Abstract
Recurrent gene fusions involving EWSR1 with members of the cAMP response element binding protein (CREB) family (ATF1 and CREB1) have been reported in a diverse group of tumors including angiomatoid fibrous histiocytoma (AFH), soft tissue and gastrointestinal clear cell sarcoma (CCS), primary pulmonary myxoid sarcoma (PPMS) and hyalinizing clear cell carcinoma of salivary gland. We have recently encountered a group of 5 myxoid mesenchymal tumors positive for EWSR1 fusions with one of the CREB family member (ATF1, CREB1 and CREM), with histologic features distinct from any of the previously described pathologic entities. Tumors occurred in children or young adults (12–23 years; mean 18), with equal gender distribution. All except one were intracranial (intra-axial, 2; meningeal, 2) while one was perirectal. Histologically, the tumors were well-circumscribed, often lobulated, composed of uniform ovoid to round cells, and arranged in cord-like or reticular structures in a myxoid background. All except one displayed unique sunburst amianthoid fibers. Immunohistochemically, tumors were positive for EMA (5/5; 4 focal, 1 diffuse) and desmin (3/5). A novel EWSR1-CREM fusion was identified by RNA sequencing in the peri-rectal tumor, which was further confirmed by FISH and RT-PCR. A 2nd case with similar EWSR1-CREM fusion was identified by RT-PCR and FISH in a meningeal tumor. The remaining cases studied by FISH showed the presence of EWSR1-CREB1 fusion in 2 cases and EWSR1-ATF1 in one. In conclusion, we report a distinct group of myxoid mesenchymal neoplasms occurring in children or young adults with a predilection for intracranial locations. Although the immunoprofile (EMA, desmin) and the fusion type raise the possibility of a myxoid AFH, none of the typical histologic findings of AFH were present, suggesting a novel entity.
Keywords: EWSR1, CREM, CREB1, ATF1, myxoid, brain, meninges
INTRODUCTION
About one third of all soft tissue tumor entities are characterized by specific recurrent chromosomal translocations, usually encoding aberrant chimeric transcription factors. The prevalence of the fusion transcripts in most sarcomas is such that they define these entities and can be used as highly specific molecular diagnostic markers in the right clinical and pathologic context. However, more recently it has become apparent that some gene fusions are not entirely histo-type specific and can be shared among sarcoma types otherwise unrelated clinically or immunophenotypically. One such example is the presence of EWSR1-CREB1 and/or EWSR1-ATF1 fusions in a diverse group of tumors, including angiomatoid fibrous histiocytoma (AFH), soft tissue and GI clear cell sarcoma (CCS), primary pulmonary myxoid sarcoma (PPMS) and hyalinizing clear cell carcinoma of salivary gland.1 CREB1 and ATF1 both belong to the cAMP response element binding protein (CREB) transcription factor gene family and are functionally related. Although CREB1 and ATF1 are interchangeable gene partners, there is a striking propensity for EWSR1-CREB1 to occur in AFH and GICCS, and for EWSR1-ATF1 in CCS.2–6 To date, only EWSR1-ATF1 fusion has been detected in hyalinizing clear cell carcinoma,7 while only EWSR1-CREB1 was reported in PPMS. Apart from the gene fusion overlap, these tumors show different clinical presentations, morphologic features, immunoprofiles and patient outcomes. The mechanism for the shared fusion genes generating different phenotypes remains unclear, but it may be dependent on the cellular context in which the translocation occurs and/or the additional secondary genetic/epigenetic changes.
In this study, we describe a group of tumors displaying a unique histomorphology and harboring fusions involving EWSR1 with one the CREB gene family (CREB1, ATF1 or CREM).
MATERIALS AND METHODS
Index case
The index case was a 20 year-old female presenting with a 9 cm peri-rectal mass for which she underwent surgical resection. Microscopically, the tumor had a multilobulated growth with monomorphic ovoid cells arranged in a reticular pattern in a diffusely myxoid stroma. Focal areas with increased cellularity were noted, with interspersed amianthoid collagen fibers. The patient received post-op external beam radiation due to the marginal resection, followed by re-excision, with no residual tumor. Subsequently, she developed 3 local recurrences, 2, 4, and 10 years after initial diagnosis, respectively, which were resected. The local recurrences were small in size, with the second being the largest, measuring 1.5 cm in diameter. No metastatic disease developed during the 13-year follow-up. Fresh frozen tissue from the last local recurrence was available for fusion discovery by RNA sequencing, which identified an EWSR1-CREM fusion. As the morphologic appearance and the non-specific immunoprofile (see below) did not fit with any known pathologic entities, we searched the consultation files of the senior authors (CRA and MKR) for additional cases with similar histology and related EWSR1 gene fusions (involving members of CREB family). The hematoxylin and eosin slides were reviewed, as well as the immunostains, whenever available. The study was approved by the Institutional Review Board.
RNA sequencing
Total RNA was extracted from frozen tissue the index case #1 using RNeasy Plus Mini (Qiagen), followed by mRNA isolation with oligo(dT) magnetic beads and fragmentation by incubation at 94°C in fragmentation buffer (Illumina) for 2.5 minutes. After gel size-selection (350–400bp) and adapter ligation, the library was enriched by PCR for 15 cycles and purified. Paired-end RNA sequencing at read lengths of 50 or 51 bp was performed with the HiSeq 2000 (Illumina). All reads were independently aligned with STAR (ver 2.3)8 against the human reference genome (hg19). The reads were then analyzed by FusionSeq for potential gene fusions.9
Reverse transcription-polymerase chain reaction (RT-PCR) and long-range DNA PCR
For case 1, RT-PCR for EWSR1-CREM was performed to validate the fusion transcript found by RNA sequencing. An aliquot of the RNA material extracted above for RNA sequencing was reverse transcribed by SuperScript III First-Strand Synthesis System (Invitrogen) into complementary DNA. PCR was performed by Advantage 2 PCR kit (Clontech, Mountain View, CA). PCR primers for EWSR1 (exon 14) and CREM (exon 8) were designed according to the reads from RNA sequencing (Supplementary table 1). In addition, we employed long range PCR to investigate the DNA intronic breakpoint in the index case 1. Long range PCR was carried out using QIAGEN LongRange PCR Kit (QIAGEN, Germantown, MD). The primer sequences, annealing temperature and cycle number were also listed in Supplementary table 1.
Fluorescence in situ hybridization (FISH)
Formalin-fixed paraffin-embedded tissues were available in each case for FISH analysis. FISH for EWSR1 was performed on all cases and followed by subsequent FISH for various fusion partners, including CREM, CREB1, and ATF1. FISH was performed on 4 μm-thick formalin-fixed paraffin-embedded (FFPE) tissue sections. Custom probes were made by bacterial artificial chromosomes (BAC) clones (Supplementary Table 2) flanking the target genes, according to UCSC genome browser (http://genome.ucsc.edu) and obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (Oakland, CA; http://bacpac.chori.org). DNA from each BACs was isolated according to the manufacturer’s instructions. The BAC clones were labeled with fluorochromes by nick translation and validated on normal metaphase chromosomes. The slides were deparaffinized, pretreated, and hybridized with denatured probes. After overnight incubation, the slides were washed, stained with DAPI, mounted with an antifade solution, and then examined on a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany) controlled by Isis 5 software (Metasystems).
RESULTS
A novel EWSR1-CREM fusion is identified in an unclassified myxoid mesenchymal neoplasm with amianthoid type fibers
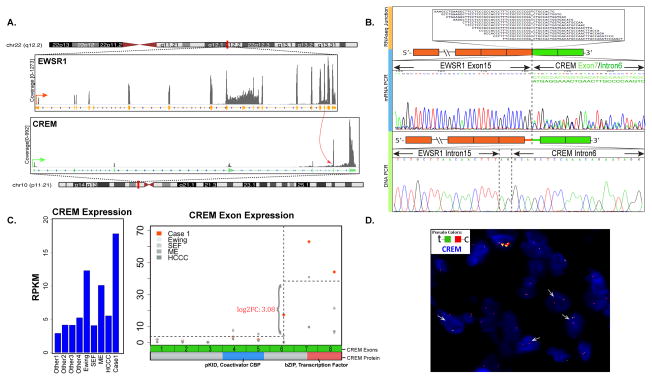
RNA sequencing of the index case revealed a candidate fusion transcript involving the EWSR1 (22q12.2) and CREM (10p11.21), which was further confirmed by RT-PCR and subsequent Sanger sequencing (Fig. 1A–B). Alternative fusion transcripts were detected by both RNA sequencing and RT-PCR, including EWSR1 exons 14 or 15 fused to either CREM exon 7 or intron 6, indicating alternative splicing events. The fusion involving EWSR1 exon 15 with either CREM transcripts resulted in in-frame products, while EWSR1 exon 14 resulted in a frameshift regardless of the CREM variant. Long range PCR using genomic DNA confirmed the DNA breakpoint located in EWSR1 intron 15 and CREM intron 6 (Fig. 1B). Gene expression analysis of RNA sequencing data showed overexpression of the CREM distal exons represented in the fusion, compared to the upstream CREM exons (Fig. 1C). A number of other samples were investigated as control showing a low level of CREM expression, except for an Ewing sarcoma with EWSR1-ERG fusion and a myoepithelial tumor with EWSR1-PBX3 fusion (Fig. 1C). FISH analysis also confirmed the break-apart of both EWSR1 and CREM genes (Fig. 1D).
Figure 1. A novel EWSR1-CREM gene fusion was identified by transcriptome sequencing in index case 1.
(A) Fusion junction reads of paired-end RNAseq identified a candidate t(10;22) translocation resulting in an in-frame fusion between EWSR1 exon 15 and CREM exon 7. (B) This result was further confirmed by RT-PCR, with an alternative fusion transcript being identified secondary to alternative splicing. DNA breakpoints in EWSR1 intron 15 and CREM intron 6 was also identified by long range DNA PCR. (C) CREM exons 7 and 8, included in the fusion transcript, showed up-regulated expression compared to the CREM 5′ exons and other control samples with EWSR1 fusions (Ewing sarcoma, sclerosing epithelioid fibrosarcoma, myoepithelial tumor, hyalinizing clear cell carcinoma of salivary gland) or not (other 1–4). Among the control samples, one Ewing sarcoma with EWSR1-ERG fusion and one myoepithelial tumor with EWSR1-PBX3 fusion also showed elevated downstream CREM exons expression, possibly due to preferential expression of ICER isoform. CREM exons 7 and 8 encode the bZIP domain responsible for dimerization and DNA binding. pKID mediates interaction with the coactivator CBP. (D) Break-apart FISH assay showed CREM rearrangement with telomeric deletion.
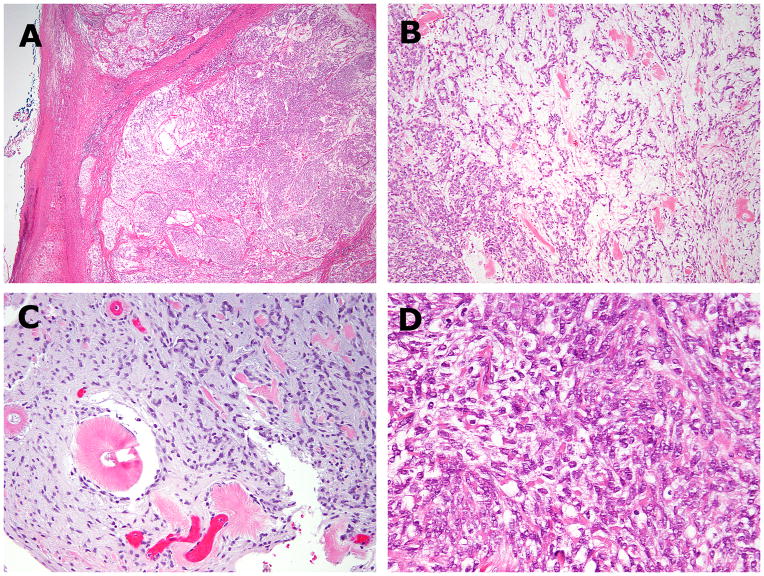
Microscopically, the tumor was well-circumscribed with a multilobulated growth pattern (Fig. 2A). The tumor lobules were composed of variably myxoid and solid areas of uniform round to ovoid cells, accompanied by scattered coarse amianthoid collagen fibers forming small rosette-type structures (Fig. 2B–C). The tumor cells had indistinct cell borders, palely eosinophilic cytoplasm, and bland ovoid nuclei with small nucleoli (Fig. 2D). Immunohistochemically, the tumor was focally positive for EMA and CD99, but negative for CK, S100, GFAP, p63, SMA and desmin.
Figure 2. Histologic findings of EWSR1-CREM positive index case 1.
(A,B) The pelvic tumor showed circumscribed borders and a lobulated growth, composed of alternating hypo and hyper-cellular areas. (C) The hypocellular myxoid areas showed uniform round to ovoid cells arranged in cord-like or reticular patterns. Scattered amianthoid fibers were seen in both the primary (B) and recurrent tumors (C). (D) Cellular areas showed densely packed cells with indistinct cell border and palely eosinophilic cytoplasm.
EWSR1 fusions with CREB family members define a unique myxoid mesenchymal neoplasm occurring with an intracranial predilection in young patients
Four additional cases were identified based on the shared genetic abnormalities, i.e. EWSR1 fusions with CREM, CREB1 or ATF1, and showing a similar morphology of an unclassified myxoid mesenchymal neoplasm (Table 1). Interestingly, all these 4 cases presented intra-cranially, with 2 cases each either dural-based/meningeal or intraparenchymal in the brain (frontal lobe). In case 5, the patient presented with one episode of generalized tonic-clonic seizure 2 months before diagnosis and subsequent occasional episodes of tongue jittering. A left frontal brain tumor was found by MRI scan. In addition, café-au-lait lesions were noted on the skin. There were no palpable skin tumors or confirmed diagnosis of neurofibromatosis.
Table 1.
Clinicopathologic characteristics of myxoid mesenchymal tumor with EWSR1 and CREB gene family fusion
| Age/Sex | Location | Histology | IHC | Fusion type | ||||
|---|---|---|---|---|---|---|---|---|
| Myxoid stroma | Amianthoid fibers | Mitosis (/10HPFs) | EMA | Desmin | ||||
| 1 | 20/F | Pelvic /Peri-rectal | 80% | Y | 2 | F+ | - | EWSR1-CREM |
| 2 | 15/F | Meninges | 95% | Y | 0 | F+ | D+ | EWSR1-CREM |
| 3 | 23/F | Meninges (occipital) | 95% | Y | 1 | F+ | D+ | EWSR1-CREB1 |
| 4 | 20/M | Brain (frontal) | 75% | Y | 4 | F+ | F+ | EWSR1-CREB1 |
| 5 | 12/M | Brain (frontal) | 20% | N | 1 | D+ | - | EWSR1-ATF1 |
M, male; F, female; Y, yes; N, no; HPF, high power field; F+, focal positive; D+, diffuse positive; −, negative
Together with the index case, all patients were children or young adults, ranging from 12 to 23 years old (mean, 18) with no obvious gender predilection (F:M ratio, 3:2). A wide spectrum of diagnoses were entertained in the differential for these cases, either based on morphology or the EWSR1 gene rearrangement, including: myoepithelial tumor (n=4), extraskeletal myxoid chondrosarcoma (n=3), angiomatoid fibrous histiocytoma (n=2), meningioma (n=2), ossifying fibromyxoid tumor (n=2) and Ewing sarcoma (n=1).
Histologically, the additional 4 cases showed circumscribed borders with a lobulated or diffuse sheet-like growth pattern (Fig. 3A, E, J). Small aggregates of lymphocytes were seen at the periphery only in one meningeal tumor (Case 2, Fig. 3A). Similar to the index case, all four additional cases showed a myxoid or fibromyxoid stroma surrounding round to ovoid tumor cells, arranged in cord-like, reticular or adenoid structures (Table 1, Fig. 3D, F, L). Amianthoid collagen fibers were also seen in all except one brain lesion (Table 1, Fig. 3B). In the more solid areas the cells were arranged in a syncytial growth pattern with indistinct cell borders (case 2, Fig. 3C), whorl-like structures (case 4, Fig. 3G) or dense sheets of cells with vacuolated cytoplasm (case 5, Fig. 3K). In all cases, tumor cells exhibited relatively uniform ovoid to round nuclei, low mitotic activity (0–4/10HPFs), and scant palely eosinophilic cytoplasm. Additional histologic findings included rare non-psammomatous microcalcifications (case 2), focal microcystic change (cases 1,3,4) and focal rhabdoid cell features (case 4). No blood-filled spaces reminiscent of AFH were seen in any of the cases.
Figure 3. Morphologic spectrum of intra-cranial tumors.
(A–C) Case 2 showed a myxoid neoplasm with a lobulated growth and focal peripheral lymphoid cuff, being composed of loosely arranged uniform round to ovoid cells with occasional amianthoid fibers. (D) Case 3 similarly showed a predominant myxoid stroma with reticular growth pattern. (E–I). Case 4 had a lobulated pattern, with hypocellular myxoid areas exhibiting reticular to adenoid architecture, while hypercellular regions showed whorl-like growth. Immunohistochemically, the cells were weakly positive for EMA (H) and focal strongly positive for desmin (I). (J–L) Case 5 revealed a pushing border into adjacent brain tissues; with solid growth of uniform round cells with occasional cytoplasmic vacuoles. The tumor also showed a myxoid stroma with reticular growth pattern.
Immunohistochemically, similar to the index case, all cases were positive for EMA, with the staining pattern ranging from diffuse to focal/weak (Fig. 3H). Desmin stain was positive in 3 cases, either diffusely (2 cases) or focally (1 case)(Fig. 3I). Other markers, including S100, GFAP, and SMA were negative in cases 2, 3 and 5.
The additional 4 cases harbored similar gene fusions involving EWSR1 with CREM/CREB1/ATF1 as demonstrated by break-apart FISH assays (Table 1, Supplementary Figure 1). A second case with EWSR1-CREM fusion was identified in a 15 year-old girl with a meningeal tumor (case 2). This result was also confirmed by RT-PCR (data not shown). An EWSR1-CREB1 fusion was found in a meningeal tumor of a 23 year-old female and in a frontal lobe lesion of a 20 year-old male. EWSR1-ATF1 was identified also in a frontal lobe tumor of a 12 year-old male, showing a predominantly solid growth and vacuolated cytoplasm, and lacking amianthoid fibers.
Follow-up information was available in case 2, who remained free of disease by clinical and MRI imaging evaluations at 17 months after diagnosis.
DISCUSSION
Most gene fusions in soft tissue tumors are strongly associated with a particular histotype and thus can be used as reliable molecular diagnostic markers and, in some cases, the resulting chimeric proteins constitute excellent therapeutic targets. However, some of the fusions are not histotype specific and are shared among two or more completely different pathologic entities, either within the same lineage or spanning multiple lines of differentiation. Two such remarkable examples are the EWSR1-ATF1 and/or EWSR1-CREB1 fusions identified in tumor types otherwise unrelated clinically or immunophenotypically. However, in certain cases the histologic overlap and the non-specific immunoprofile raise the possibility of a single pathologic entity characterized by morphologic diversity and shared genetics, rather than distinct tumor types with overlapping gene fusions. One such example is the PPMS, an entity that is mainly defined based on its pulmonary location and predominantly reticular architecture.10 Despite desmin negativity and lack of lymphoplasmacytic cuff, PPMS show some histologic overlap and similar genetic features with AFH, raising the possibility of a pulmonary myxoid variant of AFH.11,12 A somewhat similar scenario is the GI clear cell sarcoma, a term initially proposed by us in keeping with a variant of soft tissue CCS lacking melanocytic differentiation,3 but refuted by others, who suggested instead a novel entity designated as malignant gastrointestinal neuroectodermal tumor (GNET), possibly originating from a GI neuroectodermal precursor cell which has lost or does not have the potential to differentiate along the melanocytic lineage.13 Some of these controversies stem from the uncertainty on how translocation events drive phenotypic variations, and larger datasets are needed to clarify their status as independent pathologic entities. Emphasizing the importance of the cell type in which the mutation occurs, Straessler et al. demonstrated that conditional expression of EWSR1-ATF1 in a mouse model resulted in slightly different tumor phenotypes depending on the cell type, and its differentiation stage, in which the fusion was expressed.14
CREB1 (cyclic AMP [cAMP]–responsive element binding protein), ATF1, and CREM (cAMP response element modulatory protein) constitute a subfamily of the basic leucine zipper (bZIP) superfamily of transcription factors. The 3 members have a high degree of homology in the C-terminal bZIP domain, which is retained in the fusion protein and responsible for DNA binding and dimerization (Supplementary Figure 2).15 The bZIP domain binds to its target cAMP response element (CRE), which has been identified in the regulatory regions of over a hundred putative target genes,16,17 reflecting the functional diversity of the CREB family of transcription factors, including neuronal development, synaptic plasticity, glucose homeostasis, spermatogenesis, and cytokine regulation.15–19 Tissue-specific consequences of CREB activation suggest that cell type is at least one of the factors involved.17 Further understanding of the regulation of CREB gene family functions may provide insight as to how similar translocation events transform different cell types.
In this study, we report a group of 5 myxoid mesenchymal neoplasms characterized by similar morphology, immunoprofile and a remarkable predilection for the intracranial compartment in young patients. Their genetic hallmark was the presence of EWSR1 fusions with members of the CREB (cAMP response element binding protein) gene family, including CREM, CREB1 and ATF1. The distinct histologic features as well as the predilection for meninges and brain suggest a novel pathologic entity. However, similar to the other pathologic entities harboring EWSR1-CREB gene family fusions, the present cohort of tumors showed some overlapping histologic features with AFH, PMMS, and soft tissue myoepithelial tumors. The tumors had a rather non-specific immunoprofile, but the co-expression for EMA and desmin, although in some cases only focal and weak, raised the possibility of a relationship to AFH. Indeed a myxoid variant of AFH has been previously described, typically involving soft tissues of the extremities (none being reported in the brain), containing at least focal components of classic AFH (with fibrous pseudocapsules, peripheral lymphoid cuffing, and blood-filled cystic spaces) and carrying either EWSR1-CREB1 or -ATF1 fusions.20–24 In contrast, none of our cases showed the angiectatic or cystic hemorrhagic spaces, and lymphoid cuffing was only observed very focally in one meningeal tumor. In addition, the presence of amianthoid fibers identified in all but one of our cases was not described in myxoid AFH. In order to exclude the possibility that a subset of AFH harbor EWSR1-CREM fusions, we have investigated the fusion partner of 24 EWSR1-positive AFH cases from our files, but none showed CREM gene rearrangements (20 being positive for CREB1 and 4 for ATF1 by FISH). Our findings do not refute the existence of intracranial AFH, as two well-documented examples have been reported in the literature showing EWSR1-ATF1 fusion in the occipital lobe of a 25 year-old man and an EWSR1 rearrangement in the temporal lobe of a 35 year-old man.25,26 We have also identified an additional brain AFH from our files, with classic morphologic features and EWSR1-ATF1 fusion occurring in the temporal lobe of a 59 year-old man.
PPMS is another tumor with EWSR1-CREB1 fusion that may show histologic overlap, being predominantly myxoid with polygonal, spindle, or stellate cells arranged in a reticular pattern.10 PPMS usually arises in the lung of adult patients (mean, 45), often with an endobronchial growth. The tumor lacks desmin immunoreactivity and no amianthoid fibers have been reported to date.10,27,28 In keeping with the reported literature, a FISH analysis of 2 PPMS from our files showed the presence of EWSR1-CREB1 fusion.
Extraskeletal myxoid chondrosarcoma (EMC) and soft tissue myoepithelial tumors are also frequently considered in the differential diagnosis of myxoid neoplasms with EWSR1 gene rearrangements. The most common presentation of EMC is in the deep soft tissue of extremities or limb girdles of middle-aged adults.29 Although EMC are composed of monomorphic epithelioid to oval cells arranged in a reticular pattern, they usually show stromal hemorrhage and not amianthoid fibers. Furthermore, EMC lack desmin reactivity and the overwhelming majority harbor NR4A3 gene fusions with or without the EWSR1 partner.30 Similarly, myoepithelial tumors may show histologic overlap, but they consistently display the myoepithelial immunoprofile with EMA and/or cytokeratin and S100 protein expression.31,32 None of our cases were positive for S100, GFAP, SMA, or p63 immunostains. EWSR1 fusions have been identified in half of soft tissue myoepithelial tumors with various fusion partners, including POU5F1, PBX1, PBX3, ZNF444, and KLF17.31,33,34 Only one myoepithelial tumor from a pelvic tumor of a 57 year-old man was reported with an EWSR1-ATF1 fusion, but the tumor had a typical myoepithelial immunophenotype.35
In 1988, Kepes et al. reported a series of 7 meningeal tumors in young patients (8–19 years old) with “chordoid” features, peritumoral lymphoplasmacytic infiltrates and systemic manifestations (most commonly, anemia).36 In addition to microcystic anemia, one patient also developed other manifestations of Castleman syndrome, including hepatosplenomegaly, dysgammaglobulinemia, and retarded growth. In all patients, the anemia and/or other symptoms improved after resection of the meningeal tumors and resumed upon local recurrences in two patients. Histologically, the tumors showed areas resembling meningothelial meningioma with characteristic whorling pattern, but also nests and cords of cells with cytoplasmic vacuoles, reminiscent of physaliferous cells of chordoma. The surrounding lympho-plasmacytic infiltrate often formed lymphoid follicles or germinal centers. Although the age range at presentation was similar, none of our cases showed areas of typical meningioma, well-formed lymphoid follicles, physaliferous cells or chordoid features. Furthermore, one prototypical example of chordoid meningioma tested was negative for EWSR1 and FUS gene rearrangements by FISH.
An EWSR1-CREM fusion was previously reported by RNA sequencing in two melanoma cell lines (CHL-1, COLO 699), which, based on the SNP data, most likely derived from the same patient.37,38 Considering the histologic and immunohistochemical overlap between melanoma and CCS, it is possible that these cell lines may have originated from a CCS. The reported breakpoints included EWSR1 exon 6-CREM exon 7 in CHL-1 and EWSR1 exon 7-CREM exon 7 in COLO 699. The exon 7 CREM break was similar to our index case 1, but the EWSR1 breakpoints were different (EWSR1 exon 15–CREM exon 7). RNAi targeting of the CREM 3′end of the fusion transcript in the CHL-1 cell line resulted in decreased cell proliferation and invasion, with increased numbers of senescent cells.37 More recently, a FUS exon 8-CREM exon 7 fusion was reported (in an abstract form) in a CCS from the thigh of a 55 year-old man by anchored multiplex PCR and confirmed by FISH for FUS and RT-PCR.39 This limited data suggest that CREM may substitute for CREB1 and ATF1 as an alternative fusion partner to EWSR1 in CCS. Interestingly, in index case 1 the EWSR1-CREM fusion resulted in mRNA overexpression of CREM downstream exons, right after the exon 7 breakpoint (Fig. 1C). However, a slightly increased level of CREM expression downstream of exon 6 was noted in 2 unrelated EWSR1-fusion positive neoplasms lacking CREM gene abnormalities, most likely due to a preferential expression of ICER (inducible cAMP early repressor), an isoform of CREM that contains only these downstream exons, including exon 6 (Fig. 1C). In our previous studies using Affymetrix microarray analysis there was no up-regulation of CREB1 and ATF1 expressions in either AFH with EWSR1-CREB1 fusion or CCS with EWSR1-ATF1 fusion.2 Additional studies are needed to investigate whether this 3′CREM up-regulation is restricted to CREM-related fusions or occurs only in this group of tumors. Furthermore, no significant fusion transcript variability was noted among the different pathologic entities with EWSR1-ATF1/CREB1 fusions, with consistent breakpoints occurring in a given gene regardless of the tumor phenotypes, except possibly for HCCC, where the breakpoints occurred in EWSR1 exon 11 and ATF1 exon 3, which are different from other tumor types (Supplementary Figure 2).
Long-term follow-up was available in the index case 1, the patient following a protracted clinical course with late and small sized local recurrences. No metastatic event is seen in this patient after a 13-year follow-up. Of the intracranial cases, only case 2 had available follow-up information, which was uneventful for a 17-month period after surgery.
In conclusion, we report a group of myxoid mesenchymal neoplasms with distinct morphologic features and clinical presentation in children and young adults, preferentially involving intracranial locations. Collectively, they show a unique spectrum of histologic features and are genetically characterized by recurrent fusions between EWSR1 and CREB gene family of transcription factors. Additional studies are needed to establish if these represent a novel and unique pathologic entity.
Supplementary Material
FISH analysis demonstrated CREB1 (A) and ATF1 (B) break-apart in case 4 and case 5, respectively (arrows). CREB1 showed unbalanced rearrangement, with centromeric deletion.
Diagrammatic representation of the protein domains of CREM, CREB1 and ATF1 showing the common C-terminus bZIP (basic leucine zipper) domain, consistently retained in all chimeric proteins. bZIP domain is responsible for dimerization and DNA binding to target genes. The proximal pKID (phosphorylated kinase-inducible domain), mediating CBP/p300 interaction upon phosphorylation, is usually not retained in the chimeric protein. All reported CREM breakpoints involve exon 7 fused with either EWSR1 (exon 6 or 7 in the melanoma cell lines and exon 15 in our index case) or FUS (exon 8 in one CCS case).37–39 CREB1 breakpoint hotspot occurs at exon 7 across different tumor types being fused to EWSR1 (exon 7),2,3,5,10. ATF1 breakpoints occur at exon 4 or 5 being fused to EWSR1 exons 7, 8 or 10, irrespective of different types of mesenchymal tumors.2,4,5,35,40,41 HCCC harbors different exon combinations (EWSR1 exon 11-ATF1 exon 3) from other mesenchymal tumors.7,42
Acknowledgments
Supported in part by: P50 CA140146-01 (CRA); P30-CA008748 (CRA); Kristen Ann Carr Foundation (CRA); Cycle for Survival (CRA)
Footnotes
Conflicts of interest: none
References
- 1.Thway K, Fisher C. Tumors with EWSR1-CREB1 and EWSR1-ATF1 fusions: the current status. Am J Surg Pathol. 2012;36:e1–e11. doi: 10.1097/PAS.0b013e31825485c5. [DOI] [PubMed] [Google Scholar]
- 2.Antonescu CR, Dal Cin P, Nafa K, et al. EWSR1-CREB1 is the predominant gene fusion in angiomatoid fibrous histiocytoma. Genes Chromosomes Cancer. 2007;46:1051–1060. doi: 10.1002/gcc.20491. [DOI] [PubMed] [Google Scholar]
- 3.Antonescu CR, Nafa K, Segal NH, et al. EWS-CREB1: a recurrent variant fusion in clear cell sarcoma--association with gastrointestinal location and absence of melanocytic differentiation. Clin Cancer Res. 2006;12:5356–5362. doi: 10.1158/1078-0432.CCR-05-2811. [DOI] [PubMed] [Google Scholar]
- 4.Antonescu CR, Tschernyavsky SJ, Woodruff JM, et al. Molecular diagnosis of clear cell sarcoma: detection of EWS-ATF1 and MITF-M transcripts and histopathological and ultrastructural analysis of 12 cases. J Mol Diagn. 2002;4:44–52. doi: 10.1016/S1525-1578(10)60679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hisaoka M, Ishida T, Kuo TT, et al. Clear cell sarcoma of soft tissue: a clinicopathologic, immunohistochemical, and molecular analysis of 33 cases. Am J Surg Pathol. 2008;32:452–460. doi: 10.1097/PAS.0b013e31814b18fb. [DOI] [PubMed] [Google Scholar]
- 6.Thway K, Fisher C. Angiomatoid fibrous histiocytoma: the current status of pathology and genetics. Arch Pathol Lab Med. 2015;139:674–682. doi: 10.5858/arpa.2014-0234-RA. [DOI] [PubMed] [Google Scholar]
- 7.Antonescu CR, Katabi N, Zhang L, et al. EWSR1-ATF1 fusion is a novel and consistent finding in hyalinizing clear-cell carcinoma of salivary gland. Genes Chromosomes Cancer. 2011;50:559–570. doi: 10.1002/gcc.20881. [DOI] [PubMed] [Google Scholar]
- 8.Dobin A, Gingeras TR. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics. 2015;51:11 14 11–19. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sboner A, Habegger L, Pflueger D, et al. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thway K, Nicholson AG, Lawson K, et al. Primary pulmonary myxoid sarcoma with EWSR1-CREB1 fusion: a new tumor entity. Am J Surg Pathol. 2011;35:1722–1732. doi: 10.1097/PAS.0b013e318227e4d2. [DOI] [PubMed] [Google Scholar]
- 11.Smith SC, Palanisamy N, Betz BL, et al. At the intersection of primary pulmonary myxoid sarcoma and pulmonary angiomatoid fibrous histiocytoma: observations from three new cases. Histopathology. 2014;65:144–146. doi: 10.1111/his.12354. [DOI] [PubMed] [Google Scholar]
- 12.Thway K, Nicholson AG, Wallace WA, et al. Endobronchial pulmonary angiomatoid fibrous histiocytoma: two cases with EWSR1-CREB1 and EWSR1-ATF1 fusions. Am J Surg Pathol. 2012;36:883–888. doi: 10.1097/PAS.0b013e31824b1ee0. [DOI] [PubMed] [Google Scholar]
- 13.Stockman DL, Miettinen M, Suster S, et al. Malignant gastrointestinal neuroectodermal tumor: clinicopathologic, immunohistochemical, ultrastructural, and molecular analysis of 16 cases with a reappraisal of clear cell sarcoma-like tumors of the gastrointestinal tract. Am J Surg Pathol. 2012;36:857–868. doi: 10.1097/PAS.0b013e31824644ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straessler KM, Jones KB, Hu H, et al. Modeling clear cell sarcomagenesis in the mouse: cell of origin differentiation state impacts tumor characteristics. Cancer Cell. 2013;23:215–227. doi: 10.1016/j.ccr.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 16.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 17.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 18.Don J, Stelzer G. The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis. Mol Cell Endocrinol. 2002;187:115–124. doi: 10.1016/s0303-7207(01)00696-7. [DOI] [PubMed] [Google Scholar]
- 19.Rauen T, Hedrich CM, Tenbrock K, et al. cAMP responsive element modulator: a critical regulator of cytokine production. Trends Mol Med. 2013;19:262–269. doi: 10.1016/j.molmed.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer IM, Fletcher CD. Myxoid variant of so-called angiomatoid “malignant fibrous histiocytoma”: clinicopathologic characterization in a series of 21 cases. Am J Surg Pathol. 2014;38:816–823. doi: 10.1097/PAS.0000000000000172. [DOI] [PubMed] [Google Scholar]
- 21.Moura RD, Wang X, Lonzo ML, et al. Reticular angiomatoid “malignant” fibrous histiocytoma--a case report with cytogenetics and molecular genetic analyses. Hum Pathol. 2011;42:1359–1363. doi: 10.1016/j.humpath.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Chen G, Folpe AL, Colby TV, et al. Angiomatoid fibrous histiocytoma: unusual sites and unusual morphology. Mod Pathol. 2011;24:1560–1570. doi: 10.1038/modpathol.2011.126. [DOI] [PubMed] [Google Scholar]
- 23.Kao YC, Lan J, Tai HC, et al. Angiomatoid fibrous histiocytoma: clinicopathological and molecular characterisation with emphasis on variant histomorphology. J Clin Pathol. 2014;67:210–215. doi: 10.1136/jclinpath-2013-201857. [DOI] [PubMed] [Google Scholar]
- 24.Justin Wong SB, Wee A, Puhaindran ME, et al. Angiomatoid Fibrous Histiocytoma With Prominent Myxoid Stroma: A Case Report and Review of the Literature. Am J Dermatopathol. 2015;37:623–631. doi: 10.1097/DAD.0000000000000263. [DOI] [PubMed] [Google Scholar]
- 25.Dunham C, Hussong J, Seiff M, et al. Primary intracerebral angiomatoid fibrous histiocytoma: report of a case with a t(12;22)(q13;q12) causing type 1 fusion of the EWS and ATF-1 genes. Am J Surg Pathol. 2008;32:478–484. doi: 10.1097/PAS.0b013e3181453451. [DOI] [PubMed] [Google Scholar]
- 26.Ochalski PG, Edinger JT, Horowitz MB, et al. Intracranial angiomatoid fibrous histiocytoma presenting as recurrent multifocal intraparenchymal hemorrhage. J Neurosurg. 2010;112:978–982. doi: 10.3171/2009.8.JNS081518. [DOI] [PubMed] [Google Scholar]
- 27.Matsukuma S, Hisaoka M, Obara K, et al. Primary pulmonary myxoid sarcoma with EWSR1-CREB1 fusion, resembling extraskeletal myxoid chondrosarcoma: Case report with a review of Literature. Pathol Int. 2012;62:817–822. doi: 10.1111/pin.12014. [DOI] [PubMed] [Google Scholar]
- 28.Jeon YK, Moon KC, Park SH, et al. Primary pulmonary myxoid sarcomas with EWSR1-CREB1 translocation might originate from primitive peribronchial mesenchymal cells undergoing (myo)fibroblastic differentiation. Virchows Arch. 2014;465:453–461. doi: 10.1007/s00428-014-1645-z. [DOI] [PubMed] [Google Scholar]
- 29.Meis-Kindblom JM, Bergh P, Gunterberg B, et al. Extraskeletal myxoid chondrosarcoma: a reappraisal of its morphologic spectrum and prognostic factors based on 117 cases. Am J Surg Pathol. 1999;23:636–650. doi: 10.1097/00000478-199906000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Agaram NP, Zhang L, Sung YS, et al. Extraskeletal myxoid chondrosarcoma with non-EWSR1-NR4A3 variant fusions correlate with rhabdoid phenotype and high-grade morphology. Hum Pathol. 2014;45:1084–1091. doi: 10.1016/j.humpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hornick JL, Fletcher CD. Myoepithelial tumors of soft tissue. A clinicopathologic and immunohistochemical study of 101 cases with evaluation of prognostic parameters. Am J Surg Pathol. 2003;27:1183–1196. doi: 10.1097/00000478-200309000-00001. [DOI] [PubMed] [Google Scholar]
- 33.Huang SC, Chen HW, Zhang L, et al. Novel FUS-KLF17 and EWSR1-KLF17 fusions in myoepithelial tumors. Genes Chromosomes Cancer. 2015;54:267–275. doi: 10.1002/gcc.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agaram NP, Chen HW, Zhang L, et al. EWSR1-PBX3: a novel gene fusion in myoepithelial tumors. Genes Chromosomes Cancer. 2015;54:63–71. doi: 10.1002/gcc.22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flucke U, Mentzel T, Verdijk MA, et al. EWSR1-ATF1 chimeric transcript in a myoepithelial tumor of soft tissue: a case report. Hum Pathol. 2012;43:764–768. doi: 10.1016/j.humpath.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Kepes JJ, Chen WY, Connors MH, et al. “Chordoid” meningeal tumors in young individuals with peritumoral lymphoplasmacellular infiltrates causing systemic manifestations of the Castleman syndrome. A report of seven cases. Cancer. 1988;62:391–406. doi: 10.1002/1097-0142(19880715)62:2<391::aid-cncr2820620226>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 37.Giacomini CP, Sun S, Varma S, et al. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9:e1003464. doi: 10.1371/journal.pgen.1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klijn C, Durinck S, Stawiski EW, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 39.Chang K, Goytain A, Wang X, et al. Identification of novel gene fusion FUS-CREM in clear cell sarcoma of soft tissue by anchored multiplex polymerase chain reaction. Mod Pathol [Abstract #44] 2016;29:12–27. [Google Scholar]
- 40.Tsukamoto Y, Nakata Y, Futani H, et al. A rare case of clear cell sarcoma with 4 types of EWSR1-ATF1 fusions detected not in primary site but in metastatic site. Pathol Res Pract. 2013;209:803–807. doi: 10.1016/j.prp.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Lyle PL, Amato CM, Fitzpatrick JE, et al. Gastrointestinal melanoma or clear cell sarcoma? Molecular evaluation of 7 cases previously diagnosed as malignant melanoma. Am J Surg Pathol. 2008;32:858–866. doi: 10.1097/PAS.0b013e31815b8288. [DOI] [PubMed] [Google Scholar]
- 42.Weinreb I. Hyalinizing clear cell carcinoma of salivary gland: a review and update. Head Neck Pathol. 2013;7(Suppl 1):S20–29. doi: 10.1007/s12105-013-0466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FISH analysis demonstrated CREB1 (A) and ATF1 (B) break-apart in case 4 and case 5, respectively (arrows). CREB1 showed unbalanced rearrangement, with centromeric deletion.
Diagrammatic representation of the protein domains of CREM, CREB1 and ATF1 showing the common C-terminus bZIP (basic leucine zipper) domain, consistently retained in all chimeric proteins. bZIP domain is responsible for dimerization and DNA binding to target genes. The proximal pKID (phosphorylated kinase-inducible domain), mediating CBP/p300 interaction upon phosphorylation, is usually not retained in the chimeric protein. All reported CREM breakpoints involve exon 7 fused with either EWSR1 (exon 6 or 7 in the melanoma cell lines and exon 15 in our index case) or FUS (exon 8 in one CCS case).37–39 CREB1 breakpoint hotspot occurs at exon 7 across different tumor types being fused to EWSR1 (exon 7),2,3,5,10. ATF1 breakpoints occur at exon 4 or 5 being fused to EWSR1 exons 7, 8 or 10, irrespective of different types of mesenchymal tumors.2,4,5,35,40,41 HCCC harbors different exon combinations (EWSR1 exon 11-ATF1 exon 3) from other mesenchymal tumors.7,42