice. Indeed, naive T cells from TNFR1−/− mice still remain responsive to PGRN (Supplemental Fig. 4A, B), whereas naive T cells from TNFR2−/− mice were unresponsive to PGRN, but maintained sensitivity to TGF-β in terms of Foxp3 activation and IL-10 production (Fig. 5G, H). Thus, PGRN induces Treg cells and facilitates IL-10 production in a TNFR2-dependent mechanism.
Foxo4 and Stat3 jointly govern IL-10 expression in response to PGRN
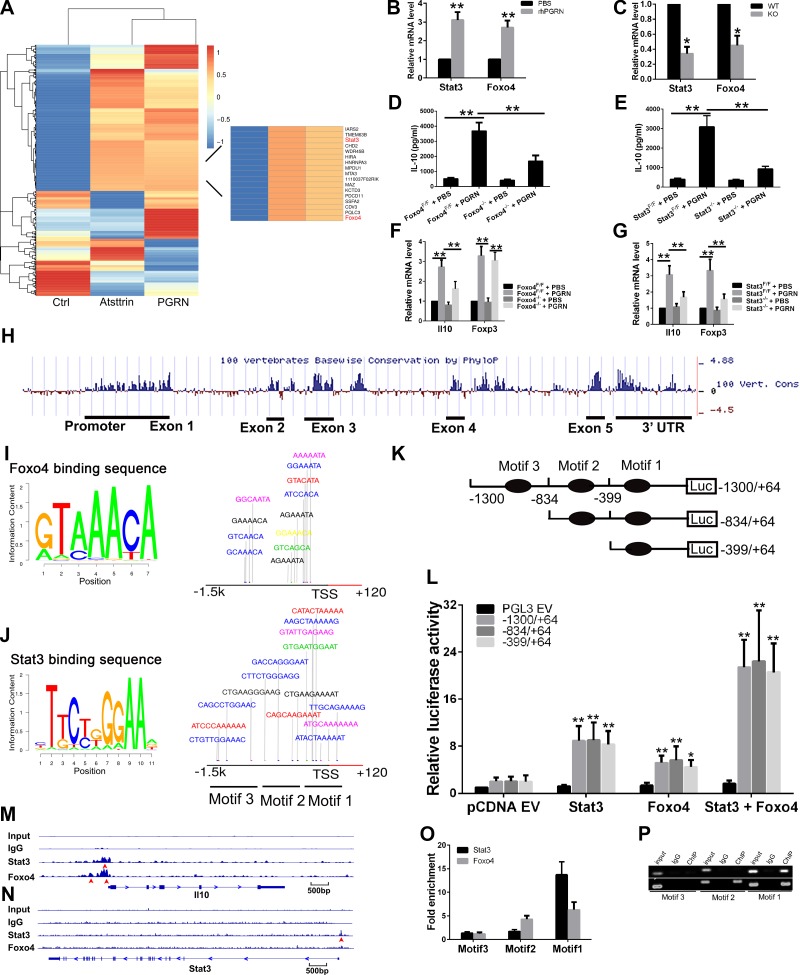
Our results suggest that Treg cells are the major new source of IL-10 production in response to PGRN, which raises the question of how PGRN could regulate Il10 transcription in this specific cell lineage. To answer this question, we performed gene expression profiling of control, PGRN-, or atsttrin (an engineered protein composed of 3 TNFR-binding fragments of PGRN)-treated (11) CD4+ T cells. Results revealed 639 differentially expressed genes when analyzed at a 5% false-discovery rate. Among them, transcription factors Foxo4 and Stat3 are of particular interest, as both are up-regulated significantly in response to both PGRN and Atsttrin (Fig. 6A), as confirmed by real-time PCR in independent samples (Fig. 6B). In addition, expression of Foxo4 and Stat3 were decreased in CD4+ T cells that were isolated from PGRN-deficient mice that were subjected to activation by anti-CD3e and anti-CD28 (Fig. 6C). Among the 3 Foxo genes that were expressed in T cells (37), the transcript of Foxo4 is specifically up-regulated in CD4+ T cells in response to PGRN. In contrast, the transcripts of other 2 Foxo genes, Foxo1 and Foxo3, which are expressed in T cells, were not responsive to PGRN treatment (Supplemental Fig. 5).
Figure 6.
Foxo4 and Stat3 govern IL-10 expression in response to PGRN. A) Genome microarry analysis of TCR-activated CD4+ T cells that were isolated from spleens of healthy WT mice that were incubated with PBS, PGRN, or atsttrin for 24 h. Differential hierarchical clustering was performed to show the distinguishable gene expression profiling among samples. Each row corresponds to a gene, and each column corresponds to an experimental sample. B) PGRN increased the expression of Foxo4 and Stat3. CD4+ T cells that were isolated from WT mice were activated with anti-CD3e and anti-CD28 in presence or absence of PGRN for 24 h, and the mRNA levels of Foxo4 and Stat3 were measured by quantitative PCR. C) PGRN deficiency decreased expression of Foxo4 and Stat3 in CD4+ T cells. D, E) Effect of Foxo4 on IL-10 expression. CD4+ T cells were isolated from Foxo4F/F mice, and then infected with Ad-GFP or Ad-Cre-GFP for 48 h, followed by incubation with or without PGRN for 4 d to determine IL-10 level in the supernatant by ELISA (D) or for 24 h to determine the mRNA level of the indicated gene by quantitative PCR (E). F, G) Effect of Stat3 on IL-10 expression. CD4+ T cells were isolated from Stat3F/F mice, then infected with Ad-GFP or Ad-Cre-GFP for 48 h, followed by incubation with or without PGRN for 4 d to determine IL-10 level in the supernatant by ELISA (F) or for 24 h to determine the mRNA level of the indicated gene by quantitative PCR (G). H) Bioinformatic analysis of Il10 gene. The degree of mammalian sequence conservation is high, with highest conservation over the 5′ proximal promoter, the 5 exons and the 3′-untranslated region. I) Predicted conserved consensus Foxo4 DNA binding sequence and potential binding site on Il10 promoter region. J) Predicted conserved consensus Stat3 DNA-binding sequence, and potential binding site on Il10 promoter region. K) Schematic diagram of luciferase reporter constructs. L) Both Stat3 and Foxo4 activate luciferase activity in Jurkat cells. Transfection into Jurkat T cells with empty pGL3 vector (pGL3 EV) or truncated constructs of Il10 promoter luciferase reporter construct, and Foxo4 and/or Stat3 expression vector. M) ChIP-Seq analysis of Stat3- or Foxo4-bound regions in Il10 locus. Red arrowhead indicates the location of binding peak. N) ChIP-Seq analysis of Stat3-bound region in Stat3 locus. Red arrowhead indicates the location of binding peak. O, P) Both Stat3 and Foxo4 bind to Il10 promoter region, as analyzed by ChIP-qPCR (O) and ChIP-PCR (P). CD4+ T cells that were isolated from WT mice were activated with anti-CD3e and anti-CD28 in presence of PGRN for 24 h, and Foxo4 or Stat3 enrichment in different motifs was analyzed by ChIP-quantitative PCR and ChIP-PCR. KO, knockout. Results are means ± sd from 3 independent experiments. *P < 0.05; **P < 0.01 vs. control group.
To further investigate effect of Foxo4 on IL-10 expression, CD4+ T cells were isolated from Foxo4F/F mice, infected with Ad-GFP or Ad-Cre-GFP for 48 h, followed by activation with anti-CD3e and anti-CD28 in the absence or presence of PGRN, and it was found that Foxo4 deletion results in significantly reduced Il10 mRNA expression and IL-10 protein level in culture supernatant in response to PGRN (Fig. 6D, E). However, the reduction in neither Il10 mRNA, nor IL-10 protein level is absolute, which suggests that other transcription factors could also contribute to govern IL-10 expression in response to PGRN. Of note, both the mRNA level of Il10 and the basal level of IL-10 from the supernatant of Foxo4−/− CD4+ T cell culture are slightly lower than that from Foxo4F/F CD4+ T cells. However, expression of Foxo4 showed no effect on Foxp3 expression (Fig. 6E). These data provide strong evidence that Foxo4 may play an important role in regulation of IL-10 expression in T-cell receptor (TCR)–activated CD4+ T cells, especially during treatment with PGRN.
To determine whether Stat3, also isolated as an up-regulated molecule by PGRN and Atsttrin (Fig. 6A), is also involved in PGRN-induced IL-10 expression, both chemical Stat3 inhibitor and Stat3−/− CD4+ T cells were used (Fig. 6F, G and Supplemental Fig. 6A, B). Both chemical blockade of Stat3 and Stat3 deficiency abrogated IL-10 expression partially, but significantly, as measured at mRNA and protein level, which is similar to observations recorded from Foxo4-deficient mice. However, in contrast to Foxo4, both chemical blockade of Stat3 and Stat3 deficiency could also regulate expression of Foxp3 (Fig. 6G). These observations indicate that Stat3 also contributes to regulation of IL-10 expression in TCR-activated CD4+ T cells in response to PGRN.
Our microarray analysis and real-time PCR data indicated that PGRN could up-regulate Stat3 and Foxo4 expression. To investigate whether both Foxo4 and Stat3 are necessary for PGRN-induced IL-10 production in CD4+ T cells, we use Foxo4F/F mice, Stat3F/F mice, and a Stat3 inhibitor. Single deletion of Foxo4 or Stat3 or treatment with a Stat3 inhibitor alone substantially reduced IL-10 expression induced by PGRN in TCR-activated CD4+ T cells. Blockade of Stat3 concomitant with Foxo4 deficiency in activated CD4+ T cells almost completely abrogated IL-10 expression at both mRNA and protein levels (Supplemental Fig. 6C, D). These results suggest that Foxo4 and Stat3 work together to achieve maximal IL-10 production in PGRN-treated TCR-activated CD4+ T cells.
To investigate further, the UCSC Genome Browser was used to identify the conserved noncoding regions that surround the Il10 locus among 100 vertebrates (Fig. 6H). We found that the 5′ proximal promoter region is highly conserved among mammals. We further searched the transcription factor binding sites on the basis of position weight matrices acquired from the Jaspar Core database (38). Figure 6I, J indicates the binding motifs of Foxo4 and Stat3 from the database and their potential binding sites on the promoter region of the murine Il10 gene with a cutoff of 85% similarity to the consensus binding sites. There are 12 putative Foxo4 binding sites and 18 putative Stat3 binding sites, and they were grouped into 3 motifs: −1020 bp/−835 bp (motif 3), −700 bp/−400 bp (motif 2), and −320 bp/−48 bp (motif 1). To determine which of the motifs was responsible for Foxo4- and Stat3-mediated Il10 promoter activity, we generated luciferase reporter constructs of the −1300 bp/+67 bp promoter by sequentially deleting each motif and testing their activity by transient transfection into Jurkat cells in response to cotransfected Foxo4 and/or Stat3 expression plasmid. Forced expression of either Foxo4 or Stat3 resulted in increased luciferase activity in all 3 constructs. Deletion of motif 3 seemed to slightly enhance, whereas further deletion of motif 2 slightly reduced the reporter activity in response to Foxo4; however, these differences did not meet statistical significance. In contrast, in response to Stat3, deletion of motif 3 enhanced the reporter activity, whereas deletion of motif 2 had no effect on the reporter activity. Of note, the effects of Stat3 on reporter activity were greater than those observed with Foxo4; cotransfection of both Foxo4 and Stat3 had synergistic effects on reporter activity (Fig. 6K, L).
For identification of Stat3 or Foxo4 DNA binding sites in TCR-activated CD4+ T cells in response to PGRN, we performed ChIP-Seq. ChIP-Seq analysis resulted in 3177 peaks of Stat3 binding and 8165 peaks of Foxo4 binding at a 0.1% false-discovery rate. Only approximately 8.5 and 7.2% of these peaks for Stat3 and Foxo4, respectively, were in promoter regions of RefSeq genes, which indicated that, although either Stat3 or Foxo4 directly binds the promoter regions of several genes, the majority of their regulation might rely on binding to longer-range DNA elements in CD4+ T cells in response to PGRN (Supplemental Fig. 7A, B). Of interest, limited target genes that had Stat3 and/or Foxo4 binding peaks in the proximal promoter regions (Supplemental Fig. 7C), among them, only Il10, Sspn, Jup, Mcmbp, and Pclb4 had both Stat3 and Foxo4 binding peaks. Moreover, on the Il10 locus, there were only Stat3 or Foxo4 binding peaks located in the proximal promoter regions in TCR-activated CD4+ T cells in response to PGRN. The binding peak for Stat3 is located 138 bp upstream from the transcription start site of Il10 gene, whereas the stronger Foxo4 binding peak is located 113 bp upstream from the transcription start site. In addition, there was a weaker binding peak for Foxo4 located 464 bp upstream from transcription start site (Fig. 6M). Of note, the Stat3 binding peak observed in the proximal promoter region of Stat3 locus suggests the possibility of Stat3 positive self-feedback, which could account for increased Stat3 expression in PGRN-treated CD4+ T cells (Fig. 6N). To our surprise, there is a clear Stat3 binding peak observed in the exon region of Grn locus (Supplemental Fig. 7D), which raises the possibility that PGRN and Stat3 could build a positive feedback loop that may eventually enhance the anti-inflammatory effects of PGRN by inducing its expression in CD4+ T cells. The underlying mechanisms still warrant further investigation.
Binding of Stat3 or Foxo4 at the aforementioned motifs in response to PGRN was further confirmed by ChIP-qPCR analysis of CD4+ T cells. We detected Foxo4 binding to motifs 1 and 2, and strong Stat3 binding to motif 1 (Fig. 6O, P), which is agreement with our ChIP-Seq results. Of particular interest is motif 1, located proximal to the Il10 transcription start site, which showed strong Stat3 enrichment in our assay—a finding that is consistent with a previous report of Stat3 binding site within 149 bp upstream of the transcription start site (39). Overall, these data clearly demonstrate that both Foxo4 and Stat3 are responsible for governing IL-10 expression in PGRN-treated CD4+ T cells. Taken together, these findings imply that both Stat3 and Foxo4 are required for IL-10 expression in TCR-activated CD4+ T cells in response to PGRN.
DISCUSSION
It was previously reported that PGRN was therapeutic against inflammatory arthritis and that IL-10 level was selectively elevated in PGRN-treated CIA mouse model (11), but the immunological mechanism that underlies the induction of IL-10 by PGRN in inflammatory diseases remains unknown. In this study, we examined the role of IL-10 in PGRN-mediated anti-inflammation and the cell types and signals responsible for production of IL-10 upon PGRN treatment by employing genetically modified mice under physiological and inflammatory conditions. We observed that IL-10 was critical for PGRN’s anti-inflammation, and Treg cells were the predominant IL-10–producing cells. Components of the TNFR2 signaling pathway, including TNFR2 and JNK, were required for expansion and activation of Treg cells and production of IL-10. Of more importance, PGRN signaling is TNFR2 dependent as blockade and deletion of TNFR2 compromised Treg expansion and activation and abolished IL-10 production completely. In addition, genome-wide microarray analysis revealed that transcription factors Foxo4 and Stat3, which bind to conserved promoter regions in the Il10 locus in CD4+ T cells, are capable of regulating IL-10 production in these cells (see proposed working model in Supplemental Fig. 7E).
Pathogenic autoimmunity is controlled in healthy individuals by a specialized subset of T cells, called Treg cells, (40), whose differentiation and function are driven by the transcription factor Foxp3 (41, 42). A large body of evidence suggests the involvement of Treg cells in both human rheumatoid arthritis and murine models of inflammatory arthritis. Treg cells are capable of inhibiting inflammatory response and damage, but are also important in both initiation and perpetuation of experimental arthritis in mice (43). Our findings that CD4+CD25+Foxp3+ cell numbers are increased after PGRN treatment in vitro and in vivo is in line with our earlier work on PGRN-deficient mice, which showed that PGRN deficiency caused a marked reduction in Treg cells during the course of inflammatory arthritis (44). Of more importance, the observation that the proportion of CD4+CD25+Foxp3+IL-10+ Treg cells is elevated in CIA mice receiving PGRN relative to those mice that received PBS indicates that Treg cells could be an important cell source of IL-10 after PGRN treatment during inflammation.
IL-10 is a typical immunosuppressive cytokine, and high and low expression levels of IL-10 are associated with amelioration and exacerbation of inflammatory arthritis, respectively (45). PGRN-deficient macrophages produce more proinflammatory cytokines and less anti-inflammatory IL-10 in response to LPS than do WT macrophages (10). In our previous study, we also found that IL-10 level is selectively elevated in the sera from PGRN-treated mice in which CIA was induced compared with that from PBS-treated mice with CIA induction (11). To gain insight into the mechanisms of the well-established therapeutic effects of PGRN, it is advantageous to monitor PGRN-induced IL-10 production during the course of CIA intervention. Given the importance of IL-10 in limiting inflammation, it is perhaps not unexpected that there are many cellular sources of this immune modulator, including macrophages, dendritic cells, B cells, and T cells (46). To our surprise, we have found that CD4+ T cells, particularly Treg cells, are the lone cell type responsible for the increased production of IL-10 upon PGRN treatment in healthy mice and during inflammation in the context of CIA induction, which suggests a role for CD4+ cells, and especially Treg-cell–associated IL-10, in control disease pathology. Treg cells exert their immunoregulatory functions via a variety of effector mechanisms, such as consumption of IL-2 (47) and production of immunomodulatory cytokines, such as IL-10 (48). Similarly, IL-10 production by Treg cells is essential for control of local immune response in the lungs and intestinal tract. In line with these reports, our studies demonstrate that Treg-cell–derived IL-10, increased in response to PGRN, ameliorates CIA. In addition, the studies we have presented here have shown that activation of JNK is required for induction of Treg cells and production of Treg-cell–derived IL-10, and that these events are TNFR2 dependent. Taken together, our results identify a previously unknown pathway that promotes the production of IL-10 in response to PGRN within the context of inflammation. Our finding that the therapeutic effect of PGRN on CIA is largely abolished in IL-10−/− mice provides additional evidence to support the conclusion that IL-10 is a target of interest for modulation of inflammatory diseases.
Although there are many cellular sources of IL-10, there are a limited number of studies that have defined the lineage-specific requirements for Il10 transcription. In macrophages, the transcription factor C/EBPα is involved in LPS-induced IL-10 production (49). In CD4+ T cells, the transcription factor GATA-3 is associated with the remodeling and stabilization of Il10 locus required for Il10 transcription (50). In lupus T cells, Stat3 promotes IL-10 expression via transactivation and chromatin remodeling of the Il10 locus (39). In addition, in human Treg cells, it was reported that IL-2–activated STAT5 binds to a STAT-responsive intronic enhancer in the Il10 locus (51), and in mouse Treg cells, both IRF4 and Blimp1 seem to be required for Il10 expression (52). Instead, our data presented here link Foxo4 and Stat3 to PGRN-mediated induction of IL-10 in CD4+ T cells. Involvement of Foxo4 and Stat3 is of special interest because both of these transcription factors have been shown to be increased in CD4+ T cells in response to PGRN, and of note, for the first time, Foxo4 is reported to mediate molecular events that instruct Il10 expression in CD4+ T cells. Applying reporter constructs, we demonstrated the synergistic effects of Stat3 and Foxo4 on Il10 transcriptional activation. In addition, involvement of Stat3 and Foxo4 in governing Il10 is underscored by results from Stat3- and Foxo4-deficient cells. Both ChIP-seq and ChIP-quantitative PCR indicate that there is strong Foxo4 binding to evolutionarily conserved regions, with consensus motifs located at Il10 proximal promoter regions. In addition, our findings indicate that Stat3 also acts as direct transcriptional regulator of Il10 in Treg cells in response to PGRN. This observation is consistent with the presence of Stat binding sites in the Il10 promoter (39, 53, 54). Our findings imply that both Stat3 and Foxo4 are synergistic for maximal IL-10 expression by CD4+ T cells, thus illuminating previously unidentified molecular events, contributory to increased IL-10 expression in CD4+ T cells in response to PGRN.
Even though Treg-cell–derived IL-10 is the major mediator of the therapeutic effect of PGRN on CIA, we cannot exclude the possibility that other cell types in addition to Treg cells are also involved in PGRN’s effect, as our data indicate that T helper (Th)17 cells decreased significantly after PGRN treatment and that IL-10 deficiency largely, but not completely, abolished PGRN-mediated effect on CIA. Th17 cells are a subtype of T cells that express the transcription factor retinoic acid receptor–related orphan nuclear receptor C and secrete the proinflammatory cytokine IL17. Treg and Th17 cells are reciprocally related cell populations with opposite roles in the immune response (55). Patients with active rheumatoid arthritis have increased numbers of Th17 cells and reduced numbers of Treg cells compared with healthy controls (56, 57). Resistance of individuals to anti-TNF therapy is associated with a further increase in Th17 cells (58). This Th17/Treg imbalance may be involved in the pathogenic response in rheumatic arthritis; therefore, the reduction in Th17 cells observed in PGRN-treated CIA may also contribute to the therapeutic effect of PGRN. We observed significant reductions in intracellular and serum level of IL-17, the end product of Th17, in mice that were treated with PGRN compared with animals treated with vehicle, which correlated with arthritis development. Of interest, it was reported that Treg cells from patients with rheumatoid arthritis who experienced a response to adalimumab, an anti-TNF therapy, were able to constrain Th17 responses via inhibition of monocyte-derived IL-6; Treg cells from patients with active rheumatoid arthritis or healthy controls could not inhibit Th17 responses (59). Our data show that PGRN could reduce IL-6, which, in turn, could account for the reduced Th17-cell number. Therefore, whether PGRN plays a role in offsetting Th17/Treg imbalance warrants further investigation.
In conclusion, our data provide novel evidence that PGRN can effectively ameliorate chronic inflammatory disorders via Foxo4- and Stat3-dependent IL-10 production in Treg cells. In brief, we have shown that not only the number of Treg cells but also their acquisition of IL-10 production contributes to the control of immune response. In addition, we have identified extrinsic signals that are crucial for regulation of this cell population. These findings are clinically important as modulation of Treg function is of great interest in many medical conditions (60). In addition, the efficacy of PGRN treatment in mouse inflammatory arthritis model supports IL-10 regulation as a promising approach for the treatment of chronic inflammatory diseases.
ACKNOWLEDGMENTS
The authors thank the U.S. National Institutes of Health, National Institute of Arthritis and Musculoskeletal and Skin Diseases for Grants R01-AR062207, R01-AR061484, and R56-AI100901, as well as W81XWH-16-1-0482 from the U.S. Department of Defense.
Glossary
- CFA
complete Freund's adjuvant
- ChIP-Seq
chromatin immunoprecipitation sequencing
- CIA
collagen-induced arthritis
- Foxo4
forkhead box protein O4
- Foxp3
forkhead box protein P3
- GFP
green fluorescent protein
- H&E
hematoxylin and eosin
- PGRN
progranulin
- Stat3
signal transducer and activator of transcription 3
- TCR
T-cell receptor
- Th
T helper
- TNFR
TNF receptor
- Treg
regulatory T
- WT
wild-type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
W. Fu designed and performed the experiments, analyzed data, and wrote the manuscript; W. Hu performed the flow cytometry analysis, microarray, and ChIP-Seq data analysis; L. Shi contributed to the purification of rhPGRN protein; J. J. Mundra performed microarray assay; G. Xiao and M. L. Dustin participated in experimental design, data analysis, and edited the paper; and C. Liu designed and supervised this study, analyzed data, and wrote and edited the manuscript.
REFERENCES
- 1.Bateman A., Bennett H. P. (2009) The granulin gene family: from cancer to dementia. BioEssays 31, 1245–1254 [DOI] [PubMed] [Google Scholar]
- 2.Feng J. Q., Guo F. J., Jiang B. C., Zhang Y., Frenkel S., Wang D. W., Tang W., Xie Y., Liu C. J. (2010) Granulin epithelin precursor: a bone morphogenic protein 2-inducible growth factor that activates Erk1/2 signaling and JunB transcription factor in chondrogenesis. FASEB J. 24, 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker M., Mackenzie I. R., Pickering-Brown S. M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A. D., Rollinson S., Cannon A., Dwosh E., Neary D., Melquist S., Richardson A., Dickson D., Berger Z., Eriksen J., Robinson T., Zehr C., Dickey C. A., Crook R., McGowan E., Mann D., Boeve B., Feldman H., Hutton M. (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919 [DOI] [PubMed] [Google Scholar]
- 4.Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J. J., van Duijn C., Peeters K., Sciot R., Santens P., De Pooter T., Mattheijssens M., Van den Broeck M., Cuijt I., Vennekens K., De Deyn P. P., Kumar-Singh S., Van Broeckhoven C. (2006) Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442, 920–924 [DOI] [PubMed] [Google Scholar]
- 5.Petkau T. L., Leavitt B. R. (2014) Progranulin in neurodegenerative disease. Trends Neurosci. 37, 388–398 [DOI] [PubMed] [Google Scholar]
- 6.Daniel R., He Z., Carmichael K. P., Halper J., Bateman A. (2000) Cellular localization of gene expression for progranulin. J. Histochem. Cytochem. 48, 999–1009 [DOI] [PubMed] [Google Scholar]
- 7.He Z., Ong C. H., Halper J., Bateman A. (2003) Progranulin is a mediator of the wound response. Nat. Med. 9, 225–229 [DOI] [PubMed] [Google Scholar]
- 8.Zhu J., Nathan C., Jin W., Sim D., Ashcroft G. S., Wahl S. M., Lacomis L., Erdjument-Bromage H., Tempst P., Wright C. D., Ding A. (2002) Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell 111, 867–878 [DOI] [PubMed] [Google Scholar]
- 9.Kessenbrock K., Fröhlich L., Sixt M., Lämmermann T., Pfister H., Bateman A., Belaaouaj A., Ring J., Ollert M., Fässler R., Jenne D. E. (2008) Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J. Clin. Invest. 118, 2438–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin F., Banerjee R., Thomas B., Zhou P., Qian L., Jia T., Ma X., Ma Y., Iadecola C., Beal M. F., Nathan C., Ding A. (2010) Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J. Exp. Med. 207, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang W., Lu Y., Tian Q. Y., Zhang Y., Guo F. J., Liu G. Y., Syed N. M., Lai Y., Lin E. A., Kong L., Su J., Yin F., Ding A. H., Zanin-Zhorov A., Dustin M. L., Tao J., Craft J., Yin Z., Feng J. Q., Abramson S. B., Yu X. P., Liu C. J. (2011) The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science 332, 478–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egashira Y., Suzuki Y., Azuma Y., Takagi T., Mishiro K., Sugitani S., Tsuruma K., Shimazawa M., Yoshimura S., Kashimata M., Iwama T., Hara H. (2013) The growth factor progranulin attenuates neuronal injury induced by cerebral ischemia-reperfusion through the suppression of neutrophil recruitment. J. Neuroinflammation 10, 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawase R., Ohama T., Matsuyama A., Matsuwaki T., Okada T., Yamashita T., Yuasa-Kawase M., Nakaoka H., Nakatani K., Inagaki M., Tsubakio-Yamamoto K., Masuda D., Nakagawa-Toyama Y., Nishida M., Ohmoto Y., Nishihara M., Komuro I., Yamashita S. (2013) Deletion of progranulin exacerbates atherosclerosis in ApoE knockout mice. Cardiovasc. Res. 100, 125–133 [DOI] [PubMed] [Google Scholar]
- 14.Wei F., Zhang Y., Jian J., Mundra J. J., Tian Q., Lin J., Lafaille J. J., Tang W., Zhao W., Yu X., Liu C. J. (2014) PGRN protects against colitis progression in mice in an IL-10 and TNFR2 dependent manner. Sci. Rep. 4, 7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jian J., Konopka J., Liu C. (2013) Insights into the role of progranulin in immunity, infection, and inflammation. J. Leukoc. Biol. 93, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu K., Zhang Y., Ilalov K., Carlson C. S., Feng J. Q., Di Cesare P. E., Liu C. J. (2007) Cartilage oligomeric matrix protein associates with granulin-epithelin precursor (GEP) and potentiates GEP-stimulated chondrocyte proliferation. J. Biol. Chem. 282, 11347–11355 [DOI] [PubMed] [Google Scholar]
- 17.Guo F., Lai Y., Tian Q., Lin E. A., Kong L., Liu C. (2010) Granulin-epithelin precursor binds directly to ADAMTS-7 and ADAMTS-12 and inhibits their degradation of cartilage oligomeric matrix protein. Arthritis Rheum. 62, 2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konopka J., Richbourgh B., Liu C. (2014) The role of PGRN in musculoskeletal development and disease. Front. Biosci. (Landmark Ed.) 19, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y. P., Liu B., Tian Q. Y., Wei J. L., Richbourgh B., Liu C. J. (2015) Progranulin protects against osteoarthritis through interacting with TNF-α and β-catenin signalling. Ann. Rheum. Dis. 74, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams A., Wang E. C., Thurner L., Liu C. J. (2016) Review: novel insights into TNF receptor, DR3 and progranulin pathways in arthritis and bone remodeling. Arthritis Rheumatol. 68, 2845–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brand D. D., Latham K. A., Rosloniec E. F. (2007) Collagen-induced arthritis. Nat. Protoc. 2, 1269–1275 [DOI] [PubMed] [Google Scholar]
- 22.Camps M., Rückle T., Ji H., Ardissone V., Rintelen F., Shaw J., Ferrandi C., Chabert C., Gillieron C., Françon B., Martin T., Gretener D., Perrin D., Leroy D., Vitte P. A., Hirsch E., Wymann M. P., Cirillo R., Schwarz M. K., Rommel C. (2005) Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat. Med. 11, 936–943 [DOI] [PubMed] [Google Scholar]
- 23.Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K. (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finnegan A., Kaplan C. D., Cao Y., Eibel H., Glant T. T., Zhang J. (2003) Collagen-induced arthritis is exacerbated in IL-10-deficient mice. Arthritis Res. Ther. 5, R18–R24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye L., Wen Z., Li Y., Chen B., Yu T., Liu L., Zhang J., Ma Y., Xiao S., Ding L., Li L., Huang Z. (2014) Interleukin-10 attenuation of collagen-induced arthritis is associated with suppression of interleukin-17 and retinoid-related orphan receptor γt production in macrophages and repression of classically activated macrophages. Arthritis Res. Ther. 16, R96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asquith D. L., Miller A. M., McInnes I. B., Liew F. Y. (2009) Animal models of rheumatoid arthritis. Eur. J. Immunol. 39, 2040–2044 [DOI] [PubMed] [Google Scholar]
- 30.Zhu X., Yang P., Zhou H., Li B., Huang X., Meng Q., Wang L., Kijlstra A. (2007) CD4+CD25+Tregs express an increased LAG-3 and CTLA-4 in anterior chamber-associated immune deviation. Graefes Arch. Clin. Exp. Ophthalmol. 245, 1549–1557 [DOI] [PubMed] [Google Scholar]
- 31.Huber S., Schramm C. (2006) TGF-beta and CD4+CD25+ regulatory T cells. Front. Biosci. 11, 1014–1023 [DOI] [PubMed] [Google Scholar]
- 32.Madan R., Demircik F., Surianarayanan S., Allen J. L., Divanovic S., Trompette A., Yogev N., Gu Y., Khodoun M., Hildeman D., Boespflug N., Fogolin M. B., Gröbe L., Greweling M., Finkelman F. D., Cardin R., Mohrs M., Müller W., Waisman A., Roers A., Karp C. L. (2009) Nonredundant roles for B cell-derived IL-10 in immune counter-regulation. J. Immunol. 183, 2312–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart C. A., Metheny H., Iida N., Smith L., Hanson M., Steinhagen F., Leighty R. M., Roers A., Karp C. L., Müller W., Trinchieri G. (2013) Interferon-dependent IL-10 production by Tregs limits tumor Th17 inflammation. J. Clin. Invest. 123, 4859–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Z., Ismail A., Kriazhev L., Sadvakassova G., Bateman A. (2002) Progranulin (PC-cell-derived growth factor/acrogranin) regulates invasion and cell survival. Cancer Res. 62, 5590–5596 [PubMed] [Google Scholar]
- 35.Kamrava M., Simpkins F., Alejandro E., Michener C., Meltzer E., Kohn E. C. (2005) Lysophosphatidic acid and endothelin-induced proliferation of ovarian cancer cell lines is mitigated by neutralization of granulin-epithelin precursor (GEP), a prosurvival factor for ovarian cancer. Oncogene 24, 7084–7093 [DOI] [PubMed] [Google Scholar]
- 36.Jian J., Zhao S., Tian Q., Gonzalez-Gugel E., Mundra J. J., Uddin S. M., Liu B., Richbourgh B., Brunetti R., Liu C. J. (2013) Progranulin directly binds to the CRD2 and CRD3 of TNFR extracellular domains. FEBS Lett. 587, 3428–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouyang W., Beckett O., Ma Q., Paik J. H., DePinho R. A., Li M. O. (2010) Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 11, 618–627 [DOI] [PubMed] [Google Scholar]
- 38.Mathelier A., Zhao X., Zhang A. W., Parcy F., Worsley-Hunt R., Arenillas D. J., Buchman S., Chen C. Y., Chou A., Ienasescu H., Lim J., Shyr C., Tan G., Zhou M., Lenhard B., Sandelin A., Wasserman W. W. (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 42, D142–D147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hedrich C. M., Rauen T., Apostolidis S. A., Grammatikos A. P., Rodriguez Rodriguez N., Ioannidis C., Kyttaris V. C., Crispin J. C., Tsokos G. C. (2014) Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc. Natl. Acad. Sci. USA 111, 13457–13462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakaguchi S. (2004) Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev. Immunol. 22, 531–562 [DOI] [PubMed] [Google Scholar]
- 41.Fontenot J. D., Gavin M. A., Rudensky A. Y. (2003) Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 [DOI] [PubMed] [Google Scholar]
- 42.Hori S., Nomura T., Sakaguchi S. (2003) Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 [DOI] [PubMed] [Google Scholar]
- 43.Monte K., Wilson C., Shih F. F. (2008) Increased number and function of FoxP3 regulatory T cells during experimental arthritis. Arthritis Rheum. 58, 3730–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei F., Zhang Y., Zhao W., Yu X., Liu C. J. (2014) Progranulin facilitates conversion and function of regulatory T cells under inflammatory conditions. PLoS One 9, e112110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan F., Beech J. (2007) Update on cytokines in rheumatoid arthritis. Curr. Opin. Rheumatol. 19, 296–301 [DOI] [PubMed] [Google Scholar]
- 46.Saraiva M., O’Garra A. (2010) The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181 [DOI] [PubMed] [Google Scholar]
- 47.Pandiyan P., Zheng L., Ishihara S., Reed J., Lenardo M. J. (2007) CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat. Immunol. 8, 1353–1362 [DOI] [PubMed] [Google Scholar]
- 48.Lu L. F., Rudensky A. (2009) Molecular orchestration of differentiation and function of regulatory T cells. Genes Dev. 23, 1270–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan W., Ding A., Kim H. J., Zheng H., Wei F., Ma X. (2016) Progranulin controls sepsis via C/EBPalpha-regulated Il10 transcription and ubiquitin ligase/proteasome-mediated protein degradation. J. Immunol. 197, 3393–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shoemaker J., Saraiva M., O’Garra A. (2006) GATA-3 directly remodels the IL-10 locus independently of IL-4 in CD4+ T cells. J. Immunol. 176, 3470–3479 [DOI] [PubMed] [Google Scholar]
- 51.Tsuji-Takayama K., Suzuki M., Yamamoto M., Harashima A., Okochi A., Otani T., Inoue T., Sugimoto A., Toraya T., Takeuchi M., Yamasaki F., Nakamura S., Kibata M. (2008) The production of IL-10 by human regulatory T cells is enhanced by IL-2 through a STAT5-responsive intronic enhancer in the IL-10 locus. J. Immunol. 181, 3897–3905 [DOI] [PubMed] [Google Scholar]
- 52.Cretney E., Xin A., Shi W., Minnich M., Masson F., Miasari M., Belz G. T., Smyth G. K., Busslinger M., Nutt S. L., Kallies A. (2011) The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 12, 304–311 [DOI] [PubMed] [Google Scholar]
- 53.Ziegler-Heitbrock L., Lötzerich M., Schaefer A., Werner T., Frankenberger M., Benkhart E. (2003) IFN-alpha induces the human IL-10 gene by recruiting both IFN regulatory factor 1 and Stat3. J. Immunol. 171, 285–290 [DOI] [PubMed] [Google Scholar]
- 54.Stumhofer J. S., Silver J. S., Laurence A., Porrett P. M., Harris T. H., Turka L. A., Ernst M., Saris C. J., O’Shea J. J., Hunter C. A. (2007) Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 8, 1363–1371 [DOI] [PubMed] [Google Scholar]
- 55.Bettelli E., Carrier Y., Gao W., Korn T., Strom T. B., Oukka M., Weiner H. L., Kuchroo V. K. (2006) Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 [DOI] [PubMed] [Google Scholar]
- 56.Wang W., Shao S., Jiao Z., Guo M., Xu H., Wang S. (2012) The Th17/Treg imbalance and cytokine environment in peripheral blood of patients with rheumatoid arthritis. Rheumatol. Int. 32, 887–893 [DOI] [PubMed] [Google Scholar]
- 57.Niu Q., Cai B., Huang Z. C., Shi Y. Y., Wang L. L. (2012) Disturbed Th17/Treg balance in patients with rheumatoid arthritis. Rheumatol. Int. 32, 2731–2736 [DOI] [PubMed] [Google Scholar]
- 58.Alzabin S., Abraham S. M., Taher T. E., Palfreeman A., Hull D., McNamee K., Jawad A., Pathan E., Kinderlerer A., Taylor P. C., Williams R., Mageed R. (2012) Incomplete response of inflammatory arthritis to TNFα blockade is associated with the Th17 pathway. Ann. Rheum. Dis. 71, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 59.McGovern J. L., Nguyen D. X., Notley C. A., Mauri C., Isenberg D. A., Ehrenstein M. R. (2012) Th17 cells are restrained by Treg cells via the inhibition of interleukin-6 in patients with rheumatoid arthritis responding to anti-tumor necrosis factor antibody therapy. Arthritis Rheum. 64, 3129–3138 [DOI] [PubMed] [Google Scholar]
- 60.Campbell D. J., Koch M. A. (2011) Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 11, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]