Abstract
Pre-mRNA splicing is an essential component of eukaryotic gene expression. Many metazoans, including humans, regulate alternative splicing patterns to generate expansions of their proteome from a limited number of genes. Importantly, a considerable fraction of human disease causing mutations manifest themselves through altering the sequences that shape the splicing patterns of genes. Thus, understanding the mechanistic bases of this complex pathway will be an essential component of combating these diseases. Dating almost to the initial discovery of splicing, researchers have taken advantage of the genetic tractability of budding yeast to identify the components and decipher the mechanisms of splicing. However, budding yeast lacks the complex splicing machinery and alternative splicing patterns most relevant to humans. More recently, many researchers have turned their efforts to study the fission yeast, Schizosaccharomyces pombe, which has retained many features of complex splicing, including degenerate splice site sequences, the usage of exonic splicing enhancers, and SR proteins. Here, we review recent work using fission yeast genetics to examine pre-mRNA splicing, highlighting its promise for modeling the complex splicing seen in higher eukaryotes.
Keywords: Pre-mRNA splicing, Schizosaccharomyces pombe, SR proteins, Alternative splicing, Exon definition
Eukaryotic genes often contain non-coding introns which must be spliced out of the pre-mRNA by the spliceosome to create a translatable mRNA. At its core, the spliceosome is composed of five highly conserved small nuclear RNA–protein complexes (snRNPs). Together with as many as 200 auxiliary proteins (Wahl et al. 2009; Will and Lührmann 2011), the spliceosome assembles anew upon each intron in a highly dynamic process. The mechanisms by which the spliceosome faithfully assembles upon and activates the correct splice sites among the vast sequence space of the transcriptome is a complex question which will remain only partially understood for the foreseeable future. It is clear that the spliceosome requires guidance from many sources, including sequence elements within the pre-mRNA, the local chromatin environment, and the influence of auxiliary splicing components which assemble upon the pre-mRNA (Matlin et al. 2005; Lee and Rio 2015). Adding to the complexity and functional importance of splicing, the use of alternative splice sites in different cellular contexts can change the coding sequence of mRNAs, thus the alternative activation of splice sites in different cell types functions as a critical control point for regulating gene expression. Recent RNA-seq datasets suggest >95 % of intron-containing human genes undergo alternative splicing (Barbosa-Morais et al. 2012; Bradley et al. 2012). Conversely, budding yeast (S. cerevisiae) has few examples of alternative splicing (Juneau et al. 2009; Marshall et al. 2013; Kawashima et al. 2014), most of which reflect intron-retention, and few of which are exon-skipping events, the type most prevalent in humans. Accordingly, the budding yeast contains a relatively reduced splicing apparatus, absent of many auxiliary components that control alternative splicing in humans (Käufer and Potashkin 2000). Nevertheless, the identification of many conserved core spliceosome components was accomplished through the use of the genetically tractable yeast model system. For example, the PRP genes were identified by screening libraries of temperature sensitive yeast strains for pre-mRNA processing (PRP) mutant phenotypes (Lustig et al. 1986; Vijayraghavan et al. 1989; Noble and Guthrie 1996). Further biochemical studies using purified proteins from these mutants in in vitro splicing reactions have played a pivotal role in delineating the mechanisms of splicing catalysis (Meyer and Vilardell 2009; Hossain and Johnson 2014).
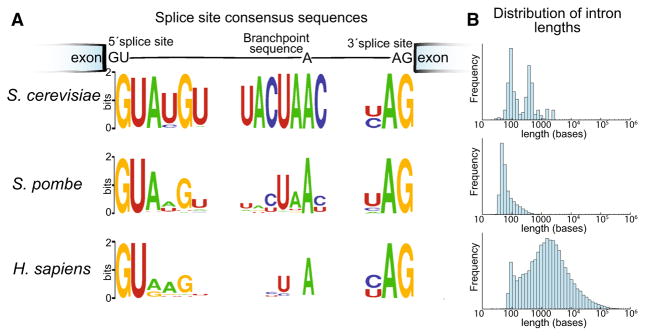
To more fully understand the complexity of splicing in higher eukaryotes, our lab and others have turned to the fission yeast, Schizosaccharomyces pombe, a distantly related ascomycete yeast which is similarly tractable to budding yeast as a model organism yet in many ways is more similar to humans from a splicing perspective. Some of these similarities become clear by comparing the genes and introns present in the genomes of budding yeast, fission yeast, and humans. Fission yeast genes are intron-dense compared to budding yeast (average of 0.9 introns per gene compared to 0.05 in budding yeast), though not as intron-dense as human genes (average of 8 introns per gene). Many fission yeast genes contain two or more introns, a pre-requisite for exon-skipping, some of which are interrupted by extremely short microexons reminiscent of those found in human genes (Scheckel and Darnell 2015). Moreover, a high degree of degeneracy is seen in the fission yeast splice site sequences that direct the spliceosome, more closely reflecting those seen in human transcripts (Fig. 1). The fission yeast genome also contains many orthologs of human splicing proteins which are not present in budding yeast (Käufer and Potashkin 2000; Kuhn and Käufer 2003), including two bona fide orthologs of human SR proteins, a class of auxiliary splicing proteins thought to be master regulators of alternative splicing in plants and animals. Additionally, two recent publications (Bitton et al. 2015; Stepankiw et al. 2015) both concluded that the fission yeast spliceosome makes use of a previously unappreciated number of alternative splice sites. Both of these studies estimate that about 2–3 % of splicing events genome-wide contain an alternative (unannotated) splice site, though the alternative mRNA isoforms are generally unstable and only detectable in RNA decay mutants. While this is a higher usage of alternative splice sites than the <1 % observed in similar RNA decay mutants in budding yeast (Kawashima et al. 2014), it pales in comparison to the extent of alternative splicing in humans.
Fig. 1.
Comparison of intron features in yeasts and humans. a Sequence logos depict the consensus sequence of splice site sequences. The total height at each position is proportional to the conservation at that position (Crooks et al. 2004). b The distribution of intron lengths for each species is shown as a histogram. Intron lengths and splice site sequences were obtained from (Kent et al. 2002; Wilhelm et al. 2008). Branch point sequences were obtained from (Clark et al. 2002; Gao et al. 2008; Stepankiw et al. 2015)
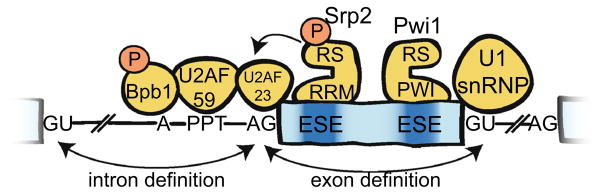
One insight as to why there isn’t more alternative splicing in fission yeast is that the yeast spliceosomes are constrained to select splice sites within a relatively short distance of each other. The natural distribution of intron lengths in both yeasts, which favor shorter introns than humans, supports this notion (Fig. 1). The prevalence of short introns in the yeast genome but long introns in humans has led to the consideration of two often juxtaposed models for the initial identification of splice site pairs: the intron-definition model and exon-definition model (Berget 1995; De Conti et al. 2013). Importantly, the intron-definition and exon-definition models are not mutually exclusive means to explain spliceosome assembly; rather both may impact assembly upon the same splice sites. In the intron-definition model, which is certainly prevalent in yeasts (Romfo et al. 2000), splice sites are identified by pairing a 5′ splice site to the nearest downstream branch point and 3′ splice site. Because the splice sites are initially paired across the intron, the intron length is under selection to remain short to maintain these interactions. In the exon-definition model, splice sites are initially identified through interactions between a 5′ splice and an upstream 3′ splice site, across the exon (Fig. 2). Thus, exons are under selection to remain short while introns may be long. These initial cross-exon interactions later need to be exchanged for cross-intron interactions for proper splicing catalysis. The molecular mechanisms by which the requisite cross-intron interactions are formed after the initial cross-exon interactions are established remain a critical, yet unanswered question in the field (De Conti et al. 2013; Hollander et al. 2016).
Fig. 2.
Mammalian-like mechanisms of splice site definition in fission yeast. Exonic splicing enhancer (ESE) sequences recruit SR and SR-related proteins, which recruit or stabilize early spliceosome assembly. Failure of these components to assemble across the exon may result in exon-skipping. Mutations in Bpb1, U2AF-59, and U2AF-23 result in exon-skipping, which can be suppressed by overexpression of Srp2. Naturally skipped exons have particularly non-consensus downstream 5′ splice sites, suggesting cross-exon interactions may contribute to recognition of upstream 3′ splice sites. Phosphorylation of Bpb1 is necessary for efficient splicing of introns with non-consensus branch point sequences. Pwi1 is an SR-related protein which is required for splicing of introns with non-consensus 5′ splice sites in fission yeast. Though the PWI domain can bind RNA, it is not clear whether it directly binds the ESE during ESE-dependent splicing
In higher eukaryotes, it is clear that the initial establishment of cross-exon interactions is facilitated by members of the SR family of proteins. These modular proteins bind to exonic splicing enhancer (ESE) sequences on the pre-mRNA through their RNA-recognition motif (RRM) and recruit the spliceosome to nearby splice sites through their RS domain (Graveley 2000; Long and Caceres 2009). The RS domain is rich in arginine-serine dipeptides and the phosphorylation status of the serine residues is important for activating splicing. Although ESE-dependent splicing does not necessarily require cross-exon interactions, it is interesting to note that when mammalian-derived purine-rich ESE sequences were placed into fission yeast exons, they still functioned to recruit the yeast SR protein SRP2 to aid in identification of weak upstream 3′ splice sites (Webb et al. 2005). RNA sequencing experiments have identified low-frequency alternative splicing events in the form of exon-skipping in fission yeast, the predominant form of alternative splicing predicted by the exon-definition model (Awan et al. 2013; Bitton et al. 2015; Stepankiw et al. 2015). Analysis of the splice site sequences surrounding the skipped exons revealed relatively consensus upstream branch point sequences, but weak downstream 5′ splice sites (Stepankiw et al. 2015), reminiscent of the observation that placement of a 5′ splice site sequence enhances recognition of upstream branch points in mammals (Robberson et al. 1990). Some of these alternative splicing events are particularly sensitive to environmental cues, suggesting their splicing may be regulated.
Together these results suggest that some elements of exon definition existed in the ancestral spliceosome (Ram and Ast 2007), and that these mechanisms persist to some extent in fission yeast, wherein SRP2 can function as a regulator of ESE-dependent splicing. Whereas loss-of-function studies on SR and SR-related proteins in humans are hampered by the high level of functional redundancy between the many SR proteins present in vertebrates, often resulting in relatively weak in vivo phenotypes (Pandit et al. 2013), this level of redundancy is not present in fission yeast. Yeast researchers are now in a strong position to tackle important questions about mammalian splicing mechanisms. What factors impact alternative splice site choice and early spliceosome assembly? How do SR proteins function to recruit the spliceosome? How do other nuclear processes such as transcription mechanistically affect the splicing process?
Several recent publications have made progress toward these goals. Haraguchi and co-workers used a reporter construct linked to a selectable auxotrophic marker in a classic forward genetic screen to identify factors which, when mutated, promote the production of exon-skipped splicing isoforms (Haraguchi et al. 2007; Sasaki-Haraguchi et al. 2015). This screen yielded point mutations in the essential proteins Bpb1, U2AF-59, and U2AF-23 (orthologs of the human SF1, U2AF-65, and U2AF-35 proteins, respectively), which bind to the branch point, polypyrimidine tract (PPT), and 3′ splice site, respectively, during early spliceosome assembly. The mechanisms by which these proteins are influenced by other auxiliary splicing proteins to selectively bind to bona fide branch point/3′ splice sites are fundamental to alternative splice site choice (Shao et al. 2014). In humans, U2AF-35 mediates protein–protein interactions involved in ESE-dependent splicing (Zuo and Maniatis 1996). Fission yeast U2AF-23 physically interacts with Srp2 (Webb and Wise 2004). Interestingly, the phenotypes of all three of the mutations identified by Haraguchi et al. could be suppressed by overexpression of Srp2, consistent with a model wherein Srp2 contacts U2AF-23 to recruit Bpb1/U2AF-59/23 to the branch point/3′ splice site (Fig. 2). Each of these proteins contains arginine-serine-rich regions with reasonable homology to the RS domains found in SR proteins (Mazroui et al. 1999; Graveley 2000; Kielkopf et al. 2001). Lipp and co-workers offered additional insights into the function of the RS domains, first by identifying relevant phosphorylation sites by mass spectrometry, then by making serine to alanine mutations at those sites (Lipp et al. 2015). Using splicing-sensitive microarrays to probe splicing genome-wide, they observed a moderate defect when the Srp2 phosphorylation sites were mutated: wherein some introns were spliced less efficiently, others were spliced more efficiently. A much more striking phenotype was observed when they mutated sites in the RS-like domain of Bpb1. The dependence of Bpb1 on splicing was relieved when a non-consensus branch point sequence was mutated to a more optimal sequence, suggesting phosphorylation is functionally important to recruiting or activating Bpb1/SF1 at suboptimal branch points (Fig. 2).
More general high throughput genetic approaches have also revealed information valuable to the splicing community. For example, Vo and co-workers recently published a proteome-wide map of protein–protein interactions using arrays of yeast-two-hybrid strains (Vo et al. 2016). This map revealed a physical interaction network between proteins containing RS domains, specifically between Pwi1, Srp1, and Srp2 (orthologs of the human SRM160, SRSF2, and SRSF4 proteins, respectively). Although the yeast-two-hybrid interactions were performed with fission yeast proteins in the cellular context of budding yeast and may not have functional relevance during splicing, these results reinforce the hypothesis that RS domains primarily interact with other RS domains. An interaction between Srp2 and Sap14 (a component of the U2 snRNP, and orthologous to human SF3b2), was also identified, further strengthening the argument for Srp2 assisting in early spliceosome assembly at the branch point. Patrick and co-workers took a complementary approach, using an epistatic miniarray profile (E-MAP) to identify loss-of-function mutations which genetically interact with splicing factor mutations (Patrick et al. 2015). Strong negative genetic interactions were found between the SWI/SNF chromatin remodeling complex and U2 snRNP components such as sap145 (SF3b145 in humans). Additional ChIP experiments showed that SWI/SNF recruits (directly or indirectly) Cdc28 (orthologous to Prp2p in budding yeast, DHX16 in humans), the helicase responsible for displacing SF3 from the branch point prior to the first catalytic step of splicing. These results highlight the interconnections between spliceosome assembly and other nuclear processes, such as nucleosome remodeling, during co-transcriptional splicing.
Recently, we published the results of a quantitative screen for mutant strains which display splicing defects (Larson et al. 2016). Using a sequencing-based approach, we were able to precisely and systematically measure the splicing efficiency of two endogenous transcripts in the background of ~3000 strains, each containing a deletion of a single, non-essential S. pombe gene. In addition to identifying known splicing factors, we found that various chromatin remodeling factors, including but not limited to SWI/SNF, resulted in as much as four-fold higher levels of unspliced pre-mRNA when deleted. Among the notable factors identified through this screen is the SR-related gene pwi1 (SRM160 in humans). This protein contains a canonical RS domain but has an atypical RNA-binding domain that contains a conserved PWI amino acid motif (Szymczyna et al. 2003). Mutation of this factor resulted in a strong genome-wide splicing defect, wherein introns with weak 5′ splice sites were particularly affected (Vo et al. 2016). This suggests that pwi1 may function similarly to its human ortholog by participating in ESE-dependent splicing that requires interactions with the U1 snRNP (Fig. 2) (Eldridge et al. 1999; Blencowe et al. 2000).
Moving past understanding splice site selection, the recent publications of atomic-resolution spliceosome structures in fission and budding yeast (Nguyen et al. 2015; Yan et al. 2015; Nguyen et al. 2016) will motivate enlightening experiments toward better understanding splicing catalysis. Much of our current understanding of splicing catalysis comes from biochemical approaches using mammalian or budding yeast extracts to purify splicing intermediates or reconstitute splicing reactions in vitro. Unfortunately, the lack of a successful in vitro splicing system has hampered biochemical studies in fission yeast. Hopefully the advancement of structural knowledge will help researchers understand why fission yeast splicing cannot be reconstituted in vitro and possibly lead to a solution, enabling additional mechanistic insights. For the time being, however, structure–function mapping experiments in fission yeast must be done using in vivo methods for assessing splicing-related phenotypes of mutations. The identification and characterization of splicing factor alleles containing discrete point mutations will be a key in these types of experiments. We have recently used our quantitative screening approach to screen through a library of thousands of temperature sensitive strains to identify alleles with major in vivo splicing defects at the restrictive temperature (Fair, Larson and Pleiss, manuscript in prep.). This quantitative screening assay could also be used to assess thousands of variants within a protein of interest. This strategy, often referred to as saturating mutagenesis or deep mutational scanning (Araya and Fowler 2011; Fowler and Fields 2014; Gao et al. 2014), has not yet been applied to study the function of spliceosomal components but could yield unanticipated findings. Surely the improvement of genetic technologies, including but certainly not limited to CRISPR, will allow the splicing field to perform previously difficult genetic experiments without the need for a model organism. Perhaps with a cleverly designed experiment, deep mutational scanning experiments could be performed in human cells. However, if one were interested in deep mutational scanning analysis of an SR protein, fission yeast may still be the ideal organism to avoid the redundancy of the plethora of SR proteins present in humans. In an experimentally useful way, the fission yeast spliceosome sits at an intermediate point between the ultra-reductionist budding yeast spliceosome and the ultra-complex human spliceosome. So despite the coming of age for effective genetic manipulations in human cells, for many reasons, the splicing community will still benefit from using fission yeast as a stepping stone to decipher the human spliceosome’s complexity.
Acknowledgments
Authors wish to thank Tokio Tani and members of the Pleiss lab for helpful comments on this manuscript. This work was funded by National Institutes of Health Grant GM098634.
References
- Araya CL, Fowler DM. Deep mutational scanning: assessing protein function on a massive scale. Trends Biotechnol. 2011;29:435–442. doi: 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan AR, Manfredo A, Pleiss JA. Lariat sequencing in a unicellular yeast identifies regulated alternative splicing of exons that are evolutionarily conserved with humans. Proc Natl Acad Sci. 2013;110:12762–12767. doi: 10.1073/pnas.1218353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Berget SM. Exon Recognition in Vertebrate Splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Bitton DA, Atkinson SR, Rallis C, et al. Widespread exon-skipping triggers degradation by nuclear RNA surveillance in fission yeast. Genome Res. 2015:884–896. doi: 10.1101/gr.185371.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Baurén G, Eldridge AG, et al. The SRm160/300 splicing coactivator subunits. RNA N Y N. 2000;6:111–120. doi: 10.1017/S1355838200991982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RK, Merkin J, Lambert NJ, Burge CB. Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLoS Biol. 2012;10:e1001229. doi: 10.1371/journal.pbio.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA. 2013;4:49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- Eldridge AG, Li Y, Sharp PA, Blencowe BJ. The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc Natl Acad Sci. 1999;96:6125–6130. doi: 10.1139/o99-903j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008;36:2257–2267. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Kan F, Wagnon JL, et al. Rapid, efficient and precise allele replacement in the fission yeast Schizosaccharomyces pombe. Curr Genet. 2014;60:109–119. doi: 10.1007/s00294-013-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. RNA N Y N. 2000;6:1197–1211. doi: 10.1017/S1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi N, Andoh T, Frendewey D, Tani T. Mutations in the SF1-U2AF59-U2AF23 complex cause exon skipping in Schizosaccharomyces pombe. J Biol Chem. 2007;282:2221–2228. doi: 10.1074/jbc.M609430200. [DOI] [PubMed] [Google Scholar]
- Hollander D, Naftelberg S, Lev-Maor G, et al. How are short exons flanked by long introns defined and committed to splicing? Trends Genet. 2016:1–11. doi: 10.1016/j.tig.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Hossain MA, Johnson TL. Using Yeast Genetics to Study Splicing Mechanisms. Methods Mol Biol. 2014;1126:285–298. doi: 10.1007/978-1-62703-980-2_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K, Nislow C, Davis RW. Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics. 2009;183:185–194. doi: 10.1534/genetics.109.105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käufer NF, Potashkin J. Analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. Nucleic Acids Res. 2000;28:3003–3010. doi: 10.1093/nar/28.16.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Douglass S, Gabunilas J, et al. Widespread use of non-productive alternative splice sites in Saccharomyces cerevisiae. PLoS Genet. 2014 doi: 10.1371/journal.pgen.1004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/S0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Käufer NF. Pre-mRNA splicing in Schizosaccharomyces pombe: regulatory role of a kinase conserved from fission yeast to mammals. Curr Genet. 2003;42:241–251. doi: 10.1007/s00294-002-0355-2. [DOI] [PubMed] [Google Scholar]
- Larson A, Fair BJ, Pleiss JA. Interconnections between RNA-processing pathways revealed by a sequencing-based genetic screen for pre-mRNA splicing mutants in fission yeast. G3 Bethesda Md. 2016;6:1513–23. doi: 10.1534/g3.116.027508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015:1–33. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp JJ, Marvin MC, Shokat KM, Guthrie C. SR protein kinases promote splicing of nonconsensus introns. Nat Struct Mol Biol. 2015;22:611–617. doi: 10.1038/nsmb.3057. [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Lustig AJ, Lin RJ, Abelson J. The yeast RNA gene products are essential for mRNA splicing in vitro. Cell. 1986;47:953–963. doi: 10.1016/0092-8674(86)90810-x. [DOI] [PubMed] [Google Scholar]
- Marshall AN, Montealegre MC, Jiménez-López C, et al. Alternative splicing and subfunctionalization generates functional diversity in fungal proteomes. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CWJ. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Mazroui R, Puoti A, Krämer A. Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA N Y N. 1999;5:1615–1631. doi: 10.1017/S1355838299991872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Vilardell J. The quest for a message: budding yeast, a model organism to study the control of pre-mRNA splicing. Brief Funct Genomic Proteomic. 2009;8:60–67. doi: 10.1093/bfgp/elp002. [DOI] [PubMed] [Google Scholar]
- Nguyen THD, Galej WP, Bai X, et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THD, Galej WP, Fica SM, et al. CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Curr Opin Struct Biol. 2016;36:48–57. doi: 10.1016/j.sbi.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, et al. Genome-wide analysis reveals sr protein cooperation and competition in regulated splicing. Mol Cell. 2013;50:223–235. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick KL, Ryan CJ, Xu J, et al. Genetic interaction mapping reveals a role for the SWI/SNF nucleosome remodeler in spliceosome activation in fission yeast. PLoS Genet. 2015;11:e1005074. doi: 10.1371/journal.pgen.1005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Ast G. SR proteins: a foot on the exon before the transition from intron to exon definition. Trends Genet. 2007;23:5–7. doi: 10.1016/j.tig.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Robberson BL, Cote GJ, Berget SM. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/MCB.10.1.84.Updated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romfo CM, Alvarez CJ, van Heeckeren WJ, et al. Evidence for splice site pairing via intron definition in Schizosaccharomyces pombe. Mol Cell Biol. 2000;20:7955–7970. doi: 10.1128/mcb.20.21.7955-7970.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Haraguchi N, Ikuyama T, Yoshii S, et al. Cwf16p associating with the nineteen complex ensures ordered exon joining in constitutive Pre-mRNA splicing in fission yeast. PLoS One. 2015;10:1–16. doi: 10.1371/journal.pone.0136336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheckel C, Darnell RB. Microexons—tiny but mighty. EMBO J. 2015;34:273–274. doi: 10.15252/embj.201490651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Yang B, Wu T, et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol. 2014;21:997–1005. doi: 10.1038/nsmb.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepankiw N, Raghavan M, Fogarty EA, et al. Widespread alternative and aberrant splicing revealed by lariat sequencing. Nucleic Acids Res. 2015;43:8488–8501. doi: 10.1093/nar/gkv763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczyna BR, Bowman J, McCracken S, et al. Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes Dev. 2003;17:461–475. doi: 10.1101/gad.1060403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- Vo TV, Das J, Meyer MJ, et al. A proteome-wide fission yeast interactome reveals network evolution principles from yeasts to human. Cell. 2016;164:310–323. doi: 10.1016/j.cell.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Webb CJ, Wise JA. The splicing factor U2AF small subunit is functionally conserved between fission yeast and humans. Mol Cell Biol. 2004;24:4229–4240. doi: 10.1128/MCB.24.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Romfo CM, van Heeckeren WJ, Wise JA. Exonic splicing enhancers in fission yeast: functional conservation demonstrates an early evolutionary origin. Genes Dev. 2005;19:242–254. doi: 10.1101/gad.1265905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:1–2. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]