Abstract
Restless legs syndrome (RLS) is a complex disorder involving sensory and motor systems. The major pathophysiology of RLS is low iron concentration in the substantia nigra, containing the cell bodies of dopamine neurons that project to the striatum, an area important for modulating movement. People who have RLS often present with normal iron values outside the brain and recent studies implicate several genes involved in the syndrome. Like most complex diseases, animal models usually do not faithfully capture the full phenotypic spectrum of “disease” which is a uniquely human construct. Nonetheless, animal models have proven useful in helping to unravel the complex pathophysiology of diseases such as RLS and suggest novel treatment paradigms. For example, hypothesis-independent genome-wide association studies (GWAS) have identified several genes as increasing risk for RLS, including BTBD9. Independently, the murine homolog, Btbd9 was identified as a candidate gene for iron regulation in the midbrain in mice. The relevance of the phenotype of another of the GWAS identified genes, MEIS1, has also been explored. The role of Btbd9 in iron regulation and RLS-like behaviors has been further evaluated in mice carrying a null mutation of the gene, and in fruit flies when the BTBD9 protein is degraded. The BTBD9 and MEIS1 stories took origin from human GWAS research, supported by work in a genetic reference population of mice (forward genetics), and further verified in mice, fish flies and worms. Finally, the role of genetics is further supported by an inbred mouse strain that displays many of the phenotypic characteristics of RLS. The role of animal models of RLS phenotypes is also extended to include periodic limb movements.
Keywords: mice, flies, worms, fish, forward genetics, reverse genetics
1.0 Introduction
Restless Legs Syndrome/Willis Ekbom disease (RLS/WED) is a highly complex disorder that includes sensory and motor symptoms. The primary sensory/behavioral phenotypes emerge during the evening and nighttime sensations due to circadian influences. The result includes urges to move the legs, leading to behaviors of reduced periods of rest without activity and to some extent increased activity. During sleep, increased non-volitional periodic limb movements (PLMS) occur in nearly all patients and represent an important endophenotype whose frequency is also strongly associated with many of the -alleles (most notably, in MEIS1 and BTBD9 genes) previously identified by genome-wide association studies (GWAS) that confer risk for RLS sensory symptoms. There may be several pathophysiological pathways that influence emergence of symptoms, but one major feature is lack of iron in the substantia nigra and subsequent compromise of synaptic dopamine signaling in its major target, viz., the striatum. Increased synaptic striatal dopamine has also been observed. Thus the biological phenotypes would be decreased brain iron and increased striatal dopamine tissue concentrations of dopamine with decreased/impaired synaptic reuptake. The precise mechanisms underlying RLS are currently intractable to experimental methods in humans, and there is great reliance on experimental animals or in vitro methods to elucidate mechanisms operational at both molecular and systems anatomical levels.
Developing an animal model of RLS is not an easy undertaking. There are differences of opinion as to what essential features comprise a valid animal model. Some believe that the phenotypes should be reproduced as closely as possible, whereas others assert that such an isomorphic model may be nearly impossible. Animal models can also be used to evaluate various aspects of the etiology and manifestations in order to elucidate underlying disease mechanisms and their response to conventional and novel treatments. In this contribution, both sides are presented showing how the different approaches are complementary.
There have been two efforts that contribute to the main part of this article. On the human side, Winkelmann and colleagues [1] conducted genome-wide (RLS) association studies (GWAS) and identified a handful of genes (including BTBD9 and MEIS1 – among others) whose variants are associated with increased risk for developing RLS (and in most instances PLMs). As concerns animal models, the Jones lab studied iron content variation in 20+ recombinant inbred mouse strains. Two compelling observations were that Btbd9 regulates iron homeostasis in the ventral midbrain (2008), and that brain and peripheral iron responses to low iron diet and behavioral responses in one specific inbred strain, viz. BXD40, mimicked several features of RLS (vide infra).
This summary of the work begins with studies aimed at revealing possible disease producing mechanisms. Winkelmann and colleagues’ studies have advanced from human GWAS to interrogating the role of Btbd9 and MEIS1 in fish, flies and worms. The work of Li, extends this by showing that that the murine null mutant of Btbd9 produces several RLS-like phenotypes. Donelson also shows that Btbd9 is important for iron regulation in Drosophila, and the work of Rouleau demonstrates how MEIS1 is important for iron regulation in the nematode, Caenorhabditis elegans. Finally two positions presented here include both physiological and behavioral phenotypes that resemble RLS. The work of Allen and colleagues describes an inbred mouse strain that shows both the basic circadian activity/rest phenotype and also the biological phenotype of RLS, and Marconi demonstrates development of the PLMS behavioral phenotype of RLS.
2.0 How genome-wide association studies shape animal research: The role of MEIS1, a gene nominated by GWAS, in behaviors mimicking RLS
Restless legs syndrome (RLS) was the first common disorder of sleep where genetic risk loci were identified by means of genome-wide association studies (GWAS). Common variants per definition with a minor allele frequency (MAF) of ≥ 5% in the population usually have a low effect size and convey only a mild risk to develop the disease. Generally, the molecular mechanism and function behind low-effect-size common genetic variants identified in complex genetic diseases provide a formidable challenge. The strongest RLS association signal identified delineates a 32-kb linkage disequilibrium (LD) block in intron 8 of MEIS1 (2p14) [3–5] MEIS1 is a transcription factor and belongs to the family of highly conserved TALE homeobox genes and interacts with PBX and HOX proteins to increase the affinity and specificity of HOX proteins [6] as well as CREB1 [2] in DNA binding. In Xenopus laevis, MEIS1 is involved in neural crest development [3]. Murine MEIS1 is essential for proximal-distal limb patterning [4] and plays a role in the Hox transcriptional regulatory network that specifies spinal motor neuron pool identity and connectivity [5]. In the CNS of the adult mouse, it is known to be expressed in cerebellar granule cells, the forebrain and the substantia nigra. While MEIS1 was initially identified in the context of acute myeloid leukemia [6], in recent years, a role in murine heart development has also been recognized [7] and SNPs in intron 8 (but in weak LD with the known RLS SNPs) play a role in determining atrioventricular conduction velocity as reflected by the length of the PR interval of the electrocardiogram in both Europeans and African Americans [8, 9].
The identified RLS locus in MEIS1 contains a large number of highly conserved noncoding regions (HCNRs) potentially functioning as cis-regulatory modules. Analyses of these HCNRs for allele-dependent enhancer activity in zebrafish and mice found that the risk allele of the lead SNP reduces it’s enhancer activity in the MEIS1 expression domain of the murine embryonic ganglionic eminences (GE). CREB1 binds to this enhancer and the identified RLS-SNP affects its binding in vitro. In addition, MEIS1 target genes suggest a role in the specification of neuronal progenitors in the GE. Thus, in vivo and in vitro analysis of a common SNP with small effect size showed allele-dependent function in the prospective basal ganglia representing the first neurodevelopmental region implicated in RLS. In addition, several rare non-synonymous variants in MEIS1 have been identified in RLS patients. However, coding variants in MEIS1 are very rare in general, possibly owing to the fact that MEIS1 represents one of the most highly conserved genes in the human genome. MEIS1 p.R272H has been identified in more than one RLS proband but also in controls [10]. Its location within the first amino acid of the homeobox domain could fuel discussion about a possible disruption of DNA binding. The pathogeneicity has been demonstrated in a large scale sequencing approach in RLS patients and controls followed by an in vivo complementation assay in the zebrafish [11]. Meis1−/− mice develop ocular and vascular defects, fail to produce megakaryocytes and display extensive hemorrhaging. They also die by embryonic day 14.5 [12]. Therefore, heterozygous Meis1-deficient mice have been investigated on a behavioral level and exhibit hyperactivity, resembling the RLS phenotype [13]. A second, independent association signal is located in an intergenic region approximately 1.3 Mb downstream of MEIS1 and potentially possesses long-range regulatory function with MEIS1 and ETAA1 as potential target genes [1, 14].
Furthermore, functional studies found a significant decrease in MEIS1 mRNA and protein expression in lymphoblastoid cell lines and brain tissue (pons and thalamus) from homozygous carriers of the risk haplotype when compared to homozygous carriers of the non-risk haplotype [15]. In a second study, knock-down of the [16] Meis1 orthologue unc-62 by RNA interference in Caenorhabditis elegans was related to increased ferritin expression and an extended lifespan. Samples from the thalamus but not the pons of RLS patients homozygous for the MEIS1 risk haplotype (n=9) compared to RLS patients carrying the protective haplotype (n=7),showed increased ferritin light and heavy chain as well as divalent metal transporter 1 (DMT1) mRNA and protein expression.. The authors argue that these data are in support of a disruption of physiological iron transport into the brain and—in conjunction with the also observed decrease of MEIS1 expression in in vitro cell models of iron deprivation—provide a functional link between the RLS gene, MEIS1 and the iron metabolism, which is believed to play a role in RLS pathogenesis [16].
Two different, but complementary approaches to studying the genetics of RLS in animals, forward and reverse genetic analysis, are presented here. In the case of forward genetics, investigators first examine the phenotypes and then search for the genes involved. Alternatively, reverse genetics starts with gene modifications, transgenic, knockout, etc., and then proceeds to the study of related phenotypes.
3.0 Impact of Btbd9 modification on RLS-like phenotypes in mice
Recently, two independent genome-wide association studies linked single nucleotide polymorphisms (SNPs) in four genes to increased risk for having RLS. The genes identified were MEIS1, MAP2K5, PTPRD, and BTBD9 [14, 17]. As SNPs in BTBD9 were found to carry an increased susceptibility to RLS in both studies, it made for an excellent gene to study RLS-like phenotypes in mice. The function of BTBD9 has not been elucidated, but has been suggested to be involved in iron homeostasis and protein ubiquitination. BTBD9 is expressed almost ubiquitously during development and adulthood. To advance the understanding of the function of BTBD9 and its potential role in RLS, a line of Btbd9 (the mouse homolog of human BTBD9) knockout (KO) mice was generated. The direct application of standard diagnostic crtieria for RLS all involve a report of a sensory event, i.e. an urge to move the legs {Allen, 2014 #3894}. These cannot be evaluated, in mice. Instead the Btbd9 mutant mice were thoroughly evaluated for similar, relevant phenotypes [20]. The test battery employed measured activity levels, sensory perception, sleep architecture, the dopaminergic system, and iron metabolism.
Activity levels were measured by two different tests. The first test, open field test, measures total spontaneous locomotor activity. The Btbd9 KO mice had increased locomotor activity compared to wild type (WT) mice. The other test used was the wheel running experiment. It uses a standard size home cage for a mouse that is equipped with a wheel with a sensor attached to a computer. The Btbd9 KO mice had a significant increase in activity compared to WT mice. Furthermore, there was no significant alteration in any of the circadian parameters of the Btbd9 KO mice.
Next, alterations in perception to a warm stimuli were analyzed using the tail flick test. The Btbd9 mutant mice have no sensory deficit during the middle of the night (the active phase), but have a sensory deficit during the middle of the day (the rest phase). Furthermore, mice were injected with 0.1 mg/kg of ropinirole. Ropinirole is a D2 dopamine receptor-like agonist with FDA approval for the treatment of RLS patients. Following this injection, there was no statistical difference between the Btbd9 KO mice and WT mice for warm stimuli sensations..
Additionally, sleep architecture was measured for the mutant mice using polysomnography (PSG). The uncontrollable urge to move often leads RLS patients to wake up during sleep in order to move around, thereby causing fragmentation of sleep. To analyze sleep architecture in free-moving mice, the mice were implanted with a wireless telemetry system (Data Sciences International). The Btbd9 KO mice had decreased sleep time and increased wake time, but no change in the amount of rapid-eye movement (REM) sleep during the rest phase.
Next, dopamine and other monoamine levels were measured in the striatum of the brain. There was no gross alteration in total dopamine or serotonin in the striatum of the brain, but there was alteration in 5-HIAA, a metabolite of serotonin, in the Btbd9 KO mice compared to WT mice. In a group of RLS patients, there is a significant decrease of CSF 5-HIAA compared to controls [21]. Additionally, there was a preferential increase in circling behavior in the counterclockwise direction in the Btbd9 KO mice compared to WT mice. Preferential changes in circling behavior have previously been linked to alterations in the dopaminergic system primarily in the striatum [22, 23].
Iron-deficiency anemia is a commonly reported symptom in RLS patients [24]. Using a colorimetric assay, total iron levels were measured in the serum of the Btbd9 KO mice and WT mice. Interestingly, the Btbd9 KO mice had elevated levels of iron in the serum. While this result is opposite of expected, not all RLS patients have iron alterations [25] and there is a subpopulation of RLS patients that have iron hemochromatosis, also known as iron overload [26, 27].
Taken together, these results suggest that the loss of Btbd9 in mice results in behavioral and iron abnormalities that have particular relevance to RLS, including locomotor activity, sensory functioning, and levels of monoamine and iron. Moreover, these results taken together suggest that BTBD9 is involved in iron metabolism and RLS.
4.0 The role of Btbd9 in iron regulation in Drosophila
The genetic toolbox, short lifespan, and roust sleep behavior of Drosophila make it an ideal research model for investigating the complexity of sleep disruptive disorders, including RLS [28]. Flies have a single BTBD9 homolog (the gene CG1826) that is 53.6% identical to the human protein [29]. The work by Freeman et al. [30, 31] has done much to investigate the relationship of BTBD9 and iron regulation using the Drosophila model, from which we’ll highlight their findings. The putative 3D structure of CG1826 conforms to a family of proteins that bind with Cullin-3 (e.g. CG1826 contains a canonical 3-box) further establishing this as a bona fide BTBD9 homolog. This is supported by the observation that Drosophila BTBD9 also binds to Cullin-3 in vivo. Flies that are deleted in Btbd9 are completely viable with a mildly shortened lifespan, but demonstrate reasonable phenotypes of RLS viz., increased locomotor activity, significant sleep fragmentation and responsiveness to dopamine agonists such as Pramipexole. Btbd9 in flies is widely expressed. However, add-back of wild type Btbd9 exclusively in the nervous system in a Btbd9 null animal restored sleep architecture to wild type status, suggesting a nervous system specific role for BTBD9 in sleep regulation.
Given the clinical correlation of RLS with iron handling and/or storage further investigation was made exploring the role for BTBD9 in iron storage and uptake. These experiments used mammalian HEK cell lines, which have been used previously to examine iron metabolism. Ferritin expression was used as a marker for measuring the status of iron storage in these cells. As expected, increased extracellular iron (through administration of FAC) caused an elevation in cellular Ferritin. By contrast, Btbd9 knockdown prevented this iron overload induced increase in Ferritin expression (stabilization by IRP2). Consistent with this observation, forced expression of Btrbd9 led to elevated Ferritin even in the presence of the iron chelator DFO. This suggested that BTBD9 is necessary and sufficient for the cell’s regulation of response to changes in iron. Additionally, this effect of BTBD9 on Ferritin expression (and thus, iron storage) was mediated through IRP2 (Iron-regulatory protein 2) – a protein that has been mechanistically linked to Ferritin mRNA stabilization [33, 34]. Consistent with this model, increased BTBD9 led to a decrease in cellular IRP2 levels. Finally, we measured free iron in flies that were mutated for Btbd9. Similar to observation in Btbdf9 mutant mice, we found that iron levels were reduced in mutants as compared to controls, while levels of Mn, Cu and Zn were unchanged [31, 35]. Taken together, these results strongly suggest a role for BTBD9 in the regulation of sleep architecture through specific iron regulatory functions in the nervous system, most likely limited to DA neurons.
5.0 Forward genetics analysis identifies a mouse strain that mimics many RLS-related phenotypes
RLS has both a well-documented biological phenotype of reduced brain iron particularly in the substantia nigra despite normal peripheral iron [35–38]. It also has a primary environmental factor of reduced peripheral iron related both to severity of RLS [39], and when severe with increased RLS prevalence [42] that can be corrected by reversing the iron deficiency [40]. One approach to establishing an animal model of RLS other than disrupting known genes would be to find an inbred mouse strain whose genetics produce the biological findings of RLS when exposed to the primary environmental factor, i.e. iron deprivation. This strain could then be examined for similarity to other behavioral and biological phenotypes of RLS. If the strain demonstrates the behavioral phenotypes of RLS then it can be evaluated for response to RLS treatments and if appropriately responsive may serve as a model for screening new medications. One of the major stumbling blocks in this chain is defining the behavioral phenotypes. RLS as a sensory disorder produce a behavior of an inability to stay at rest that is worse in the evening and night. The animal model can match these RLS behavioral phenotypes by showing increased activity or more significantly decreased rest time during the last part of active cycle. This approach has been taken over the past several years by the RLS research group at Johns Hopkins and Penn State University, mostly by Drs. Erica Unger and Byron Jones. The following summarizes this approach as another promising method for establishing an animal model of RLS.
Establishing an inbred murine strain with brain iron biology phenotype of RLS:
The BXD 40 recombinant murine strains were selected because previous work showed that among a panel of 22 BXD recombinant inbred strains, following being fed an iron deficient diet, this strain showed the greatest loss of iron in the ventral midbrain while showing minimal effect on hemoglobin. Twenty-two of the BXD strains and the 2 progenitor strains were evaluated for brain iron after 100 days post-weaning on an iron deficient diet containing 3–5 ppm iron compared to same strain controls on a normal diet of 240 ppm iron. The iron levels were evaluated for the ventral mid-brain (VMB) containing the substantia nigra for both ID and controls. The VMB iron concentrations for controls varied by about 25% over the strains while the difference between control and ID for each strain varied between 0 and 35% [41]. Moreover there was no correlation between the ID effects on VMB and hemoglobin or other peripheral measures of iron (data from a prior study [42] (see Figure 1). One mouse strain (strain 40 females) showed the biological phenotype of response to iron deficiency producing reduced VMB iron with minimal change in hemoglobin.
Figure 1.
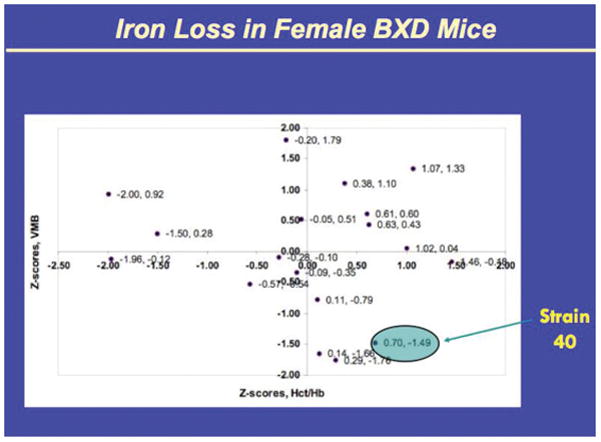
(provided by Dr. Byron Jones). The Z scores represent the difference between ID and controls presented here for 18 murine strains
5.1 Evaluating behavioral phenotypes of BXD40 females
Dr. Erica Unger (personal communication) has completed an analysis of the 24-hour activity levels of the BXD 40 compared to 5 other BXD strains, all females. The BXD 40 mice showed with dietary iron deficiency (ID) increased activity in the last part of the active cycle and the first part of rest cycle. No other strain showed this circadian change in behavior. The increased activity and decreased rest time in the last 2 hours of the active period both differed significantly for the ID compared to normal diet, but there was no similar difference for the rest period. Thus the BXD40 strain showed the two features of the behavioral phenotypes of RLS: decreased resting and critical circadian times.
5.2 Evaluating RLS medication response of BXD40 females
Preliminary data from Dr. Unger (personal communication) showed that treating the BXD40 females with either levodopa 25 mg or quinpirole 12 mg given at 3 hours before the end of the active cycle significantly reversed the lower rest time and decreased the increased activity produced by the ID diet. The effect lasted for about 2–3 hours covering the circadian time of the ID induced behavior phenotype of RLS.
5.3 Conclusions
A genetic animal model chosen for matching the brain iron biological phenotype of RLS also showed both the full behavioral RLS phenotype and also the expected response to the medications treating RLS. It should be noted that the strain was selected without any consideration of the dopamine system or any alteration of genetics. This provides a genetically determine animal model of RLS (BXD40 females) selected for brain and peripheral iron regulation matching RLS that also shows both the behavioral phenotype of RLS and the expected response to dopaminergic treatment.
6.0 The PLMS motor sign of RLS in animal models of RLS?
Four main strategies to mimic the RLS syndrome have been tested: 1) pharmacological (anti-dopaminergic) [43]; 2) lesion (spinal or cerebral-A11 region) [44, 45]; 3) genetic [13]; 4) iron deficiency [46, 47]. A limitation in these trials is the absence of specific objective markers to recognize an experimental RLS phenotype [48]. Among the possible outcome measures usually considered by the authors, the following are the most targeted: locomotor activity (open field or wheel running activity) [49]; sleep (disruption/fragmentation) [46]; spinal pain reflex [17]. But a significant increase in locomotor activity in human RLS has not been demonstrated, and insomnia might be documented for the majority but not for all RLS patients. A consensus agreement on the choice of the endpoints/outcome measures to validate a RLS behavior in animals is an important step that needs to be taken to produce trustable results. Beside insomnia, periodic limb movements (PLM) in sleep represent the only objective marker in human RLS. However, PLM are highly sensitive for RLS but much less specific, occurring up to 30% of adult general population [49]. However, PLM may anticipate RLS, and the correlation between specific polymorphisms is higher with PLM than with RLS [17]. Moreover, PLM are highly sensitive to dopamine-agonists, thus very suitable for rescue experiments [50]. Nevertheless, few steps are necessary before PLM can be routinely integrated in animal experiments. First, there are no present standard procedures to record limb movements (LMs) in animals, no scoring rules to recognize and quantify them, and overall no information on physiological limb movement activity during sleep in normal animals. Recently, Silvani et al. validated a technique to record tibialis anterior muscles activity during sleep and wake, in a polysomnographic context, in freely moving rats and mice (figure 1) [51]. Further, the same authors elaborated a computerized algorithm to identify automatically LMs, which accelerated significantly their computation. Finally, they proposed and validated new visual scoring rules for LMs. This allowed the authors to depict the physiological time structure of the EMG events and compared it to that of humans [52]. Rodents produced significantly more LMs during NREM sleep and more bilateral than unilateral LMs than humans. However, since a significant part of animal LMs occurred in only one limb, it is recommend to record both tibialis anterior muscles also in animals. The mean duration of LMs was similar, around 1 s in rodents and 2 s in humans. The resulted time structure of physiological EMG activity during NREM sleep in mice, rats and humans was very similar, with a single mode distribution of the time intervals between LMs peaking between 2 and 4 s and extending up to 10 s. This means that in healthy animals and in normal young humans the peak located around 20 s. In both animals and humans LMs separated by an inter-movement interval lower than 10 s occur in short sequences of 2 and 3 events, with a tendency for longer sequences in mice. The above described EMG pattern was stable across the same night. These results paved the way to a future routine implementation of the tibialis anterior activity during sleep among the other outcome measures in experimental genetic mouse models and pharmacological/lesion rat models of RLS.
8.0 Conclusions
Animal models of complex diseases such as RLS are often met with skepticism as to how faithfully they capture the human correlate. Progress is often slow and knowledge accrues iteratively from complementary lines of inquiry and different methodologies before converging upon a more comprehensive picture that ultimately leads to diagnostic and treatment advances. In this review, we show how animal models derived largely from our appreciation of the principal components of the disease phenotype and human genetics, have contributed to the understanding of bio-behavioral complexity and pharmacological and molecular basis of RLS. Also, the genetic-based mechanisms appear to be remarkably conserved amongst species. In fact, a genetic homolog to MEIS1 found in nematodes has been shown to regulate ferritin [53]. Finally, animal models using genetic reference populations can serve as the material for examination of genes and biochemical pathways identified by epidemiological and especially GWAS. Indeed, Ermann and Glimcher [54] made such a proposal in their article, “After GWAS, Mice to the Rescue? Recall from above that Winkelmann and colleagues [14} and Steffanson and colleagues [17] identified BTBD9 as one of several candidate genes related to RLS and periodic limb movements. In their study of iron regulation in the ventral midbrain of recombinant inbred mice, Jones and colleagues [55], identified Btbd9 as candidate for the regulation of iron in this tissue. Li and colleagues then followed up to show how manipulation of this gene in mice could replicate several of the RLS-like phenotypes [20].
9.0 Future work
The animal model research described here has focused on the biobehavioral and underlying genetic features of RLS. While the discussion featured the genes, MEIS1 and BTBD9, several other genes, including MAP2K5 and PTPRD have also been linked to RLS. As human genetic studies progress, it is likely that more genes will be identified as well as additional pathophysiological pathways. It is therefore likely that we will observe hitherto unobserved RLS-related behaviors. Advanced genetic studies in humans will reveal gene networks that can be studied for mechanisms in animals, showing that there is still much work to be done and that the value of model organisms will be integral to that effort.
Figure 2.
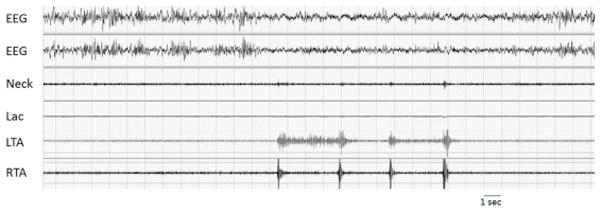
A brief series of bilateral pseudoperiodic limb movements during NREM sleep in a healthy rat recorded by intramuscular electrodes in both tibialis anterior in a polysomnographic context. Lac: locomotor activity, LTA and RTA: left and right tibialis anterior.
Highlights.
Animal models of RLS varying by species and complexity are presented
Forward and reverse genetic models are described and compared
Animal models in genetic reference populations complement human GWAS studies.
Acknowledgments
This work was supported in part by USPHS Grants P01-AG21190 and R01 NS075184 (RPA & BCJ), R01 NS082244 and Restless Legs Syndrome Foundation (YL).
Footnotes
Gene/protein notations:
BTBD9, Btbd9: Human, animal gene
BTBD9. Btbd9: Human, animal protein
Unless specified, assume protein.
References
- 1.Winkelmann J, Czamara D, Schormair B, et al. Genome-wide association study identifies novel restless legs syndrome susceptibility Loci on 2p14 and 16q12. 1. PLoS Genet. 2011;7:e1002171. doi: 10.1371/journal.pgen.1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Iwasaki M, Ficara F, et al. GSK-3 promotes conditional association of CREB and its coactivators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. Cancer Cell. 2010;17:597–608. doi: 10.1016/j.ccr.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda R, Mood K, Jones TL, Aruga J, Buchberg AM, Daar IO. Xmeis1, a protooncogene involved in specifying neural crest cell fate in Xenopus embryos. Oncogene. 2001;20:1329–42. doi: 10.1038/sj.onc.1204250. [DOI] [PubMed] [Google Scholar]
- 4.Mercader N, Leonardo E, Azpiazu N, et al. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402:425–9. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- 5.Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–91. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–43. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stankunas K, Shang C, Twu KY, et al. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ Res. 2008;103:702–9. doi: 10.1161/CIRCRESAHA.108.175489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–9. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JG, Magnani JW, Palmer C, et al. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7:e1001304. doi: 10.1371/journal.pgen.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vilarino-Guell C, Chai H, Keeling BH, et al. MEIS1 p. R272H in familial restless legs syndrome. Neurology. 2009;73:243–5. doi: 10.1212/WNL.0b013e3181ae7c79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulte EC, Kousi M, Tan PL, et al. Targeted resequencing and systematic in vivo functional testing identifies rare variants in MEIS1 as significant contributors to restless legs syndrome. Am J Hum Genet. 2014;95:85–95. doi: 10.1016/j.ajhg.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hisa T, Spence SE, Rachel RA, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–9. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spieler D, Kaffe M, Knauf F, et al. Restless Legs Syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. 2014;24:592–603. doi: 10.1101/gr.166751.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkelmann J, Schormair B, Lichtner P, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–6. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 15.Xiong L, Catoire H, Dion P, et al. MEIS1 intronic risk haplotype associated with restless legs syndrome affects its mRNA and protein expression levels. Hum Mol Genet. 2009;18:1065–74. doi: 10.1093/hmg/ddn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catoire H, Dion PA, Xiong L, et al. Restless legs syndrome-associated MEIS1 risk variant influences iron homeostasis. Ann Neurol. 2011;70:170–5. doi: 10.1002/ana.22435. [DOI] [PubMed] [Google Scholar]
- 17.Stefansson H, Rye DB, Hicks A, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–47. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 18.Allen RP, Picchietti D, Hening WA, Trenkwalder C, Walters AS, Montplaisi J. Restless legs syndrome: diagnostic criteria, special considerations, and epidemiology. A report from the restless legs syndrome diagnosis and epidemiology workshop at the National Institutes of Health. Sleep Med. 2003;4:101–19. doi: 10.1016/s1389-9457(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 19.Allen RP, Picchietti DL, Garcia-Borreguero D, et al. Restless legs syndrome/Willis-Ekbom disease diagnostic criteria: updated International Restless Legs Syndrome Study Group (IRLSSG) consensus criteria--history, rationale, description, and significance. Sleep Med. 2014;15:860–73. doi: 10.1016/j.sleep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 20.DeAndrade MP, Johnson RL, Jr, Unger EL, et al. Motor restlessness, sleep disturbances, thermal sensory alterations and elevated serum iron levels in Btbd9 mutant mice. Hum Mol Genet. 2012;21:3984–92. doi: 10.1093/hmg/dds221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earley CJ, Hyland K, Allen RP. CSF dopamine, serotonin, and biopterin metabolites in patients with restless legs syndrome. Mov Disord. 2001;16:144–9. doi: 10.1002/1531-8257(200101)16:1<144::aid-mds1009>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 22.Kim DS, Szczypka MS, Palmiter RD. Dopamine-deficient mice are hypersensitive to dopamine receptor agonists. J Neurosci. 2000;20:4405–13. doi: 10.1523/JNEUROSCI.20-12-04405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viggiano D, Vallone D, Ruocco LA, Sadile AG. Behavioural, pharmacological, morpho-functional molecular studies reveal a hyperfunctioning mesocortical dopamine system in an animal model of attention deficit and hyperactivity disorder. Neurosci Biobehav Rev. 2003;27:683–9. doi: 10.1016/j.neubiorev.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Allen RP, Earley CJ. The role of iron in restless legs syndrome. Mov Disord. 2007;22:S440–S8. doi: 10.1002/mds.21607. [DOI] [PubMed] [Google Scholar]
- 25.Matthews WB. Letter: Iron deficiency and restless legs. Br Med J. 1976;1:898. doi: 10.1136/bmj.1.6014.898-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haba-Rubio J, Staner L, Petiau C, Erb G, Schunck T, Macher JP. Restless legs syndrome and low brain iron levels in patients with haemochromatosis. J Neurol Neurosurg Psychiatry. 2005;76:1009–10. doi: 10.1136/jnnp.2003.030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaughnessy P, Lee J, O’Keeffe ST. Restless legs syndrome in patients with hereditary hemochromatosis. Neurology. 2005;64:2158. doi: 10.1212/01.WNL.0000165954.42289.03. [DOI] [PubMed] [Google Scholar]
- 28.Donelson NC, Sanyal S. Use of Drosophila in the investigation of sleep disorders. Experimental Neurology. 2015;274:72–9. doi: 10.1016/j.expneurol.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 29.Geer LY, Marchler-Bauer A, Geer RC, et al. The NCBI BioSystems database. Nucleic Acids Research. 2009;38:D492–D6. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freeman A, Pranski E, Miller RD, et al. Sleep fragmentation and motor restlessness in a Drosophila model of Restless Legs Syndrome. Curr Biol. 2012;22:1142–8. doi: 10.1016/j.cub.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman AA, Mandilaras K, Missirlis F, Sanyal S. An emerging role for Cullin-3 mediated ubiquitination in sleep and circadian rhythm: insights from Drosophila. Fly (Austin) 2013;7:39–43. doi: 10.4161/fly.23506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freeman A, Pranski E, Miller RD, et al. Sleep Fragmentation and Motor Restlessness in a Drosophila Model of Restless Leg Syndrome. Current Biology. 2012;22:1142–8. doi: 10.1016/j.cub.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salahudeen AA, Thompson JW, Ruiz JC, et al. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science. 2009;326:722–6. doi: 10.1126/science.1176326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vashisht AA, Zumbrennen KB, Huang X, et al. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science. 2009;326:718–21. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen RP, Barker PB, Wehrl FW, Song HK, Earley CJ. MRI measurement of brain iron in patients with restless legs syndrome. Neurology. 2001;56:263–5. doi: 10.1212/wnl.56.2.263. [DOI] [PubMed] [Google Scholar]
- 36.Earley CJ, Connor JR, Beard JL, Malecki EA, Epstein DK, Allen RP. Abnormalities in CSF concentrations of ferritin and transferrin in restless legs syndrome. Neurology. 2000;54:1698–700. doi: 10.1212/wnl.54.8.1698. [DOI] [PubMed] [Google Scholar]
- 37.Godau J, Schweitzer KJ, Liepelt I, Gerloff C, Berg D. Substantia nigra hypoechogenicity: definition and findings in restless legs syndrome. Mov Disord. 2007;22:187–92. doi: 10.1002/mds.21230. [DOI] [PubMed] [Google Scholar]
- 38.Rizzo G, Manners D, Testa C, et al. Low brain iron content in idiopathic restless legs syndrome patients detected by phase imaging. Mov Disord. 2013;28:1886–90. doi: 10.1002/mds.25576. [DOI] [PubMed] [Google Scholar]
- 39.Sun ER, Chen CA, Ho G, Earley CJ, Allen RP. Iron and the restless legs syndrome. Sleep. 1998;21:371–7. [PubMed] [Google Scholar]
- 40.Mehmood T, Auerbach M, Earley CJ, Allen RP. Response to intravenous iron in patients with iron deficiency anemia (IDA) and restless leg syndrome (Willis-Ekbom disease) Sleep Med. 2014;15:1473–6. doi: 10.1016/j.sleep.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 41.Jellen LC, Unger EL, Lu L, et al. Systems genetic analysis of the effects of iron deficiency in mouse brain. Neurogenetics. 2012;13:147–57. doi: 10.1007/s10048-012-0321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin L, Unger EL, Jellen LC, et al. Systems genetic analysis of multivariate response to iron deficiency in mice. Am J Physiol Regul Integr Comp Physiol. 2012;302:R1282–96. doi: 10.1152/ajpregu.00634.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baier PC, Winkelmann J, Hohne A, Lancel M, Trenkwalder C. Assessment of spontaneously occurring periodic limb movements in sleep in the rat. J Neurol Sci. 2002;198:71–7. doi: 10.1016/s0022-510x(02)00078-3. [DOI] [PubMed] [Google Scholar]
- 44.Esteves AM, de Mello MT, Lancellotti CL, Natal CL, Tufik S. Occurrence of limb movement during sleep in rats with spinal cord injury. Brain Res. 2004;1017:32–8. doi: 10.1016/j.brainres.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 45.Ondo WG, He Y, Rajasekaran S, Le WD. Clinical correlates of 6-hydroxydopamine injections into A11 dopaminergic neurons in rats: a possible model for restless legs syndrome. Mov Disord. 2000;15:154–8. doi: 10.1002/1531-8257(200001)15:1<154::aid-mds1025>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 46.Manconi M, Hutchins W, Feroah TR, Zucconi M, Ferini-Strambi L. On the pathway of an animal model for restless legs syndrome. Neurol Sci. 2007;28(Suppl 1):S53–60. doi: 10.1007/s10072-007-0738-8. [DOI] [PubMed] [Google Scholar]
- 47.Qu S, Le W, Zhang X, Xie W, Zhang A, Ondo WG. Locomotion is increased in a11-lesioned mice with iron deprivation: a possible animal model for restless legs syndrome. J Neuropathol Exp Neurol. 2007;66:383–8. doi: 10.1097/nen.0b013e3180517b5f. [DOI] [PubMed] [Google Scholar]
- 48.Clemens S, Hochman S. Conversion of the modulatory actions of dopamine on spinal reflexes from depression to facilitation in D3 receptor knock-out mice. J Neurosci. 2004;24:11337–45. doi: 10.1523/JNEUROSCI.3698-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haba-Rubio J, Marti-Soler H, Marques-Vidal P, et al. Prevalence and Determinants of Periodic Limb Movements in the General Population. Ann Neurol. 2015 doi: 10.1002/ana.24593. [DOI] [PubMed] [Google Scholar]
- 50.Manconi M, Ferri R, Zucconi M, et al. First night efficacy of pramipexole in restless legs syndrome and periodic leg movements. Sleep Med. 2007;8:491–7. doi: 10.1016/j.sleep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 51.Silvani A, Martire VL, Salvade A, et al. Physiological time structure of the tibialis anterior motor activity during sleep in mice, rats and humans. J Sleep Res. 2015;24:695–701. doi: 10.1111/jsr.12319. [DOI] [PubMed] [Google Scholar]
- 53.Catoire H, Dion PA, Xiong L, et al. Restless legs syndrome-associated MEIS risk variant influences iron homeostasis. Ann Neurol. 2011;70:170–175. doi: 10.1002/ana.22435. [DOI] [PubMed] [Google Scholar]
- 52.Ferri R. The time structure of leg movement activity during sleep: The theory behind the practice. Sleep Med. 2012;13:433–41. doi: 10.1016/j.sleep.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 54.Ermann J, Glimcher LH. After GWAS: mice to the rescue? Curr Opin Immunol. 2012;24:564–570. doi: 10.1016/j.coi.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones LC, Earley CJ, Allen RP, Jones BC. Of mice and men, periodic limb movements and iron: how the human genome informs the mouse genome. Genes Brain Behav. 2008;7:513–514. doi: 10.1111/j.1601-183X.2008.00400.x. [DOI] [PubMed] [Google Scholar]


