Abstract
Background
IL-1β is an essential factor of inflammation initiation, and it also promotes malignant transformation, indicating its tumorigenic property. We aimed to investigate the correlation between IL-1β and miR-144-3p as well as their prognostic values in LUAD and LUSC patients.
Results
The IL-1β level in both LUAD and LUSC patients was significantly higher than that of healthy donors (P < 0.001). In both populations, patients with low IL-1β level had better prognosis than high IL-1β level (P < 0.001 and P = 0.010, respectively). In A549 cells, miR-144 showed the biggest expression change (−4.38 fold) after IL-1β exposure. In LUAD patients, a negative correlation was detected between IL-1β and miR-144-3p (r = –0.540, P < 0.001) and the high miR-144-3p group had better prognosis (P = 0.003), which was validated by TCGA data. Clinical stage, IL-1β and miR-144-3p were independent risk factors in LUAD patients. In vitro, IL-1β and miR-144-3p antagomir could enhance proliferation and miR-144-3p mimics would attenuate the promoting effect of IL-1β.
Materials and Methods
ELISA and qRT-PCR were applied respectively to detected cytokines and miR-144-3p in 129 LUAD, 54 LUSC and 40 healthy donors. Moreover, miRNA array was carried out for miRNA profiling. TCGA database was employed for validation, and follow up data were collected for prognosis evaluation. MTT assay and western-blot were carried out for proliferation evaluation.
Conclusions
In LUAD patients, the serum IL-1β level was correlated with miR-144-3p may affect miR-144-3p at transcriptional level. Both of them were independent risk factors for LUAD prognosis. In addition, IL-1β and miR-144-3p might mediate inflammation-promoted tumorigenesis in LUAD patients.
Keywords: LUAD, LUSC, IL-1β, miR-144-3p, prognosis
INTRODUCTION
Lung cancer remains the leading cause of cancer-related death worldwide. It accounts for approximately 1.58 million deaths in 2015 in USA, and such deaths are projected to continue rising [1]. According to histological characteristics, lung cancer can be categorized into non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC). In NSCLC, lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) account for 60% and 25% of this population, respectively [2].
Besides histological differences, many aspects, including genetics, pathogenesis, biological behavior, treatment and prognosis, significantly vary between LUAD and LUSC [3–7]. As a result, it is highly recommended by both scientists and clinical doctors to distinguish them as two individual diseases.
It has been well established that chronic inflammation and infection are pivotal factors of carcinogenesis [8]. Interleukin-1 beta (IL-1β), which is produced by activated macrophages as a proprotein and processed to its active form by Caspase 1, is a member of the IL-1 cytokine family [9]. IL-1β is an important mediator of the inflammatory response and involved in a variety of cell behaviors. The induction of cyclooxygenase-2 (PTGS2/COX2) by IL-1β in the central nervous system (CNS) is found to contribute to inflammatory pain hypersensitivity [10]. Besides, IL-1β promotes malignant transformation and tumor aggressiveness in oral cancer [11], indicating its tumorigenic role.
Recently, mounting evidence has shown that microRNAs (miRNAs) are a novel class of inflammation-associated molecules and involved in multi-stages of tumor development, including initiation, tumor progression, invasion and metastasis [12–14]. Tumor-related miRNAs can be regulated/influenced by various inflammatory stimuli, including cytokines [15–17]. Lin et al. demonstrated that IL-1β upregulates Lin28B by downregulating miR-101 and promotes proliferation and migration of NSCLC cells, defining an IL-1β/miR-101/Lin28B pathway in NSCLC [18].
As NSCLC is a tissue specific tumor, its corresponding treatment strategies are dissimilar widely. Oncologists and scientists currently consider that LUAD and LUSC are two individual diseases with completely different genetic background [3]. In this study, we detected cytokines in both LUAD and LUSC patients and found that the IL-1β levels in both LUAD and LUSC patients were significantly higher than that of healthy donors. Moreover, in both LUAD and LUSC, low IL-1β cases had better prognosis that high IL-1β ones. In addition, miR-144-3p was a potential downstream miRNA correlated with and influenced by IL-1β in LUAD. LUAD patients with high miR-144-3p level had better prognosis. Based on TCGA database, the miR-144-3p level in tumor tissue was crucially higher than that in adjacent area in both LUAD and LUSC. However, miR-144-3p was associated with prognosis only in LUAD population, which was consistent with our results. IL-1β and miR-144-3p suppression could enhance proliferation of LUAD cells. Therefore, our findings suggested a potential link between IL-1β and miR-144-3p, which might be associated with LUAD prognosis, providing new insights into inflammation-promoted tumorigenesis.
RESULTS
Comparison of various cytokines in serum from LUAD, LUSC and healthy donors
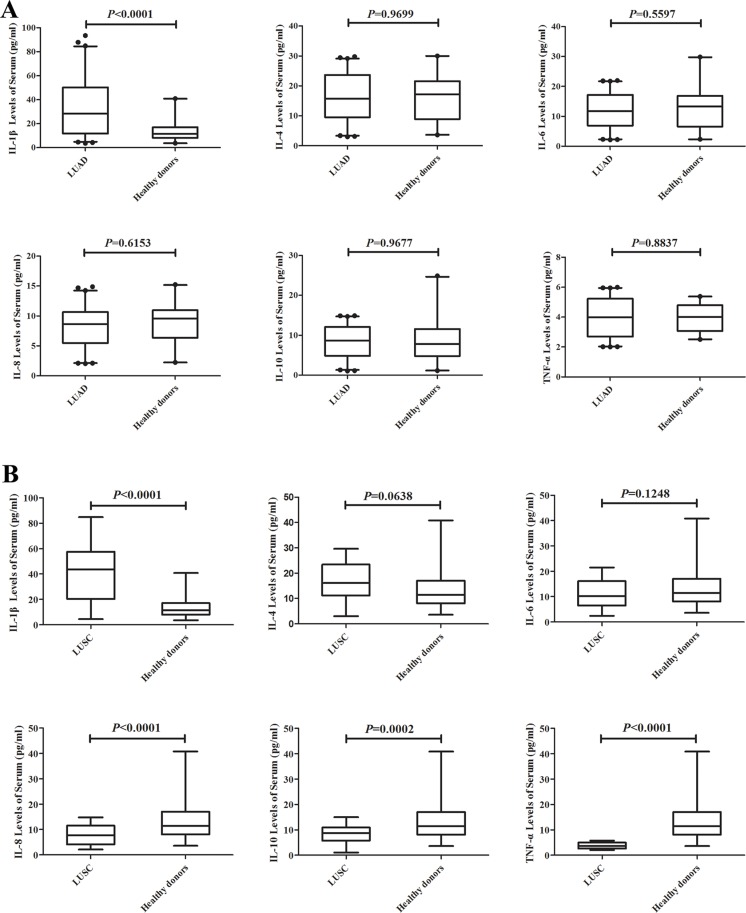
A total of six cytokines were detected using ELISA method in LUAD, LUSC patients and healthy donors. The results showed that the IL-1β level in LUAD patients was significantly higher than thatof healthy donors (P < 0.001), whereas levels of IL-4, IL-6, IL-8, IL-10 or TNF-α slightly varied between LUAD patients and healthy donors (Figure 1A). The IL-1β level in LUSC patients was significantly higher than that of healthy donors, while levels of IL-8, IL-10 and TNF-α were lower in LUSC than healthy donors (P < 0.001). However, IL-4 and IL-6 levels slightly varied between the two groups (Figure 1B).
Figure 1. Comparison of various cytokines in serum from a set of 129 LUAD, 54 LUSC and 40 healthy donors.
IL-1β level in LUAD patients was significantly higher than Healthy donors (P < 0.001), whereas IL-4, IL-6, IL-8, IL-10 or TNF-α levels varied slightly between LUAD and healthy donors (A); IL-1β level in LUSC patients were significantly higher than healthy donors, while IL-8, IL-10 and TNF-α were lower in LUSC than healthy donors (P < 0.001), however, IL-4 and IL-6 levels varied slightly between the two groups (B).
Association between IL-1β and clinical features in LUAD and LUSC patients
The IL-1β level was defined as “high” and “low” using median (28.3992 pg/mL and 43.5495 pg/mL in LUAD and LUSC, respectively) as threshold. IL-1β level of LUAD and LUSC patients was analyzed according to age, gender, smoking status and pathological stage, as well as target therapy status in LUAD patients. The results showed that in LUSC patients, the IL-1β level was associated with pathological stage (P < 0.05). However, no significant associations were observed between IL-1β level and other above-mentioned clinical features in either LUAD or LUSC population (Table 1).
Table 1. Association between IL-1β and clinical features in LUAD and LUSC patients.
| Characteristics | LUAD | LUSC | ||||||
|---|---|---|---|---|---|---|---|---|
| All patients | IL-1β Low (< mediana) | IL-1β High (≥ mediana) | p | All patients | IL-1β Low (< medianb) | IL-1β High (≥ medianb) | p | |
| n | 129 | 64 (49.60%) | 65 (50.40%) | 54 | 27 (50.00%) | 27 (50.00%) | ||
| Age (years) | 0.842 | 0.695 | ||||||
| Median (range) | 65 (48–85) | 65 (49–84) | 65 (48–85) | 64 (50–85) | 64 (50–83) | 64 (51–85) | ||
| Gender | 0.401 | 0.761 | ||||||
| Male | 93 (72.09%) | 44 (34.11%) | 49 (37.98%) | 39 (%72.22) | 20 (37.05%) | 19 (35.17%) | ||
| Female | 36 (27.91%) | 20 (15.50%) | 16 (12.40%) | 15 (27.78%) | 7 (12.97%) | 8 (14.81%) | ||
| Smoke | 0.334 | 0.248 | ||||||
| Yes | 61 (47.29%) | 33 (25.58%) | 28 (21.71%) | 36 (66.67%) | 20 (37.04%) | 16 (29.63%) | ||
| No | 68 (52.71%) | 31 (24.03%) | 37 (28.68%) | 18 (33.33%) | 7 (12.96%) | 11 (20.37%) | ||
| Pathological stagec | 0.330 | 0.023 | ||||||
| IA | 17 (13.18%) | 11 (8.53%) | 6 (4.65%) | 11 (20.37%) | 8 (14.81%) | 3 (5.56%) | ||
| IB | 34 (26.36%) | 20 (15.50%) | 14 (10.85%) | 10 (18.52%) | 8 (14.81%) | 2 (3.71%) | ||
| IIA | 23 (17.83%) | 12 (9.30%) | 11 (8.53%) | 11 (20.37%) | 5 (9.26%) | 6 (11.11%) | ||
| IIB | 32 (24.81%) | 13 (10.08%) | 19 (14.73%) | 13 (24.07%) | 5 (9.26%) | 8 (14.81%) | ||
| IIIA | 11 (8.53%) | 4 (3.10%) | 7 (5.43%) | 8 (14.81%) | 1 (1.85%) | 7 (12.96%) | ||
| IIIB | 12 (9.30%) | 4 (3.10%) | 8 (6.20%) | 1 (1.85%) | 0 (0.00%) | 1 (1.85%) | ||
| Target therapy | 0.537 | NTd | ||||||
| Yes | 65 (50.39%) | 34 (26.36%) | 31 (24.03%) | NTd | NTd | NTd | ||
| No | 64 (49.61%) | 30 (23.26%) | 34 (26.36%) | NTd | NTd | NTd | ||
Mediana = 28.3992 pg/ml Medianb = 43.5495 pg/ml NTd = Not Tested
Pathological stageC was based on NCCN Guidelines Version 3.2011 NSCLC.
Prognosis evaluation between high and low cytokine levels in LUAD and LUSC patients
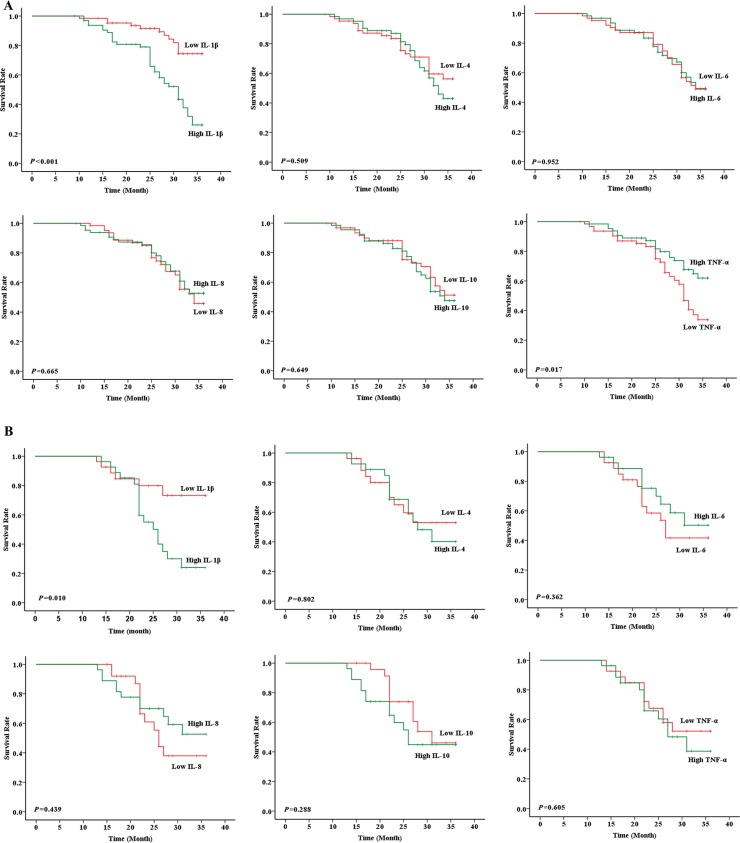
Levels of cytokines were defined as “high” and “low” using medians as thresholds. In both LUAD (Figure 2A) and LUSC (Figure 2B) populations, patients with low IL-1β level showed better prognosis than those with high IL-1β level (P < 0.001 and P = 0.010, respectively). In LUAD population, patients with high TNF-α level exhibited better prognosis than those with low TNF-α level (P = 0.017). No significant differences in prognosis were observed between patients with high and low IL-4, IL-6, IL-8 or IL-10 levels (Figure 2).
Figure 2. Prognosis evaluation between high and low cytokine levels in LUAD and LUSC patients.
In LUAD, patients with low IL-1β and high TNF-α levels showed better prognosis than those with high IL-1β (P < 0.001) and low TNF-α (P = 0.017) levels, respectively, whereas no significant differences in prognosis were observed between patients with high and low IL-4 (P = 0.509), IL-6 (P = 0.952), IL-8 (P = 0.665) or IL-10 (P = 0.649) levels (A); In LUSC, patients with low IL-1β level had better prognosis than high IL-1β (P = 0.010) level. However, no significant prognostic differences were detected between patients with high and low IL-4 (P = 0.802), IL-6 (P = 0.362), IL-8 (P = 0.439), IL-10 (P = 0.288) or TNF-α (P = 0.605) levels (B).
The prognostic value of miR-144-3p and its correlation with IL-1β
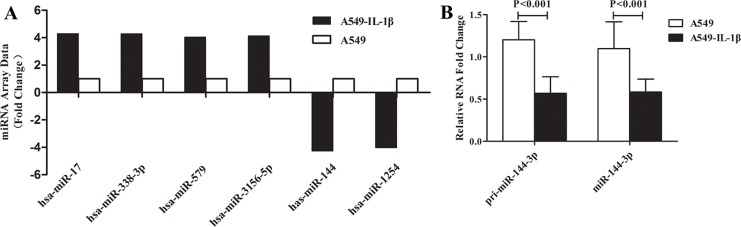
Figure 3A reveals that hsa-miR-17, hsa-miR-338-3p, hsa-miR-579, hsa-miR-3156-5p, hsa-miR-144 and hsa-miR-1254 ranked as the top six mostly altered miRNAs after IL-1β exposure in A549 cells, with an expression change greater than ± 4 fold-change, among which hsa-miR-144 showed the biggest change (−4.38 fold). Real-time PCR was carried out to validate the array data, and results showed that both pri-miR-144-3p and miR-144-3p were down-regulated in A549-IL-1β group (P < 0.05), indicating that IL-1β could regulate the transcription of miR-144-3p.
Figure 3. IL-1β was involved in regulation of miR-144-3p at the transcriptional level.
Via miRNA array, a total of six miRNAs varied greater than 4 folds (including miR-144-3p) in A549 cells after high IL-1β exposure. (A); Real-time PCR validated the result of miRNA array and showed that both pri-miR-144-3p and miR-144-3p were down-regulated in A549-IL-1β group (P < 0.05, B).
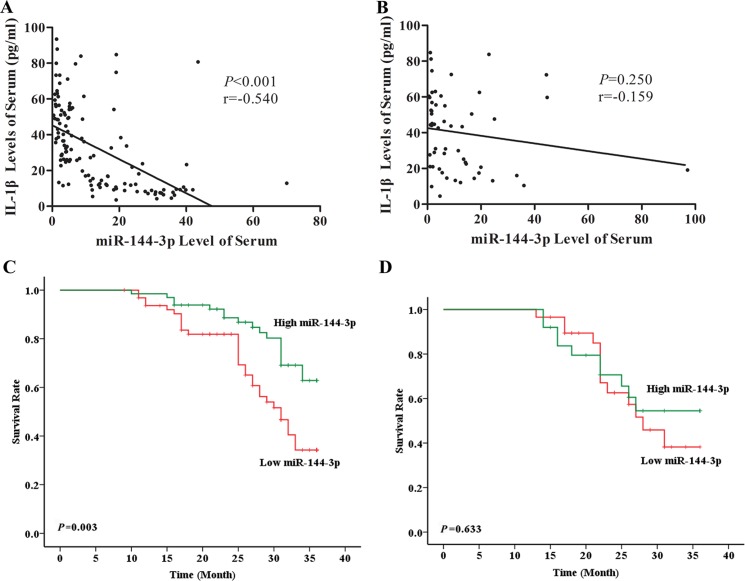
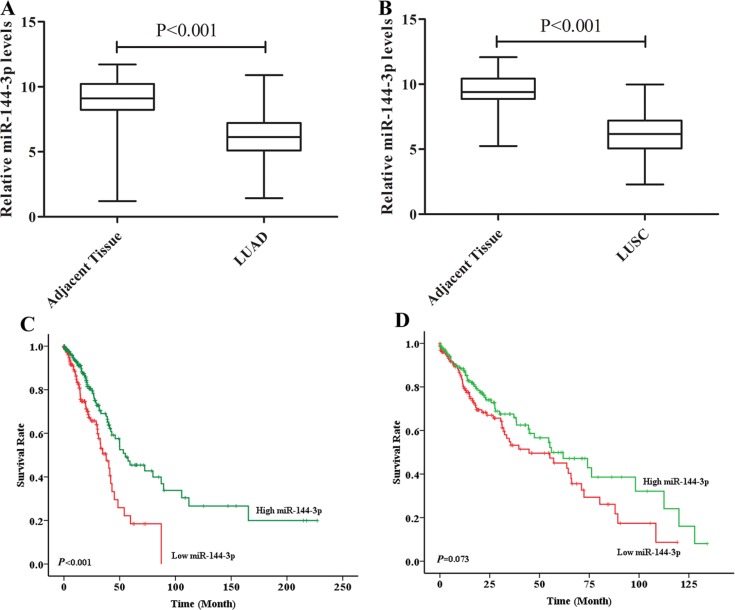
In LUAD patients, a negative correlation was detected between IL-1β and miR-144-3p (r = −0.540, P < 0.001, Figure 4A), and the high miR-144-3p group had better prognosis than low miR-144-3p group (P = 0.003, Figure 4C). In contrast, miR-144-3p was neither correlated with IL-1β (r = −0.159, P = 0.250, Figure 4B) nor prognosis (P = 0.633, Figure 4D) in LUSC patients. TCGA data displayed that miR-144-3p was significantly higher in both tumor tissues compared with adjacent area in both LUAD (Figure 5A) and LUSC (Figure 5B) populations, but its expression was only associated with prognosis in LUAD patients (Figure 5C and 5D). By χ2 test, we confirmed that miR-144-3p was correlated with IL-1β only in LUAD patients (Table 2).
Figure 4. Serum miR-144-3p level was correlated with IL-1β level and associated with prognosis in LUAD.
Via Pearson correlation analysis, serum level of miR-144-3p was negatively correlated with IL-1β (r = −0.540, P < 0.001) (A) and LUAD patient with high miR-144-3p level showed significantly better prognosis than the ones with low miR-144-3p level (P = 0.003) (C); In the contrary, in LUSC patients, serum miR-144-3p level was neither correlated with IL-1β (r = −0.159, P = 0.250) (B) nor associated with prognosis (P = 0.633) (D).
Figure 5. TCGA data validated the correlation of miR-144-3p and prognosis.
Based on TCGA database, miR-144-3p level in tumor tissue was crucially higher than that in adjacent area in both LUAD (P < 0.001) (A) and LUSC (P < 0.001) (B) patients; in LUAD, patients with high miR-144-3p had significantly longer OS than low miR-144-3p group (P < 0.001) (C), while no distinct prognostic difference was found in LUSC population (P = 0.073) (D).
Table 2. Correlation between miR-144-3p and clinical features in LUAD and LUSC patients.
| LUAD | LUSC | |||||||
|---|---|---|---|---|---|---|---|---|
| Characteristics | All patients | miR-144-3p Low (< mediana) | miR-144-3p High (≥ mediana) | p | All patients | miR-144-3p Low (< medianb) | miR-144-3p High (≥ medianb) | p |
| n | 129 (100%) | 64 (49.61%) | 65 (50.39%) | 54 (100%) | 27 (50.00%) | 27 (50.00%) | ||
| Age (years) | 0.982 | 0.817 | ||||||
| Median (range) | 65 (48–85) | 65 (48–85) | 65 (49–84) | 64 (50–85) | 64 (51–85) | 64 (50–83) | ||
| Gender | 0.736 | 0.362 | ||||||
| Male | 93 (72.09%) | 47 (36.43%) | 46 (35.66%) | 39 (72.22%) | 21 (38.89%) | 18 (33.33%) | ||
| Female | 36 (27.91%) | 17 (17.18%) | 19 (14.73%) | 15 (27.78%) | 6 (11.11%) | 9 (16.67%) | ||
| Smoke | 0.425 | 1.000 | ||||||
| Yes | 68 (52.71%) | 36 (27.91%) | 32 (24.80%) | 36 (66.67%) | 18 (33.33%) | 18 (33.33%) | ||
| No | 61 (47.29%) | 28 (21.71%) | 33 (25.58%) | 18 (33.33%) | 9 (16.67%) | 9 (16.67%) | ||
| Pathological stagec | 0.726 | 0.141 | ||||||
| IA | 17 (13.18%) | 10 (7.75%) | 7 (5.43%) | 11 (20.37%) | 3 (5.56%) | 8 (14.81%) | ||
| IB | 34 (26.35%) | 14 (10.85%) | 20 (15.50%) | 10 (18.52%) | 6 (11.11%) | 4 (7.41%) | ||
| IIA | 23 (17.83%) | 11 (8.53%) | 12 (9.30%) | 11 (20.37%) | 8 (14.81%) | 3 (5.56%) | ||
| IIB | 32 (24.81%) | 17 (13.18%) | 15 (11.63%) | 13 (24.07%) | 7 (12.96%) | 6 (11.11%) | ||
| IIIA | 11 (8.53%) | 7 (5.43%) | 4 (3.10%) | 8 (14.81%) | 2 (3.70%) | 6 (11.11%) | ||
| IIIB | 12 (9.30%) | 5 (3.88%) | 7 (5.43%) | 1 (1.85%) | 1 (1.85%) | 0 (0.00%) | ||
| IL-1β Level | < 0.001 | 0.057 | ||||||
| Low | 64 (49.61%) | 11 (8.52%) | 53 (41.09%) | 27 (50.00%) | 10 (18.52%) | 17 (31.48%) | ||
| High | 65 (50.39%) | 53 (41.09%) | 12 (9.30%) | 27 (50.00%) | 17 (31.48%) | 10 (18.52%) | ||
| Target therapy | 0.660 | NTd | ||||||
| Yes | 64 (49.61%) | 33 (25.58%) | 31 (24.03%) | NTd | NTd | NTd | ||
| No | 65 (50.39%) | 31 (24.03%) | 34 (26.36%) | NTd | NTd | NTd | ||
Medianaa = 8.1731 Medianb = 5.8994 NTd = Not Tested
Pathological stageC was based on NCCN Guidelines Version 3.2011 NSCLC.
NCCN, National Comprehensive Cancer Network; NSCLC, non-small-cell lung cancer.
In LUAD patients, univariate analysis showed that age, clinical stage, target therapy, IL-1β, TNF-α and miR-144-3p were significant prognostic factors, while gender and smoking status showed no statistical differences (Table 2). As an association was determined between IL-1β and miR-144-3p, two separate multivariate models (IL-1β and miR-144-3p) were run to avoid problems of multicollinearity. Multivariate analyses demonstrated that clinical stage, IL-1β and miR-144-3p were independent risk factors for OS (Table 3). In LUSC patients, no independent risk factor was verified (Table 4).
Table 3. Univariate and multi-variate analysis of the clinicopathological and molecular features for OS in LUAD.
| Clinicopathological parameters | Comparison reference | Univariate analysis | Multi-variate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95%CI) | P value | Hazard ratio (95%CI) | P value | ||
| Age | < 65 | Ref | Ref | ||
| ≥ 65 | 1.906 (1.079–3.366) | 0.026 | 1.566 (0.838–2.926)a | 0.159a | |
| Gender | Male | Ref | |||
| Female | 1.176 (0.640–2.160) | 0.602 | |||
| Smoke | No | Ref | |||
| Yes | 1.577 (0.891–2.790) | 0.118 | |||
| Clinical Stage | IA | Ref | < 0.001 | Ref | < 0.001a |
| IB | 1.058 (0.177–6.338) | 0.951 | 0.907 (0.150–5.499)a | 0.916a | |
| IIA | 4.515 (0.934–21.817) | 0.061 | 4.513 (0.919–22.164)a | 0.064a | |
| IIB | 6.102 (1.397–26.660) | 0.016 | 4.278 (0.966–18.945)a | 0.056a | |
| IIIA | 19.666 (4.234–91.348) | < 0.001 | 13.101 (2.796–61.386)a | 0.001a | |
| IIIB | 10.963 (2.415–49.760) | 0.002 | 6.050 (1.307–27.996)a | 0.021a | |
| Target therapy | Yes | Ref | Ref | ||
| No | 2.000 (1.129–3.542) | 0.018 | 1.697 (0.934–3.083)a | 0.083a | |
| IL-1β level | Low | Ref | Ref | ||
| High | 3.818 (1.986–7.339) | < 0.001 | 3.166 (1.592–6.297)a | 0.001a | |
| TNF-α level | Low | Ref | Ref | ||
| High | 1.954 (1.101–3.466) | 0.022 | 1.332 (0.730–2.431)a | 0.350a | |
| miR-144-3p level | Low | Ref | Ref | ||
| High | 0.427 (0.238–0.765) | 0.004 | 0.365 (0.196–0.681)b | 0.002b | |
| Age | < 65 | Ref | |||
| ≥ 65 | 0.649 (0.280–1.503) | 0.307 | |||
| Gender | Male | Ref | |||
| Female | 0.913 (0.375–2.224) | 0.842 | |||
| Smoke | No | Ref | |||
| Yes | 0.909 (0.384–2.153) | 0.828 | |||
| Clinical Stage | IA | Ref | 0.039 | Ref | 0.221 |
| IB | 2.830 (0.255–31.346) | 0.397 | 3.088 (0.275–34.686) | 0.361 | |
| IIA | 5.361 (0.626–45.921) | 0.125 | 4.805 (0.550–41.949) | 0.156 | |
| IIB | 9.739 (1.186–79.956) | 0.034 | 8.430 (0.995–71.386) | 0.050 | |
| IIIA | 17.498 (2.100–145.829) | 0.008 | 13.287 (1.442–122.460) | 0.022 | |
| IIIB | 19.248 (1.157–320.344) | 0.039 | 14.606 (0.812–262.349) | 0.069 | |
| IL-1β level | Low | Ref | Ref | ||
| High | 3.106 (1.218–7.917) | 0.018 | 1.531 (0.503–4.657) | 0.453 | |
| TNF-α level | Low | Ref | |||
| High | 0.810 (0.357–1.839) | 0.615 | |||
| miR-144-3p level | Low | Ref | |||
| High | 1.216 (0.533–2.777) | 0.642 | |||
aMultivatiate model including Age, Clinical Stage, Target therapy, IL-1β & TNF-α.
bMultivatiate model including Age, Clinical Stage, Target therapy, miR-144-3p & TNF-α.
Table 4. Univariate and multi-variate analysis of the clinicopathological and molecular features for OS in LUSC.
| Clinicopathological parameters | Comparison reference | Univariate analysis | Multi-variate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95%CI) | P value | Hazard ratio (95%CI) | P value | ||
| Age | < 65 | Ref | |||
| ≥ 65 | 0.649 (0.280–1.503) | 0.307 | |||
| Gender | Male | Ref | |||
| Female | 0.913 (0.375–2.224) | 0.842 | |||
| Smoke | No | Ref | |||
| Yes | 0.909 (0.384–2.153) | 0.828 | |||
| Clinical Stage | IA | Ref | 0.039 | Ref | 0.221 |
| IB | 2.830 (0.255–31.346) | 0.397 | 3.088 (0.275–34.686) | 0.361 | |
| IIA | 5.361 (0.626–45.921) | 0.125 | 4.805 (0.550–41.949) | 0.156 | |
| IIB | 9.739 (1.186–79.956) | 0.034 | 8.430 (0.995–71.386) | 0.050 | |
| IIIA | 17.498 (2.100–145.829) | 0.008 | 13.287 (1.442–122.460) | 0.022 | |
| IIIB | 19.248 (1.157–320.344) | 0.039 | 14.606 (0.812–262.349) | 0.069 | |
| IL-1β level | Low | Ref | Ref | ||
| High | 3.106 (1.218–7.917) | 0.018 | 1.531 (0.503–4.657) | 0.453 | |
| TNF-α level | Low | Ref | |||
| High | 0.810 (0.357–1.839) | 0.615 | |||
| miR-144-3p level | Low | Ref | |||
| High | 1.216 (0.533–2.777) | 0.642 | |||
The regulatory effect of IL-1β and miR-144-3p on LUAD cell proliferation
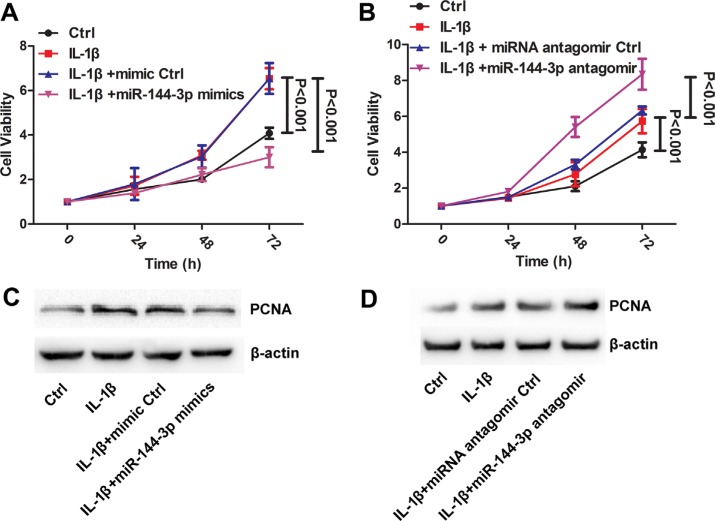
In A549 cells, IL-1β exposure and miR-144-3p mimics enhanced and suppressed cell proliferation, respectively (P < 0.001, Figure 6A). The combination of IL-1β exposure and miR-144-3p silencing promoted proliferation more significantly than mono-treatment (P < 0.0001, Figure 6B). Western-blot validated MTT result that PCNA protein level was increased after IL-1β exposure (Figure 6C and 6D), while it was increased and decreased after miR-144-3p mimics and antagomir transfection, respectively.
Figure 6. L IL-1β and miR-144-3p were involved in regulation of cell proliferation.
In A549, IL-1β exposure and miR-144-3p mimics enhanced and suppressed cell proliferation, respectively (P < 0.001) (A). The combination of IL-1β exposure and miR-144-3p silencing promoted proliferation more significantly than mono-treatment (P < 0.0001) (B). Western-blot validated MTT result that PCNA protein level was increased after IL-1β exposure (C and D), while it was increased and decreased after miR-144-3p mimics and antagomir transfection, respectively.
DISCUSSION
In the present study, we found that the IL-1β level was significantly higher in LUAD and LUSC patients. In LUAD patients, serum IL-1β level was correlated with miR-144-3p, both of which were associated with prognosis. Moreover, IL-1β was involved in regulation of miR-144-3p at the transcriptional level; IL-1β and miR-144-3p over-expression both could promote the cell proliferation.
Cytokines play key roles in initiation and development of cancer [19, 20]. IL-1β has been approved to be an essential factor in the inflammation initiation [9]. Gui et al. demonstrated that IL-1β can trigger HIF-1 through NF-κB/COX-2 signaling pathway, suggesting that HIF-1 is a bridge linking inflammation and tumor and IL-1β possesses a cancer promoting property [21]. IL-1β-induced COX-2 would mediate the evolution from inflammation to cancer through HIF-1, and such a process could be inhibited or suppressed by nonsteroidal anti-inflammatory drugs (NSAIDs), such as Aspirin and Celecoxib, leading to apoptosis of cancer cells [22]. Nevertheless, a large portion of NSCLC patients can not benefit from NSAIDs but suffer tumor progression quickly in clinic [23]. A few studies have approved that Celecoxib can promote epithelial-mesenchymal transition (EMT) of cancer cells, leading to increased metastatic risk [24, 25]. The conflict results indicated that COX-2/HIF1 is probably not the only cancer promoting pathway in downstream of IL-1β. In general, the existing evidence is not convincing enough to elucidate carcinogenetic mechanism of IL-1β, and new breakthroughs are urgently desired.
Despite under the same catalog of NSCLC, LUAD and LUSC greatly differ in genetics, histology, treatment and prognosis. Recently, Campbell JD et al. identified distinct patterns of somatic genome alterations in LUAD and LUSC. The genetics of the two major types of NSCLC significantly vary, which can be defined as individual diseases in certain level. Although targeted therapies for LUAD and LUSC are largely distinct, immunotherapy may aid in treatment for both types [3]. In the present study, we found that only in LUAD, serum IL-1β level was correlated with miR-144-3p, and both of them were independent risk factors for prognosis. However, no such significant prognostic value of IL-1β or miR-144-3p was obtained in LUSC. Our result was validated by TCGA database. Therefore, we suggested to investigate LUAD and LUSC separately to avoid major confounding factors in the future studies.
In this study, IL-8, IL-10 and TNF-α levels in LUSC were lower than those of healthy donors (P < 0.001). However, no prognostic value was observed. Although a number of studies have demonstrated the correlation between cytokine levels and prognosis in NSCLC patients, the population size was limited in the present study. The potential reason and mechanism remain unclear, and further investigation is necessary.
Although great efforts have been made to improve diagnosis and effective treatment, the prognosis of lung cancer remains poor due to lack of fine biomarkers. Accumulating evidence has confirmed that miRNAs are involved and play pivotal role in the occurrence, development and prognosis of tumor [26, 27]. Our results indicated that IL-1β probably promoted cell proliferation via suppression of miR-144-3p. Tumor miRNA is ubiquitous and stable in peripheral blood, and it is convenient to detect by real-time PCR or miRNA sequencing [28–30], making it a potential marker with synergetic diagnostic value. miRNA participates in every step of “inflammation--premalignancy--malignancy” process, including cytokine release and immune response [14]. So far, it remains unclear how miRNA mediates and regulates “inflammation-cancer transduction” in lung cancer. Therefore, such an investigation is important for the patients. Based on our findings, we hypothesized that IL-1β might regulate miR-144-3p transcription and control various biological behaviors, and its expression was associated with prognosis in LUAD patients. However, it still remains undefined how miR-144-3p acts and what the cross-talk between miR-144-3p and COX-2/HIF-1 pathway is.
Taken together, we demonstrated that in LUAD patients, the serum IL-1β level was correlated with miR-144-3p, and it could affect miR-144-3p at the transcriptional level. Moreover, both of them were associated with prognosis and independent risk factors for LUAD patients. In addition, IL-1β might promote cell proliferation via suppression of miR-144-3p. As the limit on the sample size, more elaborate studies are necessary for further exploration of the link between IL-1β and miR-144-3p and their roles in tumorigenesis.
MATERIALS AND METHODS
Patients, samples and cell line
The human NSCLC cell line A549 was obtained from American Type Culture Collection (ATCC, Manassas, USA) and cultured according to the guideline.
The study was performed on a total of 183 randomly selected NSCLC patients (129 LUADs and 54 LUSCs). Specimens were collected during biopsies and surgery from the Third Affiliated Hospital of Soochow University (Changzhou, China), and freshly collected samples were immediately fixed in formaldehyde. Each patient participated in the study after informed consent was provided. All specimens were histologically and blindly classified by two professional pathologists according to the national NCCN guidelines for NSCLC. Serum samples were collected from the same 183 patients and 40 extra healthy donors and stored in liquid nitrogen.
Enzyme linked immunosorbent assay (ELISA)
Serum samples from patients and healthy donors were obtained before biopsy or surgery from the Third Affiliated Hospital of Soochow University (Changzhou, China). The levels of cytokines, including IL-1β, IL-4, IL-6, IL-8, IL-10 and TNF-α, were determined using ELISA assay kits according to manufacturerwere determined using ELISA assay kits according tos instructions (R&D, Shanghai, China).
miRNA isolation
Total RNA was isolated from 400 μL serum of each patient using the miRNeasy Mini Kit (Qiagen, Shanghai, China) according to the manufacturer's instructions. Purified total RNA was used for further real-time PCR and miRNA array test.
miRNA array
Serum miRNA profile was generated using RiboArray TM miDETECT TM Human Array 1 × 12K (Ribobio, Guangzhou, China) according to the manufacturer's instructions.
Quantitative reverse transcription PCR (qRT-PCR)
The level of miR-144-3p was quantified by qRT-PCR using Bulge-Loop™ miRNA qRT-PCR Primer Set (Ribobio, Guangzhou, China), and results were analyzed and expressed by ΔΔCT method.
The cancer genome atlas (TCGA) database
LUAD and LUSC samples with miR-144-3p expression and clinical information were obtained from TCGA database, yielding a total of 452 LUADs and 343 LUSCs patients. The K-means cluster method (K = 2) was carried out to cluster the patients into two groups based on their miR-144-3p expression. The log-rank test and Kaplan-Meier curve were performed to evaluate the statistical significance and prognosis of the two groups.
Cell culture, miR-144-3p mimics and antagomir transfection
LUAD cell line A549 was employed for in vitro experiments. For treatment, cells were treated with 10 ng/mL IL-1β (PrimeGene, Shanghai, China). Mimics and antagomir were purchased from Ribobio Co., Ltd. (Guangzhou, China) in order to induce and suppress the miR-144-3p expression, respectively.
MTT assay
Cell proliferation was determined using Vybrant MTT Cell Proliferation Assay Kit (Invitrogen, Shanghai, China). Cells were collected at 0, 24, 48 and 72 h after IL-1β exposure and miR-144-3p mimics/antagomir transfection. Absorbance at a wavelength of 570 nm was determined on a spectrometer. The experiment was performed in triplicates.
Protein extraction and Western-blot analysis
Total proteins were extracted using CHAPS lysis buffer. Proteins were separated by sodiem dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride (PVDF) membrane. Blots were blocked in 4% dry milk at room temperature for 1 h, and immuno-stained with primary antibodies (anti-PCNA, 1:2,000; Cell signaling Technology, Shanghai, China and β-actin 1:10,000; Dako, Glostrup, Denmark) at 4°C overnight. The results were visualized via a chemiluminescent detection system (Pierce ECL Substrate Western-blot detection system; Thermo, Rockford, IL) and exposed in Molecular Imager ChemiDoc XRS System (Bio-Rad, Hercules, CA).
Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 5.01; GraphPad Software, Inc., La Jolla, USA) and SPSS (version 17, SPSS Inc., Chicago, USA) statistical softwares. The Student's t test and paired t test were used to analyze significance between independent groups and paired materials, respectively. The correlation test was used to analyze the correlation between IL-1β and miR-144-3p. The χ2 test was used to test the significance of observed differences in proportions except when the cell size was less than 5 (Fisher's exact tests). Survival curves were made using the Kaplan-Meier method and compared using the log-rank test. All variables that achieved a significance at P ≤ 0.05 in univariate analyses were enrolled in multivariate Cox's proportional hazard model. P ≤ 0.05 was considered statistically significant unless otherwise specified.
Acknowledgments
This work was financially supported by the National Key Technology R&D Program (No. 2015BAI12B12), the National Natural Science Foundation of China (No. 81171653; 81302047; 31428005), Natural science fundation of Jiangsu Province (BE2016660; BK20130243), Changzhou Science and Technology Project (Applied Based Research, No. CJ20159021) and Research support plan for post-doctor of Jiangsu Province (1601088C).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Authors’ contributions
Chen Wu designed experiments, analyzed clinical data and wrote the manuscript; Bin Xu and You Zhou analyzed data; Mei Ji provided patient samples and analyzed clinical data. Dachuan Zhang performed research and analyzed data. Jingting Jiang and Changping Wu conceived the study, supervised experiments and revised the manuscript.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. The New England journal of medicine. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, Imielinski M, Hu X, Ling S, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nature genetics. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang T, Li J, Zhang C, Hong Q, Jiang D, Ye M, Duan S. Distinguishing Lung Adenocarcinoma from Lung Squamous Cell Carcinoma by Two Hypomethylated and Three Hypermethylated Genes: A Meta-Analysis. PloS one. 2016;11:e0149088. doi: 10.1371/journal.pone.0149088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan C, Yan L, Wang L, Sun Y, Wang X, Lin Z, Zhang Y, Shi Y, Jiang W, Wang Q. Identification of immunohistochemical markers for distinguishing lung adenocarcinoma from squamous cell carcinoma. Journal of thoracic disease. 2015;7:1398–1405. doi: 10.3978/j.issn.2072-1439.2015.07.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottschling S, Jauch A, Kuner R, Herpel E, Mueller-Decker K, Schnabel PA, Xu EC, Muley T, Sultmann H, Bender C, Granzow M, Efferth T, Hoffmann H, et al. Establishment and comparative characterization of novel squamous cell non-small cell lung cancer cell lines and their corresponding tumor tissue. Lung Cancer. 2012;75:45–57. doi: 10.1016/j.lungcan.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinarello CA. A clinical perspective of IL-1beta as the gatekeeper of inflammation. European journal of immunology. 2011;41:1203–1217. doi: 10.1002/eji.201141550. [DOI] [PubMed] [Google Scholar]
- 10.Vardeh D, Wang D, Costigan M, Lazarus M, Saper CB, Woolf CJ, Fitzgerald GA, Samad TA. COX2 in CNS neural cells mediates mechanical inflammatory pain hypersensitivity in mice. The Journal of clinical investigation. 2009;119:287–294. doi: 10.1172/JCI37098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee CH, Chang JS, Syu SH, Wong TS, Chan JY, Tang YC, Yang ZP, Yang WC, Chen CT, Lu SC, Tang PH, Yang TC, Chu PY, et al. IL-1beta promotes malignant transformation and tumor aggressiveness in oral cancer. Journal of cellular physiology. 2015;230:875–884. doi: 10.1002/jcp.24816. [DOI] [PubMed] [Google Scholar]
- 12.Guo J, Feng Z, Huang Z, Wang H, Lu W. MicroRNA-217 functions as a tumour suppressor gene and correlates with cell resistance to cisplatin in lung cancer. Molecules and cells. 2014;37:664–671. doi: 10.14348/molcells.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer research. 2012;72:5576–5587. doi: 10.1158/0008-5472.CAN-12-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bu P, Wang L, Chen KY, Srinivasan T, Murthy PK, Tung KL, Varanko AK, Chen HJ, Ai Y, King S, Lipkin SM, Shen X. A miR-34a-Numb Feedforward Loop Triggered by Inflammation Regulates Asymmetric Stem Cell Division in Intestine and Colon Cancer. Cell stem cell. 2016;18:189–202. doi: 10.1016/j.stem.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng HY, Jiang SS, Hsiao JR, Hsiao M, Hsu YM, Wu GH, Chang WM, Chang JY, Jin SC, Shiah SG. IL-8 induces miR-424–5p expression and modulates SOCS2/STAT5 signaling pathway in oral squamous cell carcinoma. Molecular oncology. 2016 doi: 10.1016/j.molonc.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qayum AA, Paranjape A, Abebayehu D, Kolawole EM, Haque TT, McLeod JJ, Spence AJ, Caslin HL, Taruselli MT, Chumanevich AP, Baker B, Oskeritzian CA, Ryan JJ. IL-10-Induced miR-155 Targets SOCS1 To Enhance IgE-Mediated Mast Cell Function. J Immunol. 2016;196:4457–4467. doi: 10.4049/jimmunol.1502240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCoy CE, Sheedy FJ, Qualls JE, Doyle SL, Quinn SR, Murray PJ. O’Neill LA. IL-10 inhibits miR-155 induction by toll-like receptors. The Journal of biological chemistry. 2010;285:20492–20498. doi: 10.1074/jbc.M110.102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Zhang LF, Wu J, Xu SJ, Xu YY, Li D, Lou JT, Liu MF. IL-1beta-mediated repression of microRNA-101 is crucial for inflammation-promoted lung tumorigenesis. Cancer research. 2014;74:4720–4730. doi: 10.1158/0008-5472.CAN-14-0960. [DOI] [PubMed] [Google Scholar]
- 19.Shi M. 6P Expression of PD-L1 in lung cancer cells regulates Treg cell differentiation. Journal of thoracic oncology. 2016;11:S59. [Google Scholar]
- 20.Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, Loi S. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nature reviews Clinical oncology. 2016;13:228–241. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 21.Jung YJ, Isaacs JS, Lee S, Trepel J, Neckers L. IL-1beta-mediated up-regulation of HIF-1alpha via an NFkappaB/COX-2 pathway identifies HIF-1 as a critical link between inflammation and oncogenesis. FASEB journal. 2003;17:2115–2117. doi: 10.1096/fj.03-0329fje. [DOI] [PubMed] [Google Scholar]
- 22.Hou Z, Falcone DJ, Subbaramaiah K, Dannenberg AJ. Macrophages induce COX-2 expression in breast cancer cells: role of IL-1beta autoamplification. Carcinogenesis. 2011;32:695–702. doi: 10.1093/carcin/bgr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groen HJ, Sietsma H, Vincent A, Hochstenbag MM, van Putten JW, van den Berg A, Dalesio O, Biesma B, Smit HJ, Termeer A, Hiltermann TJ, van den Borne BE, Schramel FM. Randomized, placebo-controlled phase III study of docetaxel plus carboplatin with celecoxib and cyclooxygenase-2 expression as a biomarker for patients with advanced non-small-cell lung cancer: the NVALT-4 study. Journal of clinical oncology. 2011;29:4320–4326. doi: 10.1200/JCO.2011.35.5214. [DOI] [PubMed] [Google Scholar]
- 24.Liu R, Zheng J, Li C, Pang Y, Zheng Q, Xu X, Liu P. Celecoxib induces epithelial-mesenchymal transition in epithelial ovarian cancer cells via regulating ZEB1 expression. Archives of gynecology and obstetrics. 2015;291:1361–1369. doi: 10.1007/s00404-014-3555-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang ZL, Fan ZQ, Jiang HD, Qu JM. Selective Cox-2 inhibitor celecoxib induces epithelial-mesenchymal transition in human lung cancer cells via activating MEK-ERK signaling. Carcinogenesis. 2013;34:638–646. doi: 10.1093/carcin/bgs367. [DOI] [PubMed] [Google Scholar]
- 26.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature reviews Drug discovery. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunological reviews. 2013;253:167–184. doi: 10.1111/imr.12050. [DOI] [PubMed] [Google Scholar]
- 28.Jin P, Wang E, Ren J, Childs R, Shin JW, Khuu H, Marincola FM, Stroncek DF. Differentiation of two types of mobilized peripheral blood stem cells by microRNA and cDNA expression analysis. Journal of translational medicine. 2008;6:39. doi: 10.1186/1479-5876-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar L, Kulshreshtha R, Bhattacharya A. Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC genomics. 2010;11:288. doi: 10.1186/1471-2164-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber DG, Casjens S, Rozynek P, Lehnert M, Zilch-Schoneweis S, Bryk O, Taeger D, Gomolka M, Kreuzer M, Otten H, Pesch B, Johnen G, Bruning T. Assessment of mRNA and microRNA Stabilization in Peripheral Human Blood for Multicenter Studies and Biobanks. Biomarker insights. 2010;5:95–102. doi: 10.4137/bmi.s5522. [DOI] [PMC free article] [PubMed] [Google Scholar]