Abstract
Specific components of the intestinal microbiota are capable of influencing immune responses such that a mutualistic relationship is established. In mice, colonization with segmented filamentous bacteria (SFB) induces Th17 cell differentiation in the intestine, yet the effector functions of IL-17A in response to SFB remain incompletely understood. Here, we report that colonization of mice with SFB-containing microbiota induced IL-17A- and CXCR2-dependent recruitment of neutrophils to the ileum. This response required adaptive immunity as Rag-deficient mice colonized with SFB-containing microbiota failed to induce IL-17A, CXCL1 and CXCL2, and displayed defective neutrophil recruitment to the ileum. Interestingly, neutrophil depletion in wild-type mice resulted in significantly augmented Th17 responses and SFB expansion, which correlated with impaired expression of IL-22 and antimicrobial peptides. These data provide novel insight into a dynamic IL-17A-CXCR2-neutrophil axis during acute SFB colonization and demonstrate a central role for neutrophils in limiting SFB expansion.
INTRODUCTION
The mammalian intestine is colonized with hundreds of microbial species that provide many advantages to the host but must also be properly contained to maintain heath. Achieving this coexistence requires proper development of mucosal immune responses and their controlled activation.1-3 An important part of this balance is predicated on sensing of specific bacteria, which triggers responses required for maintaining homeostasis between host and microbiota.4,5 It is now well appreciated that individual bacterial species can profoundly influence the development and function of various immune cells and different arms of the immune response both in the intestine and systemically.6 In turn, host immune responses are involved in a number of functions including the containment of microbes.
Segmented filamentous bacteria (SFB) are gram-positive, spore-forming bacteria that primarily colonize the ileum of mice and rats.7 These bacteria form intimate associations with intestinal epithelial cells, influencing both innate and adaptive immune responses.8-11 In particular, SFB promotes the robust differentiation of T-helper-17 cells (Th17), which are characterized by the production of IL-17 related cytokines including IL-17A and IL-22. Th17 responses are important in the intestine for protection against various extracellular pathogens, but if left uncontrolled may lead to pathogenic inflammation.9,12-15 IL-22 appears to be one key mediator in this process as it targets the epithelium to support barrier function through epithelial proliferation, mucus production and antimicrobial peptide secretion.16 More recently, IL-22 derived from CD4+ T cells was found to mediate barrier protection and prevent overgrowth of SFB.17 In contrast, the function of IL-17A produced from Th17 cells in the intestine is less well defined, especially in the context of controlling SFB expansion.
Evidence from other organ systems implicates IL-17A as an important activator of innate immune mechanisms, including the recruitment and survival of neutrophils.15 Neutrophils are typically the first responders to infection and injury where they help to contain and destroy invading microbes through a number of mechanisms.18 While neutrophils can induce bystander tissue destruction and contribute to pathology, recent evidence has highlighted the protective roles of neutrophils in suppressing colitis and promoting repair processes.19,20 For example, neutrophils have been shown to be a source of IL-22 in response to intestinal injury. The production of IL-22 by neutrophils was shown to be dependent on IL-23 and capable of influencing the expression of antimicrobial peptide expression, including SA100A8, SA100A9 and RegIIIβ, which are believed to contain bacteria through direct microbicidal effects. Interestingly, IL-22 producing neutrophils failed to be efficiently recruited to sites of injury in antibiotic treated mice suggesting that commensal bacteria influence this recruitment process.19
Given that genes regulated by IL-17A include neutrophil chemokines21 and that neutrophils are capable of protecting mucosal barriers,22 we examined the contribution of the IL-17A-CXCR2-neutrophil axis in the control of acute SFB expansion. Our data presented here show that neutrophil recruitment into the ileum in response to acute colonization with SFB-containing microbiota was dependent upon adaptive immune cell-derived IL-17A and CXCR2. Following neutrophil depletion in vivo, SFB levels expanded as did intestinal Th17 cell differentiation and these effects correlated with impaired IL-22 and antimicrobial peptide expression. These finding provide novel insight into the dynamic IL-17A-CXCR2-neutrophil axis during acute SFB colonization and demonstrate a central role for neutrophils in restricting SFB expansion and in the regulation of SFB-induced Th17 responses.
RESULTS
Neutrophils are recruited into the ileum of mice in response to colonization with SFB
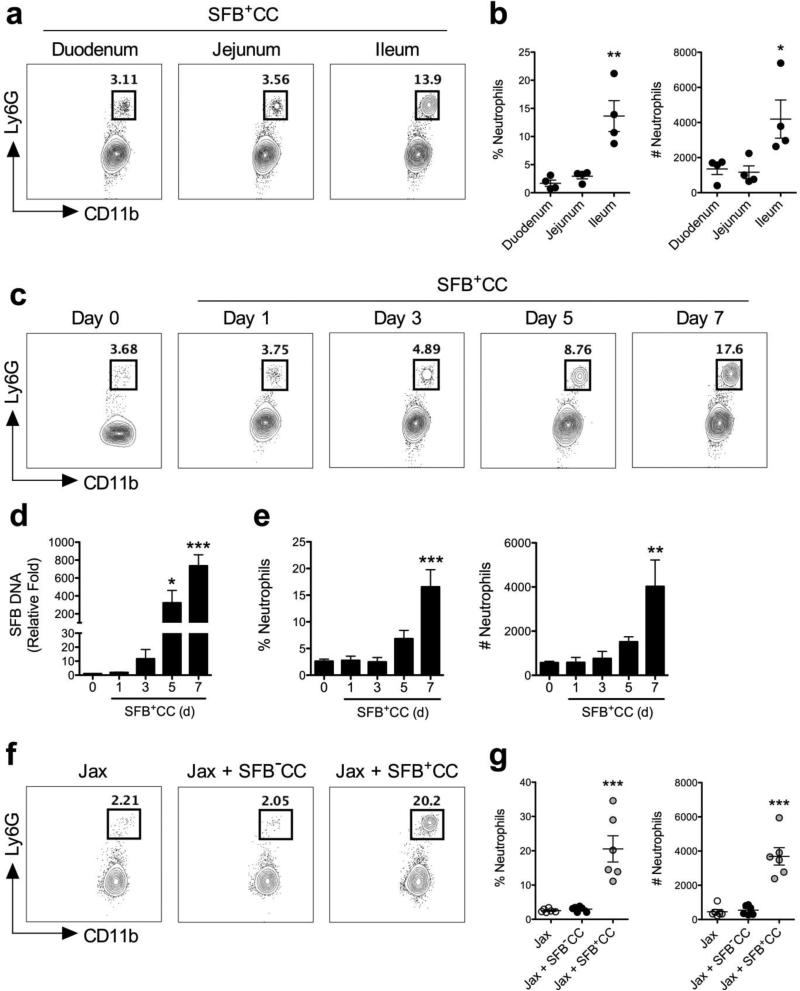
SFB colonizes the ileum making direct contact with the epithelium where it induces the differentiation of Th17 cells.9,11,23 To examine if neutrophils also respond to acute colonization with SFB, we first assessed different sections of the small intestine from SFB-void Jax mice colonized with SFB-containing cecal contents (SFB+CC). Neutrophils were as CD45+MHCII−Ly6CintCD11b+Ly6G+ cells.19 Mice that had been colonized with SFB+CC for 7 days showed the highest frequency (~14%) and number of neutrophils in the ileum where SFB is known to adhere to the epithelium, compared to the duodenum and jejunum (~3 and 4%, respectively) from the same mice (Figure 1a,b).
Figure 1.
Neutrophils are recruited into the ileum in response to colonization with SFB-containing microbiota. (a) Representative FACS plots are shown for CD11b and Ly6G expression with associated (b) frequencies and numbers representative of neutrophils in the duodenum, jejunum and ileum of SFB-void Jax mice colonized with SFB+CC for 7 days (pre-gated on CD45+MHC−Ly6Cint cells). (c) Representative FACS plots of neutrophils in the ileum of SFB-void Jax mice colonized with SFB+CC over the indicated time course. (d) SFB DNA was detected by qPCR in feces from SFB-void Jax mice colonized with SFB+CC over the indicated time course. (e) The frequencies and number of neutrophils in the ileum of SFB-void Jax mice colonized with SFB+CC over the indicated time course. Data are representative of two independent experiments with four mice per group. (f) Representative FACS plots and (g) associated frequencies and numbers of neutrophils in the ileum of SFB-void Jax mice (Jax), and SFB-void Jax mice colonized with SFB−CC or SFB+CC for 7 days. Data are representative of six mice per group. All data presented as mean ± SEM; *p<0.05, ** p<0.01, and *** p<0.001, one-way ANOVA with Tukey's multiple comparison test (b,g) or one-way ANOVA with Dunnett's multiple comparison test (d,e).
We next examined the entry of neutrophils into the ileum over the course of the first 7 days of SFB colonization. SFB-void Jax mice (day 0) and SFB-void Jax mice subsequently colonized with SFB+CC for 1, 3, 5, and 7 days were assessed for the presence of neutrophils in the ileum. FACS analysis revealed an influx of neutrophils beginning at days 3-5 and increasing further at day 7 after colonization with SFB+CC (Figure 1c). Examination of SFB DNA in feces obtained from these mice showed that SFB levels began to increase at day 3 and were significantly increased by day 5 (~300-fold increase when compared to feces from SFB-void mice) and day 7 (~800-fold increase when compared to feces from SFB-void mice) (Figure 1d). The frequency and number of neutrophils in the ileum correlated with the level of fecal SFB DNA over the course of 7 days (Figure 1e). This observed influx of neutrophils was specific to colonization with SFB as gavage of SFB-void Jax mice with SFB-void microbiota derived from separate SFB-void Jax mice (SFB−CC) failed to induce detectable neutrophil recruitment into the ileum (Figure 1f,g). Additionally, gavage of SFB- void Jax mice with fecal contents obtained from SFB-monoassociated mice resulted in neutrophil recruitment similar to that observed in Jax mice 7 days after gavage with SFB+CC, thus demonstrating that SFB was sufficient to induce neutrophil recruitment (Supplementary Figure 1). Interestingly, neutrophil accumulation in response to SFB+CC was still detectable 14 days after initial colonization indicating that neutrophil entry into the ileum was somewhat long-lasting (Supplementary Figure 2). Taken together these results show that acute colonization with SFB-containing microbiota induces the durable recruitment of neutrophils into the ileum of mice.
SFB-containing microbiota-induced neutrophil recruitment is IL-17A- and CXCR2-dependent
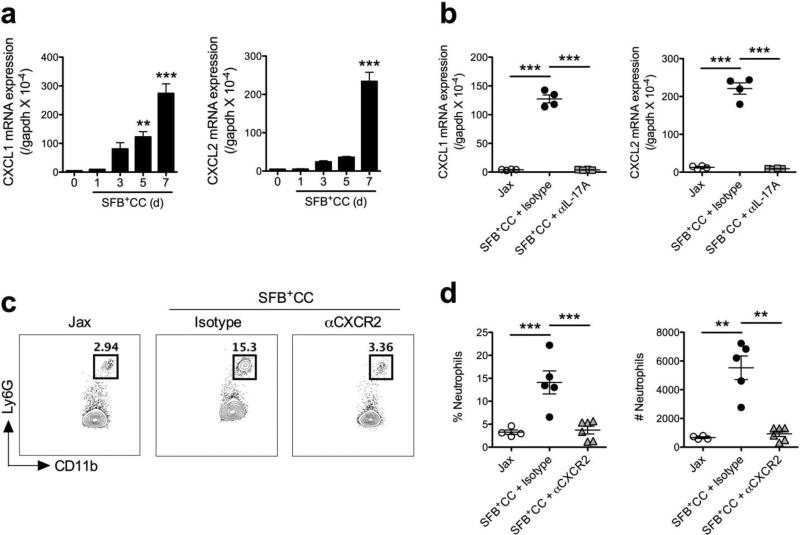
Over the time course of SFB+CC-induced neutrophil influx into the ileum, we also observed increased expression of IL-17A mRNA (Figure 2a), which coincided with increased fecal SFB DNA levels and the frequency and number of neutrophils (Figure 1c-e). Evidence from lung model systems suggests that IL-17A can function in the recruitment of neutrophils into mucosal surfaces,15 therefore we examined if daily antibody-mediated blockade of IL-17A (αIL-17A) was able to impact the entry of neutrophils into the ileum in response to acute colonization with SFB+CC. Using SFB-void Jax mice that had been colonized with SFB+CC for 7 days (a time point when robust numbers of neutrophils are present in the ileum), FACS analysis revealed that αIL-17A antibody treatment completely abolished neutrophil recruitment when compared to isotype control antibody-treated mice (Figure 2b). Indeed, the frequency and number of neutrophils in the SFB+CC-colonized ileum at day 7 was not significantly different than in SFB-void Jax mice, suggesting an important role for IL-17A in the initial recruitment of neutrophils in response to SFB-containing microbiota (Figure 2b-d).
Figure 2.
IL-17A is required for the recruitment of neutrophils into the ileum following colonization of mice with SFB-containing microbiota. (a) Expression of IL-17A mRNA was examined by qPCR in the ileum of SFB-void Jax mice colonized with SFB+CC over the indicated time course. (b) Representative FACS plots with associated (c) frequencies and numbers of neutrophils in the ileum of SFB-void Jax mice (Jax), and SFB-void Jax mice treated with isotype control antibody or αIL-17A antibody and colonized with SFB+CC for 7 days. Data are representative of at least two independent experiments with four mice per group. All data are presented as mean ± SEM; ***p<0.001, one-way ANOVA with Dunnett's multiple comparison test (a) or one-way ANOVA with Tukey's multiple comparison test (c).
One mechanism via which IL-17A may promote the recruitment of neutrophils into mucosal surfaces is through the induction of the neutrophil chemokines CXCL1 and CXCL2.15,24 Therefore, we next investigated the expression of CXCL1 and CXCL2 mRNA over the first 7 days following colonization of SFB-void Jax mice with SFB+CC. Expression of CXCL1 and CXCL2 mRNA increased in parallel to that of IL-17A mRNA (Figure 3a), correlating with increased SFB DNA levels and neutrophil numbers. We also found that αIL-17A antibody treatment during colonization with SFB+CC completely prevented the increased expression of CXCL1 and CXCL2 mRNA (Figure 3b). To directly assess the contribution of the CXCL1 and CXCL2 in the recruitment of neutrophils after colonization with SFB+CC, we blocked the receptor for these chemokines, CXCR2, with a neutralizing antibody (αCXCR2). Treatment with αCXCR2 antibody every other day during the first 7 days of colonization with SFB+CC significantly diminished the influx of neutrophils into the ileum when compared isotype control-treated mice (Figure 3c,d). Collectively, these data demonstrate that following acute colonization with SFB- containing microbiota, IL-17A is induced and leads to CXCL1 and CXCL2 expression, and subsequent recruitment of neutrophils into the ileum where SFB primarily adheres to the epithelium.
Figure 3.
CXCL1 and CXCL2 are induced by IL-17A and blocking CXCR2 reduces neutrophil influx in response to colonization with SFB-containing microbiota. (a) Expression of CXCL1 and CXCL2 mRNA were examined by qPCR in the ileum of SFB-void Jax mice colonized with SFB+CC over the indicated time course. (b) Expression of CXCL1 and CXCL2 mRNA in the ileum of SFB-void Jax mice (Jax), and SFB-void Jax mice treated with isotype control antibody or αIL-17A antibody and colonized with SFB+CC for 7 days. Data are representative of two independent experiments with four mice per group. (c) Representative FACS plots with associated (d) frequencies and numbers of neutrophils in the ileum of SFB-void Jax mice (Jax), and SFB-void Jax mice treated with isotype control antibody or αCXCR2 antibody and colonized with SFB+CC for 7 days. Data are representative of four to six mice per group. All data are presented as mean ± SEM; **p<0.01 and ***p<0.001, one-way ANOVA with Dunnett's multiple comparison test (a) or one-way ANOVA with Tukey's multiple comparison test (b,d).
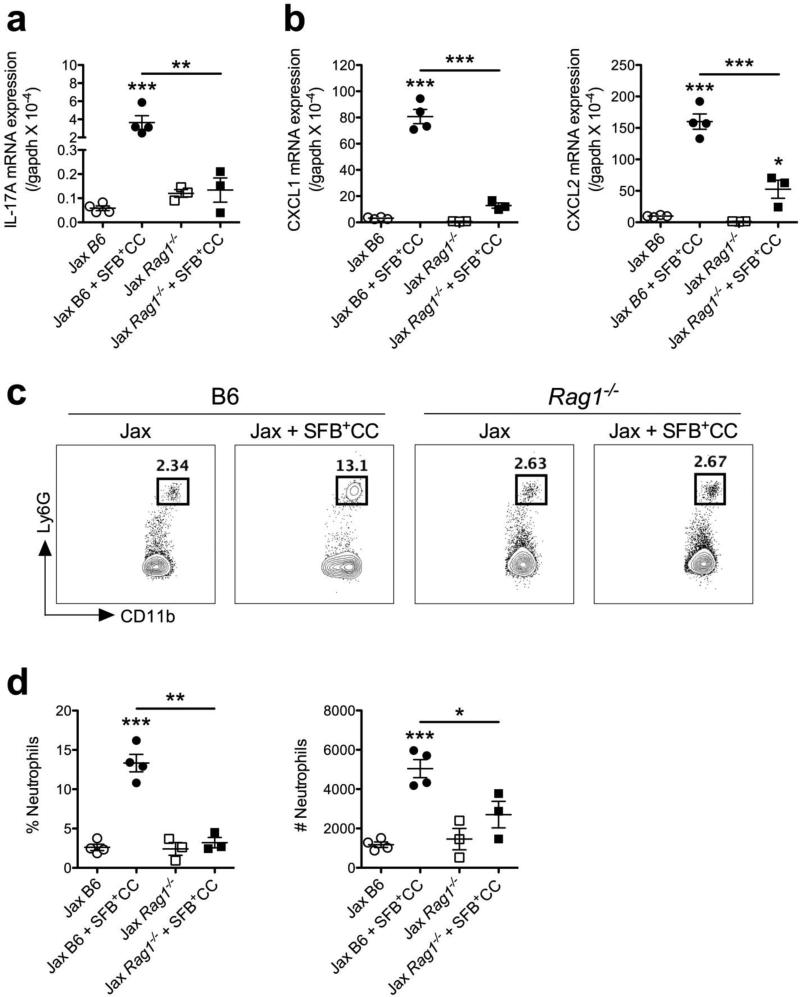
Rag-deficient mice colonized with SFB-containing microbiota fail to induce IL-17A, CXCL1, CXCL2, and display defective neutrophil recruitment into the ileum
Since CD4+ T cells and γδ T cells are important sources of IL-17A at mucosal surfaces,15,25,26 we next examined the expression of IL-17A, as well as CXCL1 and CXCL2, and neutrophil recruitment in response to colonization with SFB+CC in Rag1−/− mice (on the B6 background). Consistent with T cells being the major source of IL-17A in response to SFB+CC, colonization of SFB-void Jax Rag1−/− mice with SFB+CC for 7 days failed to stimulate expression of IL-17A mRNA when compared to colonization of SFB-void wild-type Jax B6 mice with SFB+CC for 7 days (Figure 4a). Reduced expression of IL-17 mRNA in the ileum of Rag1−/− mice colonized with SFB+CC coincided with significantly reduced expression of CXCL1 and CXCL2 mRNA as well (Figure 4b). Further, 7 days after colonizing Rag1−/− with SFB+CC, we observed significantly decreased neutrophil frequencies and numbers in the ileum when compared to wild-type B6 control mice colonized with SFB+CC (Figure 4c,d). These data indicate that adaptive immune cell-derived IL-17A is instrumental in SFB-containing microbiota-induced IL-17A, CXCL1, and CXCL2 expression, and neutrophil recruitment.
Figure 4.
Rag-deficient mice colonized with SFB-containing microbiota fail to induce IL-17A, CXCL1, and CXCL2, and display defective neutrophil recruitment into the ileum. Expression of (a) IL-17A and (b) CXCL1 and CXCL2 mRNA were examined by qPCR in the ileum of SFB-void Jax B6 and Rag1−/− mice, and SFB-void Jax B6 and Rag1−/− mice colonized with SFB+CC for 7 days. (c) Representative FACS plots with associated (d) frequencies and numbers of neutrophils isolated from the ileum of SFB-void Jax B6 and Rag1−/− mice, and SFB-void Jax B6 and Rag1−/− mice colonized with SFB+CC for 7 days. Data are representative of two independent experiments with three to four mice per group. All data are presented as mean ± SEM; *p<0.05, **p<0.01, and ***p<0.001, one-way ANOVA with Tukey's multiple comparison test.
Neutrophil depletion during colonization with SFB-containing microbiota results in augmented SFB levels and Th17 responses
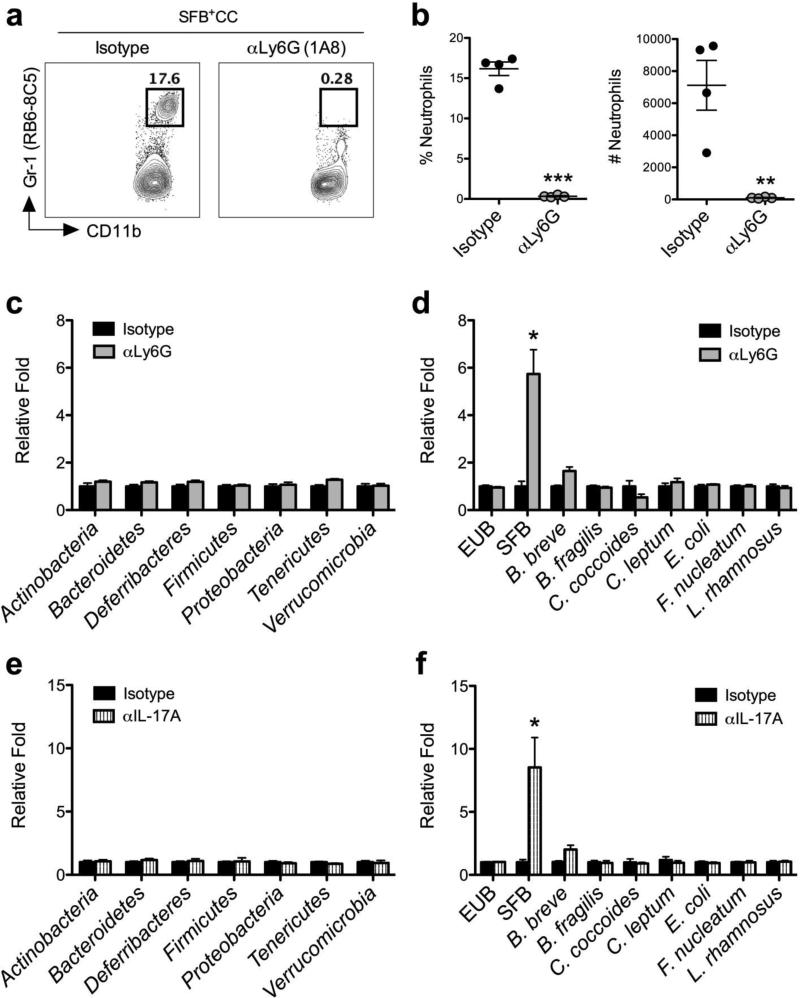
To assess the contribution of neutrophils during acute colonization of SFB-void Jax mice with SFB+CC, we examined the effect of antibody-mediated neutrophil depletion (αLy6G) on SFB levels. Upon confirming the ability of αLy6G antibody treatment to deplete neutrophils in the ileum of SFB-void Jax mice colonized with SFB+CC for 7 days (Figure 5a,b), we next quantitated the level of bacterial DNA using quantitative PCR (qPCR) in feces of αLy6G antibody-treated mice versus isotype control antibody-treated mice that had been colonized for 7 days with SFB+CC. Examination of the main bacterial phyla in feces of SFB-void Jax mice colonized with SFB+CC for 7 days revealed that αLy6G antibody treatment did not lead to detectable changes in the overall composition of microbial communities when compared to the isotype control antibody-treated group (Figure 5c). While analyses confirmed that the total amount of bacterial DNA (EUB), as well as the DNA abundance of a number of different commensal bacteria species in the feces was not affected by αLy6G antibody treatment, SFB DNA was specifically and significantly increased (approximately 6-fold) in the absence of neutrophils (Figure 5d). Treatment of SFB-void Jax mice with αLy6G antibody also resulted in augmented SFB DNA levels (~4-fold) in the ileal mucosa when compared to isotype control antibody-treated mice (Supplementary Figure 3).
Figure 5.
Neutrophils and IL-17A are required to control SFB expansion. (a) Representative FACS plots with associated (b) frequencies and numbers of neutrophils isolated from the ileum of SFB-void Jax mice treated with isotype control antibody or αLy6G antibody and colonized with SFB+CC for 7 days. The relative abundance of different (c) phyla and different (d) species of commensal bacterial DNA were examined by qPCR in feces from SFB-void Jax mice treated with isotype control antibody or αLy6G antibody and colonized with SFB+CC for 7 days. (e, f) Phyla and species analyses were performed on feces from SFB-void Jax mice treated with isotype control antibody or αIL-17A antibody and colonized with SFB+CC for 7 days. Data are representative of at least two independent experiments with four mice per group. *p<0.05, **p<0.01, and ***p<0.001. unpaired Student's t-test, two tailed (b) or Mann-Whitney test, two-tailed (c-f).
Having demonstrated a requirement for IL-17A in neutrophil recruitment during acute colonization with SFB (Figure 2,3), we next assessed the abundance of SFB DNA in the feces of SFB-void Jax mice treated with isotype control antibody or αIL-17A antibody and colonized with SFB+CC for 7 days. Similar to results with αLy6G antibody treatment (Figure 5c,d), feces obtained from αIL-17A antibody-treated mice did not show changes in the overall composition of microbial communities in the intestine as no discernable shifts in the abundance of the common bacterial phyla were observed (Figure 5e). However, αIL-17A antibody-treated mice that had been colonized with SFB+CC for 7 days showed a greater than 8-fold increase in SFB DNA levels in their feces when compared to isotype control antibody-treated mice (Figure 5f). Again, there was no significant change in the levels of total bacteria or different commensal bacteria species (Figure 5f). Interestingly, we also found that feces isolated from Rag1−/− mice contained ~8-fold more SFB DNA than B6 mice following colonization with SFB+CC (Supplementary Figure 4), which is consistent with the inability of Rag1−/− mice to induce IL-17A, CXCL1, CXCL2, and to recruit neutrophils in response to colonization with SFB-containing microbiota.
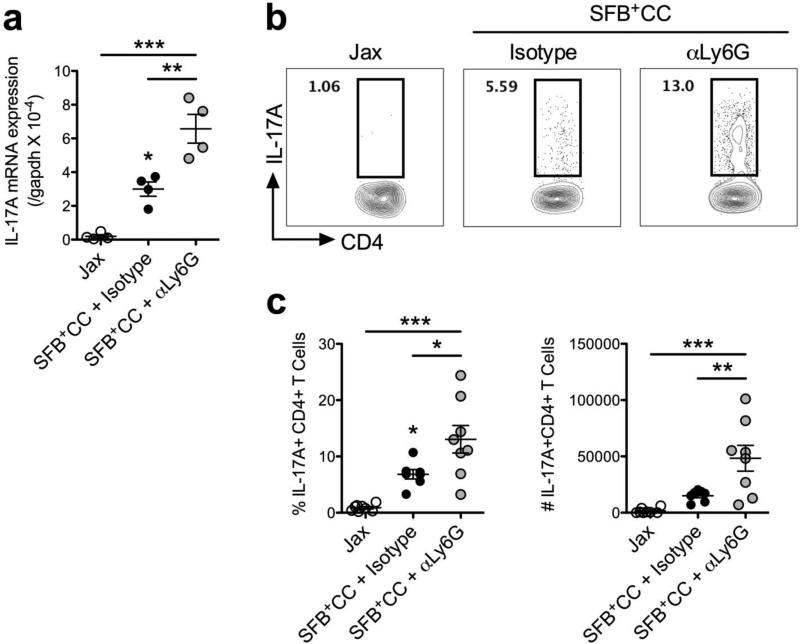
Recent evidence has demonstrated a tight relationship between levels of SFB colonization, the induction of IL-17A and Th17 differentiation.11 Therefore we sought to examine the impact that neutrophil depletion and subsequent SFB expansion had on this axis. We found that colonization with SFB+CC for 7 days induced expression of IL-17A mRNA in the ileum of SFB-void Jax mice treated with isotype control antibody, while αLy6G antibody treatment led to a significantly larger increase in the expression of IL-17A mRNA (Figure 6a). Furthermore, intracellular cytokine staining showed a >2-fold increase in the frequency and number of CD4+IL-17A+ T cells in αLy6G antibody-treated mice, when compared to isotype control antibody-treated mice (Figure 6b,c). Notably, when mice were treated with αLy6G antibody and colonized with SFB+CC for 7 days the resulting increases observed in Th17 responses did not result in histological evidence of ileal inflammation (Supplementary Figure 5). Similar to results with αLy6G antibody treatment, we also found that αIL-17A antibody treatment of SFB-void Jax mice during colonization with SFB+CC for 7 days resulted in significantly increased expression of IL-17A mRNA (Supplementary Figure 6a) and increased frequency and numbers of CD4+IL-17A+ T cells in the ileum compared to isotype control antibody-treated mice (Supplementary Figure 6b,c). Therefore, control of intestinal SFB most closely correlates with IL-17A-induced neutrophil recruitment and not IL-17A expression per se.
Figure 6.
Neutrophil depletion during colonization with SFB-containing microbiota results in augmented Th17 responses in the ileum. (a) Expression of IL-17A mRNA in the ileum of SFB-void Jax mice (Jax), and SFB-void Jax mice treated with isotype control antibody or αLy6G antibody and colonized with SFB+CC for 7 days. Data are representative of two independent experiments with four mice per group. (b) Representative FACS plots as well as cell (c) frequency and number among the indicated groups are shown for expression of IL-17A following restimulation with PMA and ionomycin (pre-gated on TCRβ+CD4+ cells). Data are pooled from two independent experiments with four mice per group. All data are presented as mean ± SEM; *p<0.05, **p<0.01, and ***p<0.001, one-way ANOVA with Tukey's multiple comparison test.
Neutrophil depletion during colonization with SFB-containing microbiota results in decreased IL-22 and antimicrobial peptide expression
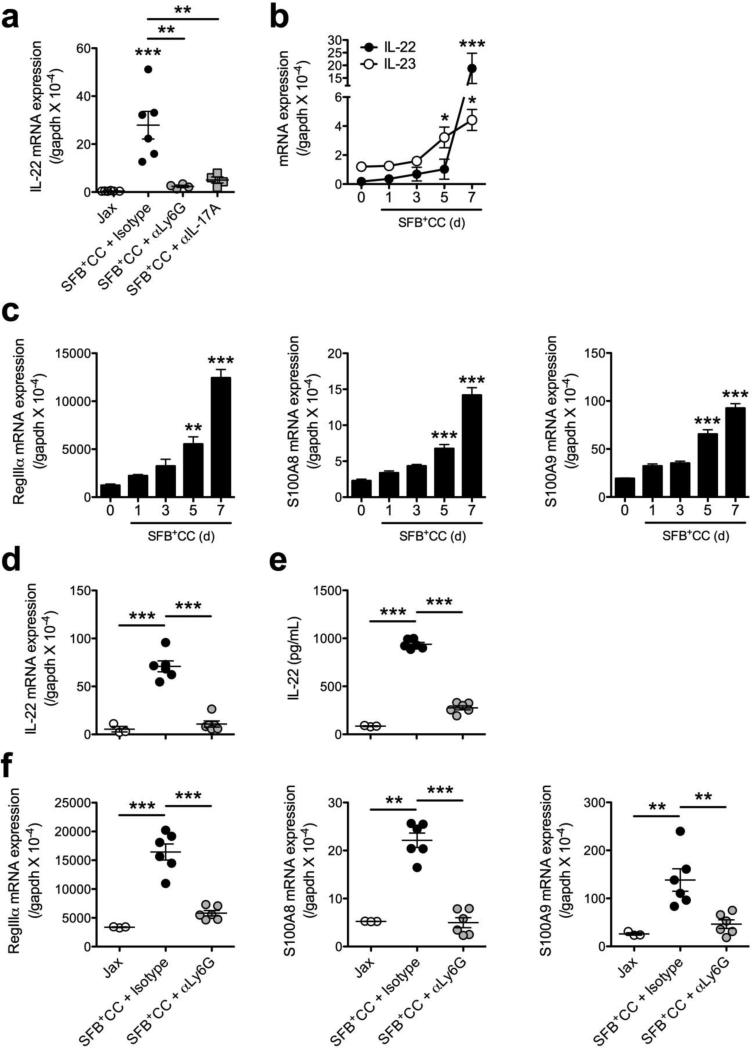
IL-22 produced by T cells and ILC3s has been demonstrated to control the abundance of SFB.17,27 This prompted us to examine if the ability of neutrophils to control SFB loads correlated with the expression of IL-22 and antimicrobial peptides. Initially, we examined IL-22 expression in the ileum of SFB-void Jax mice, and SFB-void Jax mice treated with either isotype control antibody, αLy6G antibody or αIL-17A antibody and colonized with SFB+CC for 7 days. We found that colonization with SFB-containing microbiota induced the expression of IL-22 mRNA in the ileum of isotype control-treated mice and this effect was abolished in the αLy6G antibody and αIL-17A antibody treated groups (Figure 7a). Given that IL-23 receptor signaling is required to regulate the abundance of SFB17 and that IL-23 is capable of inducing IL-22 production from colonic neutrophils during acute mucosal injury and chronic colitis,19,20 we investigated if the neutrophils that accumulate after colonization of SFB-void Jax mice with SFB+CC were able to promote IL-22 expression in response to restimulation with recombinant murine IL-23 (rmIL-23). Examination of IL-22 and IL-23 expression over the time course of colonization with SFB+CC revealed that the expression of IL-22 mRNA was significantly increased at day 5 and even more-so on day 7 (Figure 7b), a time point at which neutrophil numbers are robustly increased. Interestingly, increases in the expression of IL-23 mRNA in response to colonization with SFB+CC preceded that of IL-22 mRNA (Figure 7b). Increased mRNA expression of the antimicrobial peptides RegIIIα, S100A8, and S100A9 were also detected as early as day 5 and to a much greater extent on day 7 following colonization with SFB+CC (Figure 7c). These data indicate that IL-23 produced in response to colonization with SFB-containing microbiota may regulate neutrophil-dependent IL-22 production, which is associated with the induction of antimicrobial peptides and ultimately the control of SFB expansion.
Figure 7.
Neutrophil depletion during colonization with SFB-containing microbiota results in decreased IL-22 and antimicrobial peptide expression in the ileum. (a) Expression of IL-22 mRNA in the ileum of SFB-void Jax mice (Jax), and SFB-void Jax mice treated with isotype control antibody, αLy6G antibody, or αIL-17A antibody and colonized with SFB+CC for 7 days. (b) The expression of IL-22 and IL-23 mRNA were examined in the ileum of SFB-void Jax mice colonized with SFB+CC over the indicated time course. (c) Expression of RegIIIα, S100A8, and S100A9 mRNA in the ileum of SFB-void Jax mice colonized with SFB+CC over the indicated time course. Data are representative of four to six mice per group. Expression of IL-22 (d) mRNA and (e) protein in ileal explants isolated from indicated groups of mice and subsequently restimulated in vitro with rmIL-23 for 8 hours (for mRNA) or 24 hours (protein). (f) Expression of RegIIIα, S100A8, and S100A9 mRNA in ileal explants isolated from indicated groups of mice and subsequently restimulated in vitro with rmIL-23 for 8 hours. Data are representative of at least two independent experiments with three to six mice per group. All data are presented as mean ± SEM; *p<0.05, **p<0.01, and ***p<0.001, one-way ANOVA with Tukey's multiple comparison test (a, d, e & f) or one-way ANOVA with Dunnett's multiple comparison test (b & c).
To further investigate the contribution of neutrophils to IL-22 production in response to colonization with SFB-containing microbiota, we utilized ileal explants from SFB-void Jax mice, and SFB-void Jax mice treated with isotype control antibody or αLy6G antibody and colonized with SFB+CC for 7 days. Following colonization with SFB+CC, the expression of IL-22 mRNA and protein were induced upon restimulation with rmIL-23 (Figure 7d,e), as well as in the absence of restimulation, albeit to a lesser extent (data not shown). Remarkably, αLy6G antibody treatment of mice nearly completely abolished rmIL-23-induced expression of IL-22 mRNA and protein in ileal explants when compared to isotype control antibody-treated mice (Figure 7d,e). Similar results were also found with the RegIII, S100 and defensin families of antimicrobial peptides; rmIL-23 stimulation of ileal explants from isotype control antibody-treated mice resulted in augmented mRNA expression of these antimicrobial peptides, which was significantly reduced in explants from αLy6G antibody-treated mice (Figure 7f and Supplementary Figure 7a,b). These results suggest that IL-23 induced by SFB-containing microbiota enhances neutrophil-dependent IL-22 production, which in turn augments antimicrobial peptide expression in the ileum of mice.
DISCUSSION
In the present study, we demonstrate that colonization of mice with SFB stimulated robust neutrophil influx into the ileum of the small intestine of mice in a IL-17A- and CXCR2-dependent manner. In this context, Th17 cells were the most likely source of IL-17A production as this effect was not observed in Rag-deficient mice.9,10,28 IL-17A-CXCR2-mediated neutrophil recruitment coincided with IL-22 production and the expression of antimicrobial peptides and was fundamentally required to restrain the expansion of SFB. Together these findings highlight dynamic interplay between Th17 cells, IL-17A, CXCR2 and neutrophils that is important for controlling the expansion of SFB and potentially other bacteria that can establish intimate contact with the intestinal epithelium.
Previous findings have defined SFB as a potent inducer of antigen-specific Th17 cells in the small intestine.9,10,29 The process of Th17 differentiation can occur in the mesenteric lymph nodes and/or directly in the small intestine through the presentation of cognate antigens by MHCII+CD11c+ antigen presenting cells. 30-33 Ultimately, tight adhesion of SFB to the intestinal epithelium is capable of promoting the expression of IL-17A and IL-22. IL-22 has a direct role in barrier protection through augmenting epithelial proliferation and the generation of antimicrobial peptides and thus is capable of limiting the expansion of SFB.17 However, the function of IL-17A in controlling SFB expansion is less well understood. Here we show that IL-17A production in response to SFB colonization induced the expression of CXCL1 and CXCL2, which subsequently prompted neutrophil recruitment into the ileum and control of SFB expansion. While IL-17A is involved in the recruitment and activation of neutrophils that prevent microbial spread in response to other bacteria and fungi,34,35 and IL-22 is known to act on intestinal epithelial cells to elaborate antimicrobial peptide expression,36,37 our data link these two processes and provide key new insight into the control of SFB expansion by IL-17A-dependent neutrophil recruitment. In addition to involvement in neutrophil recruitment, IL-17A may also directly mediate barrier protective effects by regulating tight junction proteins38,39 and directly promoting the expression of α-defensin, Nox1, and Pigr via IL-17R expression on intestinal epithelial cells.40 However, it is important to highlight that under conditions of neutrophil depletion IL-17A was over-expressed, yet SFB growth was not effectively controlled. These data suggest that IL-17A and neutrophils work in concert to afford barrier protection and control of SFB expansion.
Our observation that SFB-containing microbiota-induced IL-22 expression is dependent on neutrophils is remarkable. Several different intestinal immune cell subsets can produce IL-22 including Th17 cells,41 γδ T cells,26 ILCs,41 NKT cells,42 and as recently reported, neutrophils.19,20 Given the ability of IL-22 to promote barrier function in numerous models,13,14,16 the redundancy allotted by different IL-22 producing cells may be a beneficial adaptation that provides multiple layer of host defense at barrier surfaces. In this context, the temporal production of IL-22 appears to be a highly orchestrated process unique to the specific settings. Interestingly, neutrophils have been described to package IL-22 in granules,19 which may allow for immediate release upon recruitment to the intestine following injury or microbial challenge. In response to acute introduction of adherent bacteria such as SFB, rapid IL-22-mediated barrier protection via neutrophils may complement the well-defined contributions of IL-22 production by type 3 innate lymphoid cells (ILC3s) in mediating control of SFB.11 Our current data implicate neutrophils as important responders during acute SFB colonization and show that they augment IL-22 and antimicrobial peptide production while controlling the abundance SFB. Therefore, IL-17A and Th17 cells appear to be critical mediators in controlling SFB levels through the recruitment and barrier-protective effector functions of neutrophils. In addition to contributing to IL-22-mediated barrier function, neutrophils also may play a direct role in killing of SFB via antimicrobial activities.43 Indeed a recent report demonstrated the requirement for Nox1 in controlling SFB growth in the ileum, although this was not directly linked to neutrophils.40
Notably, the induction of IL-17A and IL-22 in response to mucosal challenge or damage appears highly dependent upon the IL-23 pathway.17,19 IL-23 is produced primarily by antigen-presenting cells in response to activation of pattern-recognition receptors following barrier damage or pathogenic/adherent bacterial challenge.44 Consistent with this model, IL-23 receptor has been shown to be involved in IL-22 production, Th17 differentiation, and containment of SFB.17 IL-23R is expressed by T cells and group 3 innate lymphoid cells (ILC3s), as well as neutrophils, and their stimulation with IL-23 can potently enhance IL-22 expression.19,20 Using Rag-deficient mice, Shih et al. demonstrated that depletion of ILC3s with anti-Thy1 antibody resulted in a marked increase in SFB levels.17 These data highlight an important contribution of ILC3s in the control of SFB in the absence of adaptive immunity. It is notable that Rag-deficient hosts have an expanded population of ILCs and antimicrobial peptides45 and are more reliant on IL-23R-mediated mechanisms for control of SFB than are Rag-sufficient hosts.17 As we observed increased SFB levels in Rag-deficient mice compared to B6 mice when colonized with SFB-containing microbiota, this suggests that increased ILC and antimicrobial peptide responses in the absence of adaptive immunity and efficient neutrophil recruitment are insufficient to completely control SFB. Thus, innate and adaptive immune responses to SFB appear to be dynamically linked with at least some functions shared among distinct cell subsets depending on the timing and context. Overall, it appears that Th17 cells, IL-17A, neutrophils and ILCs collectively contribute to IL-22 mediated barrier protection in immunologically replete mice.
Recently, SFB was shown to trigger an IL-23R/IL-22 circuit that in turn promoted intestinal epithelial derived serum amyloid A and the generation of IL-17A in Th17 cells.11 These findings suggest that initial barrier protection is mediated by ILC3-derived IL-22 production that is then followed by Th17 differentiation. Our data indicate that IL-22 expression in the ileum of mice colonized with SFB-containing microbiota temporally coincides with IL-17A expression and neutrophil recruitment and that induction of IL-22 is highly dependent upon IL-17A and neutrophils. Collectively, these data suggest a model whereby IL-22-producing ILC3s are some of the earliest responders following SFB colonization and may begin to fortify the epithelial barrier. This early response may afford the host time to amplify Th17 cell differentiation and prompt IL-17A-dependent neutrophil recruitment, which plays an important and complementary role in the overall control of SFB expansion.
In addition to the expansion of SFB observed in the absence of neutrophils or IL-17A, we also noted an increase in the frequency of Th17 cells and well as IL-17A expression in the ileum. These data are consistent with a negative feedback loop whereby neutrophils suppress SFB levels thus limiting Th17 differentiation, IL-17A expression and subsequent neutrophil recruitment via CXCL1, CXCL2, and perhaps G-CSF.46 Therefore, IL-17A may be a key harbinger of danger in the intestine that is shut off only when the danger-triggered signals are effectively extinguished.47 This concept is consistent with our data demonstrating that antibody-mediated depletion of neutrophils or IL-17A neutralization both resulted in augmented levels of SFB and IL-17A expression. These findings are of interest given that overactive Th17 responses may be associated with inflammatory diseases including Crohn's disease.15 SFB has also been shown to exacerbate Th17-mediated disease in different autoimmune conditions.48,49 Thus, neutrophils represent front-line effector cells that may be required for limiting potentially detrimental Th17 responses induced by SFB and perhaps other intestinal epithelial cell adherent bacteria.
In summary, we present a model in which neutrophils are a key and previously unappreciated component of the immune response to SFB. While neutrophils are well-acknowledged as a critical component of innate immunity and host defense against enteric microbes, little is known about the contribution of these cells to the control of SFB expansion. The model presented here links SFB-induced IL-17A expression with neutrophil recruitment, barrier protection and ultimately control of SFB expansion. This mechanism may also be important in controlling the abundance of other adherent bacteria such as Citrobacter rodentium and Escherichia coli O157, which have also recently been shown to potentiate Th17 differentiation.23 Future investigations into the relative and temporal contributions of specific innate and adaptive immune cells and components in barrier protection in the intestine may provide much needed insight into the treatment of intestinal inflammatory disorders.
METHODS
Mice
Age- and sex-matched C57BL/6 (B6) or B6.129S7-Rag1tm1Mom/J (Rag1−/−) mice were purchased from The Jackson Laboratory (Jax) and verified to contain a microbiota free of SFB. To ensure these mice remained SFB-free, all experiments utilizing the transfer of SFB-containing microbiota were performed within a week of arrival of mice from Jax and mice were verified to be SFB-free by qPCR prior to colonization. SFB-positive mice were purchased from Taconic (Tac) and were similarly verified to be SFB-positive by qPCR. Mice were maintained under specific pathogen-free conditions and animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Georgia State University.
Preparation and administration of SFB-containing microbiota
Cecal contents from Tac B6 mice (SFB+CC) or Jax B6 mice (SFB−CC) were resuspended in 5 ml of sterile PBS, passed through a 100μm cell strainer, and 150 μl of the suspension was gavaged into recipient mice. Mice monoassociated with SFB were derived as previously described.50 Fecal material was sterilely collected from these mice then processed and administered in the same fashion as cecal contents at 1 ml sterile PBS per fecal pellet. Cecal contents and fecal suspensions were verified to contain SFB prior to being gavaged via qPCR. At the time-points denoted, feces were collected from mice and immediately frozen at −80. Bacterial DNA was extracted from cecal contents and feces using Qiagen DNA stool kits as per the manufacturer's instructions. SFB loads were determined via qPCR.9
Antibodies and reagents
The following antibodies (Abs) were purchased from eBioscience: CD11b (M1/70), CD45 (30-F11), Gr-1 (RB6-8C5), IL-17A (eBio17B7), IL-22 (1H8PWSR), Ly6G (1A8), Ly6C (HK1.4). Abs purchased from BD Biosciences were: TCRβ (H57-597), CD4 (RM4-5), and FcγRIII/II (2.4G2). Dead cells were identified using the fixable Aqua Live/Dead cell staining kit (Invitrogen). Isolation of LP cells and flow cytometry were performed as previously described.32
Depleting and neutralizing antibodies
Mice were injected i.p. with αIL-17A (17F3), αCXCR2 (polyclonal), αLy6G (1A8) or corresponding isotype control antibodies (αIL-17A: mouse IgG1; αCXCR2: goat IgG; αLy6G: rat IgG2a) at a concentration of 200ug/injection/daily (αIL-17A and αLy6G) or 1mg/injection/every other day (αCXCR2) 12 hours before gavage with SFB-containing cecal contents.
Intracellular cytokine staining
For intracellular cytokine analysis, cells were stimulated with PMA (50 ng/ml) and ionomycin (500 ng/ml) for 4 h in the presence of GolgiPlug (BD Pharmingen) and subsequently incubated with Live/Dead stain. Samples were then blocked with anti-FcγRIII/II and stained with αCD4 and αTCR-β. Cells were permeabilized and fixed using a Foxp3/Transcription Factor Fix/Perm kit (eBioscience) and stained by using fluorochrome-conjugated Abs according to the manufacturer's protocol.
Ileal explants
Sections of ileum were dissected from mice, opened longitudinally and immediately washed of contents. Biopsy punches (2mm; Integra Miltex) were used to excise sections of ileum, which were then placed in 96-well plates with HBSS (supplemented with 5% FBS). rmIL-23 (Miltenyi Biotec) was added to each well at 20 ng/ml. For qPCR, tissue was collected and processed for downstream application 8 hours later. For the detection of IL-22 using ELISA, supernatant from tissue was collected 24 hours after stimulation.
Real-time PCR
Total RNA was isolated from tissue sections stored in Qiagen RNAlater using the Qiagen RNeasy Mini Kit according to the manufacturer's protocol with on-column DNase digestion using the Qiagen RNase-Free DNase Set. cDNA was generated using the Superscript First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer's protocol. cDNA was used as a template for quantitative real-time PCR using SYBR Green Master Mix (Bio-Rad). PCR and analysis was performed using a StepOne PCR system (Applied Biosystems). Gene expression was calculated relative to that of gapdh.
Statistics
All statistical analyses were performed using Prism software with two-tailed unpaired Student's t test or Mann-Whitney test were appropriate and one-way ANOVA followed by the appropriate post hoc test where indicated. P-values <0.05 were considered significant (*p<0.05, **p<0.01, and ***p<0.001).
Supplementary Material
Footnotes
Author contributions
K.L.F. conceived and designed the experiments, performed the experiments, analyzed the data, and wrote the manuscript. V.L.N. and D.G. performed experiments and analyzed the data. A.H. provided reagents and technical assistance. S.A.H, N.L., A.N., C.A.P., V.G.R and N.C.B. critically read the manuscript and provided reagents. T.L.D. conceived and designed the experiments, analyzed the data, and wrote the manuscript.
CONFLICT OF INTEREST
The authors declared no conflict of interest.
REFERENCES
- 1.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis CP, Savage DC. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun. 1974;10:948–956. doi: 10.1128/iai.10.4.948-956.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 11.Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, Bonneau R, Littman DR. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 14.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: immunobiology and pathology. Annu Rev Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih VF, Cox J, Kljavin NM, Dengler HS, Reichelt M, Kumar P, Rangell L, Kolls JK, Diehl L, Ouyang W, Ghilardi N. Homeostatic IL-23 receptor signaling limits Th17 response through IL-22-mediated containment of commensal microbiota. Proc Natl Acad Sci U S A. 2014;111:13942–13947. doi: 10.1073/pnas.1323852111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 19.Zindl CL, Lai JF, Lee YK, Maynard CL, Harbour SN, Ouyang W, Chaplin DD, Weaver CT. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci U S A. 2013;110:12768–12773. doi: 10.1073/pnas.1300318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F, Cao A, Yao S, Evans-Marin HL, Liu H, Wu W, Carlsen ED, Dann SM, Soong L, Sun J, Zhao Q, Cong Y. mTOR Mediates IL-23 Induction of Neutrophil IL-17 and IL-22 Production. J Immunol. 2016;196:4390–4399. doi: 10.4049/jimmunol.1501541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen F, Hu Z, Goswami J, Gaffen SL. Identification of common transcriptional regulatory elements in interleukin-17 target genes. J Biol Chem. 2006;281:24138–24148. doi: 10.1074/jbc.M604597200. [DOI] [PubMed] [Google Scholar]
- 22.Fournier BM, Parkos CA. The role of neutrophils during intestinal inflammation. Mucosal Immunol. 2012;5:354–366. doi: 10.1038/mi.2012.24. [DOI] [PubMed] [Google Scholar]
- 23.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nunez G, Camp JG, Hattori M, Umesaki Y, Honda K. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KH. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–341. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–330. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 27.Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, Sczesnak A, Liao JJ, Torres VJ, Jenkins MK, Lafaille JJ, Littman DR. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, Ivanov II. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, Reynaud CA, Cerf-Bensussan N, Gaboriau-Routhiau V. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Geem D, Medina-Contreras O, McBride M, Newberry RD, Koni PA, Denning TL. Specific microbiota-induced intestinal Th17 differentiation requires MHC class II but not GALT and mesenteric lymph nodes. J Immunol. 2014;193:431–438. doi: 10.4049/jimmunol.1303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panea C, Farkas AM, Goto Y, Abdollahi-Roodsaz S, Lee C, Koscso B, Gowda K, Hohl TM, Bogunovic M, Ivanov II. Intestinal Monocyte-Derived Macrophages Control Commensal-Specific Th17 Responses. Cell Rep. 2015;12:1314–1324. doi: 10.1016/j.celrep.2015.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khader SA, Gaffen SL, Kolls JK. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunol. 2009;2:403–411. doi: 10.1038/mi.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14:585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnenberg GF, Fouser LA, Artis D. Functional biology of the IL-22-IL-22R pathway in regulating immunity and inflammation at barrier surfaces. Adv Immunol. 2010;107:1–29. doi: 10.1016/B978-0-12-381300-8.00001-0. [DOI] [PubMed] [Google Scholar]
- 37.Mizoguchi A. Healing of intestinal inflammation by IL-22. Inflamm Bowel Dis. 2012;18:1777–1784. doi: 10.1002/ibd.22929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JS, Tato CM, Joyce-Shaikh B, Gulan F, Cayatte C, Chen Y, Blumenschein WM, Judo M, Ayanoglu G, McClanahan TK, Li X, Cua DJ. Interleukin-23-Independent IL-17 Production Regulates Intestinal Epithelial Permeability. Immunity. 2015;43:727–738. doi: 10.1016/j.immuni.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maxwell JR, Zhang Y, Brown WA, Smith CL, Byrne FR, Fiorino M, Stevens E, Bigler J, Davis JA, Rottman JB, Budelsky AL, Symons A, Towne JE. Differential Roles for Interleukin-23 and Interleukin-17 in Intestinal Immunoregulation. Immunity. 2015;43:739–750. doi: 10.1016/j.immuni.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 40.Kumar P, Monin L, Castillo P, Elsegeiny W, Horne W, Eddens T, Vikram A, Good M, Schoenborn AA, Bibby K, Montelaro RC, Metzger DW, Gulati AS, Kolls JK. Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation. Immunity. 2016;44:659–671. doi: 10.1016/j.immuni.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paget C, Ivanov S, Fontaine J, Renneson J, Blanc F, Pichavant M, Dumoutier L, Ryffel B, Renauld JC, Gosset P, Gosset P, Si-Tahar M, Faveeuw C, Trottein F. Interleukin-22 is produced by invariant natural killer T lymphocytes during influenza A virus infection: potential role in protection against lung epithelial damages. J Biol Chem. 2012;287:8816–8829. doi: 10.1074/jbc.M111.304758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colgan SP, Ehrentraut SF, Glover LE, Kominsky DJ, Campbell EL. Contributions of neutrophils to resolution of mucosal inflammation. Immunol Res. 2013;55:75–82. doi: 10.1007/s12026-012-8350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maloy KJ, Kullberg MC. IL-23 and Th17 cytokines in intestinal homeostasis. Mucosal Immunol. 2008;1:339–349. doi: 10.1038/mi.2008.28. [DOI] [PubMed] [Google Scholar]
- 45.Korn LL, Thomas HL, Hubbeling HG, Spencer SP, Sinha R, Simkins HM, Salzman NH, Bushman FD, Laufer TM. Conventional CD4+ T cells regulate IL-22-producing intestinal innate lymphoid cells. Mucosal Immunol. 2014;7:1045–1057. doi: 10.1038/mi.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, Guttentag SH, Kolls JK, Oliver PM, Bushman FD, Worthen GS. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J Clin Invest. 2012;122:974–986. doi: 10.1172/JCI60588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubino SJ, Geddes K, Girardin SE. Innate IL-17 and IL-22 responses to enteric bacterial pathogens. Trends Immunol. 2012;33:112–118. doi: 10.1016/j.it.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015;520:99–103. doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.